Abstract
Imatinib mesylate (IM) is the first developed protein kinase inhibitor and recently it has topped consumption rates among targeted and total anticancer drugs. Although there are indications that IM possesses cyto/genotoxic activities against normal non-target cells as well, there is a lack of information regarding the underlying mechanism involved in those actions. Therefore, we aimed to evaluate the response of human circulating blood cells towards oxidative stress after IM treatment (0.0001–10 µg/mL) in vitro. Based on the results, IM had an influence on all of the oxidative stress parameters tested. Lower concentrations of IM induced an increase of glutathione level, following its decrease at higher IM concentrations indicating impairment in oxidative stress defences. Concomitant to a glutathione decrease, an increase of malondialdehyde and protein carbonyls level was observed indicating oxidative damage of lipids and proteins. The observed effects overlapped with the observed formation of oxidative base damage detected by formamidopyrimidine-DNA glycosylase modified-comet assay indicating that IM managed to induce oxidative DNA damage. Our results provide novelty in their mechanistic approach to IM-induced toxicity in non-target cells and suggest that IM can affect blood cells and induce oxidative stress.
Keywords: Imatinib mesylate, Human peripheral blood cells, Non-target cells, Oxidative stress, Oxidative DNA damage
1. Introduction
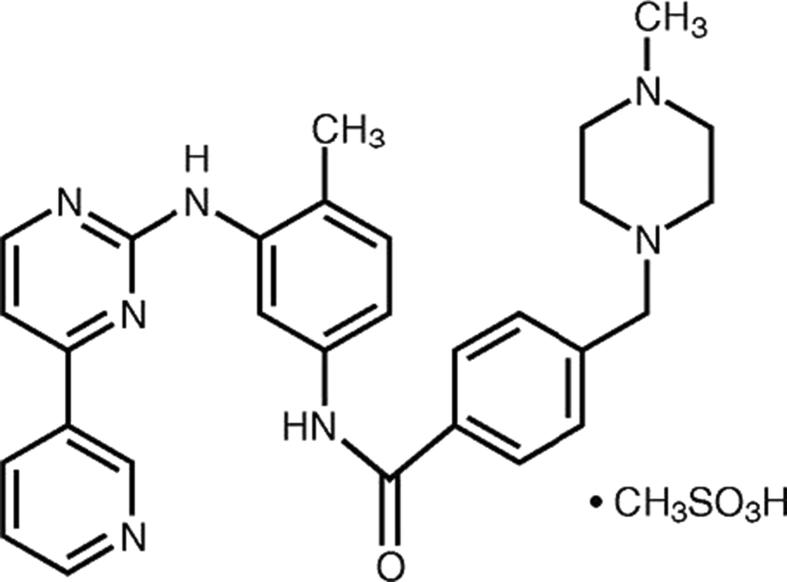
Imatinib mesylate (IM; C29H31N7O•CH3SO3H) (Fig. 1), also known by its brand name Gleevec/Glivec, is a chemotherapy medication used to treat cancer. IM is considered a pioneer protein kinase inhibitor developed for targeted cancer chemotherapy and is specifically used for the treatment of chronic myeloid leukaemia (CML) and acute lymphocytic leukaemia (ALL) that is Philadelphia chromosome-positive (Ph+). It is also used for treating certain types of gastrointestinal stromal tumours (GIST), systemic mastocytosis as well as for myelodysplastic syndrome. The target of protein kinase inhibitors is not DNA, so due to their high specificity for tumour cells they are regarded less toxic to non-target cells and a promising approach in cancer therapy (Al-Hadiya et al., 2014, Arora and Scholar, 2005, Chen and Kesselheim, 2017, Claudiani and Apperley, 2018, Hartmann et al., 2009, Longo, 2017, Manley et al., 2002, Viegas et al., 2017).
Fig 1.
Chemical structure of imatinib mesylate.
IM is a dihydrophenylaminopirimidine derivative that targets BCR-ABL kinase. Although IM is an effective BCR-ABL inhibitor, it can affect the catalytic activity of platelet-derived growth factor and c-KIT receptors. IM binds to the inactive conformation of BCR-ABL fusion protein and prevents dephosphorylation of ATP. Subsequently, the conformational changes needed for the oncoprotein’s kinase activity are suppressed. Inhibition of BCR-ABL catalytic activity initiates the proapoptotic signalling pathways and death of Ph+ leukemic cells. Nevertheless, CML stem cells are not as sensitive towards an IM-induced apoptotic effect, despite BCR-ABL activity being inhibited (Barnes and Melo, 2003, Buchdunger et al., 2000, Corbin et al., 2011, Deininger et al., 2005, Deininger et al., 1997, Goldman and Melo, 2003, Stagno et al., 2016).
Generally, IM has low rates of severe non-haematological side effects and tolerable haematological toxicity (Fabarius et al., 2007b, Wolf and Rumpold, 2009). However, recent studies on patients receiving IM revealed cytogenetic abnormalities in haematopoietic cells negative for Philadelphia chromosome (Ph–) (Wolf and Rumpold, 2009). Even though IM showed minimal toxicity and a good safety profile (Angstreich et al., 2004), still up to 8% of patients treated with IM stop their treatment due to IM’s cytotoxicity (Kulkarni et al., 2012, Smith, 2011).
Previously we performed an evaluation of IM genotoxicity in cultured human and fish cells (Novak et al., 2017). Assessing cyto/genotoxicity in vitro using human peripheral blood lymphocytes (HPBLs), human hepatoma (HepG2) cells, and zebrafish liver (ZFL) cells, we observed IM’s potential to induce cytogenetic damage. Genotoxicity was evaluated with the micronucleus test and comet assay. Results showed that ZFL cells and HPBLs were similarly sensitive to IM cytotoxicity, though HepG2 cells proved to be less sensitive. IM managed to induce primary DNA damage in HepG2 and ZFL cells at non-cytotoxic concentrations, while this was not the case for HPBLs. Higher micronuclei (MNi) frequency was detected in HPBLs and ZFL cells. Moreover, IM-treated HPBLs also had a higher frequency of nuclear buds (NBUDs) and nucleoplasmic bridges (NPBs), indicating that it has the potential to induce genome damage in normal non-target cells as well. What remained unclear are the possible mechanisms of action (MoA) responsible for its toxic effects.
Therefore, the present study aimed to investigate the response of IM-treated human peripheral blood cells (HPBCs) toward oxidative stress induction as one of the possible MoA of IM-induced cyto/genotoxicity in vitro. This was done by using concentrations relevant for therapeutic as well as occupational and environmental exposure (Novak et al., 2017, Parrillo-Campiglia et al., 2009). The contribution of oxidative stress in IM’s cytotoxicity and genotoxicity was evaluated by determining the antioxidant defence system, the lipid peroxidation (LPO), and the concentration of oxidised proteins (OXP), by using glutathione (GSH), malondialdehyde (MDA), and protein carbonyls’ (PC) as biomarkers (Augustyniak et al., 2015, Ayala et al., 2014, Domijan et al., 2012, Frijhoff et al., 2015). Moreover, modified comet assay protocol employing formamidopyrimidine-DNA glycosylase (Fpg)-enzyme was also used for the oxidative DNA damage detection (Collins, 2014, Gajski et al., 2019b, Gajski et al., 2019a). The comet assay (in vitro) is suggested as an alternative test to cytogenetic assays for screening the early genotoxicity of drug candidates (Dinter et al., 2015, Witte et al., 2007). Furthermore, the study brings up new findings on the IM impact on oxidative stress markers, which will clarify the MoA of IM-induced cyto/genotoxicity in normal human non-target cells.
2. Materials and methods
2.1. Chemicals and reagents
Imatinib mesylate (IM; CAS No 220127-57-1, MW 589.71 g/mol) was from Santa Cruz Biotechnology (Dallas, US). Fpg FLARE™ Assay Kit was from Trevigen Inc. (Gaithersburg, US); potassium dihydrogen phosphate (KH2PO4) was from Merck (Darmstadt, Germany); heparinised vacutainer tubes were from Becton Dickinson (Franklin Lakes, US); 1,1,3,3-tetramethoxy propane (TMP), 5,5′-ditiobis-2-nitrobenzoat (DTNB), ethidium-bromide (EtBr), histopaque, low melting point (LMP) agarose, and thiobarbituric acid (TBA) were from Sigma (St Louis, US). All other reagents were laboratory-grade chemicals from Kemika (Zagreb, Croatia).
2.2. Blood sampling and treatment procedure
Whole blood was taken from the antecubital vein in heparinised vacutainer tubes from two young male healthy donors ≤35 years of age (28 and 35 years of age), non-smokers, with no medical record or acute health conditions. According to a filled-in questionnaire, they had not been exposed to chemicals or diagnostic radiation that could confound the results of biomarkers assessed for at least six months. The Ethics Committee approved the project of which this study was part and the donors signed an informed consent before taking part in the study. The whole blood was exposed to IM at various concentrations (0, 0.0001, 0.001, 0.01, 0.1, 1 and 10 µg/ml) for 4 and 24 h and was used for the determination of oxidative stress responses. Each experiment was repeated at least two times according to the standard protocols listed below.
2.3. Glutathione (GSH) assay
The level of GSH in whole blood samples after IM (0.0001, 0.001, 0.01, 0.1, 1 and 10 µg/ml) treatment was determined using Ellman's reagent (Ellman, 1958). Blood samples (800 μL) were treated with an equal volume of 5% TCA to precipitate proteins and were then centrifuged for 10 min at 3500 g. After centrifugation, 500 μL of supernatant was mixed with 300 μL phosphate buffer (pH 7.4) and 50 μL DTNB (1 mM). Absorbance was measured spectrophotometrically (T70, PG Instruments Ltd., Leicestershire, UK) at 412 nm. The concentrations of GSH in the samples were calculated according to the molar absorption coefficient that was re-evaluated and calculated to be 14.15 × 103 M−1 cm−1 at 412 nm and 25 °C (Eyer et al., 2003). The results are expressed as µM.
2.4. Lipid peroxidation (LPO) assay
The level of LPO was determined by the MDA concentration in whole blood samples after IM (0.0001, 0.001, 0.01, 0.1, 1 and 10 µg/ml) treatment as described previously in detail (Domijan et al., 2015) with minor modifications. Briefly, 400 μL H3PO4 and 100 μL TBA were added to 50 μL of diluted blood sample (20×) or standard (TMP was employed as MDA standard). Samples were mixed and incubated for 30 min at 90 °C and subsequently analysed using high performance liquid chromatography (HPLC) on an HPLC (Knauer, Berlin, Germany) with an fluorescent detector (Hitachi Merck). The mobile phase consisted of 50 mM KH2PO4 and methanol, and the flow rate was set at 1 mL/min. Separation was performed on an analytical column C-18 reverse-phase (LiChrospher, Merck) with 5 μm particles (125 × 4 mm) and the fluorescent detector wavelengths were set at λex 527 nm and λem 551 nm. The MDA levels in the tested samples were calculated using the calibration curve (r2 = 0.9923) and the results were expressed as µM.
2.5. Protein carbonyl (PC) assay
The PC plasma concentration, as a measure of OXP, was determined using 2,4-dinitrophenylhydrazine (2,4-DNPH) after IM (0.0001, 0.001, 0.01, 0.1, 1 and 10 µg/ml) treatment as described in details previously (Domijan et al., 2007). The samples were analysed in duplicate and absorbance was read at 370 nm on a UV–Vis spectrophotometer (T70, PG Instruments Ltd.). The concentration of PC in the samples was calculated using a molar absorption coefficient of ε = 22 × 103 M−1 cm−1. The results are expressed as µM.
2.6. Oxidative DNA damage (Fpg-modified comet) assay
The analysis of the formation of oxidative base damage was done using a modified Fpg FLARE™ Assay Kit (Trevigen Inc.) with minor modifications as described in detail previously (Gajski et al., 2016, Gajski et al., 2008). Briefly, after the IM treatment, the whole blood was embedded in an agarose matrix and lysed at 4 °C. Next, the slides were treated with either 100 μL of Fpg-enzyme or buffer only (control) for 30 min at 37 °C, placed into alkaline solution (300 mM NaOH, 1 mM Na2EDTA, pH 12.1) for 40 min and electrophoresed for 20 min at 1 V/cm. Lastly, the slides were neutralised, stained with EtBr (10 μg/ml) and the comets were analysed at ×250 magnification by an epifluorescence microscope (Zeiss, Göttingen, Germany) linked to an image analysis system (Comet Assay II; Perceptive Instruments Ltd., Haverhill, Suffolk, UK). The level of oxidative DNA damage was expressed as percentage of DNA in the tail with a total of 100 nuclei scored from each slide.
2.7. Statistical evaluation
Analyses were performed using GraphPad Software (La Jolla, USA) and STATISTICA 13.2 (Dell Inc., Round Rock, USA). Descriptive statistics was used to assess the basic statistical parameters. The difference in oxidative stress parameters between the control and exposed samples was determined by Student t-test. The comet assay results were logarithmically transformed to equalise the variances and normalise the distribution. Afterwards, ANOVA multiple comparisons between groups were done with Newman-Keuls post hoc analysis. Statistical significance was set at the level of P < 0.05.
3. Results
3.1. Changes in glutathione (GSH) level
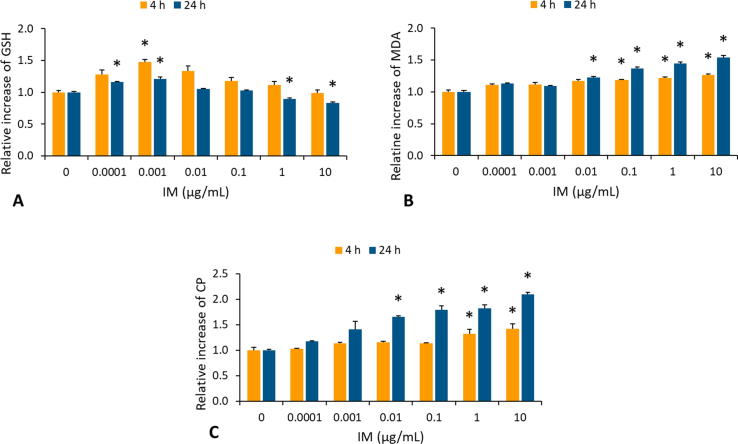
GSH level was monitored in whole blood samples after IM treatment (Fig. 2A). The results showed that treatment with lower IM concentrations (0.0001 and 0.001 µg/mL) induced an increase of GSH level, after 4 and 24 h treatment. On the contrary, higher IM concentrations resulted in a statistically significant (P < 0.05) decrease of GSH level at ≥ 1 µg/mL after 24 h exposure.
Fig 2.
The level of oxidative stress markers, glutathione (GSH) in whole blood samples (A), malondialdehyde (MDA) used as an index of lipid peroxidation (LPO) of membranes in whole blood samples (B) and protein carbonyls (PC) used as an index of oxidised proteins (OXP) in plasma samples (C) after exposure to imatinib mesylate (IM) at different concentrations (0, 0.001, 0.01, 0.1, 1 and 10 mg/mL) for 4 and 24 h evaluated using either high-performance liquid chromatography (HPLC) or spectrophotometry. A significant difference was determined with Student t-test (*P < 0.05). Data on the figures are presented as the mean values of the increase/decrease in GSH, MDA and PC in treated samples over the background level in the control samples.
3.2. Lipid peroxidation (LPO) induction
To further assess the extent of oxidative stress in IM toxicity, whole blood MDA level was assessed in treated samples (Fig. 2B). After 4 and 24 h of exposure, a statistically significant (P < 0.05) increase of MDA was detected at ≥0.1 and ≥0.01 µg/ml, respectively.
3.3. Induction of oxidised proteins (OXP)
Moreover, the plasma PC concentration (as a marker of OXP) was assessed in IM-treated samples (Fig. 2C). After 4 and 24 h of exposure, a statistically significant (P < 0.05) increase in PC was noticed at ≥1 and ≥0.01 µg/ml, respectively.
3.4. Induction of oxidative DNA damage
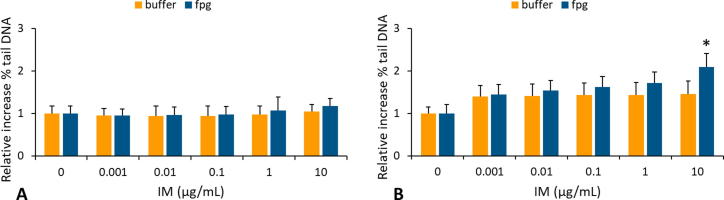
The Fpg enzyme which excises oxidised pyrimidines and purines, was used as the comet assay modification so the formation of oxidative DNA adducts in HPBCs was assessed (Fig. 3A and B). A significant change (P < 0.05) in oxidised DNA damage in HPBCs was detected only after a longer exposure period at ≥10 µg/mL (Fig. 3B).
Fig 3.
Induction of oxidative DNA damage in human peripheral blood cells (HPBCs), after exposure to imatinib mesylate (IM) at different concentrations (0, 0.001, 0.01, 0.1, 1 and 10 mg/mL) for 4 (A) and 24 (B) h evaluated by the Fpg-modified comet assay. The % of tail DNA was determined in 100 nuclei per slide. Significant difference was determined with Newman-Keuls post-test (*P < 0.05) between Fpg-digested and non-digested samples. Data on the figures are presented as the mean values of the increase in % DNA in comet tail over the background level in control cells.
4. Discussion
After we noticed in our previous study (Novak et al., 2017) that IM is capable of inducing genome damage in normal human non-target cells, we designed this study to evaluate IM-treated oxidative stress in human circulating blood cells as an underlying mechanism of its genotoxicity. Cohen et al. (2002) reported several preclinical toxicological results that were needed to approve IM use. Previously conducted studies showed that IM is not mutagenic using mouse lymphoma assay or the Ames test. Similarly, in the in vivo rat bone marrow micronucleus (MN) assay, IM did not increase MN frequency. However, in the presence of metabolic activation and at the highest concentration tested, IM was clastogenic in the Chinese hamster ovary cell assay. The potential toxicity might also be attributed to impurities of the drug that arise from manufacturing intermediates. Both intermediates were mutagenic using the Ames test and one showed genotoxic potential in vitro. Furthermore, studies also indicated that IM managed to induce DNA damage in crustaceans along with the inhibition of their reproduction. The reproduction inhibition was also observed in cyanobacteria and algae, while in higher plants, it increased MNi frequency and reduced fertility (Brezovšek et al., 2014, Cohen et al., 2002, Lutterbeck et al., 2015, Mišík et al., 2016, Novak et al., 2017, Parrella et al., 2015, Parrella et al., 2014, Pichler et al., 2014, Viegas et al., 2017).
Based on our previous results using HPBLs, IM exposure did not induce primary DNA damage assessed using the comet assay. However, IM induced genomic instability as evaluated by the CBMN assay (Novak et al., 2017). When tested in combination with 5-fluorouracil and etoposide in HPBLs in vitro, the selected mixture induced genome and cell damage at environmentally relevant concentrations. The changes in the oxidative stress parameters tested suggested impact of ROS in the cytotoxicity and genotoxicity of the IM-containing mixture (Gajski et al., 2016).
According to the results from this study, IM influenced all of the oxidative stress parameters tested. To be more precise, lower concentrations of IM induced an increase of GSH level, following its decrease at higher IM concentrations indicating impairment in oxidative stress defences. Parallel to a GSH decrease, an increase of MDA and PC level was observed indicating oxidative damage of lipids and proteins in the tested cells.
To assess the role of oxidative DNA damage in IM genotoxicity, we used the Fpg enzyme. This lesion-specific endonuclease can recognize specific oxidatively damaged DNA bases and generate additional breaks to aid in the detection of oxidative DNA damage in a modified comet assay (Davison, 2016, Schalow et al., 2011, Smith et al., 2006). Fpg catalyses the excision of a lethal lesion; 8-oxoguanine, a highly mutagenic lesion and probably the most important biological substrate of Fpg; open ring forms of 7-methylguanine, including 2,6-diamino-4-hydroxy-5-N-methylformamido-pyrimidine, and 4,6-diamino-5-formamidopyimidine, 5-hydroxycytosine and 5-hydroxyuracil; imidazole ring opened N-2-aminofluorene-C8-guanine, and the aflatoxin-bound imidazole-ring-opened guanine (Krokan et al., 1997). The removal of specifically oxidized DNA bases by Fpg enzyme leads to the formation of apurinic-apyrimidinic (AP) sites. Such DNA aberrations are then cleaved by AP-lyases creating a gap in the DNA strand. This way, the modified comet assay is able to detect those oxidation-induced gaps (Collins, 2014). Based on our results, the oxidative changes observed on the protein and lipid level coincided with the formation of oxidative damage of DNA bases.
The BCR/ABL positive human leukemic cells were susceptible to IM-induced DNA damage, assessed by the comet assay, unlike normal human lymphocytes and BCR/ABL negative leukemic cells (Czechowska et al., 2005). The same study concluded that the production of free radicals led to alkylation and oxidation of nucleic bases and formation of DNA damage. The suggested MoA was the indirect ROS-mediated DNA damage that was more probable than the direct formation of DNA strand breaks. Furthermore, Diculescu et al. (2006) investigated the interaction of DNA with the IM in bulk solution and at a dsDNA-electrochemical biosensor using differential pulse voltammetry. They found that IM could bind to dsDNA and that such interaction could induce changes in the dsDNA structure. The oxidative DNA damage and in situ electrochemical generation of IM redox metabolites can be detected using a dsDNA-biosensor. More precisely, the interaction between DNA and IM is more probable at segments with enriched adenine. This results in the generation of 2,8-dihydroxyadenine, one of the most usual IM-induced oxidation products inside the DNA double helix. Based on these results, the authors proposed that IM molecules interact with dsDNA, triggering a distortion of the double helix.
The principal mechanism of IM chemotherapy is the blocking of the ATP binding site of target tyrosine kinases (such as ABL, PDGFR, KIT). Fabarius et al., 2007a, Fabarius et al., 2005) suggested that IM genotoxicity in non-target cells could arise from the inhibition of some normal cells’ targets. The Abl kinase is linked with several stress signalling pathways and is an important protein regulating processes such as apoptosis, DNA repair, and cellular growth arrest (Fanta et al., 2008, Kharbanda et al., 1998). IM-mediated inhibition of Abl could also predispose cells to replication of damaged DNA, which, by interference with DNA repair and inhibition of cell-cycle arrest and apoptosis, can lead to genomic instability. ATP-binding domains are stereochemically conserved among different kinases; hence, inhibitors directed at the ATP binding site are not completely specific for the targeted kinases. IM also inhibits the serine-threonine kinase Raf-1 (Deininger et al., 2005), some other kinases (Karaman et al., 2008), and even non-kinase targets (Bantscheff et al., 2007). Conservation of the domain organisation of tyrosine kinase proteins is relatively high even between distantly related organisms (Shiu and Li, 2004). Therefore, IM may inhibit kinases involved in the induction of genotoxicity.
In conclusion, the results of the present study indicate that IM influenced all of the tested oxidative stress markers. Lower concentrations of IM induced an increase of GSH level, following its decrease at higher IM concentrations indicating impairment in oxidative stress defences after the treatment. At the same time, we observed an increase in PC as well as MDA concentration suggesting the induction of OXP and LPO, respectively. These changes overlapped with the formation of oxidative base damage detected by the Fpg modified-comet assay indicating oxidative DNA lesions-inducing IM potential. Our results provide up-to-date knowledge of IM-induced cyto/genotoxicity MoA. Although the IM can affect cancer cells, it is also potentially toxic to normal non-target blood cells by inducing oxidative changes to lipids, proteins, and DNA. Nevertheless, based on the accumulated evidence, IM has a favourable toxicity and side effect profile with benefits that outweighs the potential risks.
Declaration of Competing Interest
None declared.
Acknowledgements
This study received funding from the Seventh Framework Programme FP7/2007-2013 under grant agreement No. 265264, the Foundation of the Croatian Academy for Science and Arts, the Institute for Medical Research and Occupational Health, and the University of Zagreb, Faculty of Pharmacy and Biochemistry. This study was also supported by the European Cooperation in Science and Technology (CA COST Action CA15132 – hCOMET). The authors would like to thank Ms. Maja Nikolić for excellent technical assistance and Mr. Makso Herman for language editing.
Footnotes
Peer review under responsibility of King Saud University.
References
- Al-Hadiya B.M.H., Bakheit A.H.H., Abd-Elgalil A.A. Imatinib mesylate. Profiles Drug Subst. Excip. Relat. Methodol. 2014;39:265–297. doi: 10.1016/B978-0-12-800173-8.00006-4. [DOI] [PubMed] [Google Scholar]
- Angstreich G.R., Smith B.D., Jones R.J. Treatment options for chronic myeloid leukemia: imatinib versus interferon versus allogeneic transplant. Curr. Opin. Oncol. 2004;16:95–99. doi: 10.1097/00001622-200403000-00002. [DOI] [PubMed] [Google Scholar]
- Arora A., Scholar E.M. Role of tyrosine kinase inhibitors in cancer therapy. J. Pharmacol. Exp. Ther. 2005;315:971–979. doi: 10.1124/jpet.105.084145. [DOI] [PubMed] [Google Scholar]
- Augustyniak E., Adam A., Wojdyla K., Rogowska-Wrzesinska A., Willetts R., Korkmaz A., Atalay M., Weber D., Grune T., Borsa C., Gradinaru D., Chand Bollineni R., Fedorova M., Griffiths H.R. Validation of protein carbonyl measurement: a multi-centre study. Redox Biol. 2015;4:149–157. doi: 10.1016/j.redox.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala A., Muñoz M.F., Argüelles S. Lipid Peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014;2014:1–31. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., Drewes G. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- Barnes D.J., Melo J.V. Management of chronic myeloid leukemia: targets for molecular therapy. Semin. Hematol. 2003;40 doi: 10.1053/shem.2003.50002. ashem0400034. [DOI] [PubMed] [Google Scholar]
- Brezovšek P., Eleršek T., Filipič M. Toxicities of four anti-neoplastic drugs and their binary mixtures tested on the green alga Pseudokirchneriella subcapitata and the cyanobacterium Synechococcus leopoliensis. Water Res. 2014;52:168–177. doi: 10.1016/j.watres.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Buchdunger E., Cioffi C.L., Law N., Stover D., Ohno-Jones S., Druker B.J., Lydon N.B. Abl protein-tyrosine kinase inhibitor STI571 inhibits in vitro signal transduction mediated by c-kit and platelet-derived growth factor receptors. J. Pharmacol. Exp. Ther. 2000;295:139–145. [PubMed] [Google Scholar]
- Chen C.T., Kesselheim A.S. Journey of generic imatinib: a case study in oncology drug pricing. J. Oncol. Pract. 2017;13:352–355. doi: 10.1200/JOP.2016.019737. [DOI] [PubMed] [Google Scholar]
- Claudiani S., Apperley J.F. The argument for using imatinib in CML. Hematol. Am. Soc. Hematol. Educ. Progr. 2018;2018:161–167. doi: 10.1182/asheducation-2018.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M.H., Williams G., Johnson J.R., Duan J., Gobburu J., Rahman A., Benson K., Leighton J., Kim S.K., Wood R., Rothmann M., Chen G.U.K.M., Staten A.M., Pazdur R. Approval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemia. Clin. Cancer Res. 2002;8:935–942. [PubMed] [Google Scholar]
- Collins A.R. Measuring oxidative damage to DNA and its repair with the comet assay. Biochim. Biophys. Acta. 2014;1840:794–800. doi: 10.1016/j.bbagen.2013.04.022. [DOI] [PubMed] [Google Scholar]
- Corbin A.S., Agarwal A., Loriaux M., Cortes J., Deininger M.W., Druker B.J. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J. Clin. Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowska A., Poplawski T., Drzewoski J., Blasiak J. Imatinib (STI571) induces DNA damage in BCR/ABL-expressing leukemic cells but not in normal lymphocytes. Chem. Biol. Interact. 2005;152:139–150. doi: 10.1016/j.cbi.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Davison G.W. Exercise and oxidative damage in nucleoid DNA quantified using single cell gel electrophoresis: present and future application. Front. Physiol. 2016;7:249. doi: 10.3389/fphys.2016.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger M., Buchdunger E., Druker B.J. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- Deininger M.W., Goldman J.M., Lydon N., Melo J.V. The tyrosine kinase inhibitor CGP57148B selectively inhibits the growth of BCR-ABL-positive cells. Blood. 1997;90:3691–3698. [PubMed] [Google Scholar]
- Diculescu V.C., Vivan M., Brett A.M.O. Voltammetric behavior of antileukemia drug glivec. Part III. In situ DNA oxidative damage by the glivec electrochemical metabolite. Electroanalysis. 2006;18:1963–1970. [Google Scholar]
- Dinter D., Gajski G., Domijan A.-M.M.A.-M., Garaj-Vrhovac V. Cytogenetic and oxidative status of human lymphocytes after exposure to clinically relevant concentrations of antimalarial drugs atovaquone and proguanil hydrochloride in vitro. Fundam. Clin. Pharmacol. 2015;29:575–585. doi: 10.1111/fcp.12153. [DOI] [PubMed] [Google Scholar]
- Domijan A.-M., Gajski G., Peraica M., Garaj-Vrhovac V. Evaluation of oxidative status and baseline DNA damage frequency in healthy female volunteers. In: Reyes A.M., Contreras C.D., editors. Handbook on Oxidative Stress New Research. Nova Science Publishers Inc; New York: 2012. pp. 363–380. [Google Scholar]
- Domijan A.-M., Peraica M., Vrdoljak A.L., Radić B., Zlender V., Fuchs R. The involvement of oxidative stress in ochratoxin A and fumonisin B1 toxicity in rats. Mol. Nutr. Food Res. 2007;51:1147–1151. doi: 10.1002/mnfr.200700079. [DOI] [PubMed] [Google Scholar]
- Domijan A.-M.M., Ralić J., Radić Brkanac S., Rumora L., Žanić-Grubišić T. Quantification of malondialdehyde by HPLC-FL – application to various biological samples. Biomed. Chromatogr. 2015;29:41–46. doi: 10.1002/bmc.3361. [DOI] [PubMed] [Google Scholar]
- Ellman G.L. A colorimetric method for determining low concentrations of mercaptans. Arch. Biochem. Biophys. 1958;74:443–450. doi: 10.1016/0003-9861(58)90014-6. [DOI] [PubMed] [Google Scholar]
- Eyer P., Worek F., Kiderlen D., Sinko G., Stuglin A., Simeon-Rudolf V., Reiner E. Molar absorption coefficients for the reduced Ellman reagent: reassessment. Anal. Biochem. 2003;312:224–227. doi: 10.1016/s0003-2697(02)00506-7. [DOI] [PubMed] [Google Scholar]
- Fabarius A., Giehl M., Frank O., Duesberg P., Hochhaus A., Hehlmann R., Seifarth W. Induction of centrosome and chromosome aberrations by imatinib in vitro. Leukemia. 2005;19:1573–1578. doi: 10.1038/sj.leu.2403861. [DOI] [PubMed] [Google Scholar]
- Fabarius A., Giehl M., Frank O., Spiess B., Zheng C., Müller M.C., Weiss C., Duesberg P., Hehlmann R., Hochhaus A., Seifarth W. Centrosome aberrations after nilotinib and imatinib treatment in vitro are associated with mitotic spindle defects and genetic instability. Br. J. Haematol. 2007;138:369–373. doi: 10.1111/j.1365-2141.2007.06678.x. [DOI] [PubMed] [Google Scholar]
- Fabarius A., Haferlach C., Müller M.C., Erben P., Lahaye T., Giehl M., Frank O., Seifarth W., Hehlmann R., Hochhaus A. Dynamics of cytogenetic aberrations in Philadelphia chromosome positive and negative hematopoiesis during dasatinib therapy of chronic myeloid leukemia patients after imatinib failure. Haematologica. 2007;92:834–837. doi: 10.3324/haematol.11064. [DOI] [PubMed] [Google Scholar]
- Fanta S., Sonnenberg M., Skorta I., Duyster J., Miething C., Aulitzky W.E., Van Der Kuip H. Pharmacological inhibition of c-Abl compromises genetic stability and DNA repair in Bcr-Abl-negative cells. Oncogene. 2008 doi: 10.1038/onc.2008.68. [DOI] [PubMed] [Google Scholar]
- Frijhoff J., Winyard P.G., Zarkovic N., Davies S.S., Stocker R., Cheng D., Knight A.R., Taylor E.L., Oettrich J., Ruskovska T., Gasparovic A.C., Cuadrado A., Weber D., Poulsen H.E., Grune T., Schmidt H.H.H.W., Ghezzi P. Clinical Relevance of Biomarkers of Oxidative Stress. Antioxid. Redox Signal. 2015;23:1144–1170. doi: 10.1089/ars.2015.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajski G., Garaj-Vrhovac V., Oreščanin V. Cytogenetic status and oxidative DNA-damage induced by atorvastatin in human peripheral blood lymphocytes: Standard and Fpg-modified comet assay. Toxicol. Appl. Pharmacol. 2008;231:85–93. doi: 10.1016/j.taap.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Gajski G., Gerić M., Domijan A.-M.A.-M., Garaj-Vrhovac V. Combined cyto/genotoxic activity of a selected antineoplastic drug mixture in human circulating blood cells. Chemosphere. 2016;165:529–538. doi: 10.1016/j.chemosphere.2016.09.058. [DOI] [PubMed] [Google Scholar]
- Gajski G., Žegura B., Ladeira C., Novak M., Sramkova M., Pourrut B., Del Bo’ C., Milić M., Gutzkow K.B., Costa S., Dusinska M., Brunborg G., Collins A. The comet assay in animal models: from bugs to whales – (Part 2 Vertebrates) Mutat. Res. Mutat. Res. 2019;781:130–164. doi: 10.1016/j.mrrev.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Gajski G., Žegura B., Ladeira C., Pourrut B., Del Bo’ C., Novak M., Sramkova M., Milić M., Gutzkow K.B., Costa S., Dusinska M., Brunborg G., Collins A. The comet assay in animal models: from bugs to whales – (Part 1 Invertebrates) Mutat. Res. Mutat. Res. 2019;779:82–113. doi: 10.1016/j.mrrev.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Goldman J.M., Melo J.V. Chronic myeloid leukemia–advances in biology and new approaches to treatment. N. Engl. J. Med. 2003;349:1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- Hartmann J.T., Haap M., Kopp H.-G., Lipp H.-P. Tyrosine kinase inhibitors – a review on pharmacology, metabolism and side effects. Curr. Drug Metab. 2009;10:470–481. doi: 10.2174/138920009788897975. [DOI] [PubMed] [Google Scholar]
- Karaman M.W., Herrgard S., Treiber D.K., Gallant P., Atteridge C.E., Campbell B.T., Chan K.W., Ciceri P., Davis M.I., Edeen P.T., Faraoni R., Floyd M., Hunt J.P., Lockhart D.J., Milanov Z.V., Morrison M.J., Pallares G., Patel H.K., Pritchard S., Wodicka L.M., Zarrinkar P.P. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Kharbanda S., Yuan Z.M., Weichselbaum R., Kufe D. Determination of cell fate by c-Abl activation in the response to DNA damage. Oncogene. 1998;17:3309–3318. doi: 10.1038/sj.onc.1202571. [DOI] [PubMed] [Google Scholar]
- Krokan H.E., Standal R., Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325(Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni H., Göring H.H.H., Diego V., Cole S., Walder K.R., Collier G.R., Blangero J., Carless M.A. Association of differential gene expression with imatinib mesylate and omacetaxine mepesuccinate toxicity in lymphoblastoid cell lines. BMC Med. Genom. 2012;5:37. doi: 10.1186/1755-8794-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo D.L. Imatinib changed everything. N. Engl. J. Med. 2017;376:982–983. doi: 10.1056/NEJMe1700833. [DOI] [PubMed] [Google Scholar]
- Lutterbeck C.A., Kern D.I., Machado Ê.L., Kümmerer K. Evaluation of the toxic effects of four anti-cancer drugs in plant bioassays and its potency for screening in the context of waste water reuse for irrigation. Chemosphere. 2015;135:403–410. doi: 10.1016/j.chemosphere.2015.05.019. [DOI] [PubMed] [Google Scholar]
- Manley P.W., Cowan-Jacob S.W., Buchdunger E., Fabbro D., Fendrich G., Furet P., Meyer T., Zimmermann J. Imatinib: a selective tyrosine kinase inhibitor. Eur. J. Cancer. 2002;38(Supple):S19–S27. doi: 10.1016/s0959-8049(02)80599-8. [DOI] [PubMed] [Google Scholar]
- Mišík M., Filipic M., Nersesyan A., Mišíková K., Knasmueller S., Kundi M. Analyses of combined effects of cytostatic drugs on micronucleus formation in the Tradescantia. Environ. Sci. Pollut. Res. 2016;23:14762–14770. doi: 10.1007/s11356-015-5837-0. [DOI] [PubMed] [Google Scholar]
- Novak M., Žegura B., Nunić J., Gajski G., Gerić M., Garaj-Vrhovac V., Filipič M. Assessment of the genotoxicity of the tyrosine kinase inhibitor imatinib mesylate in cultured fish and human cells. Mutat. Res. 2017;814:14–21. doi: 10.1016/j.mrgentox.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Parrella A., Lavorgna M., Criscuolo E., Russo C., Fiumano V., Isidori M. Acute and chronic toxicity of six anticancer drugs on rotifers and crustaceans. Chemosphere. 2014;115:59–66. doi: 10.1016/j.chemosphere.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Parrella A., Lavorgna M., Criscuolo E., Russo C., Isidori M. Eco-genotoxicity of six anticancer drugs using comet assay in daphnids. J. Hazard. Mater. 2015;286:573–580. doi: 10.1016/j.jhazmat.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Parrillo-Campiglia S., Ercoli M.C., Umpierrez O., Rodríguez P., Márquez S., Guarneri C., Estevez-Parrillo F.T., Laurenz M., Estevez-Carrizo F.E. Bioequivalence of two film-coated tablets of imatinib mesylate 400 mg: a randomized, open-label, single-dose, fasting, two-period, two-sequence crossover comparison in healthy male South American volunteers. Clin. Ther. 2009;31:2224–2232. doi: 10.1016/j.clinthera.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Pichler C., Filipič M., Kundi M., Rainer B., Knasmueller S., Mišík M. Assessment of genotoxicity and acute toxic effect of the imatinib mesylate in plant bioassays. Chemosphere. 2014;115:54–58. doi: 10.1016/j.chemosphere.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Schalow B.J., Courcelle C.T., Courcelle J. Escherichia coli Fpg glycosylase is nonrendundant and required for the rapid global repair of oxidized purine and pyrimidine damage in vivo. J. Mol. Biol. 2011;410:183–193. doi: 10.1016/j.jmb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.-H., Li W.-H. Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol. Biol. Evol. 2004;21:828–840. doi: 10.1093/molbev/msh077. [DOI] [PubMed] [Google Scholar]
- Smith B.D. Imatinib for chronic myeloid leukemia: the impact of its effectiveness and long-term side effects. J. Natl. Cancer Inst. 2011;103:527–529. doi: 10.1093/jnci/djr073. [DOI] [PubMed] [Google Scholar]
- Smith C.C., O’Donovan M.R., Martin E.A. hOGG1 recognizes oxidative damage using the comet assay with greater specificity than FPG or ENDOIII. Mutagenesis. 2006;21:185–190. doi: 10.1093/mutage/gel019. [DOI] [PubMed] [Google Scholar]
- Stagno F., Stella S., Spitaleri A., Pennisi M.S., Di Raimondo F., Vigneri P. Imatinib mesylate in chronic myeloid leukemia: frontline treatment and long-term outcomes. Expert Rev. Anticancer Ther. 2016;16:273–278. doi: 10.1586/14737140.2016.1151356. [DOI] [PubMed] [Google Scholar]
- Viegas S., Ladeira C., Costa-Veiga A., Perelman J., Gajski G. Forgotten public health impacts of cancer – an overview. Arh. Hig. Rada Toksikol. 2017;68:287–297. doi: 10.1515/aiht-2017-68-3005. [DOI] [PubMed] [Google Scholar]
- Witte I., Plappert U., de Wall H., Hartmann A. Genetic toxicity assessment: employing the best science for human safety evaluation Part III: the comet assay as an alternative to in vitro clastogenicity tests for early drug candidate selection. Toxicol. Sci. 2007;97:21–26. doi: 10.1093/toxsci/kfl192. [DOI] [PubMed] [Google Scholar]
- Wolf D., Rumpold H. A benefit-risk assessment of imatinib in chronic myeloid leukaemia and gastrointestinal stromal tumours. Drug Saf. 2009;32:1001–1015. doi: 10.2165/11314600-000000000-00000. [DOI] [PubMed] [Google Scholar]