Abstract
Background
HIV drug resistance (HIVDR) testing can assist clinicians in selecting treatments. However, high complexity and cost of genotyping assays limit routine testing in settings where HIVDR prevalence has reached high levels.
Methods
The oligonucleotide ligation assay (OLA)-Simple kit was developed for detection of HIVDR against first-line non-nucleoside/nucleoside reverse transcriptase inhibitors and validated on 672 codons (168 specimens) from subtypes A, B, C, D, and AE. The kit uses dry reagents to facilitate assay setup, lateral flow devices for visual HIVDR detections, and in-house software with an interface for guiding users and analyzing results.
Findings
HIVDR analysis of specimens by OLA-Simple compared to Sanger sequencing revealed 99.6 ± 0.3% specificity and 98.2 ± 0.9% sensitivity, and compared to high-sensitivity assays, 99.6 ± 0.6% specificity and 86.2 ± 2.5% sensitivity, with 2.6 ± 0.9% indeterminate results. OLA-Simple was performed more rapidly compared to Sanger sequencing (<4 h vs. 35–72 h). Forty-one untrained volunteers blindly tested two specimens each with 96.8 ± 0.8% accuracy.
Interpretation
OLA-Simple compares favorably with HIVDR genotyping by Sanger and sensitive comparators. Instructional software enabled inexperienced, first-time users to perform the assay with high accuracy. The reduced complexity, cost, and training requirements of OLA-Simple could improve access to HIVDR testing in low-resource settings and potentially allow same-day selection of appropriate antiretroviral therapy.
Fund
USA National Institutes of Health R01; the Clinical and Retrovirology Research Core and the Molecular Profiling and Computational Biology Core of the UW CFAR; Seattle Children's Research Institute; UW Holloman Innovation Challenge Award; Pilcher Faculty Fellowship.
Keywords: HIV genotyping assay, NNRTI, NRT, Resistance, Regimen switching
Research in context.
Evidence before this study
In 2017, the World Health Organization HIV Drug Resistance Network (HIVRESNET) reported several point-mutation assays had been developed to detect HIV drug resistance, including the oligonucleotide ligation assay (University of Washington), PANDAA™ (Aldatu Biosciences), allele-specific primer extension (United States CDC), and multiplexed melt curve analysis (InSilixa). A more recent search on PubMed and Google Scholar using key words such as “HIV drug resistance or “HIV genotyping”, coupled with “point-of-care” up to August 30th, 2019 showed similar results. These point-mutation assays can be used in place of Sanger sequencing with a real-time thermal cycler or a standard thermal cycler and a plate reader with fluorescent or colourimetric sensing. Searching results indicate that none of these assays were implemented for routine clinical management. Evaluation of these assays using clinical specimens across various HIV subtypes and specimen types was not found. Furthermore, none of these assays were demonstrated for successful utility by non-experience users. Therefore, we concluded HIVDR testing is still an unmet public health need in LRS.
Added value of this study
A low-cost, rapid point mutation assay (OLA-Simple) was developed to enable HIVDR detection in patient specimens across HIV-subtypes (A, B, C, D, and AE) from various specimen types (DBS, PMBC DNA, and plasma) using minimal equipment (a standard thermal cycler and pipettes). Comparative drug resistance detection between OLA-Simple and Sanger sequencing or higher sensitivity assays revealed highly concordant detection of mutations but with a much shorter turnaround time (<4 h vs. 35–72 h by Sanger sequencing). Furthermore, OLA-Simple was designed to be easy-to-use. Dry reagents allowed fast and accurate assay setup, and lateral flow detection provided semi-quantitative results that can be analysed by the unaided eye or the in-house software. The in-house software provided guidance for non-trained users and maintained the records of results and reagent stocks. Untrained, first-time users processed specimens using OLA-Simple with high accuracy.
Implications of all the available evidence
Our findings highlight the potential use of the OLA-Simple for HIVDR testing in a small laboratories in LRS that may lack sophisticated equipment and trained personnel. The entire reagent cost/ specimen is <$20 and requires < 10-min hands-on time from DNA or RNA. The key equipment required in OLA-Simple is a standard thermal cycler (which is substantially more economical than a real-time thermal cycler or a sequencer) and an in-house software installed on a tablet. The rapid turnaround time of OLA-Simple could allow same-day guided ART selection in LRS.
Alt-text: Unlabelled box
1. Introduction
Effective antiretroviral therapy (ART) suppresses viral replication, allowing HIV-infected individuals to live healthy and productive lives and reduces their risk of transmitting HIV to others. However, the error-prone HIV reverse transcription generates single-base mutations conferring HIVDR. Mutations at specific locations correlate closely with in vitro susceptibility testing and with clinical treatment failure. Recently the prevalence of HIVDR has increased, especially to non-nucleoside reverse transcriptase inhibitors (NNRTI), but also to nucleoside reverse transcriptase inhibitors (NRTI) used in first-line ART in many low-resource settings (LRS) [1,2]. Individuals with transmitted HIVDR may fail to achieve suppression of HIV replication after ART initiation. Additionally, when individuals do not adhere to their daily ART regimen, virus replication may not be suppressed (i.e., treatment failure), and HIVDR variants may be selected. Testing for HIVDR can identify the appropriate therapeutic option for either clinical scenario, including prevention of unnecessary switching of ART to more expensive regimens. HIVDR testing is recommended to guide treatment decisions for HIV-infected individuals [3,4] and has proven to be cost-effective in resource-rich communities [5] but not cost-effective in LRS [6]. Commonly-used Sanger sequencing is cost-prohibitive ($150–300/test) for routine HIVDR testing in LRS [7] and is estimated to avert few Disability Adjusted Life Years [6]. Slow turnaround times for HIVDR genotypes due to shipping specimens to centralized laboratories have contributed to delays in switching ART for patients with virologic failure. Decentralizing laboratory capacity could reduce turnaround time, facilitating more timely ART switching.
The World Health Organization has prioritized expanding laboratory capacity in laboratories in LRS. Several groups have worked to improve accessibility to HIVDR testing, including development of low-cost reagents for Sanger sequencing [8,9] and development of point mutation assays that detect drug resistance associated mutations (DRMs) known to confer resistance to specific drugs [10]. Point mutation assays are potentially faster, simpler, and lower-cost alternatives to sequencing, and can detect minority-variant DRMs at frequencies <20% of an individual's virus population, often missed by Sanger sequencing [11]. Pan Degenerate Adaptation & Amplification (PANDAA™) is designed to “erase” polymorphisms surrounding DRM sites during amplification and detects six DRMs with reported sensitivity of ≥1% mutant in an individual's virus population. PANDAA™ requires manual sample preparation and a costly PCR instrument for multi-color fluorescence and high-resolution melt analysis to allow probes to differentiate single-base mismatches. The Clinical Laboratory Improvement Amendments (CLIA)-certified oligonucleotide ligation assay (OLA) is a laboratory test (CLIA-OLA) that overcomes HIV polymorphisms by annealing DNA nucleotide probes at a temperature that tolerates annealing of sequences with mismatches, while an enzyme-mediated probe ligation that requires complementary bases at the site of the targeted DRM achieves high specificity. CLIA-OLA detects DRMs at ≥2% of an individual's viral population [12] and has been implemented in several research labs in LRS [13,14]. However, the CLIA-OLA is a complex assay that requires expertise in molecular biology. The paucity of expertise and difficulty procuring molecular reagents in LRS has hindered its adoption by LRS laboratories for clinical use [15].
To address the technology gap in LRS, we have revamped the CLIA-OLA assay into a low-cost, easy-to-use “OLA-Simple” kit for detection of HIVDR to NNRTIs and NRTIs. OLA-Simple includes (i) pre-measured, dry PCR and ligation reagents with primers and probes designed to detect DRMs in the most prevalent HIV-1 subtypes (A, B, C, D, and AE), (ii) lateral flow strip detection that reports visual readout for easy interpretation of DRMs, and (iii) an in-house software to guide users, that can also capture and interpret lateral flow strip DRM results. We assessed OLA-Simple using HIV DNA and RNA from clinical specimens of various HIV subtypes and compared results to Sanger sequencing and a sensitive comparator assay, either CLIA-OLA or Illumina MiSeq. To demonstrate the usability of OLA-Simple, we also assessed the performance of inexperienced users following the step-by-step instructions from the interactive software guide, “Aquarium” [16]. With shorter time, lower cost, and easier workflow, OLA-Simple could increase the capacity of small laboratories in LRS to directly perform HIVDR from specimens near the point-of-care.
2. Materials and methods
2.1. Preparation of OLA-Simple kits
The kit includes lyophilized PCR and ligation reagents, gold mixture, and competing oligonucleotides. The 50 µL PCR was made from 5 U FastStart™ polymerase (Sigma), 0.5 mg/mL BSA, 0.2 nM dNTPs, 2 mM MgCl2, and 0.4 µM of primers (forward primer: 5′ - GRC CTA CAC CTG TCA ACA TAA TTG G - 3′ and reverse primer: 5′ - CAA AGR AAT GGA GGT TCT TTC TGA TG - 3′) in water aliquoted prior to lyophilization. The 24 µL ligation reactions was made from 4–8 mU/µL thermostable Ampligase ligase (Epicentre Technology), 12.5 mM KCl, 1 mM NAD, 1× ligase buffer (20 mM Tris–HCl, 10 mM MgCl2, 1 mM DDT), 0.1075% (v/v) Triton-× 100, 5% (w/v) trehalose, 1.5% (w/v) poly(ethylene glycol), and 3.75–60 nM probes for each DRM (K65R WT: 5′ - Digoxigenin - CTC CAR TAT TTG CYA TAA AGA A - 3′, K65R MUT: 5′ - FAM - CTC CAR TAT TTG CYA TAA AGA G - 3′, K65R COM1: 5′ Phosphorylated - RAA RGA CAG TAC TAA GTG GAG AA - Biotin 3’, K65R COM2: 5′ Phosphorylated - AAA AGA YAG YAC TAA ATG GAG RA - Biotin 3’, K103N WT: 5′ - Digoxigenin - CAT CCA GCR GGG YTA AAA AAG AAR - 3′, K103N MUT: 5′ - FAM - CAT CCA GCR GGG YTA AAA AAG AAY - 3′, K103N COM: 5′ Phosphorylated - AAA TCA GTR ACA GTA CTR GAT GTG GG - Biotin 3’, V106M/I WT: 5′ - Digoxigenin - CCA GCA GGG TTA AAA AAG AAA AAA TCA G - 3′, V106M/I MUT: 5′ - FAM - CCA GCA GGG TTA AAA AAG AAA AAA TCAA - 3′, V106M/I COM: 5′ Phosphorylated - TRA CAG TAC TRG ATG TGG GGG ATG CAT AT - Biotin 3’, Y181C WT: 5′ - Digoxigenin - AAA AAA TCC AGA AAT ART TAT YTA - 3′, Y181C MUT: 5′ FAM - AAA AAA TCC AGA AAT ART TAT YTG - 3′, Y181C COM: 5′ Phosphorylated - YCA ATA CAT GGA TGA YTT GTA TGT A - Biotin 3’, M184V WT: 5′ - Digoxigenin - ATC CAG AAA TAR TTA TCT ATA ATA YA - 3′, M184V MUT: 5′ FAM - ATC CAG AAA TAR TTA TCT ATC AAT AYG - 3′, M184V COM: 5′ Phosphorylated - TGG ATG AYT TGT ATG TAG GAT CTG A Biotin 3’, G190A WT: 5′ Digoxigenin - CAT GGA TGA YTT GTA TGT RGG - 3′, G190A MUT: 5′ FAM - CAT GGA TGA YTT GTA TGT RGC - 3′, and G190A COM: 5′ Phosphorylated - ATC TGA YTT AGA AAT AGG GCA GCA - Biotin 3’) in water aliquoted prior to lyophilization. 8uM of each competing oligo (K65R Compt: 5′ - CTC CAR TAT TTG CYA TAA AGA ARA ARG ACA GTA CTA AGT GGA GAA - 3′, K103N Compt: 5′ - CAT CCA GCR GGG YTA AAA AAG AAR AAA TCA GTR ACA GTA CTR GAT GTG GG - 3′, V106M/I Compt: 5′ - CCA GCA GGG TTA AAA AAG AAA AAA TCA GTR ACA GTA CTR GAT GTG GGG GAT GCA TAT - 3′, Y181C Compt: 5′ - AAA AAA TCC AGA AAT ART TAT YTA YCA ATA CAT GGA TGA YTT GTA TGT A - 3′, M184V Compt: 5′ - ATC CAG AAA TAR TTA TCT ATC AAT AYA TGG ATG AYT TGT ATG TAG GAT CTG A - 3′, and G190A Compt: 5′ - CAT GGA TGA YTT GTA TGT RGG ATC TGA YTT AGA AAT AGG GCA GCA - 3′) in 1×Tris-HCl pH 8.0, 0.1 mM EDTA, and water was aliquoted into 48 µL reactions prior to lyophilization. Anti-biotin antibody-coated gold nanoparticles (0.75 OD at 520 nm) was aliquoted into 860 µL reactions prior to lyophilization. The lyophilized reagents were packaged in heat-sealed foil pouches with desiccant packets and stored at 4 °C until use. For the user feasibility study, the lateral flow tests were prepared in-house. 0.5 µg anti-FAM antibodies (Southern Biotech), anti-digoxigenin antibodies (Novus Biologicals), and biotin-conjugated BSA (Sigma) were striped onto each nitrocellulose sheet (EMD Millipore) adhered onto a PVC backing card and dried overnight in desiccator. Two ends of the nitrocellulose were then assembled to the glass fiber (EMD Millipore) and absorbent pads (GE Healthcare). The assembled sheet was cut into 3.5 mm × 6 mm strips and placed into plastic cartridges (DCN Diagnostics). Assembled devices were stored in foil pouches with desiccant at 4 °C until use. Lateral flow strips used in evaluation of clinical specimens were prepared using the same protocol by InBios International, Inc.
2.2. Specimen processing
FTA dried blood spots (DBS): Briefly, 125 µL blood were collected on Whatman FTA paper (GE Healthcare Life Sciences). A 3 mm diameter disk was excised from each DBS specimen and washed following manufacturer's instructions. The HIV viral DNA copy number on each DBS punch was quantified using a nested, real-time HIV gag PCR assay [17]. DNA extraction from PBMC: PBMC separation from whole anti-coagulated blood was performed using Ficoll medium. Washed PMBC pellets were either lysed with an in-house lysis buffer or lysis buffer provided by 5 Prime DNA purification kit (Fisher Scientific). RNA extraction from plasma: RNA extraction was performed 140 µL plasma using QIAamp viral RNA mini kit (Qiagen) according to the manufacturing protocol.
2.3. Illumina MiSeq and Sanger sequencing
PCR amplicons derived from FTA DBS, PBMC DNA, RNA, and 1st-round amplicon were treated with ExoSAP-IT (Applied Biosystems, Foster City, CA) and submitted to Sanger sequencing or analysed using HIV-1 ViroSeq™ kit (Abbott) (Thai cohort only). All mutations were confirmed by bi-directional sequencing. Illumina MiSeq library preparation, sequencing and analysis was performed as previously described [2]. DRMs were identified by submitting FASTA files to the Stanford HIV Drug Resistance database [18]. GenBank accession numbers are KF544089-KF544288, MH509760-MH509936, MH935 645-MH935766, MK512771-MK513407, MH623786.1, MH623833.1, MH623884.1, MH623900.1, MH623907.1, MH623919.1, MH623924.1, MH623940.1, MH623950.1, MH623951.1, and pending release from GenBank for South African and Thailand cohort.
2.4. CLIA-OLA procedure
PCR amplification used 2 µg of purified PBMC DNA as previously described [19] or 10 µL RNA eluates reverse-transcribed and amplified using Superscript IV (Invitrogen), followed by nested PCR. Amplicons from Peruvian [20] or Thai specimens were generated as previously described [21]. For ligation, 2 µL of 1:4 diluted PCR product were added to 20 µL freshly-prepared reactions containing probes specific for K103N, V106M/I, Y181C, G190A or V184M designed for HIV subtypes B, C, or A/D/C depending on the specimen's country of origin as previously described [22]. Products from the ligation step were detected by plate-based EIA [22] and optical densities were compared to standard curves of mutant and wild-type plasmid mixtures at 0%, 2%, 5%, 10%, 25%, 50%, 75%, and 100% mutant. The proportion of mutant variant in each specimen was quantified by comparing the MUT OD / (MUT OD + WT OD) to the standard curve (mean of n = 2). Samples with negative reactions for both MUT and WT were considered IND.
2.5. OLA-Simple procedure
PCR amplification using the dry 50 µL reaction mixtures was performed directly on 3 mm washed FTA DBS punches or 1.2 µL purified DNA or 10 µL cDNA generated from 20 µL of purified RNA eluate reverse-transcribed in rehydrated dry RT mixture (Takara) at 42 °C for 1 h. Amplicons were detected in agarose gels except for eight FTA DBS. These eight amplified when repeated in 100 µL PCR to dilute potential inhibitors. Because FTA DBS amplicons were weak a second-round PCR was conducted using dry reagents. All PCR reactions underwent 4 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 55 °C, and 1 min at 72 °C, and final extension for 7 min at 72 °C. Ligation used 4.4 µL of each amplicon added to 22 µL rehydrated ligation reagents containing probes specific for one codon: K65R, K103N, V106M/I, Y181C, M184V or G190A. Ligation proceeded for 10 cycles of 30 s at 94 °C and 4 min at 37 °C. For detection, 2.4 µL of 4 µM rehydrated competing oligo were added to each ligation reaction and cycled once for 30 s at 94 °C and 4 min at 37 °C; then 24 µL of ligation products were added to the sample port of a lateral flow strip and chased by 40 µL of the detection gold mixture. The strips were scanned after 20 min. Analysis of the scanned image of each strip was independently, blindly categorized as WT, MUT, and IND by 11–13 evaluators using written instructions. Mode calls for each specimen across evaluators was reported (Supplementary Table 1a) as the final OLA-Simple result. The images were also analysed by our computer program and concordance assessed with the mode of human visual calls.
2.6. Study design and clinical specimens
The performance of OLA-Simple was validated by blindly testing archived, de-identified clinical specimens reported to have DRMs from cohorts in South Africa, Thailand, Kenya, and Peru. Each specimen was tested using OLA-Simple kit (n = 1 for PCR and n = 1 for each mutation). Specimens from South Africa consisted of sixty DBS specimens from children aged 0–5 years pre-exposed to nevirapine prophylaxis, collected in 2010–2013 and stored at room temperature (∼25 °C) [23]. Specimens from Thailand consisted of thirty-eight first-round pol amplicon derived from plasma of HIV-infected adults and children enrolled in the PHPT-GFATM cohort treatment program (Observational Cohort of HIV Infected Adults and Children in the PHPT Network Hospitals in Thailand in 2002–2008 (ClinicalTrials.gov Identifier: NCT00433030) [21]. Specimens from Kenya included twenty-three plasma and twenty-four PBMC from HIV-infected adults enrolled in studies conducted in Nairobi, Kenya in 2010–2014 (ClinialTrials.gov Identifier: NCT01898754) [2]. Specimens from Peru consisted of twenty-three first-round pol amplicons derived from plasma of HIV-infected adults initiating first-line ART collected in 2014–2015 at Hospital Nacional Dos de Mayo [20]. The genotyping results obtained by OLA-Simple were compared to those obtained by Sanger sequencing (all cohorts) and separately all cohorts were compared to CLIA-OLA, except the Kenyan PBMC DNA cohort, which was compared to Illumina MiSeq.
2.7. Feasibility study of OLA-Simple
The feasibility testing of OLA-Simple was conducted in the Biofabrication Center at the University of Washington (BIOFAB). The BIOFAB routinely processes molecular cloning jobs, but it had no prior experience running HIVDR tests. OLA-Simple was set up on three different benches based on the available space in the lab. Ten kits were stored in the available refrigerator space and restocked when the stock was low. Benches were designated as a pre-PCR bench and two post-PCR benches for ligation/detection and visual call steps. Tasks for each OLA-Simple protocol (PCR, ligation, detection, and visual calls), called “jobs,” were assigned and scheduled by two managers. A maximum of three different participants conducted different parts of the workflow in parallel. We enrolled 41 volunteer operators who completed biosafety and other related safety training prior to working in the BIOFAB, including 5 without pipetting experience, 13 unfamiliar with pipetting small volumes (<3 µL), and 23 unaware of the possibility of PCR carry-over contamination. Prior to running OLA-Simple experiments, participants without prior lab experience were provided a ten-minute overview on how to use micropipettes, mini-centrifuges, and vortex mixers. Each operator processed two specimens (n = 1 for each specimen).
2.8. Statistical analysis
Comparisons across groups were calculated using t-test with the Holm-Bonferroni correction to adjust alpha for multiple comparisons [24]. McNemar's test was used for paired comparison of the categorical data by OLA-Simple vs Sanger or OLA-Simple vs sensitive comparator assay. Concordance, sensitivity, and specificity were reported with standard errors of binomial proportion.
3. Results
3.1. Design and development of the OLA-Simple kit
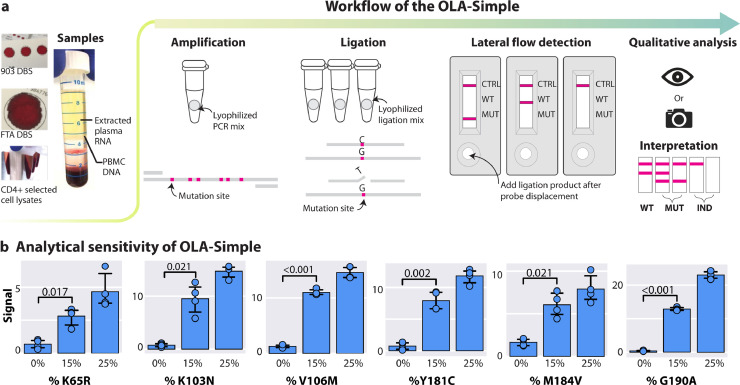
OLA-Simple (Fig. 1(a)) was designed to reduce the complexity of CLIA-OLA (Supplementary Fig. 1a) which contains four modules: sample preparation, polymerase chain reaction (PCR), enzyme-mediated probe ligation, and enzyme-linked immunosorbent assay (EIA). In OLA-Simple workflow, extracted nucleic acids from a specimen can be used to directly reconstitute the dry reagents, enabling fast set-up and accurate performance of the reactions, and visual readout of the lateral flow strips allows easy interpretation of results for the six DRMs with a total of <3.5-h wait-time (2-h PCR, 1-hour ligation, 20-min detection) and <10-min hands-on time (<1-min PCR, <3-min ligation, and ∼6-min detection). The enzymes, co-factors, and buffers used in the OLA-Simple were also optimized to eliminate the need for purification between steps. The presented OLA-Simple kit detects key DRMs in HIV pol encoding reverse-transcriptase associated with resistance to first-line NNRTI (K103N, V106M/I, Y181C, G190A), tenofovir (K65R) and lamivudine (M184V). Primers used in PCR and probes used in ligation were designed to anneal to the most prevalent global HIV-1 subtypes by incorporating degenerate bases at variable positions in the Los Alamos HIV Database. Analytical performance at each DRM site was evaluated using synthetic DNA gene fragments containing 0%, 15%, and 25% mutant (MUT) in wild-type (WT) HIV sequences; the OLA-Simple kit discriminated MUT from WT HIV in samples containing 15% mutant HIV at the six DRMs tested (Fig. 1(b)).
Fig. 1.
Workflow and analytical performance of the OLA-Simple HIV genotyping assay. (a) Workflow of OLA-Simple. DNA obtained from dried blood spots (DBS), peripheral blood mononuclear cells (PBMC), cell lysates, or cDNA reverse-transcribed from plasma RNA are amplified using pre-measured, dry PCR reagents resuspended in a single buffer. Amplicons are then tested for the presence of mutations using dry ligation reagents. Probes complementary to the wild-type and the mutant bases are reacted simultaneously at each codon. The ligated products are detected using customized lateral flow test detection strips, which are then analysed by eye or a scanner. The genotype is interpreted as mutant (MUT), wild-type (WT), or indeterminate (IND, defined as control line but no MUT or WT signal) by presence or absence of lines on each strip. (b) Analytical performance of OLA-Simple. MUT signal (means ± SD, n = 4) quantified on the lateral flow test strips for K65R, M184V, G190A, K103N, V106M, and Y181C, respectively, from control mixtures containing 1000 copies of HIV DNA input with 0%, 15%, or 25% MUT. Vertical scale is linear. Signal analysed was compared using t-test with Holm-Bonferroni adjustment.
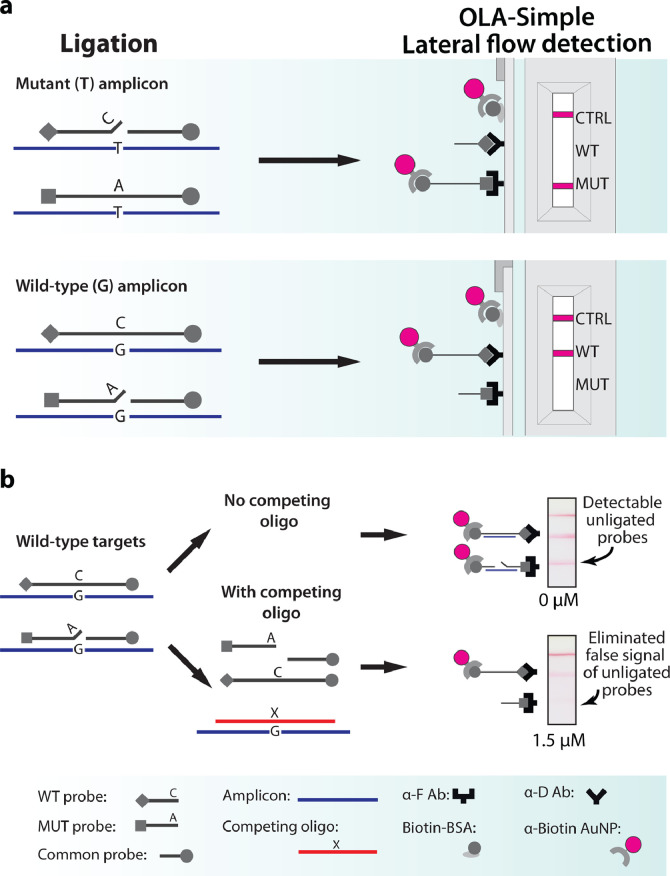
To develop dry reagents for OLA-Simple, we determined additive formulations that protect enzymes, proteins, and co-factors during the freeze-drying process (lyophilization), and serve as a water substitute to stabilize polar residues, while also maintaining buffer compatibility across all modules (see Supplementary Fig. 1 and 2 for detailed optimization). OLA-Simple's customized lateral flow devices replace a time-consuming and labor-intensive EIA in CLIA-OLA. The EIA step in CLIA-OLA involves adding and decanting multiple reagents with several incubation steps and reading colourimetric signals in series from two enzyme-tagged antibodies bound to the MUT and WT ligated probes within a single well. In contrast, OLA-Simple was designed for rapid lateral flow detection that simultaneously detects WT and MUT ligated probes (Fig. 2(a)), using: (i) multiplexed capture by immobilized antibodies, (ii) denaturation of un-ligated probes using “competing oligo,” and (iii) a shared gold nanoparticle label for both WT and MUT ligated products. (i) and (iii) were previously used to detect a mutation in the human genome which, unlike HIV, does not have neither polymorphisms nor minority mutations [25].
Fig. 2.
Detection of single-base mutations in OLA-Simple. (a) OLA-Simple detection. 5′-digoxigenin-labeled wild-type (WT) probe and 5′-FAM-labeled mutant (MUT) probe are ligated to the 3′-biotinylated, 5′-phosphorylated common probe when these anneal to WT and MUT HIV amplicon templates, respectively. A mismatched probe:template complex precludes ligation due to specificity of the ligase for complementary bases at the ligation site. MUT and WT ligated products are captured on the strip by immobilized anti-FAM antibody (α-F Ab) and anti-digoxigenin antibody (α-D Ab), respectively. Anti-biotin antibody-coated gold nanoparticles in the chase buffer bind to the ligated products as well as the biotin-conjugated BSA (Biotin-BSA) control line (CTRL), all labeled by anti-biotin antibody conjugated gold nanoparticles (α- Biotin AuNP). (b) Effects of competing oligo concentrations. Examples of scanned lateral flow strips analyzing WT HIV ligation products after treating with 0 or 1.5 µM competing oligo concentrations are shown.
For the multiplexed detection of MUT and WT ligation probes, we used EIA to quantitatively assess the performance of eight commercially-available molecular binding pairs and found three pairs bound without cross-reactivity and maintained high specificity when immobilized onto nitrocellulose test strips. Next, we investigated denaturation methods to remove unligated probe:template complexes that can cause false signals. In OLA-Simple detection, an excess of single-stranded oligonucleotide complementary to the target DNA was used as a “competing oligo” to displace both ligated and unligated probes from the amplicon template. Competing oligos at 1.5 µM eliminated the false signal (Fig. 2(b)). See detailed optimization of lateral flow detection in Supplementary Fig. 3.
3.2. OLA-Simple testing of global clinical specimens
The performance of OLA-Simple across specimen types was first evaluated by spiking synthetic HIV DNA template at 10% MUT or 0% MUT (pure WT) into nucleic acids from uninfected blood, including DNA extracted from peripheral blood mononuclear cells (PBMC), PBMC cell lysate, CD4 cell lysate, eluate from 903 dried blood spots (DBS), and from washed FTA DBS. FTA DBS slightly impaired assay performance, yet all specimen types were able to differentiate the 10% MUT sample from the 0% MUT (pure WT) sample (P < 0.05, 0% vs 10% for each specimen type) (Supplementary Fig. 4).
The robustness of OLA-Simple in analyzing DRMs in specimens across HIV-subtypes A, B, C, D, and AE was assessed using clinical specimens from multiple cohorts. A clinical panel with 168 specimens enriched for specimens with DRMs included: South African FTA DBS (n = 60), Kenyan PBMC (n = 23), Kenyan plasma (n = 23), and HIV pol PCR amplicon from Thailand (n = 38) and Peru (n = 24). Broken down by HIV subtypes, this specimen panel contained (n = 25), B(n = 26), C(n = 62), D(n = 8), AE(n = 43), AG(n = 1), G(n = 1), BC(n = 1), and CD(n = 1). OLA-Simple was performed by a trained operator “blinded” to prior genotypic testing of these specimens (Fig. 3(a)).
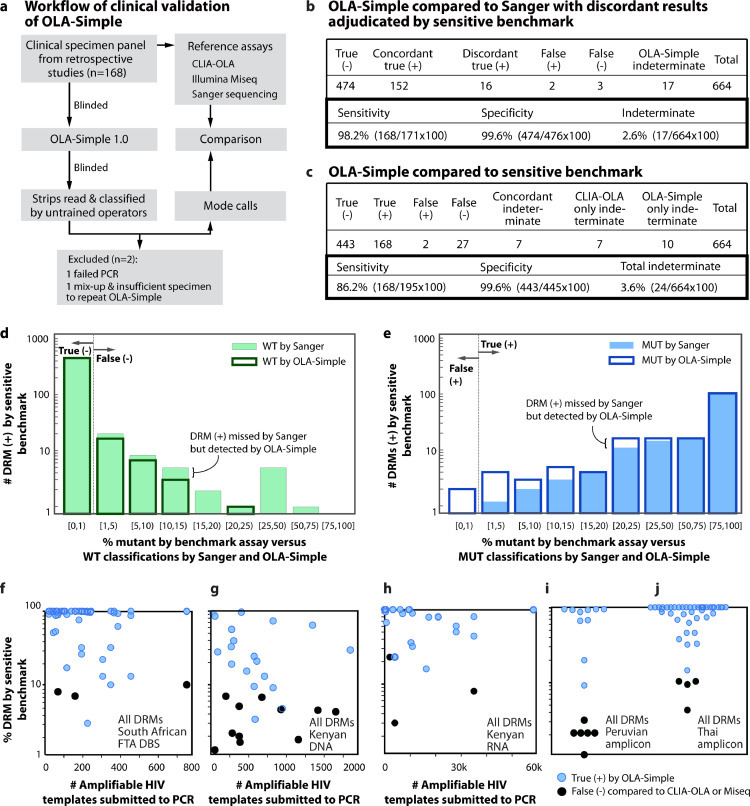
Fig. 3.
Validations of the OLA-Simple on clinical specimens, compared to other comparator assays. (a) Schema of OLA-Simple clinical specimen validation (n = 168, 672 codons). DNA, RNA, or amplicon (AMP) from specimens were re-labelled to blind the operator running the OLA-Simple test. The scanned images were sent to untrained volunteers for analysis (n = 13 for the Thai data set; n = 11 for other data sets). (b) Genotyping results of OLA-Simple compared to Sanger and (c) to a sensitive comparator assay. The comparator assay depended on the patient cohort and were used to adjudicate discordant results between the OLA-Simple and Sanger sequencing. Indeterminates were excluded from sensitivity and specificity estimations. (d) and (e) histograms of MUT frequencies in specimens identified as having DRMs by a quantitative comparator assay (CLIA-OLA or MiSeq) versus (d) WT and (e) MUT classification by Sanger and OLA-Simple. Bin spacing was based on commonly-used MUT thresholds at 1%, 5%, 10%, 15%, 20%, and 25% MUT frequencies. (f)–(j) MUT classifications of DNA samples from (f) South African FTA DBS, (g) Kenyan PBMC DNA, (h) Kenyan plasma RNA, (i) amplicon derived from Peruvian PBMC or plasma and (j) Amplicon derived from Thai plasma. CLIA-OLA (f, h–j) or MiSeq (g) were used as the comparator assay.
The OLA-Simple PCR-amplified 167/168 (99%) specimens; one Thai amplicon had a failed PCR likely due to a defective dry PCR tube rather than a low copy number or the primer sequences, as CLIA-OLA using the same primers successfully amplified this specimen. Following ligation, the OLA-Simple lateral flow detection strips were scanned and blindly classified as MUT, WT, or indeterminate (IND) by untrained evaluators (n = 13 for the Thai cohort or n = 10 for the other cohorts). Mode calls of the OLA-Simple strips were compared to classifications by Sanger sequencing and a sensitive comparator assay, MiSeq for Kenya PBMC specimens and CLIA-OLA for the remaining specimens. Comparisons were made for K103N, V106M/I, Y181C, M184V, and G190A, with V106M/I tested only on specimens from South Africa, where HIV subtype C is most prevalent. One DBS sample was excluded from the comparison due to sample mix-up (identified due to highly discordant results to both CLIA-OLA and Sanger sequencing) and insufficient specimen to repeat OLA-Simple. Fig. 3(b) shows the summary of the OLA-Simple genotyping results compared to Sanger sequencing and a sensitive comparator assay (CLIA-OLA or MiSeq).
Of the 664 codons analysed, OLA-Simple had 17 indeterminate results (IND, 2.6%), not significantly different from 14 IND (2.1%) found in the same data set by CLIA-OLA (McNemar's test, P = 0.63). Of 24 IND results by CLIA-OLA and OLA-Simple combined: 7 were concordant, 7 were CLIA-OLA only, and 10 were OLA-Simple only. Four codons with IND results by OLA-Simple also had low WT signal in CLIA-OLA. The number of HIV templates submitted to OLA-Simple and/or CLIA-OLA from the 24 specimens with IND results (median=271; interquartile range, IQR: 161–412 copies/reaction) did not differ significantly from the specimens categorized as WT (median = 270; IQR: 106–1821 copies/reaction, two-tailed Mann–Whitney U Test, P = 0.70) or MUT (median = 349; IQR: 139–1890 copies/reaction, two-tailed Mann–Whitney U Test, P = 0.32). Hence, IND results were not due to low HIV template concentrations in this study. IND results were distributed across the codons as follows: 11/166 (6.6%) at K103N, 1/59 (1.7%) at V106M/I, 3/166 (1.8%) at Y181C, 3/107 (2.8%) at M184V, and 6/166 (3.6%) at G190A. Sanger sequencing revealed that four IND codons had other mutant variants not detected by the current OLA-Simple probe sets (K103R, Y181I and G190T). A total of 83% (20/24) IND specimens had sequence mismatches likely to have caused the assay to fail at one codon, including 12/24 (50%) IND having ≥1 polymorphic bases within 3 bases of the ligation site, and 8/24 (33%) IND having multiple (≥3) polymorphic bases within the span of the probes (Supplementary Fig. 5).
Compared to Sanger sequencing, OLA-Simple had 626 concordant and 21 discordant classifications (Fig. 3(b) for combined DRM and Supplementary Table 1(b) for each DRM). OLA-Simple detected 16 MUT missed by Sanger sequencing, with 10/16 having a MUT frequency ≤20%. Compared to Sanger sequencing, and excluding 17 IND cases, OLA-Simple had a combined specificity across codons of 100% (99%, 100%, 100%, 100%, and 99% for K103N, V106M/I, Y181C, M184V, and G190A, respectively), and a combined sensitivity across codons of 98% (97%, 100%, 98%, 100%, and 100% for K103N, V106M/I, Y181C, M184V, and G190A, respectively).
Comparing OLA-Simple to a sensitive genotyping method (either 2% MUT cutoff CLIA-OLA or 1% MUT cutoff MiSeq), 93% (617/664) results were concordant, including 168 MUT, 442 WT, and 7 IND (Fig. 3(c) for combined DRM and Supplementary Table 1(c) for each DRM). Genotyping classifications by OLA-Simple and these methods were significantly different (McNemar's Test, P<0.05) but with an overall concordance of 95.5 ± 0.8% (SE). Fig. 3(d)–(e) shows the agreements and discrepancies of genotyping results across different MUT frequencies. OLA-Simple had 27 false negative (MUT misclassified as WT) and 2 false positive (WT misclassified as MUT). All false negative had ≤15% MUT except one with 23% MUT (Fig. 3(f)–(j)), while the input of HIV templates (median = 1498, IQR = 751–3710 HIV copies/reaction) of the false negative group identified by OLA-Simple was not significantly different from the group of MUT concordant to sensitive methods (median = 349; IQR = 139–2450 HIV copies/reaction, two-tailed Mann–Whitney U Test, P = 0.70). Hence, false negative results were not influenced by HIV template concentration in this study. Compared to the sensitive comparator assays and excluding 24 IND cases from both methods, OLA-Simple had a combined specificity of 100% (99%, 100%, 100%, 100% and 99% for K103N, V106M/I, Y181C, M184V, and G190A, respectively); and a combined sensitivity of 86% (81%, 100%, 87%, 82%, and 97% for K103N, V106M/I, Y181C, M184V, and G190A, respectively). These sensitivities and specificities differ depending on pre-determined% mutant cutoffs. In this data set, OLA-Simple had a maximum combined concordance of 98 ± 1% when compared to sensitive comparator assays with a cutoff of 12.5% MUT. The cutoffs with the maximum concordance for each DRM were different: 1% cutoff for G190A (99 ± 0.8% (SE)) and V106M/I (100%), 5% cutoff for M184V (99 ± 0.9%(SE)), 10% cutoff for K103N (96 ± 0.8% (SE)), and 12.5% for Y181C (99 ± 0.8% (SE)). Using results of sensitive methods as HIVDR outcomes, Receiver Operating Characteristic (ROC) curves of OLA-Simple and Sanger were compared (Supplementary Fig. 6). The ROC revealed that OLA-Simple was more sensitive than Sanger, but two false positives found in this study resulted in a similar area under the curve of 99.3%.
3.3. OLA-Simple test interpretation by untrained operators and software classification
Scanned images of OLA-Simple lateral flow strips for all clinical specimens were classified by untrained operators in large and small batch sizes. The mode human calls were then compared to a newly-developed image analysis software that automatically classified test results (Fig. 4(a)).
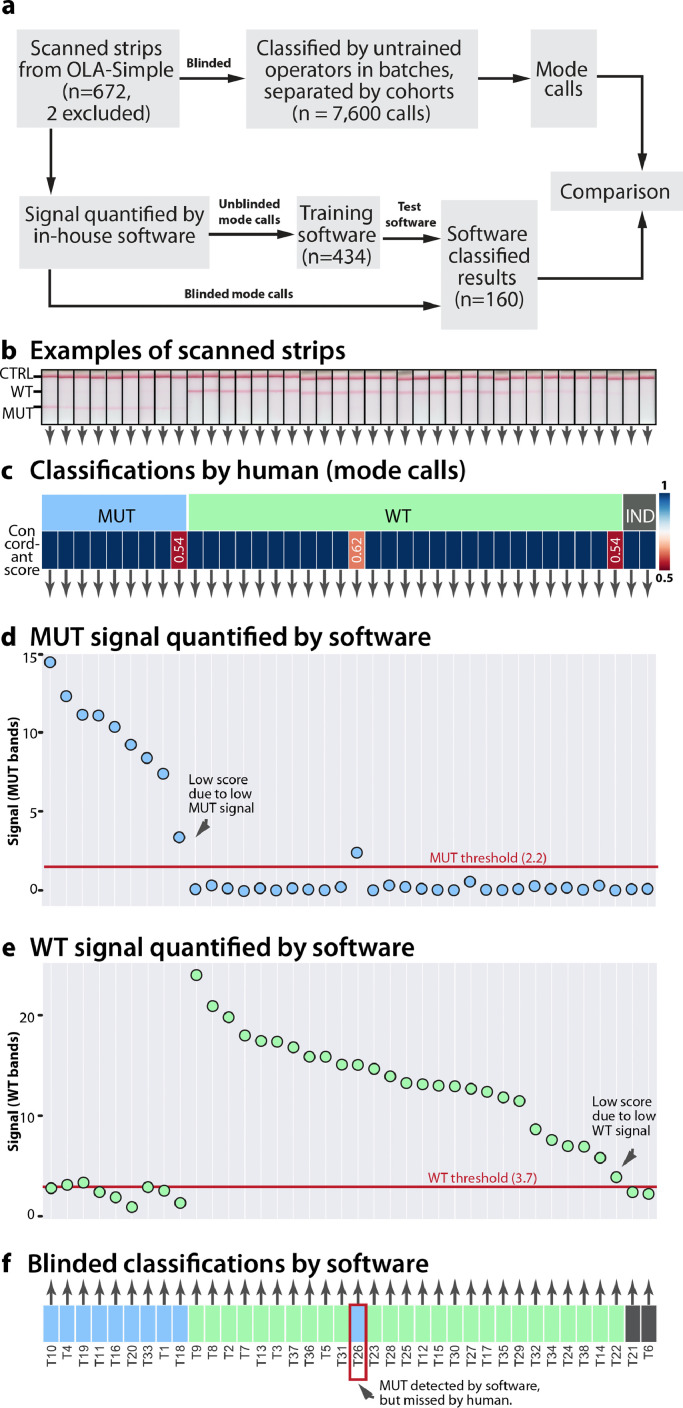
Fig. 4.
Lateral flow test results classifications determined by human evaluators and in-house software for a subset of specimens tested by OLA-Simple. (a) Schema of human analysis of OLA-Simple strip images and software analysis. (b) Scanned images of lateral flow strips for the K103N mutation in Thai specimens, with pink lines at the top, middle, and bottom of strip corresponding to flow control (CTRL), wild-type (WT), and mutant (MUT), respectively. (c) Mode genotyping classifications from 13 blinded evaluators; genotyping classification scores ranged from 0.5 to 1, where 1 = 100% agreement among evaluators. The signal from (d) MUT and (e) WT bands analysed by in-house software are plotted by grouping; highest to lowest MUT signal, highest to lowest WT signal, and indeterminates (IND). (f) Software classifications of dataset with blue, green, and gray representing MUT, WT, and IND, respectively.
For human call analysis, 672 lateral flow strip images (168 specimens excluding one PCR-failed and one mix-up) were blinded and classified by multiple evaluators (13 for Thai data; 11 for other data) in large batches, separated by specimen cohorts (92–240 strips/batch). Evaluators reported spending 2–3 min to study the classification rules and 10–20 s classifying each strip. Classifications from each evaluator were pooled and reported as the OLA-Simple mode calls and concordance scores (Supplementary Table 1(a)), with 97% (656/672) strips with >0.8 concordant scores indicating high unanimity among evaluators. All cohorts had comparable numbers of images with >0.8 concordant scores (Supplementary Fig. 7, a–f, chi-square statistic 6.31, P = 0.18). To study the effect of batch size on visual call classification, we designed an interactive online survey to assess the performance of each evaluator in a smaller dataset (Supplementary Fig. 7(g)). Each of seventy-two evaluators was presented with 20 randomly-selected lateral flow strip images from the clinical data set. First-time evaluators from the online survey spent 2.3 ± 1.1 min on the analysis of all 20 strips. Classification errors were not significantly reduced when evaluators were asked to read a smaller number of strips per batch (Supplementary Fig. 7, a and i, P = 0.74). Fig. 4(b) shows an example of scanned lateral flow strip images where most mode calls were concordant among human evaluators (scores >0.8), except for three cases with discordant calls due to either weak MUT or WT bands (Fig. 4(c), heatmap). For all 664 codons and 7600 readings (excluding one failed-PCR and one mix-up), mode calls resulted in concordance of 92 ± 1.1% (SE) compared to genotyping results by a comparator assay, while individual calls resulted in concordance of 91 ± 0.3% (SE). To overcome human variation for strips with weak signal, we developed an in-house python algorithm to calculate t-statistics between the WT, MUT, and CTRL bands and the adjacent white area of the strip. The t-statistics were normalized by the pixel count to correct for image resolution differences and were reported as signal on lateral flow tests (Supplementary Fig. 8, a–c, and examples on Fig. 4(d) and (e)).
We further trained this software to use the extracted signals to classify lateral flow test results as WT, MUT, or IND (Supplementary Fig. 8, a–c). A subset of the clinical data based on the mode call results was used as a training set (434 codons) to determine threshold signal values, and the resulting algorithm was applied to the remaining clinical data set (160 codons) to classify tests as MUT, WT, and IND (Supplementary Fig. 8, d). Only one of the 160 computer classified calls (Fig. 4(f)) did not match the mode of the human calls, however, the computer classified call was concordant with the comparator assay.
3.4. Software-guided OLA-Simple workflow for first-time users
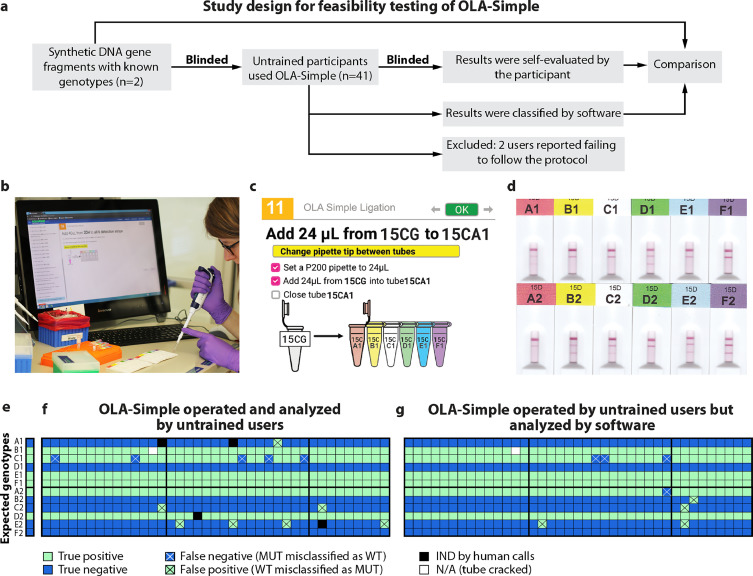
Next, we assessed if inexperienced users could successfully perform OLA-Simple. To provide detailed instructions for how to use OLA-Simple, we utilized a web-based human-in-the-loop laboratory automation application, called “Aquarium.” It integrates sample and inventory tracking, data collection, experimental protocols and workflow management [16]. In this study, Aquarium was programmed to provide interactive protocol instruction and automatic data collection for OLA-Simple using a combination of on-screen elements that included text, check boxes, and diagrams. Forty-one participants with no prior experience running OLA-Simple (Fig. 5(a)) were provided information on the location of OLA-Simple reagents, pre-PCR bench, and post-PCR bench. Without additional training, the users were then instructed to perform OLA-Simple guided by the interactive Aquarium protocols to test two blinded DNA samples containing known DRMs at the six codons assessed by OLA-Simple (K65R, K103N, V106M/I, Y181C, M184V, and G190A) (Fig. 5(b) and (c)). Scanned images of the test results (Fig. 5(d)) were visually classified by each user using guided interpretation by Aquarium as well as by the in-house software as WT, MUT, and IND. For data collection, laboratory notebooks were not provided, and instead, data such as test results, user on-screen selections, and images were automatically recorded in Aquarium.
Fig. 5.
Feasibility testing of OLA-Simple by inexperienced users guided by Aquarium software. (a) Schema of feasibility testing. 41 untrained participants were recruited to each process two blinded samples made of synthetic DNA fragments using a software guided protocol. The operators scanned the test strips and the images were used to classify each DRM by visual inspection guided by Aquarium, and by a computer algorithm. The computer classification was then compared to calls made by each participant. (b) A user performing OLA-Simple by following Aquarium. (c) Aquarium software guided users through the OLA-Simple protocol. Aquarium provided instructions as a series of interactive steps that included check boxes, timers, selection boxes, tables, and images. Aquarium automatically generates unique identifiers and reference to physical locations for all tubes and other items used in the protocols, which are then dynamically and automatically displayed in the tables, images, and text of the protocol steps shown on the computer or tablet. A permanent record is maintained for all steps, which mitigates the need for the user to record steps in a separate notebook. (d) Scanned lateral flow strip images at the end of the OLA-Simple workflow for the two samples tested (sample 1: MUT, WT, WT, MUT, WT, WT; sample 2: WT, MUT, MUT, WT, MUT, MUT for K65R, K103N, M106V, Y181C, M184V, G190A, respectively). (e) and (f) are classifications by untrained users on their own strips and by software, respectively. In one case (N/A) the ligation tube cracked during centrifugation and the specimen was lost. Gold solutions were prepared from three different manufacturing batches, which resulted in differences in signal intensities. Batches are separated by thick vertical black lines. A thick horizontal line separates results from the two samples tested (A1–F1 and A2–F2). Reduced misclassifications of genotyping results by software classifications from scanned images of strips separated by kit batches. Note that we observed some variations between the signal of images from the low-cost USB-powered scanner used in this feasibility study and the office scanner used in the clinical study, see Fig. 4, which required different thresholds for classification in this data set.
Five percent of participants (2/41) made procedural errors in performing OLA-Simple during the PCR and detection. The 39 participants who successfully completed all steps of OLA-Simple on both samples correctly genotyped them by visual call with an overall concordance for all DRMs tested of 96.8 (453/468) ±0.8% (SE) (Fig. 5(e)). Incorrect OLA-Simple results included 1 false negative and 2 IND at K65R; 2 false negative and 5 false positive at V106M/I; 1 IND at Y181C; and 4 false negative and 1 IND at M184V. V106M/I had the highest false positive rate (5/41) due to background signal causing a faint line, while M184V had the highest rate of false negative calls (4/41) due to relatively weak MUT signal. The in-house software quantification and classification of the signal (Fig. 5(f)) eliminated all four IND calls made by humans, reduced false negative calls from 5 to 4, and reduced false positive calls from 7 to 4 for an overall concordance of 98.1 ± 0.4% (SE).
4. Discussion
This work presents a unique application of cross-discipline science and engineering techniques to address the WHO's initiative to expand access to HIVDR testing. First, we developed an easy-to-use and rapid HIVDR test. The OLA-Simple workflow requires less instrumentation, lower reagent cost, and minimal hands-on time compared to Sanger sequencing or the CLIA-OLA. Second, the kits were blindly validated on representative clinical specimens from different geographic backgrounds and specimen types. Third, errors in human visual interpretations of results were reduced by use of newly-developed software to automatically quantify signal and interpret results. Last, we implemented OLA-Simple in a molecular laboratory and demonstrated that first-time users guided only by an interactive software successfully tested blinded specimens.
The workflow of OLA-Simple was built upon the established CLIA-OLA but included multiple modifications. First, single-round PCR was optimized to detect ≥10 HIV copies and replaced nested PCR in CLIA-OLA. Second, new additive formulations were developed to preserve dry reagents in OLA-Simple to minimize fluid manipulations and risks for operational errors. Third, dilution of amplicon and ligase deactivation steps performed in CLIA-OLA were eliminated. Last, multi-step EIA detection was replaced by rapid lateral flow detection with visual readout that streamlines the protocol and eliminates use of a plate washer and reader. Each component of the OLA-Simple kit was also designed for large-scale manufacturing. The kit did not include sample preparation, which allows testing of different specimen types and we show that OLA-Simple performs well on plasma, PBMC and DBS. Currently OLA-Simple maintains separate ligation reactions for each DRM, but non-overlapping probes could be multiplexed. Three multiplexed ligation reactions could be used to detect all six DRMs (K65R/Y181C, K103N/M184V, and V106M/I/G190A), but we were limited by the lack of specific hapten/antibody pairs (two per DRM) required to combine multiple DRMs in one detection strip.
OLA-Simple equipment, reagent and personnel costs are less than other existing HIVDR assays; OLA-Simple costs include a thermal cycler, a mini-centrifuge, a vortex mixer and an office scanner; $8.03 for DNA reagents ($1.12 lyophilized PCR, $1.65 lyophilized ligation, and $5.26 lateral flow devices) or $15.53 for RNA assay reagents ($8.03 DNA reagents and $6.00 for lyophilized RT reagent) and <10 min hands-on time. The total assay time excluding sample preparation has <3.5-h wait-time, mostly for single-round PCR and ligation. To further reduce the wait time, we have developed a faster two-stage PCR amplification protocol that detects ≥200 HIV copies in ∼40 min (Supplementary Fig. 9). When paired with rapid detection of virologic failure, submission of extracted RNA and DNA to OLA-Simple would add two-hour wait time to a patient's visit to inform the clinician whether the virologic failure was associated with selection of HIVDR requiring a switch to more appropriate ART, or, if OLA-Simple detects only WT, should consider strategies to enhance ART adherence.
Primer and probe sequences in OLA-Simple were designed to detect key DRMs (K65R and M184V for NRTIs; and K103N, V106M/I, Y181C, and G190A for NNRTIs), previously found in 98.8% of patients failing 1st-line NNRTI/NRTI regimens [26]. OLA-Simple reliably discriminated 15% MUT from WT sequences at each DRM using synthetic DNA fragments. This analytical detection limit is lower than the reported 20–50% MUT detection limits of Sanger sequencing [27,28]. The validation study using clinical specimens across different HIV subtypes showed that OLA-Simple detected MUT variants in 16 specimens with <20% MUT and five specimens with ≥20% MUT that were missed by Sanger sequencing. OLA-Simple had 99.6% specificity and 86 ± 1% sensitivity compared to methods with DRM detection limits of 1% for Illumina MiSeq [29] and 2% for CLIA-OLA [30]. However, the clinically-relevant% MUT cutoff remains open to debate, as treatment outcomes are also associated with other factors such as the number of DRMs, drug classes, drug adherence, and geographical backgrounds [19,31,32]. In this study, when using a 12.5% MUT cutoff for sensitive comparator assays, OLA-Simple had the highest combined concordance of 98 ± 0.6% (SE), with a sensitivity of 98 ± 1.1% (SE) and a specificity of 99 ± 0.5% (SE).
OLA-Simple genotyped efficiently across multiple HIV subtypes, with failures from 0.6% of PCR-amplifications and 2.6% of ligation reactions (IND). Polymorphisms near the ligation site (within the first two bases) have been shown to cause IND results [33,34], but this can be overcome by probe modifications to accommodate polymorphisms around the ligation site [35]. Regardless, the IND rate of OLA-Simple using universal probes tested on multiple HIV subtypes was similar to the 2.1% IND rate of CLIA-OLA using subtype-specific probes tested on corresponding HIV subtypes. It is worth noting that only 7/24 IND results were concordant between OLA-Simple and CLIA-OLA, which may be due to two factors. First, different probe sequences were used. Second, different IND classification methods were used. CLIA-OLA used a quantitative approach (EIA), and IND results were classified based on the mutant and wild-type signal (see details in Materials and Methods). On the other hand, OLA-Simple used lateral flow detection, and IND results were qualitatively classified by the mode of visual calls from multiple observers. 7/17 discordant INDs resulted from disputed user interpretation of OLA-Simple lateral flow strips, while 7/7 concordant INDs resulted from 100% agreement among users. In feasibility testing using non-clinical specimens, we found that all IND results determined by users were eliminated by software analysis (Fig. 5(g)). Thus, discordant IND results in clinical specimens should be reduced by software analysis. Moreover, ligation at each DRM is independent, and simultaneous ligation failures for multiple DRMs are rare. Thus, a specimen with IND for all DRMs indicates failure of preceding steps, and is easily differentiated from a single IND (the latter suggests a mismatched probe, which can be revised). The failure rate in OLA-Simple is lower than commercially-available HIVDR tests; FDA-approved ViroSeq™ designed for subtype B and the US CDC/ATTC HIV-1 drug resistance test kits had reported failure rates of 12–61% and 6–37%, respectively, when analyzing non-subtype B specimens [[36], [37]–38]. These failures of PCR or sequencing primers can occur due to subtype diversity. Allele-specific PCR on HIV subtype C was reported to have a similar indeterminate rate compared to the OLA-Simple applied across subtypes A, B, C, D, and AE [39].
OLA-Simple, like other point mutation assays, can only detect DRMs at pre-determined locations. Thus, point mutation assays will not replace consensus sequencing for population surveillance studies and identifying new DRMs. However, the association between DRMs and phenotypic HIVDR is well-studied and continuously-documented in a publicly-available database [40]. The DRMs assessed by CLIA-OLA have been widely-validated in patients to be predictive of phenotypic HIVDR, and probe sets have been developed and validated to detect DRMs for protease inhibitors and other NRTIs [19,22,30,34]. Moreover, in response to new WHO recommendations we have designed probe sequences for the integrase inhibitor dolutegravir. Once these probes are validated in clinical specimens, an updated version of the OLA-Simple kit can be manufactured.
OLA-Simple takes advantage of software to (i) consistently interpret results on lateral flow strips and (ii) guide users through the assay steps. Like other lateral flow tests, individual errors can occur during visual reading and interpretation of results. To address this issue, we developed and employed software to enhance result accuracy and interpretation. Automatic-calling software increased accuracy for MUT, WT, and IND classifications of the OLA-Simple results. Using interactive protocol guidance provided by Aquarium, first-time users were able to process blinded specimens and classify DRMs with 97% concordance. Aquarium has several key features that enables training on the spot, including dynamically-generated diagrams with sample identification. These features mitigate the need for domain knowledge to perform the OLA-Simple. Aquarium also forces step-wise execution of the protocols so that users cannot “lose” their place in the protocol; and by automatically collecting data in real-time, improves transparency in assay performance. Moreover, Aquarium tracks the consumption of reagent kit and therefore can help address stockout issues caused by untimely re-stocking of supplies and reagents. During our study, the refrigerator space was very limited, so we relied on Aquarium to track available kits and alert us when the supply reached a low amount and triggered re-stocking the refrigerator.
Limitations of this study include that: HIVDR testing was conducted in laboratories in the USA using archived leftover specimens, and we recognize that instructions and kits may need to be adjusted for field testing in laboratories in LRS. The probes' sequences performed well on this retrospective study, but the outcome of the tests may be different for currently-circulating HIV variants. While this study did not include specimens from a European cohort or Central Africa (subtype G and AG recombinants) due to lack of available clinical specimens for testing, the probes were designed to perform across subtypes and we expect them to perform similarly on these variants. The current workflow involves transferring amplicon during the ligation and detection steps, thus carry-over amplicon contamination is possible; however, to limit carry-over contamination the software instructs a one-way workflow from the pre- to post-PCR area, and if users need to return from post- to pre-PCR, advises users on surface decontamination and to change gloves. The dry reagents were shown to have a shelf-life of at least 4 months under refrigeration (9 months for dried PCR, 4 months for dried ligation and gold nanoparticles); however, room temperature storage and/or longer storage may be possible but requires further testing. Finally, reagent and instrument costs were based on costs to US universities; these costs may differ when manufactured in other countries.
In summary, OLA-Simple addresses challenges in implementing HIVDR testing in LRS by providing results within several hours, using low-cost reagents and equipment, and eliminating the need for extensive training. Our future plans include expanding implementation studies of OLA-Simple at international sites as well as containing the amplicon within a closed system.
Data statement
All data associated with this study are available in the main text and Supplementary Information.
Funding sources
US National Institutes of Health R01 (AI110375) and the Clinical and Retrovirology Research Core and the Molecular Profiling and Computational Biology Core of the UW CFAR (P30 AI027757) funded the study; Feasibility testing was funded by the Office of Science-Industry Partnerships at Seattle Children's Research Institute, 2018 UW Holloman Innovation Challenge Award, and BRL's Pilcher Faculty Fellowship. NP thanks 2016 Ambulatory Practice for the Future Award. NH, DM, AW, and JL thank UW Mary Gates Undergraduate Research Scholarship (2014-16). The funders of the study did not have a role in study design, data collection, data analysis, data interpretation, or writing.
Declaration of Competing Interest
U.S. Provisional Patent Application (62/672,882) was filed by the University of Washington and Seattle Children's Research Institute.
Ethic statement
All specimens were the remaining from previous studies and were transferred to Seattle with proper material transfer documents. We received picture consent from the volunteer who conducted OLA-Simple.
Acknowledgments
We thank Syamal Raychauduri, Dindo Reyes, Kim Polizzi, and Jessica Price at InBios International, Inc. for producing the lateral flow tests used in the validation study; and Aarthy Vallur and Kathryn Hjerrild of InBios International, Inc. for providing feedback on OLA-Simple prototypes. We thank Ian Andrews and Annapurni Sriram for stimulating discussion and technical support. We thank Prof. Patrick Stayton, Robert Britschgi, Selvi Srinivasan, and Rick Edmark for their training, assistance, and access to the lyophilizer; Cameron Paxton Cordray for recruiting and scheduling Aquarium experiments during the feasibility testing; seventy-two volunteers for their participation in the online visual call survey, and forty-one volunteers for feasibility testing.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.002.
Contributor Information
Lisa M. Frenkel, Email: lfrenkel@uw.edu.
Barry R. Lutz, Email: blutz@uw.edu.
Appendix. Supplementary materials
References
- 1.Gupta R.K., Gregson J., Parkin N., Haile-Selassie H., Tanuri A., Andrade Forero L. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis. 2018;18(3):346–355. doi: 10.1016/S1473-3099(17)30702-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverman R.A., Beck I.A., Kiptinness C., Levine M., Milne R., McGrath C.J. Prevalence of pre-antiretroviral-treatment drug resistance by gender, age, and other factors in HIV-Infected individuals initiating therapy in Kenya, 2013–2014. J Infect Dis. 2017;216(12):1569–1578. doi: 10.1093/infdis/jix544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luz P.M., Morris B.L., Grinsztejn B., Freedberg K.A., Veloso V.G., Walensky R.P. Cost-Effectiveness of genotype testing for primary resistance in brazil. J Acquir Immune Defic Syndr. 2015;68(2) doi: 10.1097/QAI.0000000000000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinstein M.C., Goldie S.J., Losina E., Cohen C.J., Baxter J.D., Zhang H. Use of genotypic resistance testing to guide HIV therapy: clinical impact and cost-effectiveness. Ann Interal Med. 2001;134(6):440. doi: 10.7326/0003-4819-134-6-200103200-00008. [DOI] [PubMed] [Google Scholar]
- 5.Saag M.S., Benson C.A., Gandhi R.T., Hoy J.F., Landovitz R.J., Mugavero M.J. Antiretroviral drugs for treatment and prevention of HIV infection in adults. JAMA. 2018;320(4) doi: 10.1001/jama.2018.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips A., Cambiano V., Nakagawa F., Mabugu T., Miners A., Ford D. Cost-effectiveness of HIV drug resistance testing to inform switching to second line antiretroviral therapy in low income settings. PLoS ONE. 2014;9(10) doi: 10.1371/journal.pone.0109148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levison J.H., Wood R., Scott C.A., Ciaranello A.L., Martinson N.A., Rusu C. The clinical and economic impact of genotype testing at first-line antiretroviral therapy failure for HIV-infected patients in South Africa. Clin Infect Dis. 2013;56(4):587–597. doi: 10.1093/cid/cis887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessells R.J., Stott K.E., Manasa J., Naidu K.K., Skingsley A., Rossouw T. Implementing antiretroviral resistance testing in a primary health care hiv treatment programme in rural Kwazulu-Natal, South Africa: early experiences, achievements and challenges. BMC Health Serv Res. 2014;14:116. doi: 10.1186/1472-6963-14-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inzaule S.C., Ondoa P., Peter T., Mugyenyi P.N., Stevens W.S., de Wit T.F.R. Affordable hiv drug-resistance testing for monitoring of antiretroviral therapy in sub-Saharan africa. Lancet Infect Dis. 2016;16(11):e267–ee75. doi: 10.1016/S1473-3099(16)30118-9. [DOI] [PubMed] [Google Scholar]
- 10.Duarte H.A., Panpradist N., Beck I.A., Lutz B., Lai J., Kanthula R.M. Current status of point-of-care testing for human immunodeficiency virus drug resistance. J Infect Dis. 2017;216(suppl_9):S824–S8S8. doi: 10.1093/infdis/jix413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson RM S.C., Quiñones-Mateu M.E. Next-Generation sequencing to help monitor patients infected with HIV: ready for clinical use? Curr Infect Dis Rep. 2014;16(401) doi: 10.1007/s11908-014-0401-5. [DOI] [PubMed] [Google Scholar]
- 12.Frenkel L.M., Wagner L.E., Atwood S.M., Cummins T.J., Dewhurst S. Specific, sensitive, and rapid assay for human immunodeficiency virus type 1 pol mutations associated with resistance to zidovudine and didanosine. J Clin Microbiol. 1995;33(2):342–347. doi: 10.1128/jcm.33.2.342-347.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner T.A., Kress C.M., Beck I., Techapornroong M., Wittayapraparat P., Tansuphasawasdikul S. Detection of HIV-1 drug resistance in women following administration of a single dose of nevirapine: comparison of plasma RNA to cellular DNA by consensus sequencing and by oligonucleotide ligation assay. J Clin Microbiol. 2010;48(5):1555–1561. doi: 10.1128/JCM.02062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Dyke R.B., Ngo-Giang-Huong N., Shapiro D.E., Frenkel L., Britto P., Roongpisuthipong A. A comparison of 3 regimens to prevent nevirapine resistance mutations in HIV-Infected pregnant women receiving a single intrapartum dose of nevirapine. Clin Infect Diseas Offic Publ Infect Dis Soc Am. 2012;54(2):285–293. doi: 10.1093/cid/cir798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duarte H.A., Beck I.A., Levine M., Kiptinness C., Kingoo J.M., Chohan B. Implementation of a point mutation assay for HIV drug resistance testing in Kenya. Aids. 2018 doi: 10.1097/QAD.0000000000001934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller B., Vrana J., Miller A., Newman G., Klavins E. Aquarium: the laboratory operating system (Version v2.5.0) Zenodo. 2019 http://doiorg/105281/zenodo2535715 [Google Scholar]
- 17.Micek Mark A., Blanco Ana J., Beck Ingrid A., Dross S., Matunha L., Montoya P. Nevirapine resistance by timing of hiv type 1 infection in infants treated with single‐dose nevirapine. Clin Infect Dis. 2010;50(10):1405–1414. doi: 10.1086/652151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu T.F., Shafer R.W. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42(11):1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne R.S., Silverman R.A., Beck I.A., Mckernan-Mullin J., Deng W., Sibley T.R. Minority and majority pre-treatment HIV-1 drug resistance associated with failure of 1st-line NNRTI Art in Kenyan women. AIDS. 2019;6(33):941–951. doi: 10.1097/QAD.0000000000002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soria J., Mugruza R., Levine M., Leon S.R., Arevalo J., Ticona E. Pretreatment hiv drug resistance and virologic outcomes to first-line antiretroviral therapy in Peru. AIDS Res Hum Retroviruses. 2019 doi: 10.1089/aid.2018.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ngo-Giang-Huong N., Jourdain G., Amzal B., Sang-a-gad P., Lertkoonalak R., Eiamsirikit N. Resistance patterns selected by nevirapine vs. efavirenz in HIV-infected patients failing first-line antiretroviral treatment: a bayesian analysis. PLoS ONE. 2011;6(11):e27427. doi: 10.1371/journal.pone.0027427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck I.A., Deng W., Payant R., Hall R., Bumgarner R.E., Mullins J.I. Validation of an oligonucleotide ligation assay for quantification of human immunodeficiency virus type 1 drug-resistant mutants by use of massively parallel sequencing. J Clin Microbiol. 2014;52(7):2320–2327. doi: 10.1128/JCM.00306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanthula R., Rossouw T.M., Feucht U.D., van Dyk G., Beck I.A., Silverman R. Persistence of hiv drug resistance among south african children given nevirapine to prevent mother-to-child-transmission. AIDS. 2017;31(8):1143–1148. doi: 10.1097/QAD.0000000000001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm S. A simple sequentially rejective multple test procedure. Scand J Stat. 1979;6(2):65–70. [Google Scholar]
- 25.Toubanaki D.K., Christopoulos T.K., Ioannou P.C., Flordellis C.S. Identification of single-nucleotide polymorphisms by the oligonucleotide ligation reaction: a dna biosensor for simultaneous visual detection of both alleles. Anal Chem. 2009;81:218–224. doi: 10.1021/ac801870x. [DOI] [PubMed] [Google Scholar]
- 26.Rhee S.Y., Jordan M.R., Raizes E., Chua A., Parkin N., Kantor R. HIV-1 drug resistance mutations: potential applications for point-of-care genotypic resistance testing. PLoS ONE. 2015;10(12) doi: 10.1371/journal.pone.0145772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunthard H.F., Wong J.K., Ignacio C.C., Havlir D.V., Richman D.D. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of hiv type 1 pol from clinical samples. AIDS Res Hum Retroviruses. 1998;14(10):869–876. doi: 10.1089/aid.1998.14.869. [DOI] [PubMed] [Google Scholar]
- 28.Schuurman R., Demeter L., Reichelderfer P., Tijnagel J., de Groot T., Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 1999;37(7):2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casadella M., Paredes R. Deep sequencing for HIV-1 clinical management. Virus Res. 2017;239:69–81. doi: 10.1016/j.virusres.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 30.Chung M.H., Beck I.A., Dross S., Tapia K., Kiarie J.N., Richardson B.A. Oligonucleotide ligation assay detects hiv drug resistance associated with virologic failure among antiretroviral-naïve adults in Kenya. J Acquir Immune Defic Syndr. 2014;67(3):246–253. doi: 10.1097/QAI.0000000000000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clutter D.S., Zhou S., Varghese V., Rhee S.Y., Pinsky B.A., Jeffrey Fessel W. Prevalence of drug-resistant minority variants in untreated HIV-1-Infected individuals with and those without transmitted drug resistance detected by sanger sequencing. J Infect Dis. 2017;216(3):387–391. doi: 10.1093/infdis/jix338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stella-Ascariz N., Arribas J.R., Paredes R., Li J.Z. The role of HIV-1 drug-resistant minority variants in treatment failure. J Infect Dis. 2017;216(suppl_9):S847–SS50. doi: 10.1093/infdis/jix430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck I.A., Crowell C., Kittoe R., Bredell H., Machaba M., Willamson C. Optimization of the oligonucleotide ligation assay, a rapid and inexpensive test for detection of HIV-1 drug resistance mutations, for non-North American variants. J Acquir Immune Defic Syndr. 2008;48(4):418–427. doi: 10.1097/QAI.0b013e31817ed7d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck I.A., Mahalanabis M., Pepper G., Wright A., Hamilton S., Langston E. Rapid and sensitive oligonucleotide ligation assay for detection of mutations in human immunodeficiency virus type 1 associated with high-level resistance to protease inhibitors. J Clin Microbiol. 2002;40(4):1413–1419. doi: 10.1128/JCM.40.4.1413-1419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mutsvangwa J., Beck I.A., Gwanzura L., Manhanzva M.T., Stranix-Chibanda L., Chipato T. Optimization of the oligonucleotide ligation assay for the detection of nevirapine resistance mutations in Zimbabwean human immunodeficiency virus type-1 subtype c. J Virol Methods. 2014;210:36–39. doi: 10.1016/j.jviromet.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackard J., Rosemary A., Chika O., Jonathan O., Godwin I., Georgina O. Genotyping performance evaluation of commercially available HIV-1 drug resistance test. PLoS ONE. 2018;13(6) doi: 10.1371/journal.pone.0198246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z., Wagar N., DeVos J.R., Rottinghaus E., Diallo K., Nguyen D.B. Optimization of a low cost and broadly sensitive genotyping assay for HIV-1 drug resistance surveillance and monitoring in resource-limited settings. PLoS ONE. 2011;6(11):e28184. doi: 10.1371/journal.pone.0028184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mracna M., Becker-Pergola G., Dileanis J., Guay L.A., Cunningham S., Jackson J.B. Performance of applied biosystems viroseq HIV-1 genotyping system for sequence-based analysis of non-subtype b human immunodeficiency virus type 1 from Uganda. J Clin Microbiol. 2001;39(12):4323–4327. doi: 10.1128/JCM.39.12.4323-4327.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang G., Cai F., Zhou Z., DeVos J., Wagar N., Diallo K. Simultaneous detection of major drug resistance mutations in the protease and reverse transcriptase genes for HIV-1 subtype c by use of a multiplex allele-specific assay. J Clin Microbiol. 2013;51(11):3666–3674. doi: 10.1128/JCM.01669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shafer R.W. Human immunodeficiency virus type 1 drug resistance mutations update. J Infect Dis. 2017;216(suppl_9):S843–S8S6. doi: 10.1093/infdis/jix398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.