Abstract
Background
Although various molecular subtypes of bladder cancer (BC) have been investigated, most of these studies have focused on muscle-invasive BC (MIBC). A few studies have investigated non-muscle-invasive BC (NMIBC) or NMIBC and MIBC together, but none has classified progressive NMIBC or immune checkpoint inhibitor (ICI)-based therapeutic responses in early-stage BC patients.
Methods
A total of 1,934 samples from seven patient cohorts were used. We performed unsupervised hierarchical clustering to stratify patients into distinct subgroups and constructed a classifier by applying SAM/PAM algorithms. We then investigated the association between molecular subtypes and immunotherapy responsiveness using various statistical methods.
Findings
We explored large-scale genomic datasets encompassing NMIBC and MIBC, redefining four distinct molecular subtypes, including a subgroup containing progressive NMIBC and MIBC with poor prognosis that would benefit from ICI treatment. This subgroup showed poor progression-free survival with the distinct features of high mutation load, activated cell cycle, and inhibited TGFβ signalling. Importantly, we verified that BC patients with this subtype were significantly responsive to an anti-PD-L1 agent in the IMvigor210 cohort.
Interpretation
Our results reveal an immunotherapeutic option for ICI treatment of highly progressive NMIBC and MIBC with poor prognosis.
Funding
This research was supported by the National Research Foundation of Korea grant funded by the Korean government, a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare, Republic of Korea, and a grant from the KRIBB Research Initiative Program.
Keywords: Bladder cancer, Disease progression, Subtype, Immunotherapy, Genomic signature
Research in context
Evidence before this study
Bladder cancer (BC) is one of the most prevalent cancers in the world and one of the cancers that is most difficult to treat; thus, improving patient outcomes is an important challenge. Cancer immunotherapy, including immune checkpoint inhibitors (ICIs), has emerged as an important therapeutic approach to treat BC and other malignancies, but only a fraction of patients respond to treatment. Although several potential predictive markers have been proposed, the underlying mechanisms governing the response to ICIs, which would be utilized to classify the most responsive subset of patients, have not been fully elucidated.
Added value of this study
We identified four distinct molecular subtypes, including a subgroup containing progressive non-muscle-invasive BC (NMIBC) and muscle-invasive BC (MIBC) with poor prognosis that would benefit from ICI treatment. We verified the independent utility of this subtype using a logistic regression model that included PD-L1 expression and tumour mutational burden in the IMvigor210 cohort.
Implications of all the available evidence
Our newly constructed classification system supports an immunotherapeutic option via ICI treatment for highly progressive NMIBC and MIBC with poor prognosis. We believe that our results provide a modality for developing new treatment strategies for both NMIBC and MIBC.
1. Introduction
Bladder cancer (BC) is one of the most prevalent malignancies worldwide, with an estimated 430,000 new cases and 165,000 deaths annually [1]. BC continues to be one of the most difficult malignancies to treat; thus, improving patient outcomes remains an important challenge. BC is categorized into two subtypes according to tumour stage: non-muscle-invasive BC (stage Ta or T1; NMIBC) and muscle-invasive BC (stage T2, T3, or T4; MIBC). Upon initial diagnosis, the majority of cases of BC are NMIBCs, which are treated with transurethral resection of the bladder tumour followed by intravesical Bacillus Calmette-Guérin (BCG) therapy. Despite these treatments, many patients with NMIBC, especially those with T1 high-grade tumours, experience progression to MIBC [2]. MIBCs, which are treated with cisplatin-based neoadjuvant chemotherapy followed by radical cystectomy, have poor prognosis with a five-year survival <50% and accompanying high rates of metastatic relapse after radical cystectomy [3].
Recently, immune checkpoint inhibitors (ICIs), such as anti-PD-1/PD-L1 inhibitors, have emerged as an important therapeutic approach for advanced and metastatic BC [4] and for BCG-unresponsive NMIBC [5], [6], [7]. Despite the success of immunotherapy, only a fraction of patients benefit from treatment with ICIs. Although the detection of tumour and/or immune cell PD-L1 by immunohistochemistry has been investigated as a potential biomarker for response to ICI [8], the prognostic value of PD-L1 has not been determined, and conflicting results have been reported regarding the relationship between PD-L1 protein expression and patient survival. Thus, to increase the utility of ICIs, further development of the genomic signature as a robust and predictive biomarker for potential response to ICIs in concert with immunohistochemistry-based biomarkers in BC is needed. Numerous investigations have been conducted on predictive biomarkers [9,10], but the underlying mechanisms governing the response to ICIs, which would be utilized to classify the most responsive patient subset, have yet to be fully elucidated.
Several studies have revealed that the molecular subtypes MIBC [11], [12], [13], [14] and NMIBC [15] resemble breast cancer subtypes. Although the molecular differences between MIBC and NMIBC are reportedly distinct, some studies suggest that transcriptional subtypes might be independent of conventional stages or grades [16], [17], [18]. While molecular subtypes associated with response [14] or resistance [11] to neoadjuvant chemotherapy have been suggested, distinct subtypes that can predict a potential response to ICIs in NMIBC and MIBC have not been investigated. Although a previous investigation [19] reported an association between ICI responsiveness and subtype classification using a system that addresses NMIBC and MIBC, namely, Lund taxonomy [17], this study was unable to classify progressive NMIBC or ICI-based therapeutic clues in early-stage BC patients. Considering these limitations and recent publications of large-scale BC cohorts, such as The Cancer Genome Atlas (TCGA; ref. 13) and UROMOL [15], there is a great need to explore the landscape of BC to recharacterize its molecular subtypes.
In this study, we report that BC can be sub-grouped into four major classes with different molecular and clinical characteristics, regardless of the histopathological classification system. The four subtypes of BC exhibited distinct features: (1) the presence of FGFR3 or TP53 mutations, (2) the activity of immune response pathways, (3) the expression of cell cycle genes, and (4) the epithelial-mesenchymal transition (EMT). Integrative analysis of mutation and gene expression data revealed that class 3, exhibiting similar proportions of high-grade NMIBC and MIBC, showed distinct biological features associated with response to ICIs. Using gene expression profiles from the IMvigor210 cohort [19], we also verified a significant association between class 3 and response to ICIs, suggesting an immunotherapeutic option via ICI treatment for high-risk NMIBCs with progressive disease and for a subset of MIBCs that are responsive to ICI.
2. Materials and methods
2.1. Patients and gene expression data
Microarray datasets related to gene expression in BC were obtained from the GEO database. Datasets with clinical information including follow-up time data were included in this study. Gene expression and clinical data from 165 primary BC patients from the Chungbuk National University Hospital were used as the discovery cohort (GSE13507; the CNUH cohort, n = 102 NMIBC and 63 MIBC; ref. 20). Collection and analysis of all samples was approved by the institutional review board of Chungbuk National University, and informed consent was obtained from each subject. Gene expression datasets for BC from the Yonsei University Severance Hospital (GSE120736; the YUSH cohort, n = 78 NMIBC and 61 MIBC), University Hospital of Lund (GSE19915; the UHL cohort, n = 97 NMIBC and 45 MIBC; ref. 16), and Swedish southern healthcare region (GSE32894; the SSH cohort, n = 213 NMIBC and 93 MIBC; ref. 17) were used for subtype validation. For more critical validations, RNA-seq datasets from the TCGA (GSE62944; n = 408 MIBC; ref. 21), European UROMOL consortium (E-MTAB-4321; n = 460 NMIBC and 16 MIBC; ref. 15), and IMvigor210 (n = 298 MIBC; ref. 19) were downloaded from GEO, ArrayExpress, and the Supplementary data of Mariathasan et al. [19], respectively. To characterize mutation profiles of BC patients, variant data from the TCGA and UROMOL cohorts were also collected from the UCSC Xena Browser and European Genome-Phenome Archive, respectively.
Clinical data were obtained from the Supplementary Information of the corresponding literature or were requested from the authors. Cancer-specific survival was defined as the time from surgery to death caused by cancer, and progression-free survival was defined as the time elapsed between treatment initiation and tumour progression. In this study, disease progression was defined as an increase in stage from either Ta or T1 to T2 or higher after disease relapse. Patients with available survival time data were subjected to survival analysis. The clinicopathologic characteristics of the study subjects are provided in the Supplementary Tables 1, 4, 5, 6 and 7.
2.2. Gene expression analysis
A summary of the methodology is shown in Supplementary Fig. 1. All gene expression datasets were separately log2 transformed and quantile normalized. Before clustering, we selected 3938 genes with an expression level that exhibited at least a two-fold difference relative to the median value in greater than 15% of the samples in the discovery cohort (the CNUH cohort). Cluster analysis with 3938 genes revealed four distinct BC subtypes (data not shown). To select small genes retaining molecular characteristics of the subtypes, we applied the identical procedures for gene expression data of other cohorts that were generated by the same experimental platform (Illumina) as the CNUH cohort, identifying 3235 and 2990 genes in the YUSH and SSH cohorts, respectively. Only the expression of 1627 genes was commonly varied in all three cohorts (Supplementary Fig. 2). Unsupervised clustering of the gene expression matrix consisting of 1627 genes in the discovery cohort revealed four clusters with distinct gene expression patterns. Hierarchical cluster analysis was conducted using Gene Cluster 3.0 (median centring of genes, centroid linkage) and visualized using Java TreeView. Silhouette width was calculated to determine the accuracy of clustering assignment [22]. Only samples with positive silhouette values were retained for further analysis because they best represented each subtype (R-package: Cluster).
2.3. Building a gene-based classifier
We applied significance analysis of microarrays (SAM) to identify significantly differentially expressed genes in each subtype (class 1 vs. the rest, class 2 vs. the rest, class 3 vs. the rest, and class 4 vs. the rest; R-package: SAMR). Subsequently, we selected genes with the greatest difference in expression level among the four subtypes in the CNUH cohort (239, 251, 275, and 471 genes for classes 1, 2, 3, and 4 subtypes, respectively). The resulting four gene sets were combined to yield 851 subtype-specific gene sets. Among these gene sets, genes without the “approved symbol” from the HUGO Gene Nomenclature Committee were removed. The remaining 826 genes were trained using prediction analysis of microarrays (PAM) to build a gene-based classifier (R-package: PAMR). We performed 10-fold cross-validation to select the optimal threshold for centroid shrinkage and selected the value Δ = 2.555, yielding good performance with the 786 genes (Supplementary Fig. 3).
2.4. Subtype classification on validation cohorts
For RNA-seq datasets, genes with a value of zero were set to the missing value after log2 transformation (following the addition of 1 to the FPKM values), and genes were removed if they had missing data in >30% of the samples. After quantile normalization, expression levels of each gene in each dataset were independently standardized to a mean of zero and a standard deviation of 1 and then merged to yield a large, pooled RNA-seq validation cohort consisting of 884 BC samples.
The validation cohorts were independently assigned to BC subtypes using the PAM classifier built based on the discovery cohort. YUSH and the SSH cohorts were generated by the same microarray platform as the discovery cohort (Illumina microarray platform). For each dataset, expression profiles were median centred across all samples and subjected to classification using the classifier built based on the discovery cohort.
The gene expression data from the UHL (Swegen microarray platform), TCGA, UROMOL, and IMvigor210 cohorts do not contain all of the gene symbols in the discovery cohort. Thus, we used signature genes that were contained in both the discovery and validation cohorts. We trained a classifier on the discovery cohort and used it to predict the tumour subtype in the validation cohort as mentioned above. Finally, a heat map was generated using predicted class and subtype-specific genes.
2.5. Mutation significance analysis
Using somatic variant data from the TCGA cohort, the mutation profiles of the 485 genes with total mutation frequency ≥5% and the genes involved in the oncogenic signature (64 genes) and DNA damage response and repair (DDR; 165 genes) were investigated. Likewise, the mutation profiles of FGFR3, genes involved in the oncogenic signature (64 genes) and DDR (165 genes) were investigated in the UROMOL cohort.
2.6. Analysis of an association between class 3 and response to ICIs
To investigate the association between the class 3 subtype and ICI response, we examined tumours from the IMvigor210 cohort of patients with metastatic urothelial cancer who were treated with an anti-PD-L1 agent (atezolizumab). RNA-seq data and clinical information from 298 patients with metastatic urothelial cancer were obtained from IMvigor210CoreBiologies, a fully documented software and data package for the R statistical computing environment. The four subtypes of BC were assigned as mentioned above. To assess the independent utility of BC subtype for predicting ICI response, generalized linear models were used for fitting a binary response (responder vs. nonresponder) as the dependent variable, while PD-L1 expression on tumour cells (TC), PD-L1 expression on immune cells (IC), or tumour mutation burden, and BC subtypes were used as independent variables.
2.7. Statistical analysis
The chi-squared test and the two-sample t-test were used to assess differences between groups for categorical and continuous variables, respectively. A log-rank test was used to estimate associations between subtypes and survival outcomes. Statistical analyses were performed in the R language environment and were considered to be significant with a P-value <0·05 (two-sided).
3. Results
3.1. Identification of BC subtypes by hierarchical clustering
To identify distinct molecular subtypes of BC regardless of previously known clinicopathological factors (stage or grade), we performed an unsupervised hierarchical cluster analysis using gene expression profile data from the Chungbuk National University Hospital (CNUH; GSE13507) cohort [20]. With 1627 genes commonly varied among the three microarray cohorts (Supplementary Fig. 2), the clustering analysis revealed four distinct BC subtypes (Supplementary Fig. 3a). The PAM algorithm was applied to samples reflecting the molecular characteristics of each subtype along with subtype-specific genes (Supplementary Fig. 3b and 3c) and retained a final subtype-specific signature consisting of 786 genes with the lowest prediction error (Supplementary Fig. 3d). We applied this genomic subtype predictor defined by 786 genes, namely, GSP786, to all 165 patients from the CNUH cohort and obtained consequent subtype stratifications, in which 49 (30%), 44 (27%), 43 (26%), and 29 (18%) patients were noted in classes 1, 2, 3, and 4, respectively (Fig. 1a; Supplementary Table 1).
Fig. 1.
Prediction of four molecular subtypes of BC using the GSP786 classifier in the CNUH and RNA-seq cohorts. (a) Heat map of the subtype-specific signature consisting of 786 genes and associations with clinicopathological features in the CNUH cohort. Class 1 contained many low-grade NMIBCs. Class 2 included both low-grade NMIBCs and a small number of MIBCs. Class 3 exhibited similar involvement in high-grade NMIBC and MIBC. Class 4 contained the most MIBC cases. (b) Heat map of the GSP786 classifier profiled in the RNA-seq cohort combined with TCGA and UROMOL. Samples are ordered according to the four classes with pathological features together with their previous subtype assignment information from the UROMOL, SSH, and TCGA cohorts. Class 1 included many low-stage and low-grade tumours with more MIBCs (71 out of 275 in class 1, 25%) compared with the CNUH cohort (1 out of 49 in class 1, 2%). The sample compositions of classes 2 and 3 were similar to those in the CNUH cohort. Class 4 tumours consist of high-grade invasive tumours with more NMIBCs (76 out of 215, 35%) compared with the CNUH cohort (6 out of 29, 20%). The heat map ranks genes based on fold-change, and genes with the largest fold-change appear at the top. Data are presented in a matrix format in which each row represents an individual gene and each column represents a tissue sample. Each cell in the matrix represents the expression level of a gene feature in an individual sample. The colouring in the cells reflects relativity high (red) and low (green) expression levels as indicated in the scale bar (log2 transformed scale).
To understand the biological characteristics of each subtype, we applied a functional enrichment test to each subtype-specific gene list (Supplementary Table 2). These analyses provided substantial insight into the biological understanding of subtypes (Supplementary Fig. 4; Supplementary Table 3). Tumour samples involved in class 1 mainly included low-grade NMIBCs (Fig. 1a). Class 1 is characterized by decreased expression of genes involved in cell proliferation (Supplementary Fig. 4a), signifying the less aggressive characteristics of class 1. Class 2 included both low-grade NMIBCs and a small number of MIBCs (Fig. 1a). Class 2 displayed the downregulation of immune response pathways, such as antigen processing and presentation and T cell receptor signalling pathways (Supplementary Fig. 4b). All verified human leukocyte antigen (HLA) genes, which were associated with clinical prognosis in cancer patients [23], exhibited a specifically inhibited pattern in class 2 (Supplementary Fig. 5a). We also examined activated functions in class 2, observing increased expression of the oncogenes FGFR3 and CCND1 (Supplementary Fig. 5b; ref. 24). Class 3 exhibited similar involvement of high-grade NMIBC and MIBC (Fig. 1a). In particular, most T1 high-grade tumours (11 out of 16, 69%) were classified into class 3, indicating that class 3 might be capable of detecting high-risk NMIBC with progressive disease. Class 3 displayed the activation of cell cycle-associated functions (Supplementary Fig. 4c) and the inhibition of genes involved in the Notch signalling pathway (Supplementary Fig. 5c). These processes are associated with tumour progression [25], [26], [27]. Based on our findings and the previously reported crucial role of cell cycle-related genes in BC prognosis, such as E2F1, FOXM1, CCNB1, and CCNE1 [20,27,28], we examined expression levels of these genes in NMIBC and MIBC. Increased expression of these genes in class 3 was observed in both the NMIBC and MIBC subgroups (Supplementary Fig. 5d). These results indicate that class 3 may have distinct molecular characteristics suitable for predicting the aggressive clinical behaviour of tumours, regardless of the pathological subtypes of NMIBC and MIBC. Finally, class 4, which contains the most MIBC cases, exhibited clear upregulation of genes implicated in extracellular matrix organization along with strong activation of the immune response (Supplementary Fig. 4d). Additionally, the overexpression of genes associated with EMT or myofibroblasts was noted in class 4 (Supplementary Fig. 5e), which displayed similar features to the infiltrated subtype [17].
3.2. GSP786-based subtype classification in independent cohorts
We applied our classifier to four validation cohorts to verify whether the identification of the four subtypes was reproducible. When GSP786 was applied to the RNA-seq cohort (total n = 884) combined with TCGA [13] and UROMOL [15], all four subtypes were represented (Fig. 1b; Supplementary Table 4). Despite the increased involvement of invasive tumours compared to the CNUH cohort, class 1 demonstrated consistent inhibition of genes involved in cell proliferation compared to other subtypes (Supplementary Fig. 6a). Furthermore, many of the high-grade invasive tumours in class 1 were histologically papillary tumours (36 out of 71, 51%), which exhibit a better prognosis compared with nonpapillary tumours. Moreover, class 1 tumours classified into the luminal-papillary subgroup, a subtype with good prognosis reported by the TCGA consortium [13], were more numerous compared with those in other subclasses (Supplementary Table 4), indicating that class 1 reflects the good prognostic molecular characteristics of BC. The sample compositions of classes 2 and 3 in the RNA-seq cohort were similar to those in the CNUH cohort, and the overall molecular characteristics were retained (Supplementary Fig. 6b and 6c). NMIBC tumours in class 3 were predominantly associated with UROMOL class 2, which was characterized by concomitant carcinoma in situ (CIS), high expression of late cell cycle genes, and the worst progression-free survival (PFS; ref. 15). Although UROMOL class 2 is stratified into infiltrated or genomically unstable (GU) subtypes from the Lund taxonomy [17], the infiltrated subtype might not represent a distinct cancer subtype given that the infiltrated subtype exhibited heterogeneous molecular characteristics [17], and UROMOL class 2 reportedly exhibited a solid characteristic [15]. In contrast to previous investigations, GSP786 successfully classified UROMOL class 2 tumours into two molecular subgroups. Specifically, most tumours with GU were classified into class 3, whereas many tumours with the infiltrated subtype were classified into class 4 (Fig. 1b). These results emphasize the relevance of the predictor in accurately reflecting the molecular differences between GU and the infiltrated subtypes. Class 4 displayed the homogeneous upregulation of genes involved in EMT or myofibroblasts, regardless of tumour stage or grade (Supplementary Fig. 6d), which is consistent with other discovery and validation cohorts. Moreover, the majority of MIBCs in class 4 were associated with the basal-squamous subtype, and the remaining samples corresponded to the luminal-infiltrated subtype. We also identified four subtypes in the Yonsei University Severance Hospital (YUSH; Supplementary Fig. 7a and 8a; Supplementary Table 5), University Hospital of Lund (UHL; Supplementary Fig. 7b; Supplementary Table 6), and Swedish southern healthcare region (SSH; Supplementary Fig. 7c and 8b; Supplementary Table 7) cohorts, indicating the consistency and confidence of the GSP786 classification. Taken together, our classifier demonstrated a consistent molecular classification pattern despite different sample compositions and heterogeneous distributions of histological factors across multiple patient cohorts. More details regarding subtype classification based on the GSP786 in independent cohorts are provided in the Supplementary Results.
3.3. Prognostic significance of subtypes
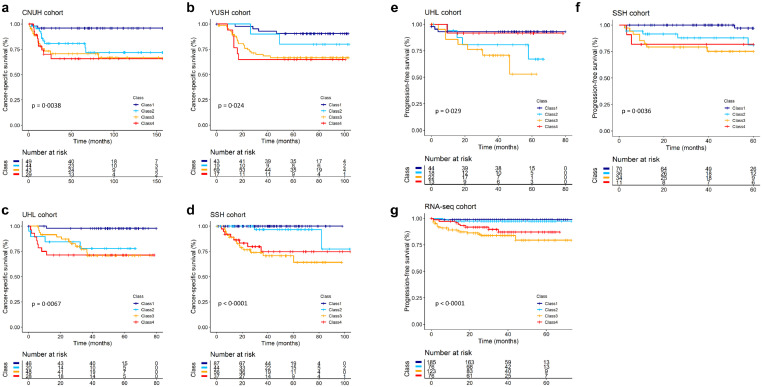
To investigate the ability of molecular subtypes to predict the clinical behaviours in BC patients, we compared patient survival rates among these subtypes in the independent cohorts. We found a statistically significant difference in cancer-specific survival among the four subtypes in the CNUH, YUSH, UHL, and SSH cohorts (each P < 0·05, log-rank test, Fig. 2a–d). In addition, we estimated the PFS rates of the NMIBC patients in three independent cohorts (the UHL, SSH and RNA-seq cohorts). As expected from our molecular characterization of class 3, the frequency of NMIBC progression was higher in class 3 compared to other subclasses (each P < 0·05, log-rank test, Fig. 2e–g). These results support a distinct molecular feature of class 3, reflecting tumour aggressiveness due to cell cycle disorder and its predictive value in classifying high-risk NMIBC patients with progressive disease.
Fig. 2.
Prognostic significance of BC subtypes. (a–d) Kaplan-Meier curves showing time to death in the CNUH, YUSH, UHL, and SSH cohorts. Class 1 exhibited a better prognosis compared with other subclasses. Class 2 exhibited an intermediate level in patient survival, whereas classes 3 and 4 patients exhibited reduced survival rates compared with other subtypes. (e–g) Kaplan-Meier curves showing time to progression in the UHL, SSH, and RNA-seq cohorts. The frequency of NMIBC progression in class 3 was significantly increased compared with the other subclasses. P-values were obtained by log-rank tests. The + symbols in the panels indicate censored data.
3.4. Mutational landscape of the four BC subtypes
We next sought to identify mutations that were specifically associated with the four subtypes using somatic variant data from the TCGA cohort. We found that the somatic mutation rate was significantly increased in class 3 compared with the other subclasses (P < 0·001, two-sample t-test, Fig. 3), suggesting that the DNA repair system is highly impaired in class 3 tumours [29]. Although we identified differences in the mutation frequency among the subtypes, we did not observe unique and recurrent mutations that were solely associated with a particular subtype. The vast majority of class 3 (70%) or class 4 (52%) patients harboured mutations in TP53, which was previously reported as the most frequently mutated gene in MIBC [12,13]. Along with these findings, RB1 mutations, which frequently co-occur with TP53 mutations [13,30] and have adverse prognostic importance [31], were profoundly enriched in classes 3 or 4, demonstrating that classes 3 and 4 exhibit more adverse biological and clinical features of MIBC. On the other hand, the mutation frequency of FGFR3, which is mutually exclusive with TP53 [13] and frequently occurs in NMIBC [32], was significantly increased in classes 1 or 2 compared with classes 3 or 4 (P < 0·001, chi-squared test, Fig. 3), indicating that classes 1 and 2 have less aggressive characteristics and more favourable prognosis. Taken together with our gene expression data, these results imply that MIBC patients could be stratified into distinct subgroups comprising more or less aggressive features.
Fig. 3.
Mutational landscape of BC subtypes in the TCGA cohort. Frequently mutated genes (≥5%, top panel), genes involved in the oncogenic signature (middle panel) and DNA damage response and repair (bottom panel). Samples are sorted according to mutation frequency within each class. Chi-squared tests for differences in frequency pattern between the classes are presented on the left together with total mutation frequency. The mutation frequency for each subtype is presented on the right. The mutation load for each sample is presented above the mutation plot.
To illustrate the mutation pattern associated with the four subtypes in patients with NMIBC, we analysed variant data from the UROMOL cohort (Supplementary Fig. 9). Consistent with our findings in MIBC, the mutation frequency of TP53 was significantly increased in classes 3 and 4 compared with classes 1 and 2. The frequency of FGFR3 was higher in class 1 (61%) than in the other subclasses, but the difference was not statistically significant. This observation may be due to the prevalence of FGFR3 mutations in NMIBC (60–80%; ref. 33). Among the genes involved in DNA damage response and repair (DDR), the frequency of BRCA1 mutations was increased in class 3 compared with the other subtypes in both TCGA and UROMOL cohorts, demonstrating that class 3 exhibits more aggressive characteristics associated with DDR.
3.5. Association between the class 3 subtype and response to ICIs
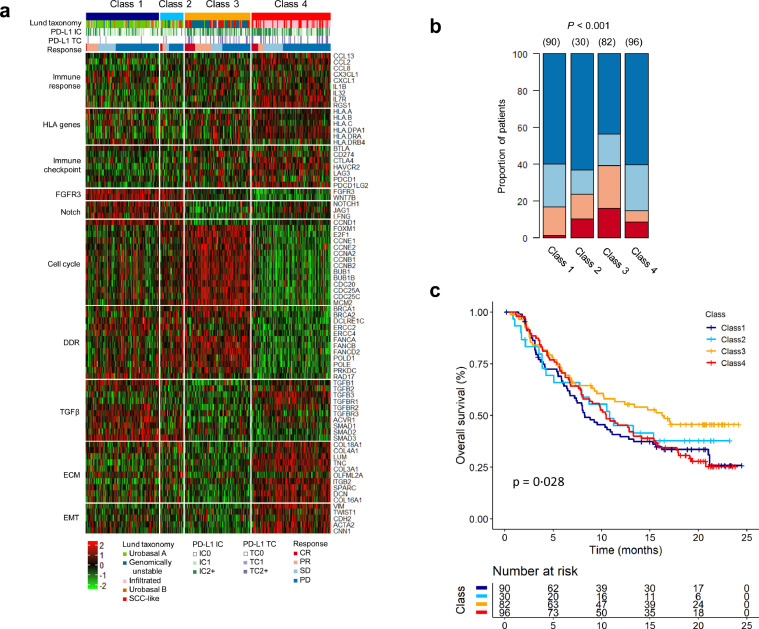
By identifying the four subtypes by GSP786 and exploring their prognostic associations in BC patients, we observed that class 3 had distinct biological features associated with potential responses to ICIs, such as a high somatic mutation rate (Fig. 3; refs. 34, 35) and alterations of genes involved in DDR (Fig. 3; Supplementary Fig. 9; refs. [36], [37], [38]). To verify an association between the class 3 subtype and response to ICIs, we applied GSP786 to the IMvigor210 cohort from 298 BC patients treated with an anti-PD-L1 agent (atezolizumab; ref. 19). As expected, a higher response rate was observed in patients with class 3 tumours, exhibiting increased expression of genes associated with the cell cycle and DDR (Figs. 4a and 4b). Importantly, the class 3 subtype displayed a better survival rate than the other subtypes (P = 0·028, log-rank test, Fig. 4c), implying that MIBC patients with class 3 might have a superior response to ICI treatment.
Fig. 4.
Class 3 subtype is associated with response to ICI. (a) Heat map of the GSP786 classifier profiled in the IMvigor210 cohort. Rows of the heat map show gene expression grouped by specific functions or pathways. IC, immune cells; TC, tumour cells; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease. (b) Response versus predicted subtype based on the GSP786 classifier, demonstrating that class 3 had a significantly increased response rate (P < 0·001, two-sided Fisher's exact test). The numbers in parentheses specify sample numbers in each class. (c) Class 3 exhibited a better survival rate compared with other subtypes (P = 0·028, log-rank test).
Although a previous investigation reported that the GU subtype from Lund taxonomy was enriched for response to ICIs [19], the GU subtype might not represent a potential biomarker for ICIs given that the GU subtype did not show a higher complete response rate than the other subtypes. Furthermore, samples in the class 3 subtype with a complete response were stratified into GU or SCC-like (Fig. 4a), and these subtypes showed similar biological features, such as activation of genes involved in the cell cycle and DDR. This observation further emphasizes the significance of our subtype as a potential biomarker for predicting response to ICIs.
In gene set enrichment analysis, class 3 was negatively associated with the TGFβ signalling pathway (Supplementary Fig. 10a), corresponding to TGFβ attenuating the response to an anti-PD-L1 agent [19]. We also observed decreased expression of TGFβ signalling pathway-associated genes in class 3 (Supplementary Fig. 10b). To assess the independent utility of BC subtypes for predicting the response to ICIs, we fit a logistic regression model to our data and used PD-L1 expression on tumour cells, PD-L1 expression on immune cells, TMB, and BC subtype as potential explanatory factors. In this test, class 3 exhibited a statistically significant relationship with response to ICIs, suggesting the independent utility of subtype for classifying the best responsive patient subset (Table 1). Taken together, these results propose a potential benefit of ICIs as a more aggressive treatment option in class 3 tumours containing high-risk NMIBC that could progress to MIBC.
Table 1.
Logistic regression analysis for prediction of response to ICI.
| Variable | Estimated β | Std. Error | Z-value | P (> Z) |
|---|---|---|---|---|
| PD-L1 expression on TC | −0·2252 | 0·2321 | −0·97 | 0·33198 |
| PD-L1 expression on IC | 0·3746 | 0·221 | 1·695 | 0·09003 |
| Tumour mutation burden | 1·3138 | 0·4517 | 2·909 | 0·00363 |
| BC subtype | 0·9663 | 0·3241 | 2·981 | 0·00287 |
Regression coefficients (Estimated β) represent the mean change in the response variable for one unit of change in the predictor variable while holding other predictors in the model constant.
TC = tumour cell; IC = immune cell; BC = bladder cancer.
4. Discussion
The clinical heterogeneity of BC suggests that biologically relevant subtypes may exist within and between NMIBC and MIBC. Using a series of unsupervised learning approaches, we demonstrated that patients with BC could be sub-grouped into four major classes with distinct molecular characteristics, regardless of previously known clinicopathological factors. By constructing the genomic subtype predictor based on 786 genes (GSP786), we demonstrate the robustness of GSP786 for classifying these four subtypes, and its reproducibility was validated in six independent cohorts with a total of 1934 samples. Furthermore, our data clearly demonstrated the prognostic significance of the four subtypes of BC, particularly the aggressive clinical behaviour of class 3 with ICI responsiveness.
One of the most prominent biological characteristics of classes 1 and 2 was significant alterations in FGFR3, including both mutations and overexpression (Fig. 3; Supplementary Fig. 5b). This finding suggests that FGFR3 is a candidate therapeutic target in these subgroups. Although many studies have reported FGFR3 dysregulation as a promising therapeutic target in multiple preclinical trials [33], further studies are needed to verify the benefit of FGFR3 inhibitors in these subgroups. The main biological difference between classes 1 and 2 was the significant downregulation of immune response pathways in class 2. We demonstrated that HLA gene expression was significantly reduced in class 2 (Supplementary Fig. 5a), and this downregulation is associated with poor prognosis in cancer [23]. Detection and further characterization of class 2 may potentially result in optimized therapy options for patients.
One of the distinct biological characteristics of class 3 tumours was the concordant activation of cell cycle-associated functions in both the NMIBC and MIBC subgroups, suggesting that a subset of patients with high-grade NMIBC or MIBC may share molecular characteristics associated with more aggressive clinical behaviour. Furthermore, class 3 also exhibited high rates of somatic mutations and alterations in DDR genes (Fig. 3; Supplementary Fig. 9). These features suggest a potential benefit for immunotherapy via treatment of class 3 patients with ICIs [37], which is also supported by our findings demonstrating an association between class 3 and ICI responses (Fig. 4). As expected, class 3 was strongly correlated with the response to ICIs. We also found that class 3, which showed poor prognosis in the discovery and validation cohorts, exhibited better survival rates compared with other subclasses in the IMvigor210 cohort.
TMB was shown to be useful in predicting the treatment response to ICIs, and the class 3 subtype has a statistically significant relationship with the response to ICIs (Table 1), suggesting that TMB is not the only contributing factor for higher response in class 3. Furthermore, whole-exome sequencing for the determination of mutation load is expensive and time-consuming, hindering its application in clinical practice, and disease-specific TMB thresholds for the effective prediction of a response in BC are not well-established [39]. Taken together, these results propose our classifier, in concert with TMB, as a potential biomarker for ICI selection across NMIBC and MIBC; however, further prospective validation studies are required to confirm these conclusions.
Although the class 3 subtype has molecular characteristics similar to the previously recognized GU subtype from the Lund taxonomy [17] with respect to activation of cell cycle-related functions, we showed that the class 3 subtype, exhibiting similar involvement of high-grade NMIBC and MIBC, had the worst PFS compared with the other subtypes. Since the urobasal B or SCC-like subtypes showed higher complete response rates than the GU subtype [19], the GU subtype might not be appropriate for identifying patients who are likely to respond to treatment with ICI. On the other hand, we demonstrated the independent utility of our classifier for predicting patient response to ICIs in the IMvigor210 cohort. These results emphasize this approach's predictive value by classifying high-risk NMIBC and MIBC patients who might benefit from ICI therapy.
In conclusion, we identified four molecular subtypes of BC that display discriminative biological and clinical features, even considering all pathological subtypes of BC. Our newly constructed classification system suggests biological activity and potential treatment guidelines for each subclass of BC, especially the prognostic and predictive significance of class 3 with ICI responsiveness. Our findings may contribute to precision medicine in BC by classifying optimal patient subgroups and determining an appropriate therapeutic course. A close relationship between the class 3 subtype and the response to ICIs may present an appropriate treatment option for highly progressive NMIBC and MIBC with a poor prognosis.
Funding sources
This research was supported by the National Research Foundation of Korea (NRF) grant (NRF-2017R1A2B2007302, 2017R1A2B2007836) funded by the Korea government (MSIP), a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (HI16C1866), and a grant from KRIBB Research Initiative Program. The funders had no role in the study design, data collection, data analysis, interpretation, writing of the report.
CRediT authorship contribution statement
Bic-Na Song: Data curation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing. Seon-Kyu Kim: Data curation, Investigation. Jeong-Yeon Mun: Data curation, Investigation. Young-Deuk Choi: Data curation, Investigation. Sun-Hee Leem: Investigation, Project administration, Funding acquisition, Supervision, Writing - original draft. In-Sun Chu: Formal analysis, Project administration, Funding acquisition, Supervision, Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgements
Not applicable.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.10.058.
Contributor Information
Sun-Hee Leem, Email: shleem@dau.ac.kr.
In-Sun Chu, Email: chu@kribb.re.kr.
Appendix. Supplementary materials
References
- 1.Ferlay J., Soerjomataram I., Dikshit R. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Jordan B., Meeks J.J. T1 bladder cancer: current considerations for diagnosis and management. Nat Rev Urol. 2019;16(1):23–34. doi: 10.1038/s41585-018-0105-y. [DOI] [PubMed] [Google Scholar]
- 3.Alfred Witjes J., Lebret T., Comperat E.M. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71(3):462–475. doi: 10.1016/j.eururo.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Balar A.V., Galsky M.D., Rosenberg J.E. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. The Lancet. 2017;389(10064):67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.US National Library of Medicine. Atezolizumab in treating patients with recurrent BCG-unresponsive non-muscle invasive bladder cancer [Internet]. [cited 2019 May 9] Available from: https://ClinicalTrials.gov/show/NCT02844816.
- 6.US National Library of Medicine. Safety and pharmacology study of atezolizumab alone and in combination with Bacille Calmette-Guérin (BCG) in high-risk non-muscle-invasive bladder cancer (NMIBC) participants [Internet]. [cited 2019 May 9] Available from: https://ClinicalTrials.gov/show/NCT02792192.
- 7.US National Library of Medicine. Phase 2 durvalumab (Medi4736) for bacillus calmette-guérin (BCG) refactory urothelial carcinoma in situ of the bladder [Internet]. [cited 2019 May 9] Available from:https://ClinicalTrials.gov/show/NCT02901548.
- 8.Yi M., Jiao D., Xu H. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17(1):129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eich M.-.L., Chaux A., Guner G. Tumor immune microenvironment in non–muscle-invasive urothelial carcinoma of the bladder. Hum Pathol. 2019;89:24–32. doi: 10.1016/j.humpath.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Zeng D, Li M, Zhou R. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol Res. 2019;7(5):737–750. doi: 10.1158/2326-6066.CIR-18-0436. canimm.0436.2018. [DOI] [PubMed] [Google Scholar]
- 11.Choi W, Porten S, Kim S. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25(2):152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson AG, Kim J, Al-Ahmadie H. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(3):540–556. doi: 10.1016/j.cell.2017.09.007. e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seiler R., Ashab H.A.D., Erho N. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol. 2017;72(4):544–554. doi: 10.1016/j.eururo.2017.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Hedegaard J, Lamy P, Nordentoft I. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell. 2016;30(1):27–42. doi: 10.1016/j.ccell.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Lindgren D., Frigyesi A., Gudjonsson S. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70(9):3463–3472. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 17.Sjodahl G., Lauss M., Lovgren K. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18(12):3377–3386. doi: 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- 18.Tan T.Z., Rouanne M., Tan K.T., Huang R.Y., Thiery J.P. Molecular subtypes of urothelial bladder cancer: results from a meta-cohort analysis of 2411 tumors. Eur Urol. 2019;75(3):423–432. doi: 10.1016/j.eururo.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 19.Mariathasan S, Turley SJ, Nickles D. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J.S., Leem S.H., Lee S.Y. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J Clin Oncol. 2010;28(16):2660–2667. doi: 10.1200/JCO.2009.25.0977. [DOI] [PubMed] [Google Scholar]
- 21.Rahman M., Jackson L.K., Johnson W.E., Li D.Y., Bild A.H., Piccolo S.R. Alternative preprocessing of RNA-Sequencing data in the cancer genome atlas leads to improved analysis results. Bioinformatics. 2015;31(22):3666–3672. doi: 10.1093/bioinformatics/btv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- 23.Hirohashi Y, Torigoe T, Mariya T, Kochin V, Saito T, Sato N. HLA class I as a predictor of clinical prognosis and ctl infiltration as a predictor of chemosensitivity in ovarian cancer. Oncoimmunology. 2015;4(5) doi: 10.1080/2162402X.2015.1005507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanli O., Dobruch J., Knowles M.A. Bladder cancer. Nat Rev Dis Primers. 2017;3:17022. doi: 10.1038/nrdp.2017.22. [DOI] [PubMed] [Google Scholar]
- 25.Nowell C.S., Radtke F... Notch as a tumour suppressor. Nat Rev Cancer. 2017;17(3):145–159. doi: 10.1038/nrc.2016.145. [DOI] [PubMed] [Google Scholar]
- 26.Rampias T., Vgenopoulou P., Avgeris M. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat Med. 2014;20(10):1199–1205. doi: 10.1038/nm.3678. [DOI] [PubMed] [Google Scholar]
- 27.Song B.N., Kim S.K., Chu I.S. Bioinformatic identification of prognostic signature defined by copy number alteration and expression of CCNE1 in non-muscle invasive bladder cancer. Exp Mol Med. 2017;49:e282. doi: 10.1038/emm.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S.K., Roh Y.G., Park K. Expression signature defined by FOXM1-CCNB1 activation predicts disease recurrence in non-muscle-invasive bladder cancer. Clin Cancer Res. 2014;20(12):3233–3243. doi: 10.1158/1078-0432.CCR-13-2761. [DOI] [PubMed] [Google Scholar]
- 29.Andor N., Maley C.C., Ji H.P. Genomic instability in cancer: teetering on the limit of tolerance. Cancer Res. 2017;77(9):2179–2185. doi: 10.1158/0008-5472.CAN-16-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J., Akbani R., Creighton C.J. Invasive bladder cancer: genomic insights and therapeutic promise. Clin Cancer Res. 2015;21(20):4514–4524. doi: 10.1158/1078-0432.CCR-14-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knowles M.A., Hurst C.D... Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15(1):25–41. doi: 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- 32.Pietzak E.J., Bagrodia A., Cha E.K. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol. 2017;72(6):952–959. doi: 10.1016/j.eururo.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babina I.S., Turner N.C... Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer. 2017;17(5):318–332. doi: 10.1038/nrc.2017.8. [DOI] [PubMed] [Google Scholar]
- 34.Goodman A.M., Kato S., Bazhenova L. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyu G.Y., Yeh Y.H., Yeh Y.C., Wang Y.C. Mutation load estimation model as a predictor of the response to cancer immunotherapy. NPJ Genom Med. 2018;3:12. doi: 10.1038/s41525-018-0051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouw K.W., Goldberg M.S., Konstantinopoulos P.A., D'Andrea A.D. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017;7(7):675–693. doi: 10.1158/2159-8290.CD-17-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teo M.Y., Seier K., Ostrovnaya I. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J Clin Oncol. 2018;36(17):1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le DT, Durham JN, Smith KN. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Havel J.J., Chowell D., Chan T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.