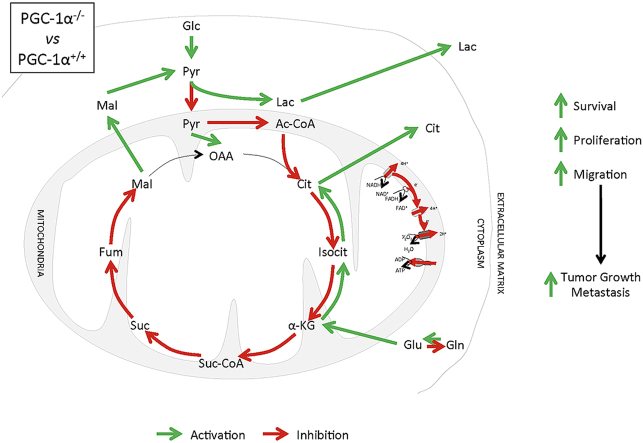
Abstract
PGC-1α controls, to a large extent, the capacity of cells to respond to changing nutritional requirements and energetic demands. The key role of metabolic reprogramming in tumor development has highlighted the potential role of PGC-1α in cancer. To investigate how loss of PGC-1α activity in primary cells impacts the oncogenic characteristics of spontaneously immortalized cells, and the mechanisms involved, we used the classic 3T3 protocol to generate spontaneously immortalized mouse embryonic fibroblasts (iMEFs) from wild-type (WT) and PGC-1α knockout (KO) mice and analyzed their oncogenic potential in vivo and in vitro. We found that PGC-1α KO iMEFs formed larger and more proliferative primary tumors than WT counterparts, and fostered the formation of lung metastasis by B16 melanoma cells. These characteristics were associated with the reduced capacity of KO iMEFs to respond to cell contact inhibition, in addition to an increased ability to form colonies in soft agar, an enhanced migratory capacity, and a reduced growth factor dependence. The mechanistic basis of this phenotype is likely associated with the observed higher levels of nuclear β-catenin and c-myc in KO iMEFs. Evaluation of the metabolic adaptations of the immortalized cell lines identified a decrease in oxidative metabolism and an increase in glycolytic flux in KO iMEFs, which were also more dependent on glutamine for their survival. Furthermore, glucose oxidation and tricarboxylic acid cycle forward flux were reduced in KO iMEF, resulting in the induction of compensatory anaplerotic pathways. Indeed, analysis of amino acid and lipid patterns supported the efficient use of tricarboxylic acid cycle intermediates to synthesize lipids and proteins to support elevated cell growth rates. All these characteristics have been observed in aggressive tumors and support a tumor suppressor role for PGC-1α, restraining metabolic adaptations in cancer.
Keywords: PGC-1α, Metabolism, Cancer, Tumor, Metastasis
Graphical abstract
1. Introduction
Since Otto Warburg first hypothesized that the metabolic alterations observed in cancer cells had a causative impact on tumor development, the role of metabolism in cancer has been the focus of intense research and lively debate. Warburg's central observation was that tumor cells increase glucose uptake and lactate (Lac) secretion under aerobic conditions [1]– a phenomenon now known as the Warburg effect, a term now generally used to refer to the reduced oxidative phosphorylation rates in cancer cells [2]. While it is generally agreed that this glycolytic switch favors tumor growth by providing high levels of the biosynthetic precursors needed for rapid proliferation [3], it remains unclear to what extent the suppression of oxidative metabolism supports or is detrimental for tumor development or to what extent general conclusions can apply. Indeed, there are many studies that support the oncogenic role of oxidative metabolism [4]. Statements like “mitochondrial respiration always work as a tumor suppressor” are no longer accepted, nor other general conclusions on the role of tumor metabolism, so the possible efficacy of therapeutic approaches that target mitochondria is being questioned. An emerging picture is that cancer cells need mitochondria even if they are largely dysfunctional, at least in part, because they require the intermediary metabolites produced by these organelles [5], and likely also because the production of reactive oxygen species (ROS) by dysfunctional mitochondria favor, or are necessary for, cell proliferation [6]. Mitochondria also provide the metabolic flexibility that allows cancer cells to revert to oxidative metabolism to survive nutrient deprivation in the restrictive tumor environment [7]. Resistance to chemotherapy agents [8] can also be boosted by mitochondria, as cells with active oxidative metabolism express high levels of stress resistance genes, including antioxidants [9]. In addition, mitochondria can provide the plasticity necessary for metastasis [10], and maintenance of the cancer stem cell reservoir [11].
All that being said, epidemiological studies stress the link between metabolic dysfunctions, such as obesity, where oxidative metabolism is reduced and increased risk of tumor development [12], and also highlight the positive effect of interventions in patients that favor oxidative metabolism such as diet [13] and exercise [14]. Accordingly, oxidative metabolism seems to play an overall protective role in cancer, although the underlying molecular mechanisms involved having been only partly elucidated.
PGC-1α is a transcriptional coactivator and master regulator of oxidative metabolism and plays a fundamental role in the metabolic plasticity of mammalian cells. It is tightly regulated by environmental cues to respond to changing energetic demands and nutritional status. PGC-1α transcriptional program directly controls the number and activity of the mitochondria [15], the antioxidant systems that prevent the build-up of oxidants derived from the activity of the electron transport chain (ETC) [9], as well as the genes involved in the metabolism of lipids and glucose [16]. The central role of PGC-1α in metabolism has led to the evaluation of its impact in tumor development. As is generally the case when considering the role of mitochondria in cancer, however, arguments can be made both in favor of a tumor suppressor role and of a tumor-promoting role [17,18], a role that it could play when working as a coactivator of ERRa [19]. In particular, in breast cancer PGC-1α levels were initially found to be reduced in aggressive tumor cell lines [20], however, later studies suggested that PGC-1α was associated with lung metastasis of breast cancer cells and to higher migratory capacity [21]. In melanoma PGC-1α antioxidant capacity was linked to chemotherapy resistance [22]. However, PGC-1α transcriptional axis was later shown to suppress melanoma metastasis by the same group [22]. On one hand PGC-1α has also been proposed to promote colon cancer growth [23] and on the other hand to be a good prognosis factor for colon cancer [24] and PGC-1α KO mice are more sensitive than WT to colon cancer induction [23]. In prostate cancer PGC-1α has been shown to prevent metastasis [25], etc.
To investigate the role of PGC-1α in cancer from a comprehensive perspective and the mechanisms involved, we analyzed the characteristics of mouse embryonic fibroblasts (MEF) from both wild-type (WT) and PGC-1α knock-out (KO) mice immortalized spontaneously in vitro following the classical 3T3 protocol. This way we could follow the history of PGC-1α deficient cells from their primary origin to their immortalized state and possibly their transformation without forcing changes in PGC-1α levels.
Molecular and functional analyses revealed that immortalized MEFs (iMEFs) maintained the features characteristic to the role of PGC-1α in metabolism and showed metabolic adaptations commonly found in advanced tumors. Examination of the oncogenic properties of the iMEFs indicated that KO cells were resistant to contact inhibition and migrated faster than WT cells. The impact of these alterations was evaluated using in vivo xenograft assays, which indicated that the loss of PGC-1α results in larger primary tumors and enhances the capacity of tumor cells to form lung metastatic nodules, overall supporting the notion that PGC-1α plays primarily a tumor suppressor role in cancer development.
2. Materials and methods
Animals. Adult male NOD/SCID and C57BL/6 mice were used (12–16 weeks old). Animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the CSIC and all procedures conformed to the Declaration of Helsinki. All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (No. 86–23 revised 1985).
Cell culture. Wild-type and PGC-1α KO MEFs were isolated and cultured as previously described [26]. Wild type (n = 4) and PGC-1α KO (n = 3) iMEF lines were obtained using the classical 3T3 protocol. Briefly, MEFs were cultured in Dulbecco's modified essential medium (DMEM) (Sigma-Aldrich) with 10% Fetal Bovine Serum (FBS) (Gibco), 2 mM glutamine (Gibco) and antibiotics (Gibco), counted every 72 h using a hemocytometer and were re-seeded at 106 cells/dish. This process was repeated until the cultures reached senescence. When cultures escaped senescence and immortalized, cells were counted and seeded again for 3 additional passages to determine post-immortalization growth rates. Since spontaneous immortalization can lead to significant genetic heterogeneity, the protocol was repeated for a total of six MEF preparations per genotype, from six independent embryos, derived from 3 different off-springs, that were passed independently, and no clonal-isolation protocol was implemented. Three WT and three KO immortalized cell lines (iMEF) were randomly selected for further analysis. Each of the immortalized cell lines derived originally from a different embrion. No specific selection procedure was implemented other that, for all the cell lines used, all the original embrions were simultaneously processed and the MEFs were simultaneously obtained and passed. HEK293T cells and B16–V5 murine melanoma cells were also cultured in DMEM with 10% FBS, 2 mM glutamine and antibiotics.
pH measurements. Cells (n = 3 per group) were seeded in 100-mm dishes and cultured to confluency. The culture medium was then replenished with fresh medium and the pH was measured 24 h later using an HI 2211 pH/ORP Meter (Hanna Instruments).
Mitochondrial energetics measurements. A total of 6 × 104 cells (n = 3 per group) were seeded per well in XF24 Cell Culture Microplates (Seahorse Biosciences) and incubated at 37 °C overnight. Simultaneously, XF24 FluxPacks (Seahorse Biosciences) were incubated with XF Calibrant Solution (Seahorse Biosciences) overnight at 37 °C in a CO2 incubator. The next day, the cell culture medium was changed to non-buffered DMEM with 5 mM galactose. The microplate and the FluxPlack were placed in an XFe24 Analyzer (Seahorse Biosciences) where the oxygen consumption rate (OCR) was measured before and after the sequential injections of oligomycin (6 μM final), FCCP (0.3 μM final) and rotenone/antimycin A (0.1 μM final of each) (all from Sigma-Aldrich). All samples were measured in triplicate.
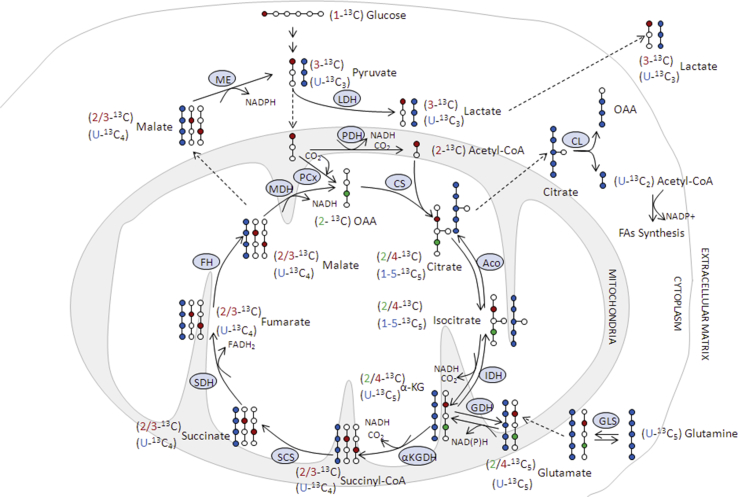
13C Metabolic flux assay. Cells were cultured to confluency, washed with PBS 1x and suspended in fresh medium containing one of the following 6 different substrate combinations: glucose (Glc) 5 mM + glutamine (Gln) 2 mM, (1–13C) Glc 5 mM, (1–13C) Glc 5 mM + Gln 2 mM, (1–13C) Glc 5 mM + (U–13C5) Gln 2 mM, Glc 5 mM + (U–13C5) Gln 2 mM or Glc 2 mM + U–13C5 Gln 5 mM, respectively. Samples (0.6 ml) of the culture medium were collected at initiation and after 24 h and cell pellets were extracted at the end of the incubation, as previously described [27] for 13C nuclear magnetic resonance (NMR) and amino acids analysis. 1H and 13C NMR spectra were obtained and analyzed as indicated below. n = 4 per group in each cell culture medium condition.
High resolution 13C NMR spectroscopy. Proton-decoupled 13C NMR spectra (22 °C, pH. 7.2) of incubation media and cellular extracts were acquired at 11,7 T in a Bruker DRX-500 spectrometer, operating at 125,13 MHz for 13C, using a commercial 1H, 13C dual probe. In brief, acquisition conditions were the following:/3 pulses, 30 kHz sweep width, 64K data table (1.7 s acquisition time), 6s total cycle time and either 3594 (incubation media), or 35944 (cell extracts) scans. The proton de-coupler was gated only during the acquisition using a broad-band composite pulse decoupling to minimize nuclear overhausser effects. Chemical shifts were calibrated with respect to an external capillary of dioxane (10% v/v, 67.4 ppm). Assignments were based on literature values and on the addition of internal standards. 13C fractional enrichments in relevant carbons were determined where indicated by comparing the intensities of the corresponding 13C resonance with that of the same resonance from a natural abundance sample (1.1% 13C) obtained from cell pellets or media obtained from incubations with unenriched substrates. See detailed description in the Supplementary Information.
Metabolomic analysis of 13C labeled cell extracts. Samples were analyzed with liquid chromatography (UPLC 1290, Agilent Technologies) coupled to a triple quadrupole mass spectrometer (ESI-QQQQ MS/MS 6420, Agilent Technologies) as described in detail in the Supplementary Information.
Glutamine deprivation survival test. Cells (n = 3 per group) were seeded in 6-well plates and cultured to confluency. Then, culture medium was removed and fresh medium without Gln was added. Every 24 h on 4 consecutive days, cells were counted with a hemocytometer.
Colony formation assay. Cells were plated in 100 mm plates at low density (2 × 103 cells per plate). After 11 days, the colonies were fixed with 4% paraformaldehyde (PFA) for 30 min, stained with crystal violet (Sigma-Aldrich) for 30 min, and washed with distilled water. The dishes were scanned and counted with ImageJ processing software (NIH), and the colonies were photographed using a binocular magnifier MZ16 F (Leica) equipped with a Nikon Digital Sight DS-L camera. 3–4 different clones from each genotype were examined and experiments were made in quintuplicate.
Soft-agar colony formation assay. Assays were performed in 6-well plates with cells (n = 3 per group) embedded in 0.3% agarose in culture medium (3 × 105 cells per well) seeded on top of a layer of 0.6% agarose. Colonies were photographed as above after 30 days.
Contact inhibition assay. Cells (n = 3 per group) were plated in 100-mm plates and allowed to grow for two weeks with a medium change every two days. Three-dimensional (3D) colony-like outgrowth structures were counted and photographed with a Nikon TS100 microscope (Nikon) equipped with a Nikon Digital Sight DS-L camera.
Wound healing assay. Cells (n = 3 per group) were seeded in 6-well plates and grown to confluency. A diagonal linear scratch was then made across the center of the well using a sterile 1 ml micropipette tip. The plate was washed three times with PBS and incubated with fresh culture medium containing 5% FBS. Images were captured with a Zeiss Cell Observer Z1 inverted microscope equipped with a Cascade 1K high resolution camera (Photometrics). Images were analyzed with TScratch software.
Boyden-chamber migration assay. 1.5 × 104 cells (n = 3 per group) were plated onto 8-μm pore size polycarbonate membranes in a Boyden-chamber (Costar) and allowed to migrate for 18 h. The cells that had entered the membrane were then fixed with cold 70% ethanol and stained with propidium iodide (Sigma-Aldrich) for 30 min at 37 °C followed by incubation at room temperature for 1 h and 30 min. The stained nuclei were imaged with a Zeiss LSM710 (Zeiss) spectral confocal microscope and analyzed using ZEN 2009 software (Zeiss) and ImageJ.
Protein extraction and western blotting. Whole cell extracts were prepared. Proteins were separated in 12% SDS-PAGE gels and transferred to Amersham Hybond PVDF membranes (GE Healthcare) by semidry transference using the TransBlot SD cell system (Bio-Rad). The following antibodies were used: PFKFB3 (Proteintech, 13763-1-AP), c-myc (Santa Cruz Biotechnology, sc-764), Malic enzyme (kindly provided by Dr. Ángela M. Valverde, IIB, CSIC-UAM, Madrid) and β-actin (A5441, Sigma). ImageJ software was used to analyze western blots.
RNA isolation and quantitative reverse transcription PCR (qRT-PCR). Total mRNA was extracted with TRI Reagent (Sigma) and retro-transcribed (RT) with M-MLV enzyme (Invitrogen). The resulting cDNA was analyzed by qPCRs using a SYBR Green Master Mix (Applied Biosystems) and an Eppendorf MasterCycler RealPlex platform. The primer sets used are listed in the Supplementary Information section.
Retroviral and lentiviral vectors. Retroviral particles were generated in HEK293T cells. pCL-Eco and pCX4-GFP were cotransfected using a calcium phosphate transfection protocol. Twenty-four hours later, the virus-containing culture medium was collected and passed through a 0.45 μm filter to remove cells in suspension, and 8 μg/ml of polybrene was added to aid infection. Lentiviral particles were similarly generated in HEK293T using VSVg, p8.9 and pHRSIN–CS–Luc-IRES-emGFP plasmids.
Viral infections. WT and PGC-1α KO iMEFs were infected with the retroviral virus-containing culture medium for 20 h, whereas B16–V5 and iMEF cultures were infected by incubation with lentivirus-containing culture media for 6 h. Infection efficacy was assessed by following the GFP expression using an inverted fluorescence microscope (Axiovert 135, Zeiss). GFP- positive cells were selected using a FACSVantage SE cell sorter (Becton Dickinson). Luciferase activity in cell extracts was evaluated with a GloMax luminometer (Promega).
Cell proliferation assay in Matrigel. GFP-positive iMEFs (2 × 106) from exponentially growing cell cultures were embedded in 200 μl a 2:1 (v/v) mixture of Matrigel (BD Biosciences) and DMEM, which was then injected into the back of C57BL/6j mice (n = 6 per group). After 60 days the Matrigel plugs were extracted and fixed in 4% PFA for 24 h, soaked in 30% sucrose (Sigma-Aldrich), embedded in Tissue-Tec OCT Compound (Sakura) and frozen. Sections were prepared in a cryostat at a thickness of 10 μm.
Xenograft tumor assay. A total of 1 × 107 cells were injected subcutaneously into the back of NOD/SCID mice (n = 6 per group). Mice were euthanized 15 days later, and tumor volume was calculated as 4/3*π*width*length*height. Tumors were then dissected and fixed in 4% PFA for histological analysis.
Experimental metastasis assay. A total of 5 × 105 each of iMEF and B16–V5 cells were co-injected into the lateral tail vein of NOD/SCID mice (n = 15 per group). Every week, mice were anaesthetized and were injected retro-orbitally with 5 μg/mg of luciferase substrate (Cayman). Lung metastasis was visualized in vivo using an IVIS lumina imaging system (PerkinElmer). At 4 weeks post-inoculation, mice were euthanized and lungs were dissected and fixed with Bouin's solution (Sigma-Aldrich) for histological analysis.
Hematoxylin and eosin staining (H&E). Tissue sections were fixed in 10% buffered formalin and embedded in paraffin, and 4 μm sections were de-waxed and hydrated. Sections were stained for 3 min with Harris hematoxylin and for 2 min with eosin. Finally, the slides were dehydrated and mounted using DPx Mountant (Sigma-Aldrich). Images were acquired with a Nikon E90i microscope equipped with a DS-Fi1 camera and analyzed with ImageJ.
Immunofluorescence (IF). Fixation, staining and analysis procedures were performed as previously described [28]. The following antibodies were used: β-catenin (Santa Cruz Biotechnology, sc-1496), GFP (Santa Cruz Biotechnology, sc-8334) and Ki67 (Thermo Scientific, RM-9106-S1). Images were acquired with a Nikon E90i microscope equipped with a DS-Q1 camera and analyzed with ImageJ.
Immunohistochemistry. Fixation and staining was performed with the Vectastain ABC (Vector Labs) and the Peroxidase Substrate Kit DAB (Vector Labs) following the manufacturer's instructions using an αKi67 antibody RM-9106-S1 (Thermo Scientific). Images were acquired with a Nikon E90i (Nikon) microscope equipped with a DS-Fi1 camera and analyzed with ImageJ.
Lipidomic analysis. Fatty acids from total lipids were transmethylated according to Lepage & Roy [7]. The methylated fatty acids were separated and quantified on a HP5890 series II gas chromatograph equipped with a flame ionization detector (Agilent Technologies, Spain). Individual fatty acids were identified comparing relative retention times with commercial standards (Nu Chek, Elysian, MN, USA). Tridecanoic acid was used as an internal standard for quantification. The average carbon chain length (ACL), the double bond index (DBI) and the peroxidizability index (PI) were determined as previously described [29].
Amino acid analysis. Amino acids were analyzed in the Protein Chemistry facility at the Center for Biological Research (CSIC), Madrid, on a Biochrom 30 amino acid analyzer. The Biochrom 30 uses classical amino acid analysis methodology based on ion-exchange liquid chromatography and postcolumn continuous reaction with ninhydrin.
Statistical analysis. Data are expressed as mean ± SD. Statistical significance was evaluated by two-tailed unpaired t-test, paired t-test or analysis of variance as required. Values were considered statistically significant at p < 0.05.
3. Results
3.1. PGC-1α KO iMEFs are more oncogenic than WT iMEFs
Aiming to analyze the role of PGC-1a in the process of cell immortalization and tumurogenesis, we generated immortalized cell lines derived from MEFs using the classical 3T3 protocol. Passage 0 for iMEFs was considered when the cell plate where the MEFs immortalized become confluent, following evaluation of cell proliferation rates for three passages, the cells were frozen and subsequently, when all the required cell lines were collected, they were simultaneously put again in culture for further analysis.
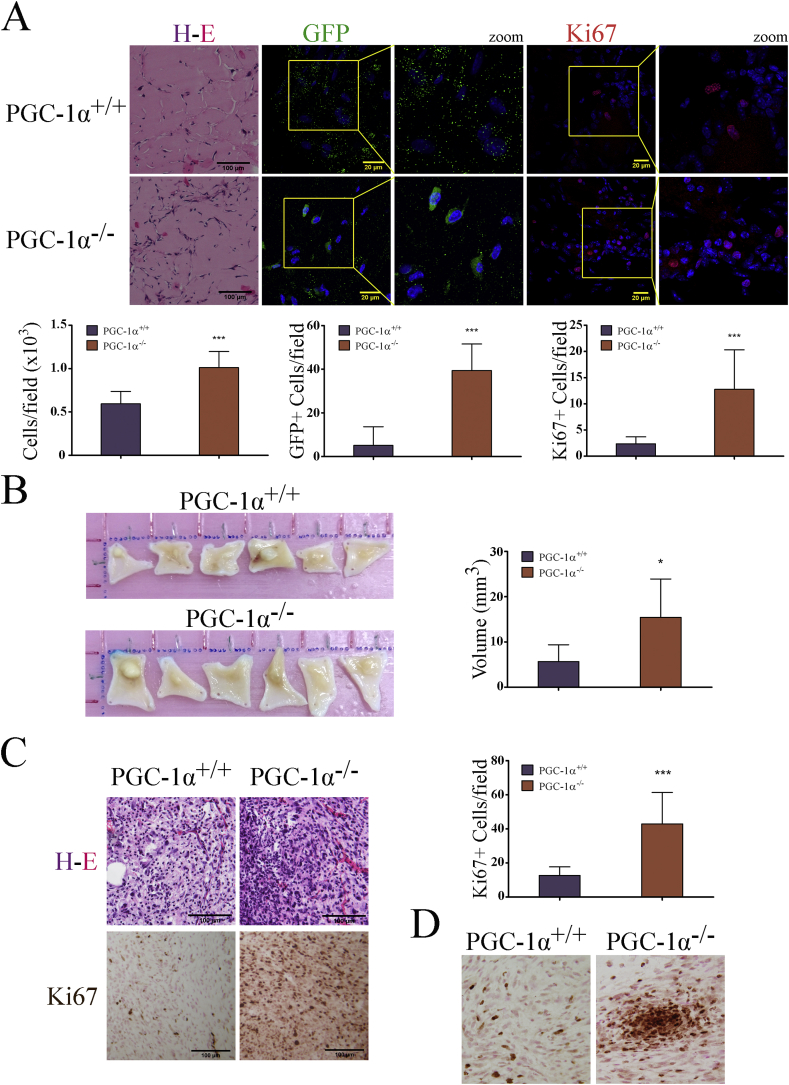
As an initial test to compare and evaluate the survival and proliferative capacity of WT and PGC-1α KO iMEFs in vivo, we infected cells with a retrovirus constitutively expressing GFP, embedded them in Matrigel, and then injected them subcutaneously into C57BL6 mice. Matrigel was used to provide a suitable environment for anchorage, growth and also to provide pro-angiogenic factors that facilitate access to nutrients. After 60 days, the mice were sacrificed and the plugs were removed, fixed and embedded in paraffin. We determined the total number of cells per section by nuclear DAPI staining, and also measured the percentage of GFP+ and Ki67+ (actively proliferating cells). We found that the Matrigel plugs containing PGC-1α KO iMEFs had significantly more cells per field than those containing WT iMEFs, and this was accompanied by a significantly higher percentage of GFP+ and Ki67+ cells (Fig. 1A).
Fig. 1.
PGC-1α KO iMEFs have a greater oncogenic potential than WT iMEFs. A) Matrigel plugs with embedded WT and KO iMEFs from injected in C56BL/6j mice. Top-left, H&E-stained sections. Top-center, immunofluorescence (IF) analysis of GFP+ cells. Top-right, IF analysis of Ki67+ cells. Bottom panels, quantitative analysis of cell counts. B) Tumors dissected from NOD-SCID mice injected with WT and KO iMEFs. Left, full size tumors. Right, tumor volume. C) Histological analysis of primary tumors. Top-left, H&E staining. Bottom left, IHQ Ki67 staining. Right, quantitative analysis of Ki67+ cells. D) Ki67 staining shows the presence of hyper-proliferative nodules in KO but not in WT iMEFs tumors. Data are mean ± standard deviation. *, p < 0.05; **, p ≤ 0.01; ***, p ≤ 0.005.
To question whether the survival/proliferative capacity of iMEFs could sustain tumor development, we injected them subcutaneously in immunodeficient NOD-SCID mice and examined tumor appearance after 60 days. Results showed that KO iMEF produced significantly larger tumors than WT iMEFs (Fig. 1B). Histological analysis and evaluation of Ki67+ proliferative cells further showed a significantly higher percentage of actively proliferating cells in the tumors produced by KO iMEFs (Fig. 1C) and also the presence of hyper-proliferative nodules, which were absent in WT iMEF-derived tumors (Fig. 1D). Overall, these results suggest that KO iMEFs have higher survival and/or proliferation capacity in vivo than WT iMEFs.
3.2. PGC-1α KO iMEFs facilitate the formation of lung metastases by B16–V5 melanoma cells
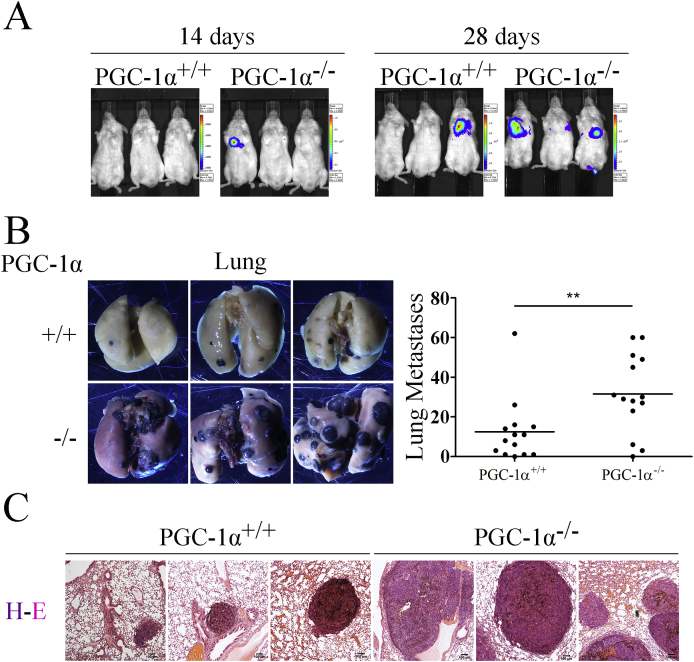
While iMEFs do not have sufficient tumor formation capacity to generate metastasis on their own, they have been shown to promote lung metastasis formation by melanoma cells, possibly by providing an optimal matrix environment [30]. We therefore tested the capacity of WT and KO iMEFs to facilitate metastasis formation by co-inoculation of iMEFs and B16–V5 melanoma cells, modified by lentiviral infection to constitutively express luciferase. Cells were injected into the lateral tail vein of NOD-SCID mice and luciferase expression in vivo was used to monitor tumor development.
Lung nodules were already detectable 2 weeks after inoculation in animals co-incoculated with KO iMEFS but not with WT iMEFs, where nodules were only evident 4 weeks after inoculation (Fig. 2A). Following lung dissection, results showed that animals co-inoculated with KO iMEFs had a significantly higher number of lung nodules than those co-inoculated with WT iMEFs (Fig. 2B). Moreover, H&E histological staining revealed that animals co-inoculated with KO iMEFs had larger nodules than those co-inoculated with WT iMEFs (Fig. 2C), suggesting that KO iMEFs have a higher capacity to promote metastasis formation than WT iMEFs.
Fig. 2.
PGC-1α KO iMEFs can boost more the metastatic potential of B16 melanoma cells than WT iMEFs. A) Bioluminiscence detection of metastatic nodules in NOD-SCID mice coinoculated with iMEFs and B16–V5 melanoma cells. B) Lungs bearing metastasis. Left, representative images of lungs. Righ, quantification of the number of nodules. C) H&E staining of lung sections. Data are mean ± standard deviation. *, p < 0.05; **, p ≤ 0.01; ***, p ≤ 0.005.
3.3. PGC-1α KO iMEFs present a transformed phenotype
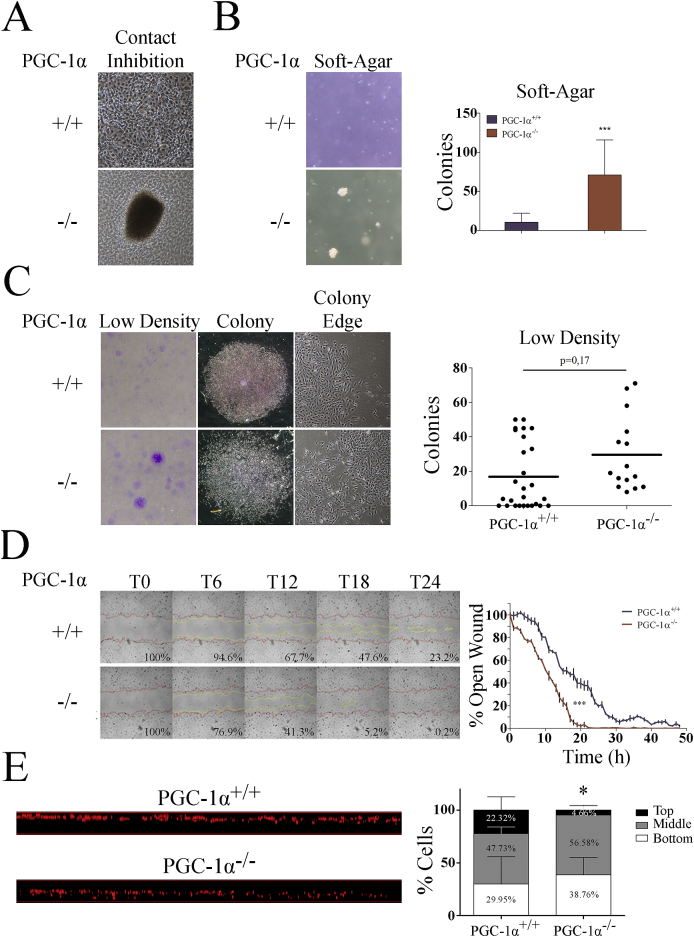
To investigate the cellular basis for the higher oncogenic and pro-metastatic potential of PGC-1α KO iMEFs, we used a panel of in vitro assays to explore the different phenotypic characteristics typically associated with transformed cells.
We first looked for differences in proliferation rates in optimal exponential culture conditions, but we failed to find any significant difference between WT and KO iMEFs [31]. We next, we tested their capacity to induce cell cycle arrest in response to cell-contact inhibition. To do this, confluent cell plates were maintained in culture with a medium change every 2 days for up to 2 weeks, at which point we evaluated the formation of 3D colony-like outgrowth structures. We found that KO iMEFs, but not WT iMEFs, formed 3D colony-like structures, suggesting that contact inhibition was impaired in KO iMEFs (Fig. 3A).
Fig. 3.
In vitro phenotypic characterization of iMEFs. A) Representative images of overgrown iMEF cell cultures. B) Formation of colonies in soft-agar. Left, representative images. Right, quantification of the number of colonies. C) Formation of colonies grown at low cell densities. Left, representative images of culture dishes stained with crystal violet. Center, representative images of single colonies. Right, representative images of colony edges. Graph shows colony number quantification. D) Wound-healing assay using WT and KO iMEFs. Left panel, representative images of the wound closure at different times, the red lines mark the edge at time 0, yellow lines mark the edge at the indicated time. Right panel, graph representing the percentage of the remaining open area at each time point. E) Migration in a Boyden chamber. Left, orthogonal view of the porous membrane of a Boyden chamber where immortalized iMEFs were allowed to migrate. Cell nuclei were stained with propidium iodide and visualized by confocal microscopy. Right, quantitative evaluation of migrated distances. The membrane span was divided into three even sections, top, middle, and bottom and the percentage of cells per section were quantified. Data are mean ± standard deviation. *, p < 0.05; **, p ≤ 0.01; ***, p ≤ 0.005. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To determine whether KO iMEFs had a general defect in the response to extracellular cues, we next examined anchorage-independent growth by evaluating colony formation in soft agar. We found that whereas KO cells lines formed colonies, this capacity was almost absent in WT cells lines (Fig. 3B).
As a complementary approach, we examined the ability of iMEFs to form colonies when cells were plated at low densities, finding a non-significant increase in the number of colonies from KO iMEFs relative to WT iMEFs (Fig. 3C). Importantly, when colonies were visualized microscopically, we observed that KO iMEFs formed rough-edged colonies wherease WT iMEFs formed compact colonies (Fig. 3C), a characteristic normally associated with faster migration rates [32].
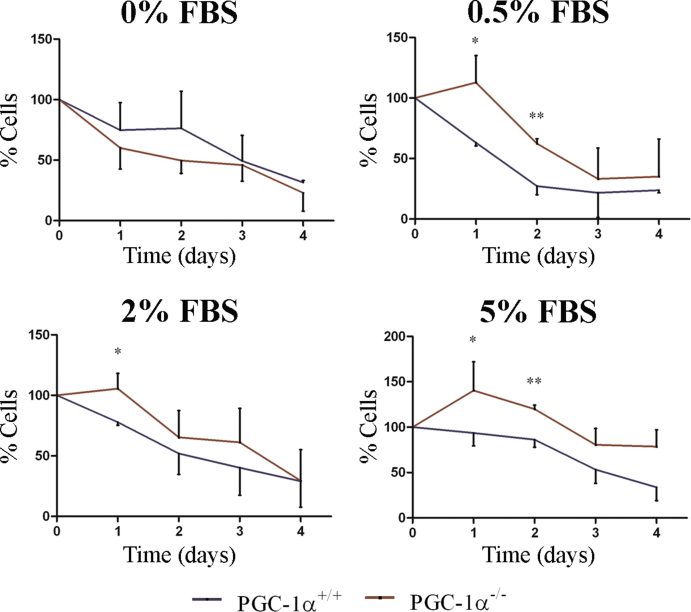
Continuing this analysis, we measured growth factor dependency by testing cell survival under reduced serum conditions (0%, 0.5%, 2% and 5% FBS). Significant differences were found at 0.5%, 2% and 5% FBS (Supp. Fig. 1) at 24 and 48 h with a higher percentage of surviving KO iMEFs.
Given the above findings and our previous observations suggesting that PGC-1α deficiency is associated with enhanced cell migration [33], we used two different assays to asses migration of iMEFs: an in vitro wound healing (scratch) assay that evaluates migration in response to loss of cell contact and a Boyden-chamber assay that measures directional migration. We found that KO iMEFs migrated faster in both assays. In scratch assays, KO iMEFs closed the wound in 20 h compared with 30 h for WT cells (Fig. 3D). Of note, the cell density at the wound closure site was lower for KO cells, suggesting a more disorganized migratory profile. To avoid the confounding effect of cell proliferation, the cells were serum-starved o/n prior to the initiation of the essay. In the Boyden chamber assay, the migrated distance after 18 h was evaluated by dividing the membrane into three Z-sections (top, middle and bottom) and quantifying the percentage of cells in each section. At 18 h, the percentage of WT cells in the top section was significantly higher wherease the percentage of KO cells in the bottom section was higher, supporting a faster migratory speed for KO iMEFs (Fig. 3E).
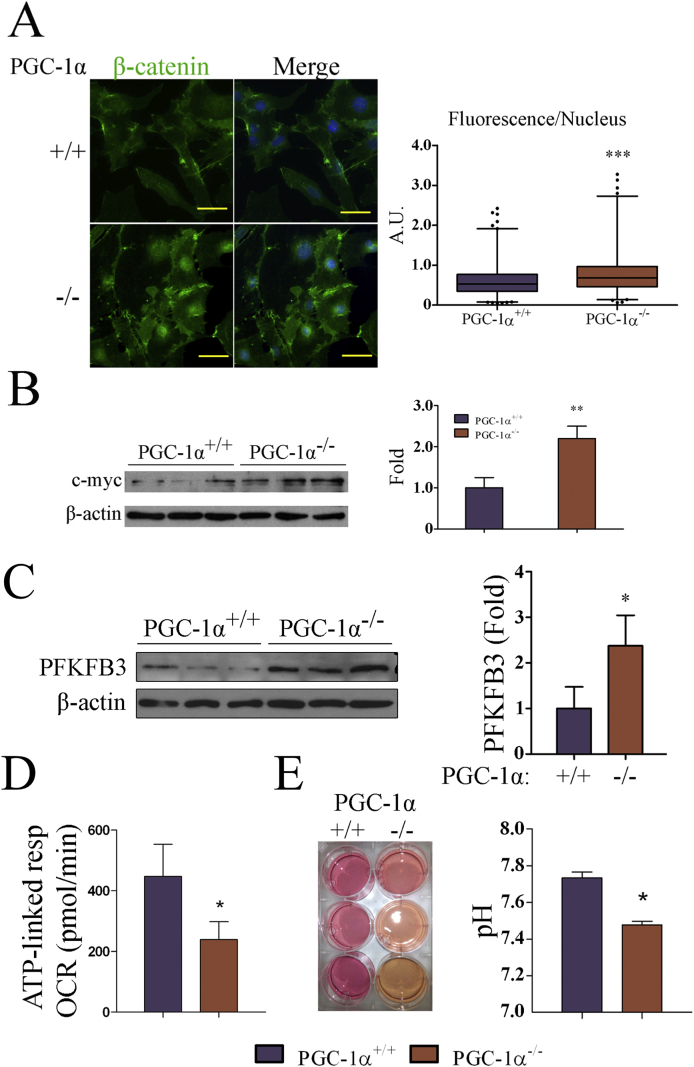
3.4. Nuclear β-catenin and c-myc levels are higher in PGC-1α KO cell lines
Loss of contact inhibition and enhanced cell migration and metastatic potential typically characterize impaired integrin and cadherin mediated cell contacts, leading to the activation and nuclear localization of the oncogene β-catenin [34].
β-catenin is present in adherens junctions in quiescent cells, but accumulates in the nucleus in proliferative and tumor cells where it regulates gene expression [35]. Nuclear translocation of β-catenin in cancer cells results in the induction of many oncongenes [36] including c-myc, an oncogene associated with most if not all the metabolic alterations found in tumors [37], and whose levels have been shown to inversely correlate with those of PGC-1α in at least two types of cancer cells [38,39].
Given that KO iMEFs displayed phenotypic characteristics reminiscent of tumor cells, we analyzed the sub-cellular distribution of β-catenin by immunoflourescence, finding that the levels of nuclear β-catenin were higher in KO cells than in WT cells (Fig. 4A). Moreover, we detected higher levels of c-myc in western blots of KO iMEFs, an observation consistent with a possible increase in β-catenin transcriptional activity (Fig. 4B).
Fig. 4.
A-B, PGC-1α KO iMEFs show elevated levels of nuclear β-catenin and c-myc. A) β-catenin subcellular localization in iMEFs. Left, representative IF images of β-catenin staining in PGC-1α WT and KO iMEFs. Right, Cellular cross sections were used to determine the levels of β-catenin nuclear staining. B) c-myc protein levels. Left, WB of c-myc in WT and KO iMEF. Right, quantification of c-myc protein levels. C-E, PGC-1α KO iMEFs show reduced oxygen consumption rates. C) PFKFB3 levels in WT and KO iMEFs. Left, PFKFB3 Western blot. Right, quantification of PFKFB3 protein levels. D) Oxygen consumption rate (OCR) coupled to ATP production. E) Media acidification by iMEF. Left, images of cell culture dishes 24 h after change of media of confluent cultures. Right, pH of culture media at 24 h. Data are mean ± standard deviation. *, p < 0.05; **, p ≤ 0.01; ***, p ≤ 0.005.
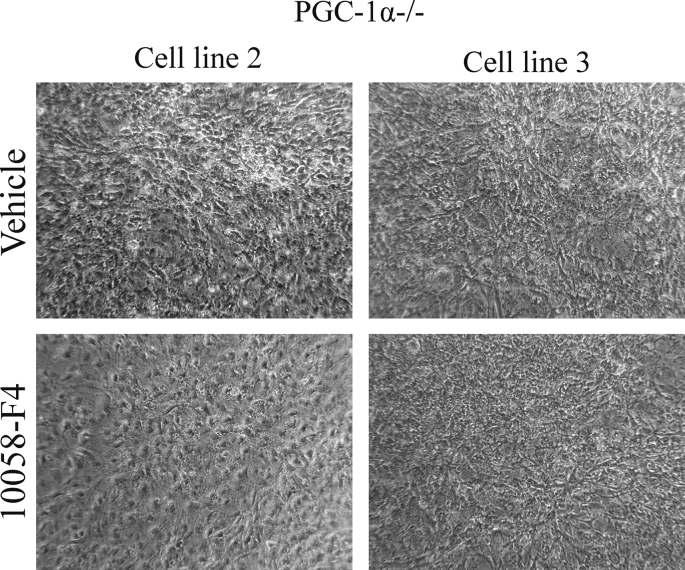
In order to test the relevance of c-myc induction in KO iMEFs, we analyzed how the response to contact inhibition in vitro of the KO iMEFs was modulated in the presence of a c-myc inhibitor, 10058-F4. One of the KO cell lines did not tolerate the treatment, the other two showed a reduction in the formation of 3D structures, suggesting a functionally relevant role of c-myc induction (Supp. Fig. 2).
3.5. PGC-1α KO iMEFs are more glycolytic than WT's
The tumor metabolic switch that suppresses oxidative metabolism and triggers glycolytic metabolism is driven to a large extent by c-myc. Considering that PGC-1α is a master regulator of oxidative metabolism and that c-myc levels were elevated in PGC-1α KO cell lines, we next sought to evaluate the metabolic adaptations of the immortalized cell lines.
To this end we first examined the protein expression levels of PFKFB3, a kinase that positively controls the glycolytic flux and is elevated in several cancers [40]. PFKFB3 was selected as a marker of glycolysis because it is a highly regulated enzyme whose activity boost the formation of fructose 2,6-bisphosphate (F2,6P2), which is, in turn, a potent activator of 6-phosphofructo-1-kinase (PFK-1), the enzyme in charge of the first committed rate-limiting step of glycolysis, which makes of PFKFB3 a potent inducer of the glycolytic flux. Western blotting showed that PFKFB3 levels were significantly higher in PGC-1α KO iMEFs than in WT counterparts, pointing to a more glycolytic metabolism in KO cells (Fig. 4C).
Elevated glycolytic rates generally result in increased lactate (Lac) secretion to the media leading to media acidification. Therefore, determination of culture medium pH can be used as a surrogate marker of non-oxidative glucose catabolism. Evaluation of the pH in confluent cultures revealed a higher acidification of the culture medium of KO cells, suggesting an elevated production of Lac (Fig. 4E).
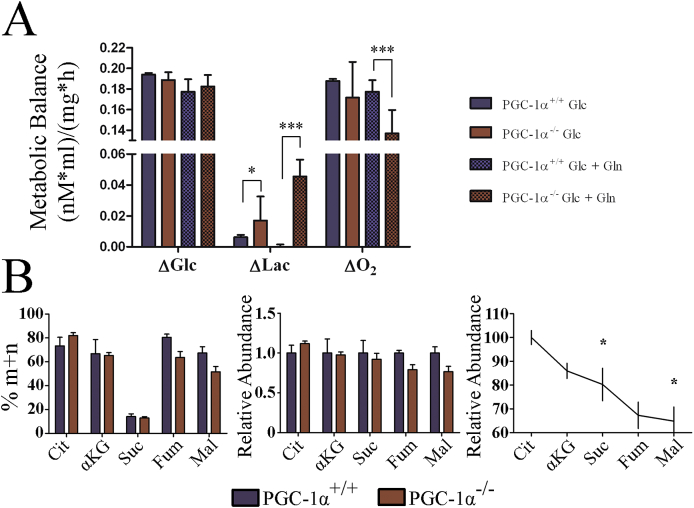
To directly evaluate the production of Lac from glucose (Glc), we used stable isotope tracing. Thus, iMEFs were incubated in culture medium containing glucose labeled with 13C in carbon 1 (1–13C) Glc. Determination of (1–13C) Glc and (1–13C) Lac levels in the culture media by NMR allowed the quantification of Glc consumption and Lac production rates at equimolar rates, since 1 M of labeled glucose would produce 1 M of labeled Lac, whereas the total Lac production from Glc would be twice that. The amount of labeled CO2 produced by Glc oxidation could be estimated from the difference between glucose consumption and Lac production. As anticipated, we observed a significantly higher production of labeled Lac in KO cell medium, while Glc consumption was similar for both genotypes, suggesting a more glycolytic metabolism in KO cells (Fig. 5A).
Fig. 5.
Progression of metabolites through the TCA cycle occurs at a lower pace in PGC-1α KO iMEFs. A) iMEFs were incubated with (1–13C) Glc with or without Gln to determine the use of Glc to produce Lac by NMR analysis of cell culture media. The graph shows the moles of used Glc, produced labeled Lac. B) MS analysis of TCA intermediates in cell extracts of iMEFs labeled with (1–13C) Glc. Left, percentage of labeled 13C TCA metabolites in WT and KO iMEFs relative to the total amount of each metabolite in the cell extract. Center, percentage of each labeled metabolite in KO iMEFs relative to WT cells. Right, percentage variation in the rate of labeled metabolite in WT vs KO iMEFs in the sequential steps of the TCA cycle. Data are mean ± standard deviation. *, p < 0.05; **, p ≤ 0.01; ***, p ≤ 0.005.
3.6. TCA cycle activity is reduced in the absence of PGC-1α
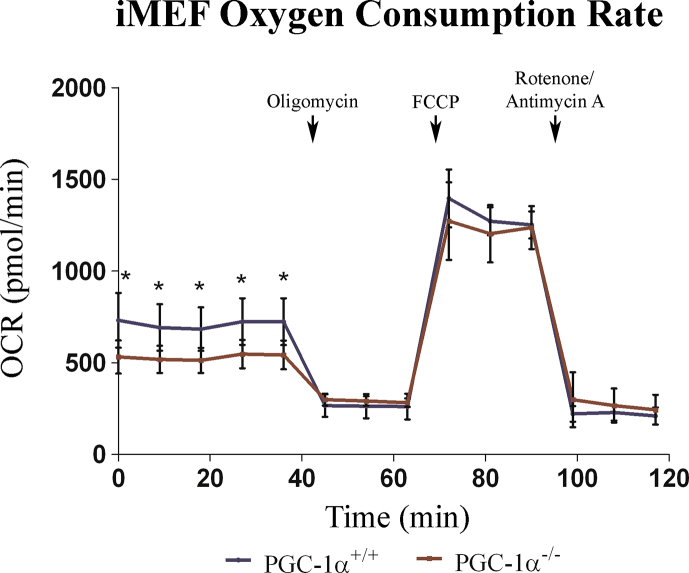
Given these results, we next evaluated the use of oxidative metabolism to generate ATP (ATP-coupled oxygen consumption). Cell lines were incubated in the presence of 5 mM galactose, and in the absence of glucose, to force oxidative metabolism [41], and respiration was measured in the SeaHorse XFe24 analyzer. Results showed that both basal and ATP-coupled oxygen consumption rates (OCR) were lower in KO iMEFs (Fig. 4D and Supp. Fig. 2), but maximal respiratory capacity, proton leakage and non-mitochondrial respiration were not significantly different between KO and WT iMEFs, as evaluated by the determination of the OCR with the sequential addition of oligomycin, an inhibitor of complex V (ATP synthase) of the Electron Transport Chain (ETC), FCCP, a membrane uncoupler, and rotenone and antimycin A, inhibitors of ETC complex I (NADH+ ubiquinone oxidoreductase) and complex III, respectively (Supp. Fig. 2).
Continuing this analysis, we measured the catabolism of Pyr in the TCA cycle (Supp. Fig. 3) using liquid chromatography-mass spectrometry (LC-MS). Specifically, we determined the levels of the labeled the TCA intermediates, citrate (Cit), αKetoglutarate/2-oxoglutarate (αKG), succinate, fumarate and malate (Supp. Fig. 3). We found that the relative levels of labeled Cit, the first metabolite of the TCA cycle following the incorporation of the carbons derived from glucose as acetyl-CoA, tended to be higher in KO iMEFs; however, this difference was nor maintained for αKG and succinate, and was reversed for fumarate and malate, which showed higher labeling in WT cells (Fig. 5B). Importantly, we found a significant difference in the step-wise relative decrease in the percentage of labeled Glc in KO iMEFs relative to equivalent WT cells (Fig. 5B, right panel) possibly suggesting that Cit progressed more slowly through the TCA cycle in KO cells than in WT cells. This result is consistent with the previously established role of PGC-1α in the control of succinate dehydrogenase (SDH) and fumarate hydratase (FH) expression levels [42].
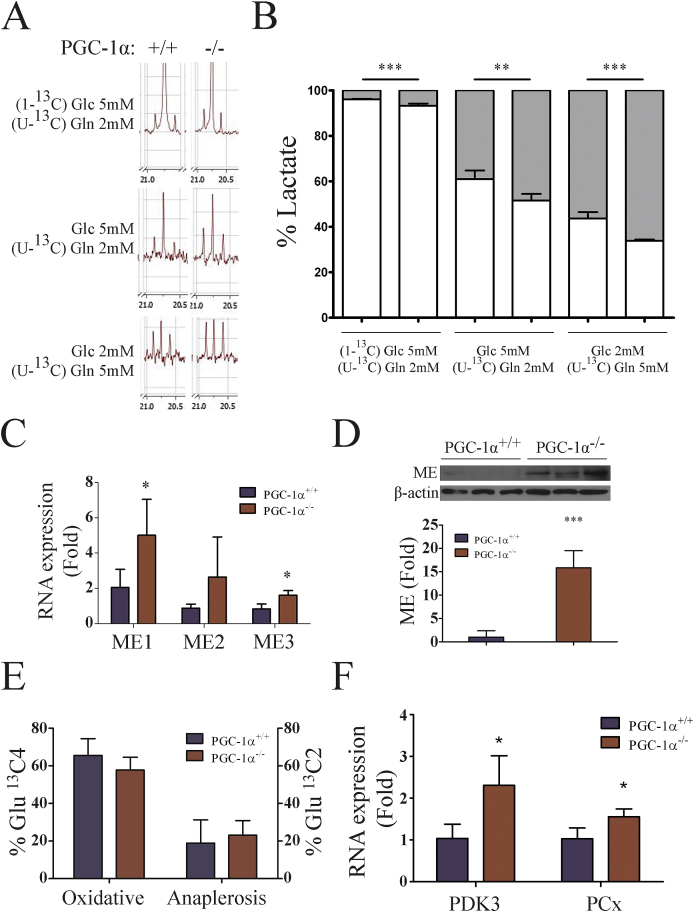
As the TCA cycle gradually deteriorates during tumor development different anaplerotic systems became prevalent. The first one to be induced is commonly malic Enzyme (ME), which catalyzes the formation of pyruvate (Pyr) from malate [43] to compensate for the deficit in malate dehydrogenase that forms oxalacetate (OAA) from malate in the TCA cycle [44] and generates NADPH+, providing an important source of reducing equivalents to maintain antioxidant systems [45].
To determine the activity of ME, we used stable isotope tracing and NMR spectrum-analysis in cells labeled with (U–13C) glutamine (Gln) and/or (1–13C) Glc. Since the only way to form (U–13C) Lac is via the activity of ME, determination of the Lac Carbon 3 resonance peaks, which double when the C2 carbon is also labeled, can be used to determine whether the origin of Pyr is directly from Glc (glycolysis) or derives from the incorporation of αKG into the TCA cycle, to form malate, which leaves the mitochondria and is catalyzed by ME in the cytosol to form Pyr. We found, that the amount of (U–13C) Lac was significantly higher in PGC-1α KO iMEFs than in WT cells (Fig. 6A and B), suggesting a higher level of ME activity. In accord with this, gene expression analysis showed increased expression ME isoforms 1 and 3 in KO cell lines (Fig. 6C), and this finding was confirmed by western blotting (Fig. 6D).
Fig. 6.
Enhanced use of anaplerotic reactions in KO iMEFs. A-D) Evaluation of ME activity in iMEFs. A) NMR spectra of Lac C3 from (U–13C) Gln and/or (1–13C) Glc labeled cells. B) Quantitative analysis of the origin of labeled Lac C3. C) RNA expression of ME1, ME2 and ME3 in WT and KO iMEFs. D) ME protein levels. Top, western blotting of ME in WT and KO iMEFs. Bottom, quantitative analysis of ME protein levels. E-F) Evaluation PCx activity in iMEFs. E) PDH (oxidative) or PCx (anaplerotic) TCA entry rates for Pyr were calculated by NMR evaluation of Glu isotopologues following incubation with (1–13C) Glc. F) RNA expression of PDK3 and PCx in WT and KO iMEFs. Data are mean ± standard deviation. *, p < 0.05; **, p ≤ 0.01; ***, p ≤ 0.005.
Another relevant anaplerotic pathway activated in tumor cells involves the induction of the enzyme pyruvate carboxylase (PCx), which makes use of pyruvate (Pyr) to synthesize OAA [46]. To determine the relative contribution of PCx and Pyr Deshidrogenase (PDH) to the catabolism of Pyr, we used stable isotope tracing of 1–13C Glc metabolism, which differentially marks TCA metabolite carbon atoms depending on which enzyme incorporates the labeled Pyr into the cycle. In particular, PCx activity is responsible for the C2 13C mark whereas PDH is responsible for the C4 13C mark on αKG. The amount of Glu formed from αKG by the action of transaminases, can be determined by NMR to evaluate the relative rate of PCx and PDH activity. We found that KO iMEFs showed more PCx and less PDH activity than WT iMEFs, although differences did not reach statistical significance (Fig. 6E). The relevance of this result was supported by gene expression analysis of PCx and Pyr deshydrogenase kinase 3 (PDK3), an inhibitor of PDH [47], by qRT-PCR. Both genes were found to be significantly upregulated in PGC-1α KO iMEFs (Fig. 6F).
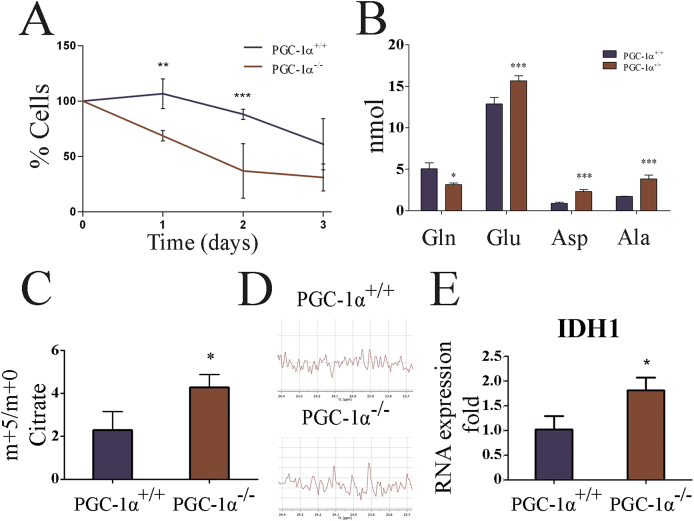
3.7. PGC-1α KO cell lines are glutamine (Gln) dependent
While pyruvate is used predominantly for Lac formation by tumor cells, the TCA cycle is actively used to provide biosynthetic precursors needed for the synthesis of amino acids, nucleic acids and fatty acids, which together support the elevated cell proliferations rates. This use of the TCA cycle is boosted by the utilization of Gln that enters the TCA cycle after deamination and transamination as αKG. As a consequence, advanced tumors are characteristically Gln dependent [48].
To evaluate the Gln dependency, iMEFs were cultured in Gln-deprived culture medium and the remaining live cells were counted every 24 h. We found that whereas cell viability was compromised in all cases following Gln deprivation, the percentage of live WT cells was significantly higher at 24 and 48 h relative to KO cells, indicating a higher Gln dependency in the latter (Fig. 7A).
Fig. 7.
KO iMEFs are dependent on Gln to produce Cit. A) Survival rates of iMEFs in the absence of Gln. B) Concentration of the amino acids, Gln, Glu, Asp and Ala in WT and KO iMEFs. Data are mean ± standard deviation. C) MS analysis of Cit in cell extracts of iMEFs labeled with (U–13C) Gln. The graph shows the ratio Cit m+5/m+0. D) NMR analysis of acetate from (U–13C) Gln labeled iMEF. E) Gene expression analysis of IDHI in iMEF. *, p < 0.05; **, p ≤ 0.01; ***, p ≤ 0.005.
In advanced tumors, Gln taken up by the amino acid transporter ASCT2, is first deaminated by glutaminase (GLS) to form Glu, both ASCT2 and GLS are targets of c-myc [49], and then a transaminase reaction transfers the second amino group to form αKG, which can be incorporated into the TCA cycle [50]. Glu can also produce aspartete (Asp) to from OAA [51]. The maintenance of the OAA and Cit pools seems to be the main molecular basis of Gln dependency in tumor cells. It has been proposed that this provides TCA components for the synthesis of biomolecules and alleviates the need for Pyr oxidation in tumor cells with compromised ETC activity [51]. Indeed, Gln supplementation increased Lac production in both WT and KO iMEFs (Fig. 5A).
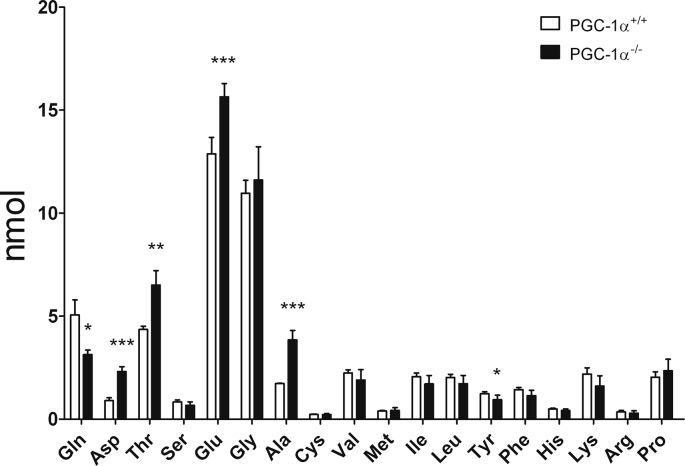
In addition to its incorporation into the TCA cycle, another important use of Gln is in the synthesis of other amino acids, in particular Glu, Asp, and alanine (Ala) [52]. As a rule, quiescent and proliferative cells have relatively different needs in terms not only of the amount but also of the types of amino acids, and Gln plays a key role in the restoration of amino acid deficit in cancer cells [53]. Thus, we analyzed the intracellular levels of amino acids using ion-exchange liquid chromatography and post-column continuous reaction with ninhydrin (Supp. Fig. 4). We found that KO iMEFs had significantly less amount of Gln and more Glu, Asp, and Ala, consistent with a strong use of Gln, at least in part to boost Glu, Asp and Ala synthesis (Fig. 7B & Supp. Fig. 4).
To directly determine how Gln was used by the TCA cycle, iMEFs were incubated with 13C Gln, (U–13C5) Gln. Were prepared cell extracts and analyzed the incorporation of labeled Gln to Cit by LC-MS. It has been proposed that the synthesis of Cit from αKG by reductive carboxylation can be induced in some aggressive tumor cell lines [54,55]. This process would directly produce Cit m+5 from Gln m+5. Analysis showed that KO cell lines showed a higher level of 13C m+5/m+0 ratio (Fig. 7C), possibly suggesting an active reductive carboxylation pathway.
To evaluate this possibility we used NMR to determine the origin of labeled acetate in cells labeled with (U–13C) Gln. Acetate is formed from Cit by the enzyme citrate lyase. In the NMR spectrum the presence of acetate signal doublets is indicative of the presence of 13C in consecutive carbons. Importantly, the presence of this signal could be observed only in KO cells, suggesting the presence of an active reductive carboxylation (Fig. 7D). However, this result is tempered by the fact that acetate can also be formed from Gln when it is used to generate Lac following the TCA cycle and then by the action of ME generating Lac from malate (Supp. Fig. 3).
To further test this hypothesis, we analyzed the expression levels of IDH1, an isocitrate deshydrogenase isoform that has been proposed to be responsible for active reductive carboxylation in tumor cells [56]. Consistently, we found that the mRNA levels of IDH1 where significantly higher in PGC-1α KO iMEFs than in WT iMEFs (Fig. 7E).
These results are in overall agreement with previously published data on the use of Gln in advanced tumors which has been suggested to be due to the use of citrate as a source of carbons for fatty acid biosynthesis [57,58].
3.8. PGC-1α KO cell lines have increased levels of polyunsatureated fatty acids (PUFAs)
Tumor cells show altered fatty acid (FA) profiles that may be related to their metabolic alterations and this impacts tumor development [59]. We thus sought to evaluate how the absence of PGC-1α impacts the cellular fatty acid profile in iMEFs. Whereas no differences were found in the ratio of unsaturated vs saturated FAs, we did find a higher amount of polyunsaturated fatty acids (PUFAs) in KO cells (Table 1). This higher level of unsaturation corresponded with a higher peroxidizability index, a higher level of double bonds and a higher n6/n3 ratio in KO cells. Among the PUFAs detected, we noted a significant increase in the levels of arachidonic acid (20:4n-6, AA) in KO iMEFs. Estimation of desaturase activities based on the levels of the different FAs suggested a higher activity of the Δ5 desaturase, which is responsible for the formation of a double bond in the acid dihomo-γ-linolenic acid (20:3n-6) (DGLA) to form AA. Importantly, elevated levels of AA and Δ5 desaturase have been found in some types of cancer and reduction of desaturase activity has been reported to increase the efficacy of chemotherapy in some cases [60,61]. The basis of this effect seems to be related to the formation of pro-inflammatory intermediates by the enzyme COX-2 [62]. Importantly, Δ5 desaturase has also been linked to migration and invasion, while its downregulation increases E-cadherin levels, which could also possibly link the pathway to β-catenin [[1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81]] activity [63]. In contrast, there is no evidence that links desaturase activity to c-myc activity although altered lipid patterns have been identified in c-myc cancer cell models [64]. The noted increases in AA levels in KO cells might therefore also play a role in the more aggressive cancer prone phenotype of the PGC-1α KO iMEFs.
Table 1.
Fatty acid profile of PGC-1α WT and KO MEF & iMEFs. KO iMEFs show higher levels of PUFAs, in particular of AA, a precursor of pro-inflammatory mediators. Unsaturated FAs (UFA), saturated FAs (SAT), polyunsaturated FAs (PUFA), peroxidizability index (PI), double bond index (DBI), araquidonic acid (20:4n-6, AA), desaturase Δ5 (n-6).a, PGC-1α+/+ MEFs vs PGC-1α−/- MEFs;b, PGC-1α+/+ MEFs vs PGC-1α+/+ iMEFs;c, PGC-1α−/- MEFs vs PGC-1α−/- iMEFs;d, PGC-1α+/+ iMEFs vs PGC-1α−/- iMEFs. *, p < 0.05; **, p ≤ 0.01; ***, p ≤ 0.005.
| PGC-1α+/+ MEF | PGC-1α−/- MEF | PGC-1α+/+ iMEF | PGC-1α−/- iMEF | |
|---|---|---|---|---|
| UFA/SAT | 1.09 ± 0.08 | 0.99 ± 0.06 | 1.10 ± 0.07 | 1.10 ± 0.03 |
| PUFA (% nmol) | 23.98 ± 0.18 | 31.10 ± 1.03a** | 24.70 ± 0.77 | 25.75 ± 3.28c* |
| PI | 1.05 ± 0.02 | 1.35 ± 0.05a* | 1.06 ± 0.06 | 1.12 ± 0.17 |
| DBI | 127.02 ± 2.40 | 149.17 ± 5.07a* | 125.67 ± 4.98 | 132.35 ± 11.35 |
| n-6/n-3 | 2.69 ± 0.21 | 3.9 ± 0.32a** | 2.75 ± 0.36 | 3.39 ± 0.18 |
| 20:4n-6 (% nmol) | 10.79 ± 0.57 | 17.98 ± 0.11a*** | 11.36 ± 0,62 | 13.47 ± 3.33c***;d*** |
| Desaturase Δ5 (C20:4n-6/C20:3n-6) | 11.79 ± 1.54 | 18.00 ± 1.60a** | 8.51 ± 6.05 | 18.04 ± 7.46d*** |
4. Discussion
Reduced oxidative metabolism and mitochondrial dysfunction are hallmarks of cancer; however, tumor cells make use mitochondria and oxidative metabolism to boost their oncogenic potential [65]. PGC-1α is a master regulator of oxidative metabolism that responds to limited nutrient availability [66]. Its activities have been demonstrated to both facilitate and inhibit tumor progression in different contexts [21,25,38]. Here, we aimed to test, how the loss of PGC-1α activity in a primary cell would impact its oncogenic potential following spontaneous immortalization, in an attempt to directly asses whether PGC-1α is a tumor suppressor or an oncogene.
We found that, following immortalization, the metabolic differences between PGC-1α KO iMEFs and WT cells that are characteristic of PGC-1α activity are maintained, including, reduced oxygen consumption, increased glycolytic flux, and reduced TCA activity (Fig. 4, Fig. 5). These changes are associated with the induction of anaplerotic systems that typically mark advanced tumors (Fig. 6, Fig. 7). Consistently, PGC-1α KO iMEFs formed larger and more proliferative primary tumors and facilitated the metastatic potential of B16 melanoma cells (Fig. 1, Fig. 2). These characteristics are likely associated with the greater mobility of the KO cells and their enhanced capacity to sustain cell growth, supporting the role of PGC-1α as a tumor suppressor gene (Fig. 3).
Whereas PGC-1α KO iMEFs do have a proliferative advantage WT iMEFs in optimal cell culture conditions [31], they respond poorly to cues that inhibit cell growth such as contact inhibition and lack of substrate anchorage. KO iMEFs are also more resistant to cell death in limiting growth factor conditions than WT iMEFs (Fig. 3, Supp. Fig. 1). These characteristics are more typical of fully transformed cell lines rather than of simply immortalized cells [67]. Generally, spontaneously immortalized MEFs do not form tumors [68] and in order to test the oncogenic potential of a particular gene using iMEFs as a test model, the co-expression an oncogene such as RAS is necessary [68]. By contrast, PGC-1α KO iMEFs were able to form primary tumors when injected subcutaneously into immunodeficient mice within highly proliferative nodules, as identified by Ki67 staining (Fig. 1). Since the in vitro data suggest that growth rates are comparable between WT and KO iMEFs [31], and the contribution of the host immune response is not relevant in the mouse model used, KO iMEFs are likely to be more oncogenic due, at least in part, to their capacity to escape growth inhibition. Even though the contribution of angiogenesis in this model cannot be ruled out, the finding that KO iMEFs embedded in Matrigel (which supplies pro-angiogenic factors), proliferate more than WT iMEFs supports the relevance of the growth-control phenotype.
In addition, KO iMEFs also display a greater migratory phenotype (Fig. 3), first identified in primary cells [33,69]. This characteristic could be associated with the capacity of PGC-1α to prevent metastasis in prostate cancer [25]. Since iMEFs could not sustain metastasis formation by themselves, we tested their capacity to promote metastasis formation through co-inoculation with the highly metastatic B16–V5 cell line. Under these conditions iMEFs have been shown to contribute to the tumor matrix and impact on tumor progression [30]. Consistently, we found that KO iMEFs promoted the formation of lung metastasis (Fig. 2). Expression of an oncogene in this model could have increased the metastatic potential of the iMEFs but, our rationale was to test PGC-1α as the sole driving force, and follow the fate of PGC-1α-deficient cells. For the same reason, rescue of the phenotype by forced PGC-1α expression was not tested, since the “history” of a cell that did not have PGC-1α from the outset could not be recapitulated by forced expression following immortalization.
Loss of PGC-1α activity in cancer cells has been linked to the down-regulation of E-cadherin, and the activation of epithelial-mesenchymal transition (ETM) [70,71], a process that could lead to the nuclear translocation of β-catenin and the induction of c-myc [72,73]. Furthermore, in two previous studies we noted that PGC-1α deficient endothelial cells had reduced levels of VE-Cadherin in the cell membrane, and also showed alterations in the organization of the actin cytoskeleton. Therefore, a possible link between PGC-1α and β-catenin could be related to the formation of the cadherin complexes [74,75]. Importantly, c-myc activity in cancer cells is associated with most, if not all, the characteristics observed for the loss of PGC-1α in iMEFs, [76]. Indeed, nuclear β-catenin and c-myc levels were higher in KO iMEFs (Fig. 4), supporting the proposed molecular basis for this phenotype. It has been shown that β-catenin nuclear translocation following Wnt signaling activation results in the transcriptional induction of c-myc [77]. Importantly, c-myc and PGC-1α expression negatively correlates with c-my in pancreatic cancer stem cells [38]. However, how PGC-1α controls the β-catenin and c-myc remains to be established.
Given that PGC-1α is a master regulator of oxidative metabolism and that metabolic dysfunction is a hallmark of cancer, we evaluated whether KO MEFs remained metabolically distinct from WT cells after immortalization, and if so, how these differences could be related to their oncogenic potential. We found that KO iMEFs showed reduced oxygen consumption and were more glycolytic than WT cells (Fig. 4, Fig. 5). KO iMEFs also exhibited reduced TCA activity for the oxidation of Pyr (Fig. 5, Fig. 6).
The reduced TCA rates observed in KO iMEFs is a common feature of advanced tumors, in which, SDH and FH activities are typically reduced [78] and further drainage of TCA intermediates to feed intermediate metabolism has to be compensated by the activation of anaplerotic pathways in the cytosol [79]. In this case, cytosol/mitochondrial shuttles that normally work to incorporate oxidized intermediates and export reduced intermediates work in reverse.
All of these characteristics are to be expected in cells that lack PCG-1α and confirm that the metabolic imprint associated with PGC-1α is not lost following immortalization. Importantly, detailed evaluation of the metabolic derangements of KO iMEFs revealed close similarities with alterations found in advanced, aggressive tumors, such as Gln dependency, and induction of anaplerotic pathways that aim to compensate for TCA deficiencies and feed biosynthetic flux (Fig. 6, Fig. 7). All of these alterations have been proposed as targets for the development of novel therapies since they considerably impact on the viability of the cancer cell but not of the normal cell [80,81]. That all of these changes can be detected in PGC-1α KO iMEFs is strong evidence for its central role as a tumor suppressor.
In sum, characterization of the oncogenic potential and metabolic adaptations of spontaneously immortalized PGC-1α KO MEF suggests that lack of PGC-1α is to a large extent a hallmark of the metabolic changes associated with tumor progression, and strongly argue in favor of a tumor suppressor role for PGC-1α.
Acknowledgements
The authors wish to acknowledge the technical support of the “Servicio de Imagen Multimodal Experimental (SIME) of the IIB (CSIC-UAM), the “Servicio Interdepartamental” of the UAM, the Metabolomics Service of IRB Lleida, and the Protein Chemistry Facility of the CIB (CSIC). We thank Dr. Pablo Hernansanz-Agustín (IIS-IP and UAM) for technical advice on the use of the SeaHorse system, Dr. Marisol Soengas (CNIO) for the B16 cell line, Dr. Antonio Rodriguez (UAM) for the Luc-GFP lentivial vector, Dr. Ángela M. Valverde (IIB) for the Malic Enzime antibody. Editorial support was provided by Dr. Kenneth McCreath.
This work was funded by grants from the Spanish “Ministerio de Ciencia, Innovación y Universidades” (MICINN) and ERDF/FEDER funds, SAF2012-37693, SAF2015-63904-R, SAF2015-71521-REDC, RTI2018-093864-B-I00 to M.M., SAF2017-83043-R and B2017/BMD-3724 to S·C., PI15/00107 to A.M.R, the University of the Basque Country UPV/EHU grant GIU16/62) to J.l.R.S. and M.B.R.L., and the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement 721236-TREATMENT to M.M.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101396.
Contributor Information
Ignacio Prieto, Email: nprieto58@gmail.com.
Carmen Rubio Alarcón, Email: ruala42@gmail.com.
Raquel García-Gómez, Email: rgarcia@iib.uam.es.
Rebeca Berdún, Email: rebecaberdun@gmail.com.
Tamara Urgel, Email: turgel@iib.uam.es.
Manuel Portero, Email: manuel.portero@mex.udl.cat.
Reinald Pamplona, Email: reinald.pamplona@mex.udl.cat.
Antonio Martínez-Ruiz, Email: amartinezruiz@salud.madrid.org.
José Ignacio Ruiz-Sanz, Email: joseignacio.ruizs@ehu.eus.
M. Begoña Ruiz-Larrea, Email: mbego.ruizlarrea@ehu.eus.
Mariona Jove, Email: mariona.jove@udl.cat.
Sebastián Cerdán, Email: scerdan@iib.uam.es.
María Monsalve, Email: mpmonsalve@iib.uam.es.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
References
- 1.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 2.Otto A.M. Warburg effect(s)-a biographical sketch of Otto Warburg and his impacts on tumor metabolism. Cancer Metabol. 2016;4:5. doi: 10.1186/s40170-016-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porporato P.E., Filigheddu N., Pedro J.M.B., Kroemer G., Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28(3):265–280. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoshan M. On mitochondrial metabolism in tumor biology. Curr. Opin. Oncol. 2017;29(1):48–54. doi: 10.1097/CCO.0000000000000336. [DOI] [PubMed] [Google Scholar]
- 6.Verbon E.H., Post J.A., Boonstra J. The influence of reactive oxygen species on cell cycle progression in mammalian cells. Gene. 2012;511(1):1–6. doi: 10.1016/j.gene.2012.08.038. [DOI] [PubMed] [Google Scholar]
- 7.Valcarcel-Jimenez L., Gaude E., Torrano V., Frezza C., Carracedo A. Mitochondrial metabolism: Yin and Yang for tumor progression. Trends Endocrinol. Metab.: TEM (Trends Endocrinol. Metab.) 2017;28(10):748–757. doi: 10.1016/j.tem.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navarro P. Targeting tumor mitochondrial metabolism overcomes resistance to antiangiogenics. Cell Rep. 2016;15(12):2705–2718. doi: 10.1016/j.celrep.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 9.Valle I., Alvarez-Barrientos A., Arza E., Lamas S., Monsalve M. PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc. Res. 2005;66(3):562–573. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 10.Porporato P.E., Payen V.L., Baselet B., Sonveaux P. Metabolic changes associated with tumor metastasis, part 2: mitochondria, lipid and amino acid metabolism. Cell. Mol. Life Sci. : CM. 2016;73(7):1349–1363. doi: 10.1007/s00018-015-2100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song I.S. Mitochondria as therapeutic targets for cancer stem cells. World J. Stem Cells. 2015;7(2):418–427. doi: 10.4252/wjsc.v7.i2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colditz G.A., Peterson L.L. Obesity and cancer: evidence, impact, and future directions. Clin. Chem. 2018;64(1):154–162. doi: 10.1373/clinchem.2017.277376. [DOI] [PubMed] [Google Scholar]
- 13.Poff A. Seminars in Cancer Biology. 2017. Targeting the Warburg effect for cancer treatment: ketogenic diets for management of glioma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann P. Cancer and exercise: Warburg hypothesis, tumour metabolism and high-intensity anaerobic exercise. Sports. 2018;6(1) doi: 10.3390/sports6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ventura-Clapier R., Garnier A., Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc. Res. 2008;79(2):208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 16.Summermatter S., Troxler H., Santos G., Handschin C. Coordinated balancing of muscle oxidative metabolism through PGC-1alpha increases metabolic flexibility and preserves insulin sensitivity. Biochem. Biophys. Res. Commun. 2011;408(1):180–185. doi: 10.1016/j.bbrc.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Gravel S.P. Deciphering the dichotomous effects of PGC-1alpha on tumorigenesis and metastasis. Front Oncol. 2018;8:75. doi: 10.3389/fonc.2018.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun S.H., Han S.H., Park J.I. Peroxisome proliferator-activated receptor gamma and PGC-1alpha in cancer: dual actions as tumor promoter and suppressor. PPAR Res. 2018;2018:6727421. doi: 10.1155/2018/6727421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deblois G., St-Pierre J., Giguere V. The PGC-1/ERR signaling axis in cancer. Oncogene. 2013;32(30):3483–3490. doi: 10.1038/onc.2012.529. [DOI] [PubMed] [Google Scholar]
- 20.Jiang W.G., Douglas-Jones A., Mansel R.E. Expression of peroxisome-proliferator activated receptor-gamma (PPARgamma) and the PPARgamma co-activator, PGC-1, in human breast cancer correlates with clinical outcomes. Int. J. Cancer. 2003;106(5):752–757. doi: 10.1002/ijc.11302. [DOI] [PubMed] [Google Scholar]
- 21.Andrzejewski S. PGC-1alpha promotes breast cancer metastasis and confers bioenergetic flexibility against metabolic drugs. Cell Metabol. 2017;26(5):778–787. doi: 10.1016/j.cmet.2017.09.006. e775. [DOI] [PubMed] [Google Scholar]
- 22.Luo C. A PGC1alpha-mediated transcriptional axis suppresses melanoma metastasis. Nature. 2016;537(7620):422–426. doi: 10.1038/nature19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhalla K. PGC1alpha promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Res. 2011;71(21):6888–6898. doi: 10.1158/0008-5472.CAN-11-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Souza-Teixeira F. PGC-1alpha as a biomarker of physical activity-protective effect on colorectal cancer. Cancer Prev. Res. 2018;11(9):523–534. doi: 10.1158/1940-6207.CAPR-17-0329. [DOI] [PubMed] [Google Scholar]
- 25.Torrano V. The metabolic co-regulator PGC1alpha suppresses prostate cancer metastasis. Nat. Cell Biol. 2016;18(6):645–656. doi: 10.1038/ncb3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olmos Y. Mutual dependence of Foxo3a and PGC-1alpha in the induction of oxidative stress genes. J. Biol. Chem. 2009;284(21):14476–14484. doi: 10.1074/jbc.M807397200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cruz F. Intracellular compartmentation of pyruvate in primary cultures of cortical neurons as detected by (13)C NMR spectroscopy with multiple (13)C labels. J. Neurosci. Res. 2001;66(5):771–781. doi: 10.1002/jnr.10048. [DOI] [PubMed] [Google Scholar]
- 28.Zambrano A. The thyroid hormone receptor beta induces DNA damage and premature senescence. J. Cell Biol. 2014;204(1):129–146. doi: 10.1083/jcb.201305084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arranz L., Naudi A., De la Fuente M., Pamplona R. Exceptionally old mice are highly resistant to lipoxidation-derived molecular damage. Age. 2013;35(3):621–635. doi: 10.1007/s11357-012-9391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mezawa Y., Orimo A. The roles of tumor- and metastasis-promoting carcinoma-associated fibroblasts in human carcinomas. Cell Tissue Res. 2016;365(3):675–689. doi: 10.1007/s00441-016-2471-1. [DOI] [PubMed] [Google Scholar]
- 31.Prieto I. Early induction of senescence and immortalization in PGC-1alpha-deficient mouse embryonic fibroblasts. Free Radic. Biol. Med. 2019;138:23–32. doi: 10.1016/j.freeradbiomed.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Muzzio N.E., Pasquale M.A., Gonzalez P.H., Arvia A.J. Influence of individual cell motility on the 2D front roughness dynamics of tumour cell colonies. J. Biol. Phys. 2014;40(3):285–308. doi: 10.1007/s10867-014-9349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borniquel S. Inactivation of Foxo3a and subsequent downregulation of PGC-1 alpha mediate nitric oxide-induced endothelial cell migration. Mol. Cell. Biol. 2010;30(16):4035–4044. doi: 10.1128/MCB.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huttenlocher A. Integrin and cadherin synergy regulates contact inhibition of migration and motile activity. JCB (J. Cell Biol.) 1998;141(2):515–526. doi: 10.1083/jcb.141.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archbold H.C., Yang Y.X., Chen L., Cadigan K.M. How do they do Wnt they do?: regulation of transcription by the Wnt/beta-catenin pathway. Acta Physiol. 2012;204(1):74–109. doi: 10.1111/j.1748-1716.2011.02293.x. [DOI] [PubMed] [Google Scholar]
- 36.Shang S., Hua F., Hu Z.W. The regulation of beta-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8(20):33972–33989. doi: 10.18632/oncotarget.15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordan J.D., Thompson C.B., Simon M.C. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12(2):108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sancho P. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metabol. 2015;22(4):590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 39.Kaminski L. PGC1alpha inhibits polyamine synthesis to suppress prostate cancer aggressiveness. Cancer Res. 2019;79(13):3268–3280. doi: 10.1158/0008-5472.CAN-18-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi L., Pan H., Liu Z., Xie J., Han W. Roles of PFKFB3 in cancer. Signal transduction and targeted therapy. 2017;2:17044. doi: 10.1038/sigtrans.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aguer C. Galactose enhances oxidative metabolism and reveals mitochondrial dysfunction in human primary muscle cells. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgess S.C. Diminished hepatic gluconeogenesis via defects in tricarboxylic acid cycle flux in peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha)-deficient mice. J. Biol. Chem. 2006;281(28):19000–19008. doi: 10.1074/jbc.M600050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murai S. Inhibition of malic enzyme 1 disrupts cellular metabolism and leads to vulnerability in cancer cells in glucose-restricted conditions. Oncogenesis. 2017;6(5):e329. doi: 10.1038/oncsis.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desideri E., Vegliante R., Ciriolo M.R. Mitochondrial dysfunctions in cancer: genetic defects and oncogenic signaling impinging on TCA cycle activity. Cancer Lett. 2015;356(2 Pt A):217–223. doi: 10.1016/j.canlet.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 45.Lahey R. Enhanced redox state and efficiency of glucose oxidation with miR based suppression of maladaptive NADPH-dependent malic enzyme 1 expression in hypertrophied hearts. Circ. Res. 2018;122(6):836–845. doi: 10.1161/CIRCRESAHA.118.312660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lao-On U., Attwood P.V., Jitrapakdee S. Roles of pyruvate carboxylase in human diseases: from diabetes to cancers and infection. J. Mol. Med. 2018;96(3–4):237–247. doi: 10.1007/s00109-018-1622-0. [DOI] [PubMed] [Google Scholar]
- 47.Sradhanjali S., Reddy M.M. Inhibition of pyruvate dehydrogenase kinase as a therapeutic strategy against cancer. Curr. Top. Med. Chem. 2018;18(6):444–453. doi: 10.2174/1568026618666180523105756. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J., Pavlova N.N., Thompson C.B. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36(10):1302–1315. doi: 10.15252/embj.201696151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dang C.V. Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harbor Symp. Quant. Biol. 2011;76:369–374. doi: 10.1101/sqb.2011.76.011296. [DOI] [PubMed] [Google Scholar]
- 50.Choi Y.K., Park K.G. Targeting glutamine metabolism for cancer treatment. Biomolecules & therapeutics. 2018;26(1):19–28. doi: 10.4062/biomolther.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeBerardinis R.J. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U.S.A. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sookoian S., Pirola C.J. Alanine and aspartate aminotransferase and glutamine-cycling pathway: their roles in pathogenesis of metabolic syndrome. World J. Gastroenterol. 2012;18(29):3775–3781. doi: 10.3748/wjg.v18.i29.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsun Z.Y., Possemato R. Amino acid management in cancer. Semin. Cell Dev. Biol. 2015;43:22–32. doi: 10.1016/j.semcdb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullen A.R. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7(5):1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mullen A.R. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dang L. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 2016;16(10):619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fendt S.M. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat. Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beloribi-Djefaflia S., Vasseur S., Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azrad M., Turgeon C., Demark-Wahnefried W. Current evidence linking polyunsaturated Fatty acids with cancer risk and progression. Front Oncol. 2013;3:224. doi: 10.3389/fonc.2013.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pender-Cudlip M.C. Delta-6-desaturase activity and arachidonic acid synthesis are increased in human breast cancer tissue. Cancer Sci. 2013;104(6):760–764. doi: 10.1111/cas.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Camps J (Oxidative Stress and Inflammation in Non-communicable Diseases - Molecular Mechanisms and Perspectives in Therapeutics p pages (cm).
- 63.Yang X. Inhibition of cancer migration and invasion by knocking down delta-5-desaturase in COX-2 overexpressed cancer cells. Redox Biol. 2017;11:653–662. doi: 10.1016/j.redox.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griffitts J. In vivo MRS assessment of altered fatty acyl unsaturation in liver tumor formation of a TGF alpha/c-myc transgenic mouse model. J. Lipid Res. 2009;50(4):611–622. doi: 10.1194/jlr.M800265-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu Y. The reverse Warburg effect is likely to be an Achilles' heel of cancer that can be exploited for cancer therapy. Oncotarget. 2017;8(34):57813–57825. doi: 10.18632/oncotarget.18175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang H., Ward W.F. PGC-1alpha: a key regulator of energy metabolism. Adv. Physiol. Educ. 2006;30(4):145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 67.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 68.Sun H., Taneja R. Analysis of transformation and tumorigenicity using mouse embryonic fibroblast cells. Methods Mol. Biol. 2007;383:303–310. doi: 10.1007/978-1-59745-335-6_19. [DOI] [PubMed] [Google Scholar]
- 69.Qu A. PGC-1alpha attenuates neointimal formation via inhibition of vascular smooth muscle cell migration in the injured rat carotid artery. Am. J. Physiol. Cell Physiol. 2009;297(3):C645–C653. doi: 10.1152/ajpcell.00469.2008. [DOI] [PubMed] [Google Scholar]
- 70.Lee H.J. Peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1alpha) upregulated E-cadherin expression in HepG2 cells. FEBS Lett. 2008;582(5):627–634. doi: 10.1016/j.febslet.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 71.Yuan Y. Mitochondrial dysfunction accounts for aldosterone-induced epithelial-to-mesenchymal transition of renal proximal tubular epithelial cells. Free Radic. Biol. Med. 2012;53(1):30–43. doi: 10.1016/j.freeradbiomed.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 72.Tian X. E-cadherin/beta-catenin complex and the epithelial barrier. J. Biomed. Biotechnol. 2011;2011:567305. doi: 10.1155/2011/567305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yochum G.S., Cleland R., Goodman R.H. A genome-wide screen for beta-catenin binding sites identifies a downstream enhancer element that controls c-Myc gene expression. Mol. Cell. Biol. 2008;28(24):7368–7379. doi: 10.1128/MCB.00744-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garcia-Quintans N. Regulation of endothelial dynamics by PGC-1alpha relies on ROS control of VEGF-A signaling. Free Radic. Biol. Med. 2016;93:41–51. doi: 10.1016/j.freeradbiomed.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 75.Garcia-Quintans N. Oxidative stress induces loss of pericyte coverage and vascular instability in PGC-1alpha-deficient mice. Angiogenesis. 2016;19(2):217–228. doi: 10.1007/s10456-016-9502-0. [DOI] [PubMed] [Google Scholar]
- 76.Pelengaris S., Khan M., Evan G. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer. 2002;2(10):764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 77.Shi L. Research of the relationship between beta-catenin and c-myc-mediated Wnt pathway and laterally spreading tumors occurrence. Eur. Rev. Med. Pharmacol. Sci. 2017;21(2):252–257. [PubMed] [Google Scholar]
- 78.King A., Selak M.A., Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25(34):4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 79.Corbet C., Feron O. Cancer cell metabolism and mitochondria: nutrient plasticity for TCA cycle fueling. Biochim. Biophys. Acta. 2017;1868(1):7–15. doi: 10.1016/j.bbcan.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 80.Anderson N.M., Mucka P., Kern J.G., Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein & cell. 2018;9(2):216–237. doi: 10.1007/s13238-017-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luengo A., Gui D.Y., Vander Heiden M.G. Targeting metabolism for cancer therapy. Cell chemical biology. 2017;24(9):1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.