Abstract
Background
Metastasis of rhabdomyosarcoma (RMS) is the primary cause of tumour-related deaths. Previous studies have shown that overexpression of the guanine nucleotide exchange factor T (GEFT) is correlated with a poorer RMS prognosis, but the mechanism remains largely unexplored.
Methods
We focused on determining the influence of the GEFT-Rho-GTPase signalling pathway and the epithelial–mesenchymal transition (EMT) or mesenchymal–epithelial transition (MET) on RMS progression and metastasis by using RMS cell lines, BALB/c nude mice and cells and molecular biology techniques.
Findings
GEFT promotes RMS cell viability, migration, and invasion; GEFT also inhibits the apoptosis of RMS cells and accelerates the growth and lung metastasis of RMS by activating the Rac1/Cdc42 pathways. Interestingly, GEFT upregulates the expression levels of N-cadherin, Snail, Slug, Twist, Zeb1, and Zeb2 and reduces expression level of E-cadherin. Thus, GEFT influences the expression of markers for EMT and MET in RMS cells via the Rac1/Cdc42-PAK1 pathways. We also found that the level of GEFT gene promoter methylation in RMS is lower than that in normal striated muscle tissue. Significant differences were observed in the level of GEFT gene methylation in different histological subtypes of RMS.
Interpretation
These findings suggest that GEFT accelerates the tumourigenicity and metastasis of RMS by activating Rac1/Cdc42-PAK signalling pathway-induced EMT; thus, it may serve as a novel therapeutic target.
Fund
This work was supported by grants from the National Natural Science Foundation of China (81660441, 81460404, and 81160322) and Shihezi University Initiative Research Projects for Senior Fellows (RCZX201447). Funders had no role in the design of the study, data collection, data analysis, interpretation, or the writing of this report.
Keywords: Rhabdomyosarcoma, GEFT, Rac1/Cdc42-PAK1 pathways, EMT, Methylation
Research in context.
Evidence before this study
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children, and those with high-risk RMS have a poor prognosis. Our previous studies have demonstrated that overexpression of guanine nucleotide exchange factor T (GEFT) is significantly correlated with lymph node metastasis, distant metastasis, and poor RMS prognosis. The epithelial–mesenchymal transition (EMT) is essential for tumour metastasis. The references searched were all available PubMed publications, and the search terms were RMS and GEFT, with an emphasis on EMT.
Added value of this study
GEFT influences the expression of markers for EMT and MET in RMS cells via the Rac1/Cdc42-PAK1 pathways, as demonstrated through a variety of in vitro and in vivo experiments. Our novel findings, along with the existing evidence regarding the resolution mechanisms in RMS, have revealed one of the mechanisms underlying RMS.
Implications of all the available evidence
Our research may lead to potential targets for RMS therapies by not only focusing on the function of GEFT in the tumourigenicity and metastasis of RMS but also by revealing its underlying molecular mechanism. Future research should try to identify the role of GEFT in the progression and metastasis of other cancers, as such studies could provide new therapeutic opportunities for these patients.
Alt-text: Unlabelled box
1. Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children. There are two major RMS histologic subtypes—embryonal RMS (ERMS) and alveolar RMS (ARMS)—and two rare variants—pleomorphic RMS and sclerosing RMS [1]. The prognosis for RMS depends on the tumour's metastasis, location, size, and staging, as well as the patient's age. Among these factors, the presence of metastasis is the strongest predictor of poor clinical outcomes [2]. The majority of human cancer-related deaths are caused by tumour metastasis [3,4]. Despite multimodal therapies, patients with high-risk RMS have a poor prognosis, with a 5-year overall survival rate of 20–30% [2]. The incomplete understanding of RMS’ molecular mechanisms and metastasis-related genes is the primary reason for its poor prognosis. Therefore, identifying new molecular therapeutic targets relevant to the pathogenesis of RMS is crucial. In our previous studies, we found that overexpression of guanine nucleotide exchange factor T (GEFT) was significantly correlated with lymph node metastasis, distant metastasis, and a poor RMS prognosis [5,6].
GEFT was first identified in a retroviral mutation screen for potential oncoproteins. It is a member of the Dbl protein family that is Rho-specific for the Rho GEFs and contains 60 distinct mammalian members [7], [8], [9]. It is highly expressed in brain, heart, and muscle tissues [8,10]. GEFT promotes the myogenesis of C2C12 cells via activating RhoA, Rac1, and Cdc42 and their downstream effector proteins [11]. However, two studies have reported that GEFT/p63RhoGEF is a RhoA-specific GEF and not a Rac/Cdc42-specific GEF [12,13]. The potential functions and pathways of GEFT in regard to RMS progression and metastasis are unclear.
Interestingly, cells with a high GEFT expression level had cell shapes that had more spindle-like appearance and fewer cell contacts, suggesting that epithelial–mesenchymal transition (EMT) or mesenchymal–epithelial transition (MET) may have occurred in RMS. We know that EMT is essential for tumour metastasis. Hallmarks of EMT include the loss of expression or function of E-cadherin and a markedly reduced number of tight junction proteins, which is accompanied by a considerable increase in mesenchymal markers, such as N-cadherin [14]. Induction of transcription factors is the key initiating step of EMT. These factors include the zinc-finger binding transcription factors, namely, Snail1 and Slug, zinc finger E-box–binding homeobox 1 (ZEB1), ZEB2, and Twist, all proteins that bind to the promoter region of genes associated with cell–cell adhesion and repress their transcription [15], [16], [17]. Thus, we investigated any changes in the hallmarks of EMT and EMT-inducing transcription factors in RMS.
We have long known that epigenetics often affects gene expression, including mechanisms involving DNA methylation, histone modification and microRNAs. Our previous investigation showed that targeting GEFT by miR-874 could impair tumour proliferation and invasion of RMS [18]. DNA methylation changes may result in altered gene expression profiles, the silencing of tumour suppressors due to promoter DNA hypermethylation, as well as oncogene upregulation due to promoter DNA hypomethylation [19]. There are close relationships between methylation status and gene mutations and the biological behaviour of RMS [20]. Sun's study showed that fusion-positive and fusion-negative rhabdomyosarcomas possess characteristic methylation profiles [21].
Herein, our study revealed that GEFT promotes RMS cell viability, migration, and invasion. GEFT also inhibits apoptosis by activating the Rac1/Cdc42 signalling pathways, and it accelerates lung metastasis, as demonstrated by the construction of a nude mouse model of rhabdomyosarcoma xenografts in order to better verify the results of the in vitro experiments. Furthermore, GEFT influences the expression of hallmarks of EMT and EMT-inducing transcription factors in RMS cells via the Rac1/Cdc42-PAK1 pathways, subsequently promoting the tumourigenicity and metastasis of RMS. We also found hypomethylation of GEFT. Thus, GEFT can potentially be a therapeutic target.
2. Materials and methods
2.1. Tissue samples
Thirty-nine formalin-fixed paraffin-embedded (FFPE) RMS and fifteen normal skeletal muscle tissue samples were selected from archives of the Department of Pathology of the First Affiliated Hospital, Shihezi University School of Medicine, China. These samples included 19 embryonal rhabdomyosarcomas (ERMS), 6 alveolar rhabdomyosarcomas (ARMS), 9 pleomorphic rhabdomyosarcomas (PRMS) and 5 sclerosing rhabdomyosarcomas (SRMS). Written informed consent was obtained from all participating patients before enrolment in the study. This study was approved by the institutional ethics committee at the First Affiliated Hospital of Shihezi University School of Medicine and conducted in accordance with the ethical guidelines of the Declaration of Helsinki.
2.2. Cell cultures and transfection
Our study utilised three cell lines derived from human RMS: RD (ERMS), PLA-802 (ARMS), and RH30 (ARMS). They were purchased from the Biological Technology Co., Ltd. (Fu Xiang, Shanghai, China). PLA-802 cells were cultured in RPMI 1640. RD and RH30 cells were cultured in DMEM medium supplemented with 4.5 g/L of glucose, 2 mM of L-glutamine, and 10% foetal bovine serum (FBS) at 37 °C with 5% CO2. Human RMS cells were grown on six-well plates for transfection with Lipofectamine 2000 (Life Technologies, USA). We performed the procedure according to the manufacturer′s instructions. Cells were harvested for quantitative real time PCR (qRT-PCR) or Western blot analysis at 48 h after transfection, and these experiments were performed in triplicate. All experiments were repeated at least three times.
2.3. Antibodies and inhibition
The primary antibodies were rabbit anti-GEFT (Abcam, ab127690), rabbit anti-Snail (Abcam, ab229701), mouse anti-Slug (Abcam, ab51772), mouse anti-Twist (Abcam, ab175430), rabbit anti-Rac1 (Abcam, ab33186), mouse anti-E-cadherin (CST, #14472), rabbit anti-N-cadherin (CST, #13116), rabbit anti-PAK1 (Abcam, ab223849), Cdc42 (Abcam, ab187643), rabbit anti-RhoA (Abcam, ab187027), mouse anti-Zeb1 (Santa Cruz, sc-81428), mouse anti-Zeb2 (Santa Cruz, sc-271984), and mouse anti-β-actin (ZSGB-BIO, China). The secondary antibody was peroxidase-conjugated goat anti-mouse/rabbit IgG (ZB-2305). Cdc42 Activation Assay Biochem Kit™ (cytoskeleton, Cat. # BK034), Rac1 Activation Assay Biochem Kit™ (cytoskeleton, Cat. # BK035) and Rho A Activation Assay Biochem Kit™ (cytoskeleton, Cat. # BK036) were used for analysis of Cdc42, Rac1 and Rho A activation. NSC23766 (Selleck, S8031), ZCL278 (Selleck, S7293), and IPA-3 (Selleck, S7093) inhibitors were acquired commercially.
2.4. Plasmid generation
Human GEFT plasmid (GenBank-ID: NM-182947) was purchased from QIAGEN (Shanghai, China). The shRNA sequence targeting GEFT was also designed by QIAGEN (Supplementary Fig. S1). We chose the shGEFT sequence with the best results to carry out the follow-up cell function experiments, except for the colony formation assay.
2.5. qRT-PCR analyses
Total RNA was extracted with an RNeasy Mini Kit (QIAGEN) following the manufacturer's protocol. GEFT gene primers were purchased from QuantiTect Primer Assays (QIAGEN). The reaction was performed in an ABI 7500 real-time PCR thermocycler (Applied Biosystems) with a Quantifast SYBR Green PCR Kit (QIAGEN). The 2−ΔΔCt method was used to quantify the expression of GEFT.
2.6. Western blotting
Transfected RMS cells were lysed in RIPA lysis buffer (Solarbio). Protein lysates were centrifuged at 4 °C for 15 min at 12,000 rpm to remove cellular debris. Equal amounts of total protein were loaded onto 15% SDS-polyacrylamide gels and transferred to PVDF membranes (Millipore) after electrophoresis. The membranes were blocked for 2 h with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 and were incubated overnight with the primary antibodies at 4 °C. Then, the membranes were incubated with the secondary antibodies for 2 h at room temperature.
2.7. Cell proliferation assays
Cell proliferation was monitored by a Cell Counting Kit-8 (CCK8) assay (Dojindo, Shanghai, China) and an EDU (KGA337, KeyGen BioTECH, China) assay. CCK8 can be used to measure both proliferation and cytotoxicity by utilising Dojindo's highly water-soluble tetrazolium salt. Tumour cells transfected with GEFT, shGEFT, Rac1, or Cdc42 were cultured in 96-well plates at a cell density of 1 × 104 cells/well. Cell proliferation was assessed every 24 h following the manufacturer's protocol. For the EdU assay, tumour cells transfected with GEFT, shGEFT, Rac1, or Cdc42 were cultured in 12-well plates at a cell density of 1 × 105 cells/well. Cell proliferation was assessed every 48 h following the manufacturer's protocol.
2.8. Colony formation assay
Tumour cells were transfected with GEFT plasmid. A total of 1000 cells were seeded per six-well plate. After 2 weeks, the colonies were fixed with 4% paraformaldehyde for 20 min and then stained with 0.1% crystal violet for 20 min.
2.9. Scratch assay
A total of 6 × 105 cells was seeded in six-well plates and cultured overnight. When the cultures reached 85% confluency, the cell layer was scratched with a sterile pipette tip, washed thrice with PBS to remove any debris, and cultured in complete medium.
2.10. Cell migration and invasion assays
Cell migration and invasion experiments were performed with Transwell® chambers (Corning Inc., USA), following the manufacturer's protocol. A total of 3 × 104 cells in serum-free media were placed in the upper chambers of the Transwells. For the invasion assays, Matrigel™ matrix was used to coat the basement membrane (BD Biosciences). The basement membrane had an 8.0 µm pore size. The migration assays did not utilise Matrigel. The assays measured how many cells moved from the upper chamber through the membrane (migration) or through the membrane plus Matrigel (invasion) to the lower chamber over 24 h (migration) or 48 h (invasion). Medium containing 20% FBS was added to the lower chamber.
2.11. Flow cytometry analysis of apoptosis
Cells were cultured for 48 h and then washed thrice with cold PBS. After digestion with Accutase (eBioscience, USA) and centrifugation, the cells were resuspended in 1 × binding buffer at a 1 × 106 cell/ml concentration. The cells were analysed by PAS flow cytometry (PARTEC, Germany) with FlowJo 7.6 software after staining with Annexin V-APC/7-AAD or Annexin V-FITC/PI.
2.12. Pull-down assay
Pull-down assays were conducted using an Activation Assay Biochem kit (Cytoskeleton, Denver, CO) according to the manufacturer's protocols. Cell lysates were incubated for 60 min at 4 °C with either Rhotekin-RBD or PAK-PBD beads to detect active RhoA or Rac1/CdC42, respectively. Proteins were separated on 15% gradient polyacrylamide gels and transferred to polyvinylidene fluoride membranes, which were probed for RhoA, Rac1, or Cdc42.
2.13. Quantitative analysis of GEFT DNA methylation by MALDI-TOF MS
DNA was isolated using a DNA Extraction Kit (QIAGEN, 56404). DNA purity and quality were evaluated by using a NanoDrop spectrophotometer (NanoDrop Technologies Inc.) and gel electrophoresis. Human GEFT methylation primers were 5′ aggaagagagAGTTTTTTTGTTTTTTGAGGATTTG 3′ (forward) and 5′ cagtaatacgactcactatagggagaaggct AAAACTTCATACTAAACCCCCACC3’ (reverse). Bisulfite was applied to treat the DNA by using the EZ DNA Methylation Kit™ (Zymo Research, D5008). Mass spectra were obtained by MassARRAY Compact MALDI-TOF MS, and the methylation proportions of individual units were generated by EpiTyper 1.0.5 (SEQUENOM).
2.14. Animal studies
RD or RH30 cells were constructed with lentiviruses according to standard protocols by QIAGEN. The animal study was approved by the Ethics Committee of the First Affiliated Hospital of Medical College, Shihezi University. The 5-week-old male BALB/c nude mice, weighing 16–20 g, were purchased from Beijing Weitonglihua Experimental Animal Technology Co., Ltd. Nine mice were randomly divided into 3 groups—the RD group, Vehicle group or RD-GEFT group—for the analysis of GEFT function. A total of 2 × 106 cells was subcutaneously injected into the upper flanks of the nude mice. The mice were monitored every other day, and the tumours were measured with callipers. Tumour size was estimated by the modified ellipsoid formula: 1/2 (length × width2). The tumour length and width were measured every 2 or 4 days.
As for the injected inhibitor groups, after the tumour length size was >5 mm, NSC23766 (50 mg/kg) or ZCL278 (50 mg/kg) treatment was started. Eighteen mice were randomly divided into 3 groups—the RH30-GEFT group, the RH30-GEFT + NSC23766 group or the RH30-GEFT + ZCL278 group—for analysis of the GEFT pathways. To establish the mouse model, the mice were injected subcutaneously with cells, and they were weighed before and during the experiment. Inhibitor or vehicle was administered intraperitoneal every 2 days. A Vernier calliper used to measure the tumour size to observe the growth of the tumours. After anaesthesia (ketamine/xylazine cocktail, 85 mg/kg ketamine, 15 mg/kg xylazine), the mice were exposed to observe in vivo GFP expression of tumour cells using the small animal in vivo imaging system IVIS-Lumina-Series-III. Mice were euthanised by CO2 inhalation at the end of the experiments. The study was conducted according to the Animal Research Reporting In Vivo Experiments (ARRIVE) requirements (Supplementary Table 1).
2.15. Statistical analysis
SPSS 20.0 was employed to conduct the statistical analysis. Data were presented as the mean ± SD. One-way ANOVA (analysis of variance) was used to compare the indexes among three groups, and 2 independent sample t-test or Wilcoxon nonparametric test was used to compare the indexes between two groups. Differences of the rate changes between the two groups were tested by chi-square test. P < 0.05 was recognised as the level of statistical significance. *P < 0.05, **P < 0.01, and ***P < 0.001 were noted.
3. Results
3.1. GEFT displays oncogene activity in RMS
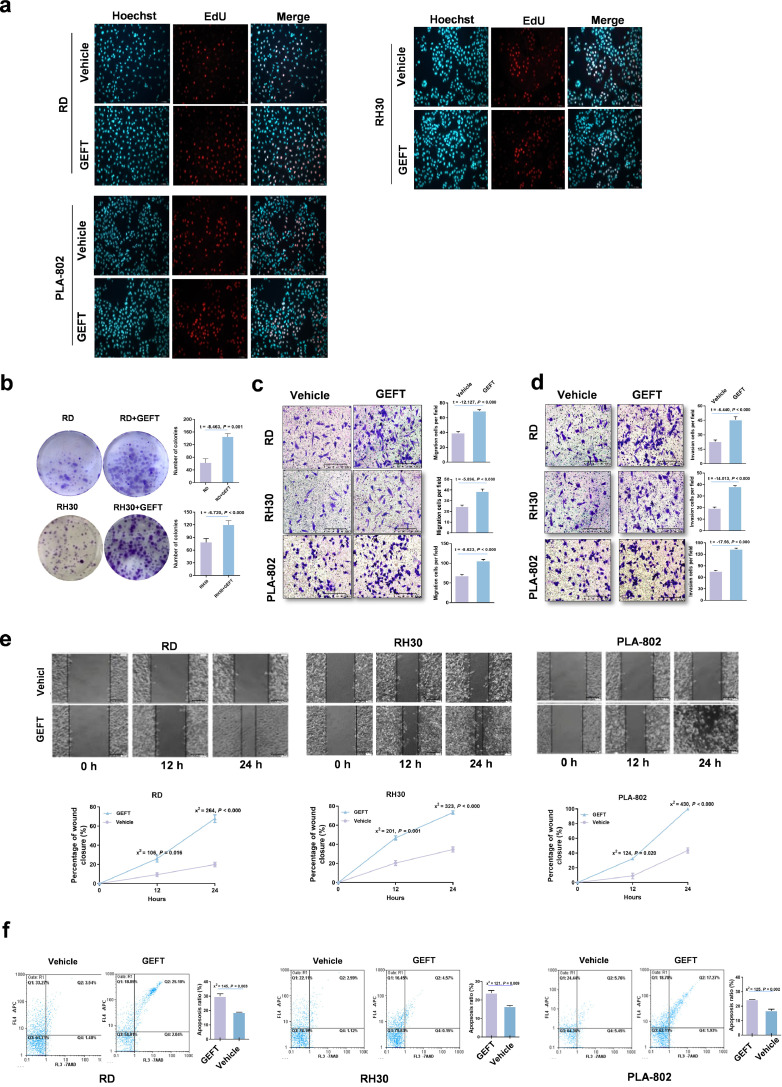
Our previous work found that overexpression of GEFT was significantly correlated with lymph node metastasis, distant metastasis, and a poor RMS prognosis; thus, GEFT may act as an oncogene in RMS [5,6]. To investigate the GEFT signalling pathways in RMS development, we first evaluated the biological role of GEFT in RMS cell lines. The results showed that RMS cell lines transfected with pCDNA-GEFT showed high GEFT expression relative to the vehicle control, as assessed by qRT-PCR (Supplementary Fig. S2a). The GEFT protein levels of cells transfected with shGEFT decreased compared with those of the vehicle control group, as shown by Western blotting (Supplementary Fig. S2b). The CCK-8 assay results showed that compared with control cells, the growth of RMS cells transfected with pCDNA-GEFT was enhanced (Supplementary Fig. S2c). EdU was used to verify the cell proliferation results, and we found that GEFT could promote cell proliferation (Fig. 1a). The number of colonies increased following GEFT overexpression in RD and RH30 cells (Fig. 1b). The number of migrating and invading tumour cells was remarkably higher in pCDNA-GEFT-treated cells than that in the control groups (Fig. 1c and d). Scratch assays with cells transfected with pCDNA-GEFT healed faster compared with the control cells (Fig. 1e). Similarly, flow cytometry analysis of RMS cells showed that GEFT inhibited apoptosis (Fig. 1f). Thus, GEFT promotes RMS cell viability, proliferation, migration, and invasion, and inhibits apoptosis; therefore, GEFT may act as an oncogene in RMS.
Fig. 1.
GEFT displays oncogene activity in RMS. (a) EdU was used to detect cell proliferation. Hoechst stained all cells (blue), EdU labelled proliferating cells (red), and GEFT promoted cell proliferation (pink). (b) Colony-forming assays. (c) The number of migrating tumour cells, as well as their migratory ability, was significantly higher in pCDNA GEFT-treated cells than that in the control groups. (d) The number of invading tumour cells, as well as their invasion ability, was significantly higher in pCDNA GEFT-treated cells than that in the control groups. (e) The migratory ability of RMS cells was investigated by wound-healing assays. (f) GEFT inhibited apoptosis of RMS cells. An independent sample t-test was used to detect differences between the two groups (b, c, d). Differences of the rate changes between the two groups were tested by chi-square test (e, f).
3.2. GEFT influences RMS cells via the Rho guanosine triphosphatase (Rho-GTPase) pathway
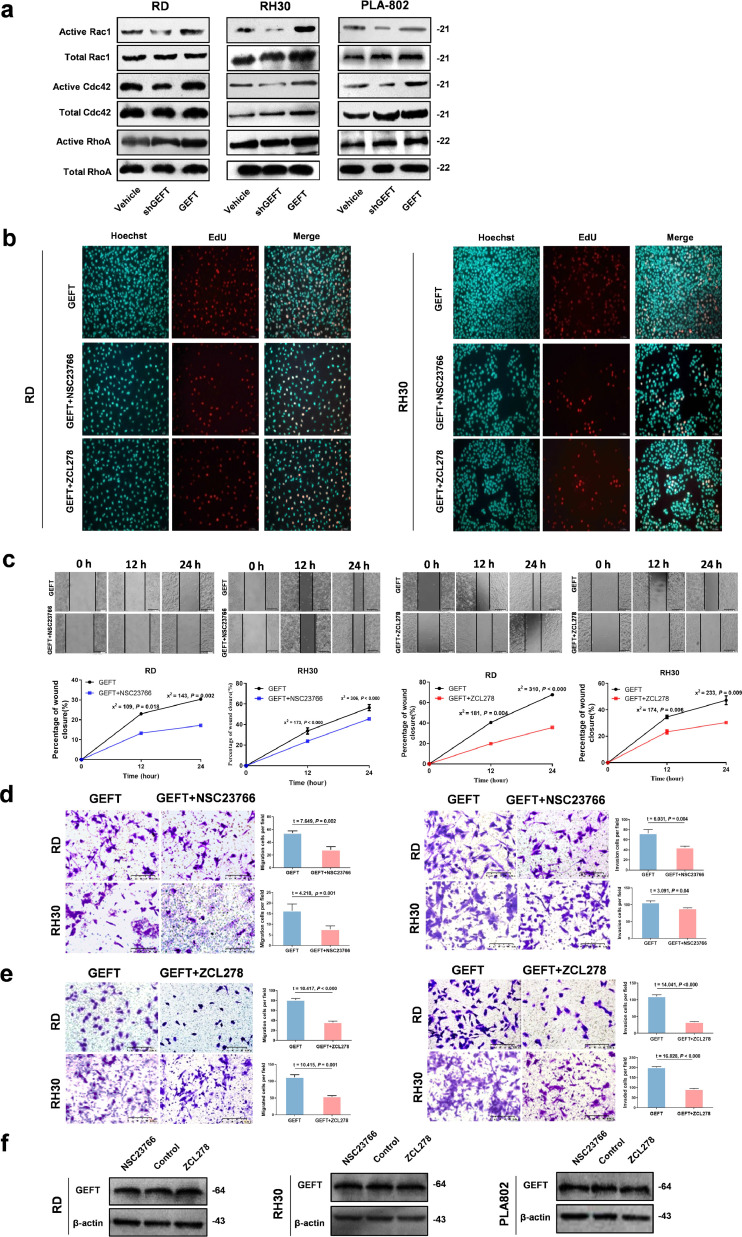
Then, we focused on the signalling pathway mediating GEFT-induced Rho-GTPase activation. Inhibiting GEFT activity reduced Rac1 and Cdc42 activation, whereas inducing GEFT activity enhanced Rac1 and Cdc42 activation in RMS cell lines, but RhoA remained unchanged (Fig. 2a). To further indicate that GEFT enhances RMS cell viability, migration, and invasion via the Rho-GTPase pathway, we treated RMS cells transfected with pCDNA-GEFT with Rac inhibitor (NSC23766) or Cdc42 inhibitor (ZCL278). EdU assays showed that NSC23766 and ZCL278 could inhibit cell proliferation (Fig. 2b). The CCK-8 assay results showed that treatment with NSC23766 inhibited the growth of RMS cells transfected with pCDNA-GEFT (Supplementary Fig. S3). The scratch assay results showed that cells transfected with pCDNA-GEFT and treated with NSC23766 or ZCL278 had slower wound healing compared with cells transfected with pCDNA-GEFT (Fig. 2c). The number of migrating and invading tumour cells was remarkably lower in NSC23766- or ZCL278-treated cells than in the control groups according to the Matrigel and Transwell assays (Fig. 2d and e). The level of GEFT protein did not change in response to NSC23766 or ZCL278 treatment in RMS cell lines (Fig. 2f). Therefore, these results indicated that GEFT as an upstream gene of Rac1 or Cdc42 via Rac1/Cdc42-dependent pathways regulates the progression of rhabdomyosarcoma.
Fig. 2.
GEFT is involved in Rho activation. (a) Rac1 and Cdc42 were activated by GEFT in RMS cells, as shown by pull-down assays, but RhoA was not changed. (b) The EdU assay shows that NSC23766 or ZCL278 can inhibit cell proliferation of RMS. (c) The migratory ability of RMS cells is inhibited by NSC23766 or ZCL278. Differences of the rate changes between the two groups were tested by chi-square test. (d and e) The number of migrating and invading tumour cells was significantly lower in NSC23766 or ZCL278-treated cells than that in the control groups. An independent sample t-test was used to detect differences between the two groups. (f) The level of GEFT protein did not change when the RMS cell lines were treated with either NSC23766 or ZCL278.
3.3. GEFT promotes RMS cell growth and metastasis via the Rac1/Cdc42 pathway in vivo
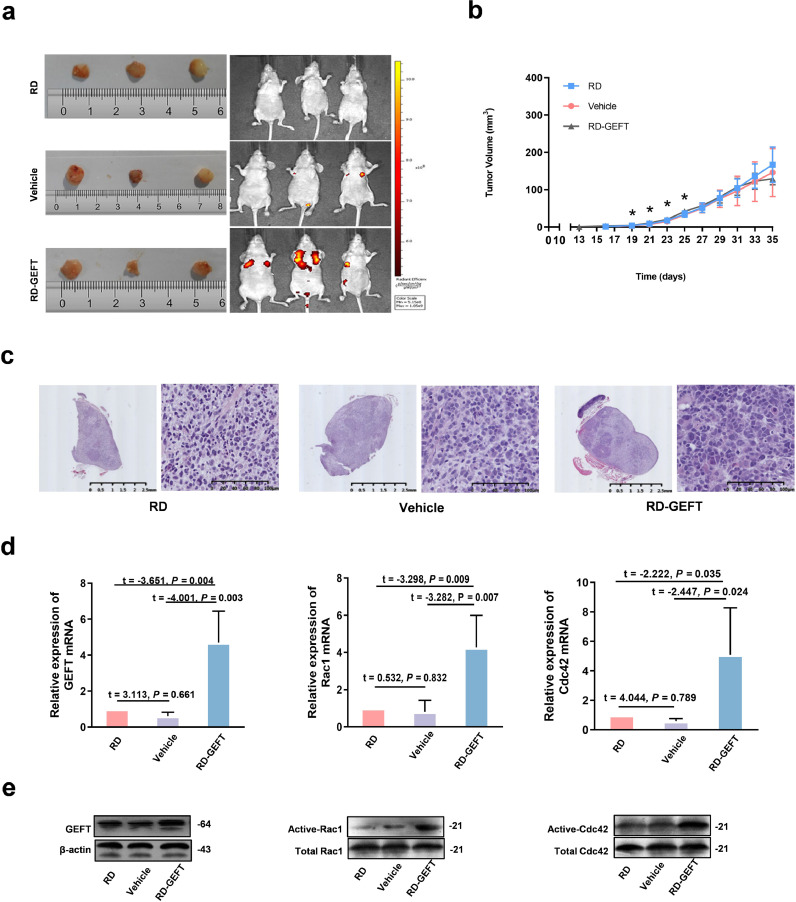
To further demonstrate GEFT promotes RMS cell growth and metastasis in vivo, RD cells stably transfected with lentivirus GEFT were injected into nude mice. The whole-body green fluorescent protein (GFP) imaging system revealed that metastasis was more extensive in the RD-GEFT group than in the two control groups (Fig. 3a). The subcutaneous tumourigenic latency of nude mice in the RD-GEFT group was shortened (13.875 ± 1.464 days); additionally, the growth rate of tumours over the first 25 days was significantly faster, and the volume of the tumours was larger. Compared with the RD group (17.286 ± 1.604 days) and Vehicle group (17.714 ± 1.604 days), the difference was statistically significant (P < 0.05, Fig. 3b). Haematoxylin and eosin (HE) staining of the tumour sections revealed that the RD-GEFT group exhibited more atypical tumour cells and had enhanced pathological mitosis (Fig. 3c). We then assessed mRNA and protein expression levels in the tumours. The mRNA expression levels of GEFT, Rac1, and Cdc42 were increased in the LV-GEFT group (P < 0.05, Fig. 3d). Additionally, the activation of Rac1and Cdc42 were increased in the RD-GEFT group (P < 0.05, Fig. 3e). These data are thus consistent with the GEFT-Rac1/Cdc42 signalling in the RMS cell lines.
Fig. 3.
Effects of GEFT overexpression on tumour growth and metastasis in vivo. (a) Excised tumour samples and whole-body GFP imaging. (b) Growth curves of the xenografted tumours. One-way ANOVA (analysis of variance) was used to compare the indexes among three groups. (c) HE-stained xenografted tumours. (d) The mRNA levels of GEFT, Rac1 and Cdc42 are increased in the RD-GEFT+ group. An independent sample t-test was used to detect differences between the two groups. (e) Pull-down assays show that Active-Rac1 and Active -Cdc42 levels are increased in the RD-GEFT group. *P < 0.05.
3.4. Rac1/Cdc42 pathway inhibitor inhibited tumour growth in vivo
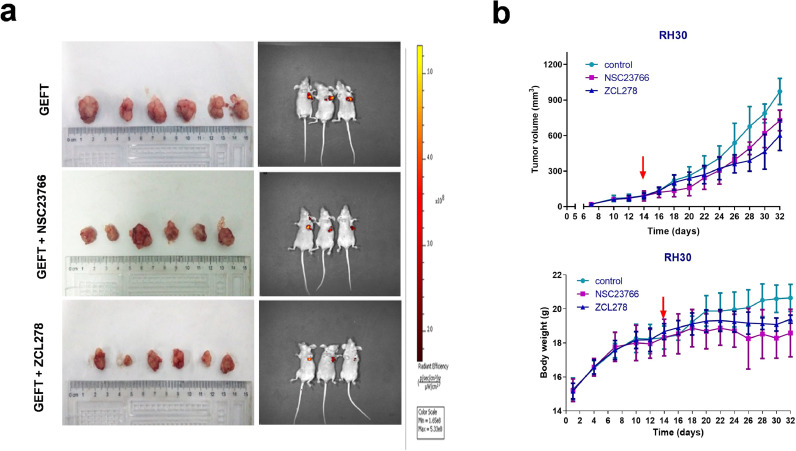
Next, we wanted to investigate whether Rac1/Cdc42 pathway inhibitors could inhibit tumour growth in vivo. RH30 cells stably transfected with lentivirus GEFT were injected into nude mice. Tumours formed after 7 days. On the 14th day, the nude mice were given intraperitoneal injections of the optimal concentrations of NSC23766 or ZCL278. Compared with the GEFT control group, the tumour growth slowed after the 14th day of NSC23766 treatment (P < 0.05) or the 12th day of ZCL278 treatment (P < 0.05). The weight of nude mice in the NSC23766 and ZCL278 groups was significantly lower than that of the control group after the 14th day (P < 0.05 and P < 0.01) (Fig. 4). These data demonstrate that Rac1/Cdc42 pathway inhibitors could inhibit RMS tumour growth in vivo.
Fig. 4.
NSC23766 and ZCL278 slowed the growth rate of the tumours in mice. On the 14th day, the nude mice were given inhibitors (50 mg/kg). (a) Excised tumour samples and whole-body GFP imaging. (b) Growth curves of the xenografted tumours. One-way ANOVA (analysis of variance) was used to compare the indexes between two groups (a, b).
3.5. GEFT influences EMT/MET via Rac1/Cdc42-PAK1 pathways
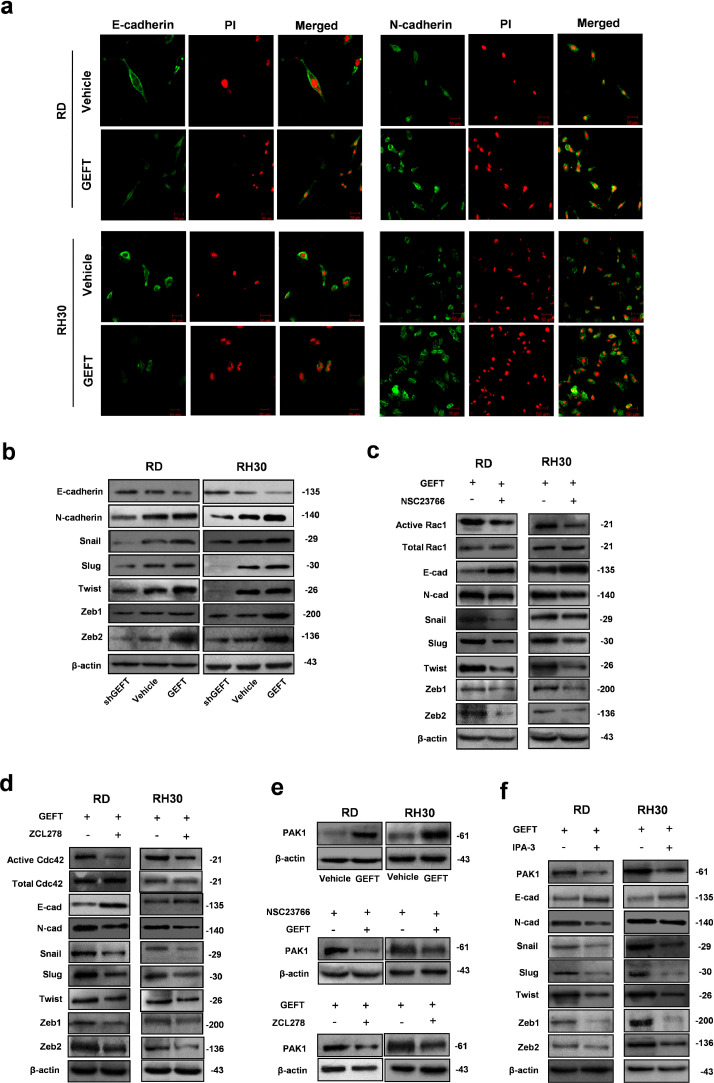
We next tested our hypothesis that GEFT influences EMT and EMT-inducing transcription factors to promote tumour invasion and metastasis by Rac1/Cdc42 signalling. First, we used immunofluorescence assays to analyse the fluorescence intensity of various EMT-related markers. In RMS cells, GEFT enhanced the fluorescence intensity of N-cadherin protein and reduced the fluorescence intensity of E-cadherin protein (Fig. 5a). Thus, the protein level of E-cadherin was reduced, whereas those of N-cadherin and EMT-inducing transcription factors (Snail, Slug, Twist, ZEB1, and ZEB2) increased in cells transfected with pCDNA-GEFT; further, the opposite effects were observed in cells transfected with shGEFT (Fig. 5b). When cells were treated with NSC23766 or ZCL278, the expression level of E-cadherin increased, but the expression levels of N-cadherin and Snail, Slug, Twist, ZEB1, and ZEB2 were reduced (Fig. 5c and d). Last, upregulation of GEFT could promote the expression of PAK1, but NSC23766 or ZCL278 could inhibit the expression of PAK1 (Fig. 5e). When treated with IPA-3 (a classic inhibitor of PAK1), the expression level of E-cadherin increased, and the expression levels of N-cadherin and EMT-inducing transcription factors were reduced (Fig. 5f). In summary, these data provide strong evidence that GEFT influences EMT/MET via the Rac1/Cdc42-PAK1 pathways to promote tumour invasion and metastasis.
Fig. 5.
GEFT influences the expression of EMT markers in RMS cells via the Rac1/Cdc42-PAK1 pathways. (a) Immunofluorescence assay confirmed that GEFT enhanced the fluorescence intensity of N-cadherin protein in RMS cells and reduced the fluorescence intensity of E-cadherin protein in RMS cells. (b) GEFT overexpression upregulated the expression levels of EMT related markers (N-cadherin, Snail, Slug, Twist, Zeb1, and Zeb2) and reduced the expression level of E-cadherin; the expression levels of EMT-related proteins were reduced by shGEFT, which also upregulated the expression level of E-cadherin. (c and d) The expression level of E-cadherin increased, whereas the expression levels of N-cadherin and Snail, Slug, Twist, ZEB1, and ZEB2 were reduced after NSC23766 or ZCL278 interference. (e) GEFT can promote the expression of PAK1, and NSC23766 or ZCL278 can inhibit the expression of PAK1. (f) The expression level of E-cadherin increased and the expression levels of N-cadherin and EMT-inducing transcription factors were reduced after IPA-3 interference.
3.6. GEFT gene methylation levels in rhabdosyosarcoma and their correlation with clinical pathological characteristics
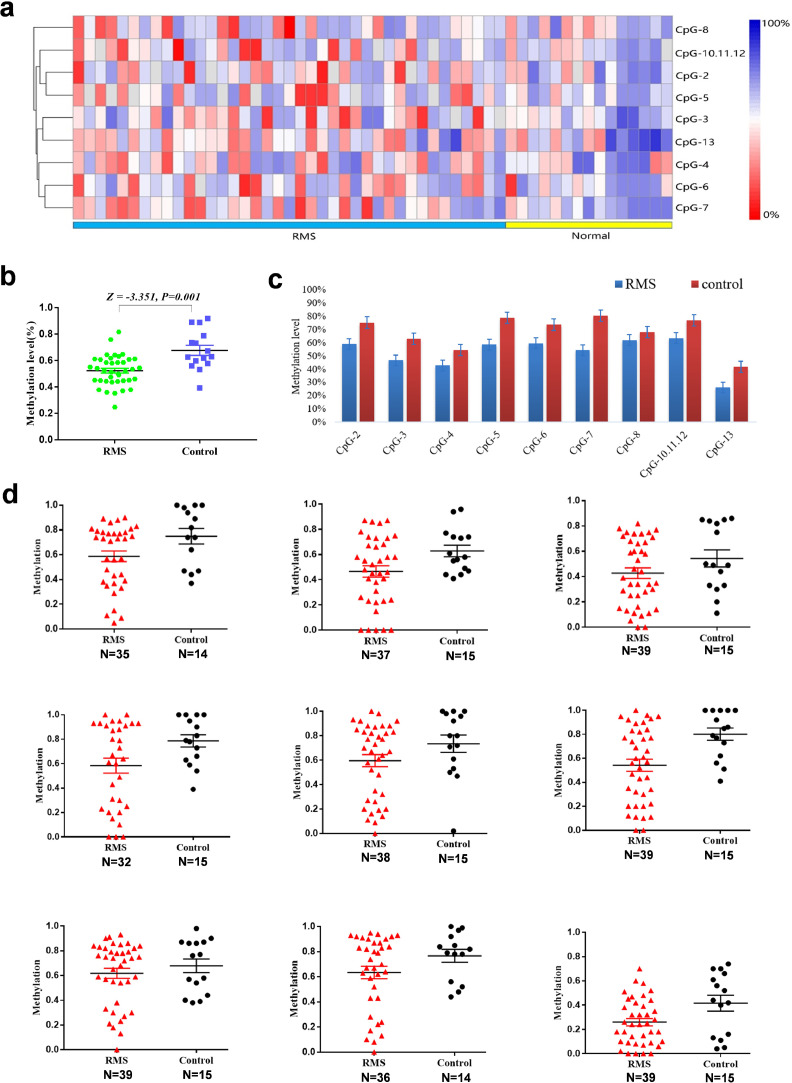
The overall methylation level of the target fragment of the GEFT promoter was statistically lower (0.5226±0.1147) in rhabdosyosarcoma than in normal skeletal muscle tissues (0.6768±0.1483, Z= −3.351, P = 0.001) (Fig. 6a and b). Importantly, the methylation level of 9 CpG units of the GEFT promoter in rhabdomyosarcoma was lower than that in normal skeletal muscle tissue, and changes at CpG-2, CpG-7, and CpG-13 were statistically significant (Fig. 6c, d and Supplementary Table 2). Furthermore, rhabdomyosarcoma patients had significant differences among different histological subtypes of rhabdomyosarcoma in regard to CpG-7 hypomethylation (χ2 = 10.886, P = 0.012; Supplementary Table 3). However, GEFT gene methylation levels showed no obvious differences among the patients with regards to their sex, age, ethnicity, or tumour location (P > 0.05).
Fig. 6.
GEFT is upregulated in rhabdomyosarcoma via aberrant promoter hypomethylation. (a) Hierarchical cluster analysis of CpG units’ methylation profiles of the GEFT promoter region in RMS (n = 39) and normal tissues (n = 15). Each vertex indicates one CpG site. Each column represents one sample. The colour gradient between red and blue indicates methylation of each GEFT CpG unit in each sample (0–100%). Grey represents inadequate or missing data. (b) Comparison of average methylation of GEFT promoter between RMS and skeletal muscle tissues. (c) Histogram analysis of the average methylation level of 9 CpG units of the GEFT promoter in RMS and skeletal muscle tissues. (d) Scatter plot of the average methylation level of 9 CpG units of the GEFT promoter in RMS and skeletal muscle tissues. Wilcoxon nonparametric test was used to compare the indexes between two groups.
We have previously used immunohistochemistry to show that in 39 cases of rhabdomyosarcoma, 37 cases showed overexpression of GEFT protein (37/39, 94.9%), while in 15 cases of normal skeletal muscle, only 1 case was weakly positive (1/15, 6.7%). Pearson bivariate correlation analysis showed that low level methylation of the CpG-2, CpG-7, and CpG-13 sites increased the expression level of the GEFT protein (P = 0.006; P = 0.041; P = 0.037) (Supplementary Table 4).
4. Discussion
Metastasis of tumour cells, rather than the primary tumour, is the major cause of death from cancer. Therefore, understanding the molecular mechanism involved in the metastasis of cancer cells is crucial. Metastasis is a complex process that requires the concerted action of many genes and signalling pathways [22,23]. In this context, Rho-GTPases play fundamental roles. Rho-GTPases not only regulate the formation of specific actin-containing structures [24] but also regulate several other cell migration-related processes, including polarity, adhesion, and substrate recognition [25]. Furthermore, Rho-GTPases cycling between GTP-bound and GDP-bound states have been widely implicated in cancer progression. The key step in their activation is the exchange of GDP for GTP, which is catalysed by RhoGEFs.
RhoGEFs are intracellular effectors of cell surface receptors; some RhoGEFs are specific for Rho, Rac, or Cdc42 [26]. Known RhoGEFs include Vav1-3, P-Rex, Tiam1, TRIO, and Ect2. Most RhoGEFs display increased abundance or activity in human tumours and can promote tumour cell migration and invasion [27], [28], [29], [30], [31], [32], [33]. Inhibiting some RhoGEFs blocks many functions associated with tumourigenesis and prevents metastasis [34,35]. However, reports about the roles of RhoGEFs in sarcomas are rare. In one study, TRIO was found to be frequently affected by high-level amplifications and abundant expression, and it may be involved in the development of soft tissue sarcomas [36]. To date, the roles of GEFT in tumours have been poorly investigated.
GEFT, as one of the RhoGEFs, comprises the Dbl and pleckstrin homology domains with short N- and C-terminal sequences. At present, the GEFT signalling pathway involved in RMS is unknown. The interaction between GEFT and the blood vessel epicardial substance controls cell shape and movement through regulating Rac1/Cdc42 activity [37]. GEFT regulation of lens differentiation and eye development occurs through a Rac-mediated mechanism [38]. GEFT selectively couples with Gαq/11 to activate RhoA in blood vessels and cultured cells; it also mediates the physiologically important Ca2+sensitisation of force induced by Gαq/11-coupled agonists [39,40]. Moreover, GEFT is an important mediator of angiotensin II-dependent RhoA activation in rat aortic smooth muscle cells [41]. GEFT limits lamellipodial protrusion to one direction via RhoA activation in breast carcinoma cells [42]. The GEFT-RhoA signalling pathway participates in the regulation of breast tumour initiation and progression [43]. GEFT activation of the Rac/Cdc42-p21-activated kinase I (PAK) signalling pathways enhances neurite outgrowth in neuroblastoma cells [10]. In our study, we confirmed that GEFT influences RMS cells via the Rho-GTPase signalling pathway.
EMT consists of disaggregating epithelial units and reshaping epithelia for movement. In transition, the epithelium loses polarity, adherens junctions, and tight junctions. This phenotypic conversion requires new molecular biochemical instructions [44]. The cellular adaptations characterising EMT are driven by growth factor signalling pathways, such as transforming growth factor b (TGF-b), Wnt, and fibroblast growth factor (FGF) [45]. Members of the TGF-b family of growth factors can initiate and maintain EMT in a variety of biological systems and during pathophysiological progression [46]. EMT involves signalling factors that induce the expression of specific transcription factors (e.g., Snail, Twist, Zeb) and post-translational regulators, many of which are involved in embryonic development, wound healing and cancer metastasis [47].
EMT has been extensively studied in the last decade, but MET has only recently become noteworthy [48]. MET may exist in sarcomas [49,50]. The MET process is associated with a better prognosis through integrated proteomics and genomics analyses in soft tissue leiomyosarcomas [51]. Snail1 is highly expressed both in ARMS samples from patients and in ARMS cell lines. The expression level of E-cadherin is downregulated in ARMS [52]. Snail is a key regulator of ARMS tumour growth and differentiation through repressing the functions of MYF5 and MYOD [53]. Therefore, MET may play an important role in mesenchymal tumours.
Rac1 promotes the EMT programme in gastric adenocarcinoma and in the acquisition of a cancer stem-like cell state [54]. Rac1, Snail1, Twist1, N-cadherin, and Vim levels are markedly elevated, whereas E-cadherin levels are substantially decreased in non-small cell lung cancer (NSCLC). Rac1 expression is positively correlated with Snail1, Twist1, N-cadherin, and Vim levels and negatively correlated with E-cadherin levels in NSCLC tissues [55]. Interferon regulatory factor 4 binding protein regulates EMT and the motility of breast cancer cells via the Rac1/Cdc42 signalling pathways [56]. In our study, GEFT overexpression upregulated the expression levels of N-cadherin, Snail, Slug, Twist, Zeb1, and Zeb2 and reduced the expression level of E-cadherin. Additionally, GEFT influenced the expression of EMT/MET markers of RMS cells via the Rac1/Cdc42-PAK1 pathways.
There was a study showing that NELL1 activation was correlated with CpG hypomethylation in patients with RMS [57]. We found that elevated GEFT expression in RMS was due to promoter hypomethylation, and CpG-2, CpG-7, and CpG-13 hypomethylation was correlated with the expression of the GEFT protein. Kurmasheva's study found that hypermethylation of the PAX3 gene decreased its protein expression in ERMS, while hypomethylation of the PAX3 gene has been found in ARMS [58]. A hypermethylated MYOD1 gene was found in ERMS but not in ARMS [59]. Our results showed that the hypomethylation of the GEFT gene promoter region was different in different histologic subtypes of RMS, which may open up new approaches for the auxiliary diagnosis of RMS.
5. Conclusion
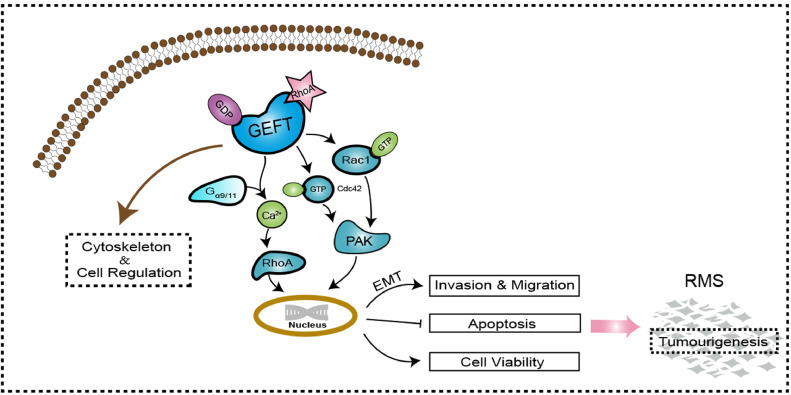
This study not only demonstrated a function of GEFT in the tumourigenicity and metastasis of RMS but also delineated its potential molecular mechanism. GEFT influences RMS cells via the Rac1/Cdc42-PAK signalling pathway and contributes to the molecular mechanism of EMT to promote the invasion and metastasis of RMS (Fig. 7). The newly identified GEFT-Rho-GTPase-EMT axis provides new insights into the invasion and metastasis of RMS and is a valuable target for RMS therapy. Future studies on its function in other cancers will provide further information about the role of GEFT in cancer progression and metastasis.
Fig. 7.
GEFT function and signalling pathway. GEFT comprises the Dbl and pleckstrin homology domains and regulates cytoskeleton and cellular processes. GEFT has specific exchange activity for Rac, Cdc42 and GST-PAK. GEFT selectively couples with Gαq/11 to activate RhoA in blood vessels and cultured cells; it also mediates Ca2+ sensitization of the force induced with Gαq/11-coupled agonists. GEFT activates the Rac1/Cdc42 signalling pathway and promotes the tumourigenicity and metastasis of RMS by influencing the expression of EMT-related proteins of RMS cells.
Collectively, our findings provide important clues about the mechanisms that GEFT uses to accelerate the tumourigenicity and metastasis of RMS by activating Rac1/Cdc42-PAK signalling pathway-induced EMT. These findings suggest that GEFT may serve as a novel therapeutic target.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgement
We thank Professor Yiting Zhou for offering expert advice concerning this study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.10.060.
Contributor Information
Chunxia Liu, Email: liuliu2239@sina.com.
Feng Li, Email: lifeng@shzu.edu.cn, lifeng7855@126.com.
Appendix. Supplementary materials
References
- 1.Doyle L.A. Sarcoma classification: an update based on the 2013 World Health Organization classification of tumors of soft tissue and bone. Cancer. 2014;120(12):1763–1774. doi: 10.1002/cncr.28657. [DOI] [PubMed] [Google Scholar]
- 2.Breneman J.C., Lyden E., Pappo A.S., Link M.P., Anderson J.R., Parham D.M. Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma – a report from the Intergroup Rhabdomyosarcoma Study IV. J Clin Oncol. 2003;21(1):78–84. doi: 10.1200/JCO.2003.06.129. [DOI] [PubMed] [Google Scholar]
- 3.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 4.Valastyan S., Weinberg R.A. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C., Li D., Jiang J., Hu J., Zhang W., Chen Y. Analysis of molecular cytogenetic alteration in rhabdomyosarcoma by array comparative genomic hybridization. PLoS One. 2014;9(4):e94924. doi: 10.1371/journal.pone.0094924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun C., Liu C., Li S., Li H., Wang Y., Xie Y. Overexpression of GEFT, a Rho family guanine nucleotide exchange factor, predicts poor prognosis in patients with rhabdomyosarcoma. Int J Clin Exp Pathol. 2014;7(4):1606–1615. [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D., Yang X., Yang D., Songyang Z. Genetic screens in mammalian cells by enhanced retroviral mutagens. Oncogene. 2000;19(52):5964–5972. doi: 10.1038/sj.onc.1203992. [DOI] [PubMed] [Google Scholar]
- 8.Guo X., Stafford L.J., Bryan B., Xia C., Ma W., Wu X. A Rac/Cdc42-specific exchange factor, GEFT, induces cell proliferation, transformation, and migration. J Biol Chem. 2003;278(15):13207–13215. doi: 10.1074/jbc.M208896200. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt A., Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16(13):1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 10.Bryan B., Kumar V., Stafford L.J., Cai Y., Wu G., Liu M. GEFT, a Rho family guanine nucleotide exchange factor, regulates neurite outgrowth and dendritic spine formation. J Biol Chem. 2004;279(44):45824–45832. doi: 10.1074/jbc.M406216200. [DOI] [PubMed] [Google Scholar]
- 11.Bryan B.A., Mitchell D.C., Zhao L., Ma W., Stafford L.J., Teng B.B. Modulation of muscle regeneration, myogenesis, and adipogenesis by the Rho family guanine nucleotide exchange factor GEFT. Mol Cell Biol. 2005;25(24):11089–11101. doi: 10.1128/MCB.25.24.11089-11101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutz S., Freichel-Blomquist A., Rumenapp U., Schmidt M., Jakobs K.H., Wieland T. p63RhoGEF and GEFT are Rho-specific guanine nucleotide exchange factors encoded by the same gene. Naunyn Schmiedebergs Arch Pharmacol. 2004;369(5):540–546. doi: 10.1007/s00210-004-0926-5. [DOI] [PubMed] [Google Scholar]
- 13.Souchet M., Portales-Casamar E., Mazurais D., Schmidt S., Leger I., Javre J.L. Human p63RhoGEF, a novel RhoA-specific guanine nucleotide exchange factor, is localized in cardiac sarcomere. J Cell Sci. 2002;115(Pt 3):629–640. doi: 10.1242/jcs.115.3.629. [DOI] [PubMed] [Google Scholar]
- 14.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peinado H., Portillo F., Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol. 2004;48(5–6):365–375. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- 16.Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez D.M., Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7(344):re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang H., Liu Y., Li Z., Liu Q., Cui W., Zhang L. MicroRNA-874 functions as a tumor suppressor in rhabdomyosarcoma by directly targeting GEFT. Am J Cancer Res. 2019;9(4):668–681. [PMC free article] [PubMed] [Google Scholar]
- 19.Liang G., Weisenberger D.J. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics. 2017;12(6):416–432. doi: 10.1080/15592294.2017.1311434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki M., Nishimura R., Yoshida K., Shimamura T., Shiraishi Y., Sato Y. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun. 2015;6:7557. doi: 10.1038/ncomms8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun W., Chatterjee B., Wang Y., Stevenson H.S., Edelman D.C., Meltzer P.S. Distinct methylation profiles characterize fusion-positive and fusion-negative rhabdomyosarcoma. Mod Pathol. 2015;28(9):1214–1224. doi: 10.1038/modpathol.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang A.C., Massague J. Molecular basis of metastasis. New Eng J Med. 2008;359(26):2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang Y., Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118(3):277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Ridley A.J., Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70(3):389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 25.Jaffe A.B., Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 26.Barrio-Real L., Kazanietz M.G. Rho GEFs and cancer: linking gene expression and metastatic dissemination. Sci Signal. 2012;5(244):pe43. doi: 10.1126/scisignal.2003543. [DOI] [PubMed] [Google Scholar]
- 27.Citterio C., Menacho-Marquez M., Garcia-Escudero R., Larive R.M., Barreiro O., Sanchez-Madrid F. The rho exchange factors vav2 and vav3 control a lung metastasis-specific transcriptional program in breast cancer cells. Sci Signal. 2012;5(244):ra71. doi: 10.1126/scisignal.2002962. [DOI] [PubMed] [Google Scholar]
- 28.Zheng M., Simon R., Mirlacher M., Maurer R., Gasser T., Forster T. TRIO amplification and abundant mRNA expression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Am J Pathol. 2004;165(1):63–69. doi: 10.1016/S0002-9440(10)63275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fortin S.P., Ennis M.J., Schumacher C.A., Zylstra-Diegel C.R., Williams B.O., Ross J.T. Cdc42 and the guanine nucleotide exchange factors Ect2 and trio mediate Fn14-induced migration and invasion of glioblastoma cells. Mol Cancer Res. 2012;10(7):958–968. doi: 10.1158/1541-7786.MCR-11-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Zapico M.E., Gonzalez-Paz N.C., Weiss E., Savoy D.N., Molina J.R., Fonseca R. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7(1):39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 31.Prieto-Sanchez R.M., Hernandez J.A., Garcia J.L., Gutierrez N.C., San Miguel J., Bustelo X.R. Overexpression of the VAV proto-oncogene product is associated with B-cell chronic lymphocytic leukaemia displaying loss on 13q. Br J Haematol. 2006;133(6):642–645. doi: 10.1111/j.1365-2141.2006.06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menacho-Marquez M., Garcia-Escudero R., Ojeda V., Abad A., Delgado P., Costa C. The Rho exchange factors Vav2 and Vav3 favor skin tumor initiation and promotion by engaging extracellular signaling loops. PLoS Biol. 2013;11(7) doi: 10.1371/journal.pbio.1001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin H., Li T., Ding Y., Deng Y., Zhang W., Yang H. Methylation status of T-lymphoma invasion and metastasis 1 promoter and its overexpression in colorectal cancer. Hum Pathol. 2011;42(4):541–551. doi: 10.1016/j.humpath.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Lazer G., Katzav S. Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy? Cell Signal. 2011;23(6):969–979. doi: 10.1016/j.cellsig.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Razidlo G.L., Magnine C., Sletten A.C., Hurley R.M., Almada L.L., Fernandez-Zapico M.E. Targeting pancreatic cancer metastasis by inhibition of Vav1, a driver of tumor cell invasion. Cancer Res. 2015;75(14):2907–2915. doi: 10.1158/0008-5472.CAN-14-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adamowicz M., Radlwimmer B., Rieker R.J., Mertens D., Schwarzbach M., Schraml P. Frequent amplifications and abundant expression of TRIO, NKD2, and IRX2 in soft tissue sarcomas. Genes Chromosomes Cancer. 2006;45(9):829–838. doi: 10.1002/gcc.20343. [DOI] [PubMed] [Google Scholar]
- 37.Smith T.K., Hager H.A., Francis R., Kilkenny D.M., Lo C.W., Bader D.M. Bves directly interacts with GEFT, and controls cell shape and movement through regulation of Rac1/Cdc42 activity. Proc Natl Acad Sci USA. 2008;105(24):8298–8303. doi: 10.1073/pnas.0802345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell D.C., Bryan B.A., Liu L., Hu X.H., Huang X.Q., Ji W.K. GEFT, a rho family guanine nucleotide exchange factor, regulates lens differentiation through a Rac1-mediated mechanism. Curr Mol Med. 2011;11(6):465–480. doi: 10.2174/156652411796268687. [DOI] [PubMed] [Google Scholar]
- 39.Momotani K., Artamonov M.V., Utepbergenov D., Derewenda U., Derewenda Z.S., Somlyo A.V. p63RhoGEF couples Galpha(q/11)-mediated signaling to Ca2+ sensitization of vascular smooth muscle contractility. Circ Res. 2011;109(9):993–1002. doi: 10.1161/CIRCRESAHA.111.248898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Momotani K., Somlyo AV. p63RhoGEF: a new switch for G(q)-mediated activation of smooth muscle. Trends Cardiovasc Med. 2012;22(5):122–127. doi: 10.1016/j.tcm.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wuertz C.M., Lorincz A., Vettel C., Thomas M.A., Wieland T., Lutz S. p63RhoGEF–a key mediator of angiotensin II-dependent signaling and processes in vascular smooth muscle cells. FASEB J. 2010;24(12):4865–4876. doi: 10.1096/fj.10-155499. [DOI] [PubMed] [Google Scholar]
- 42.Hayashi A., Hiatari R., Tsuji T., Ohashi K., Mizuno K. p63RhoGEF-mediated formation of a single polarized lamellipodium is required for chemotactic migration in breast carcinoma cells. FEBS Lett. 2013;587(6):698–705. doi: 10.1016/j.febslet.2013.01.043. [DOI] [PubMed] [Google Scholar]
- 43.Cho S.G., Wang Y., Rodriguez M., Tan K., Zhang W., Luo J. Haploinsufficiency in the prometastasis Kiss1 receptor Gpr54 delays breast tumor initiation, progression, and lung metastasis. Cancer Res. 2011;71(20):6535–6546. doi: 10.1158/0008-5472.CAN-11-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalluri R., Neilson E.G. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112(12):1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zavadil J., Bottinger E.P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 47.Moustakas A., de Herreros A.G. Epithelial-mesenchymal transition in cancer. Mol Oncol. 2017;11(7):715–717. doi: 10.1002/1878-0261.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J., Du X., Wang G., Sun Y., Chen K., Zhu X. Mesenchymal to epithelial transition in sarcomas. Eur J Cancer. 2014;50(3):593–601. doi: 10.1016/j.ejca.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Wells A., Yates C., Shepard C.R. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin Exp Metastasis. 2008;25(6):621–628. doi: 10.1007/s10585-008-9167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito T., Nagai M., Ladanyi M. SYT-SSX1 and SYT-SSX2 interfere with repression of E-cadherin by snail and slug: a potential mechanism for aberrant mesenchymal to epithelial transition in human synovial sarcoma. Cancer Res. 2006;66(14):6919–6927. doi: 10.1158/0008-5472.CAN-05-3697. [DOI] [PubMed] [Google Scholar]
- 51.Yang J., Eddy J.A., Pan Y., Hategan A., Tabus I., Wang Y. Integrated proteomics and genomics analysis reveals a novel mesenchymal to epithelial reverting transition in leiomyosarcoma through regulation of slug. Mol Cell Proteom. 2010;9(11):2405–2413. doi: 10.1074/mcp.M110.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puskulluoglu M., Lukasiewicz E., Miekus K., Jarocha D., Majka M. Differential expression of Snail1 transcription factor and Snail1-related genes in alveolar and embryonal rhabdomyosarcoma subtypes. Folia Histochemica et Cytobiologica. 2010;48(4):671–677. doi: 10.2478/v10042-010-0046-7. [DOI] [PubMed] [Google Scholar]
- 53.Skrzypek K., Kusienicka A., Trzyna E., Szewczyk B., Ulman A., Konieczny P. SNAIL is a key regulator of alveolar rhabdomyosarcoma tumor growth and differentiation through repression of MYF5 and MYOD function. Cell Death Dis. 2018;9(6):643. doi: 10.1038/s41419-018-0693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon C., Cho S.J., Chang K.K., Park D.J., Ryeom S.W., Yoon S.S. Role of Rac1 pathway in epithelial-to-mesenchymal transition and cancer stem-like cell phenotypes in gastric adenocarcinoma. Mol Cancer Res. 2017;15(8):1106–1116. doi: 10.1158/1541-7786.MCR-17-0053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Zhou Y., Liao Q., Han Y., Chen J., Liu Z., Ling H. Rac1 overexpression is correlated with epithelial mesenchymal transition and predicts poor prognosis in non-small cell lung cancer. J Cancer. 2016;7(14):2100–2109. doi: 10.7150/jca.16198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z., Yang M., Chen R., Su W., Li P., Chen S. IBP regulates epithelial-to-mesenchymal transition and the motility of breast cancer cells via Rac1, RhoA and Cdc42 signaling pathways. Oncogene. 2014;33(26):3374–3382. doi: 10.1038/onc.2013.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tombolan L., Poli E., Martini P., Zin A., Romualdi C., Bisogno G. NELL1, whose high expression correlates with negative outcomes, has different methylation patterns in alveolar and embryonal rhabdomyosarcoma. Oncotarget. 2017;8(20):33086–33099. doi: 10.18632/oncotarget.16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurmasheva R.T., Peterson C.A., Parham D.M., Chen B., McDonald R.E., Cooney C.A. Upstream CpG island methylation of the PAX3 gene in human rhabdomyosarcomas. Pediatr Blood Cancer. 2005;44(4):328–337. doi: 10.1002/pbc.20285. [DOI] [PubMed] [Google Scholar]
- 59.Chen B., Dias P., Jenkins J.J., Savell V.H., 3rd, Parham D.M. Methylation alterations of the MyoD1 upstream region are predictive of subclassification of human rhabdomyosarcomas. Am J Pathol. 1998;152(4):1071–1079. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.