Abstract
Background
The spatial distributions of different types of cells could reveal a cancer cell's growth pattern, its relationships with the tumor microenvironment and the immune response of the body, all of which represent key “hallmarks of cancer”. However, the process by which pathologists manually recognize and localize all the cells in pathology slides is extremely labor intensive and error prone.
Methods
In this study, we developed an automated cell type classification pipeline, ConvPath, which includes nuclei segmentation, convolutional neural network-based tumor cell, stromal cell, and lymphocyte classification, and extraction of tumor microenvironment-related features for lung cancer pathology images. To facilitate users in leveraging this pipeline for their research, all source scripts for ConvPath software are available at https://qbrc.swmed.edu/projects/cnn/.
Findings
The overall classification accuracy was 92.9% and 90.1% in training and independent testing datasets, respectively. By identifying cells and classifying cell types, this pipeline can convert a pathology image into a “spatial map” of tumor, stromal and lymphocyte cells. From this spatial map, we can extract features that characterize the tumor micro-environment. Based on these features, we developed an image feature-based prognostic model and validated the model in two independent cohorts. The predicted risk group serves as an independent prognostic factor, after adjusting for clinical variables that include age, gender, smoking status, and stage.
Interpretation
The analysis pipeline developed in this study could convert the pathology image into a “spatial map” of tumor cells, stromal cells and lymphocytes. This could greatly facilitate and empower comprehensive analysis of the spatial organization of cells, as well as their roles in tumor progression and metastasis.
Keywords: Deep learning, Convolutional neural network, Lung adenocarcinoma, Pathology image, Cell distribution and interaction, Prognosis
Evidence before this study
Since 2011, computer algorithms have been developed to analyze tissue pathology images for cancer diagnosis, grading and prognosis. Recently, deep learning-based algorithms have made remarkable achievements in pathology image analysis. Several deep learning models for lung cancer pathology image analysis have been proposed for lung cancer H&E-stained pathology images. Furthermore, several deep learning methods have been developed to characterize the tumor micro-environment, since the tumor micro-environment plays an important role in tumor progression and response to treatment.
The major cell types in a malignant tissue of lung include tumor cells, stromal cells, and lymphocytes. Stromal cells are connective tissue cells such as fibroblasts and pericytes, and their interaction with tumor cells plays an important role in cancer progression and metastasis inhibition. For example, the crosstalk between cancer cells and stromal cells is needed for invasive growth and metastasis. Spatial heterogeneity of TILs is associated with the tumor molecular profile and patient prognosis. How to automatically classify different types of cells is a major technical challenge in studying the tumor microenvironment.
Added value of this study
In this study, we developed a pathological image analysis and cell classification pipeline, which can perform nuclei segmentation, CNN-based cell type prediction, and feature extraction. This pipeline successfully visualizes the spatial distributions of tumor, stromal, and lymphocyte cells in the ROI of lung ADC pathology images.
Implications of all the available evidence
Quantifying distribution and interaction with tumor or stromal cells of lymphocytes can potentially provide a way to evaluate immune response status and serve as a biomarker for immunotherapy response. The analysis pipeline developed in this study could convert the pathology image into a “spatial map” of tumor cells, stromal cells and lymphocytes. This could greatly facilitate and empower comprehensive analysis of cell spatial organization, as well as its role in tumor progression and metastasis.
Hematoxylin and Eosin (H&E)-stained tissue whole-slide image (WSI) scanning is becoming a routine clinical procedure that produces massive pathology images with histological details in high resolution. Tumor pathology images contain not only essential information for tumor grade and subtype classification [1], but also information on the tumor microenvironment and the spatial distributions of different types of cells. Tumor tissues are complex structures with cancer cells and surrounding non-malignant cells (such as stromal cells and lymphocytes) that form the tumor micro-environment [2]. Understanding the interactions among these cells can provide critical insights into tumor initiation, progression, metastasis and potential therapeutic targets. For example, the crosstalk between cancer cells and stromal cells is needed for invasive growth and metastasis [3,4]. However, the major technical challenge to studying cell spatial organization is how to classify different types of cells from tumor tissues. It is impractical for a pathologist to manually recognize and localize every individual cell in a pathology slide.
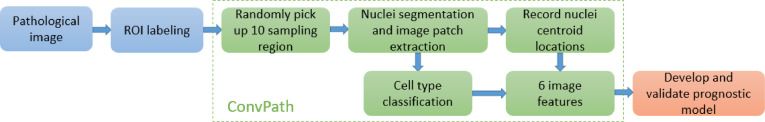
In recent years, convolutional neural networks (CNNs), one of the deep learning strategies, have made great success in image recognition tasks [5], [6], [7]. In this study, we developed a CNN model to automatically classify tumor cells, stromal cells, and lymphocytes for lung adenocarcinoma (ADC) pathology images. Furthermore, we developed an automated image analysis pipeline, ConvPath, to facilitate researchers in studying the spatial interactions of different types of cells and their roles in tumor progression and metastasis. The ConvPath pipeline is composed of nuclei segmentation, cell type recognition, microenvironment characterization, and prognosis (Fig. 1). The prognostic performance of the model was validated in two independent lung ADC cohorts.
Fig. 1.
Flow chart of ConvPath-aided pathological image analysis.
CHCAMS, National Cancer Center/Cancer Hospital of Chinese Academy of Medical Sciences, China; CI, confidence interval; HR, hazard ratio; TCGA, The Cancer Genome Atlas.
1. Methods
1.1. Datasets
H&E-stained histology images and clinical information for lung ADC patients and corresponding clinical data were collected from four independent cohorts: The Cancer Genome Atlas lung ADC project LUAD data (referred as the TCGA dataset), the National Lung Screening Trial project (the NLST dataset), the University of Texas Special Program of Research Excellence (SPORE) in Lung Cancer project (the SPORE dataset), and the National Cancer Center/Cancer Hospital of Chinese Academy of Medical Sciences, China (the CHCAMS dataset).
The TCGA data, including 1337 tumor images from 523 patients, were obtained from the TCGA image portal (https://wiki.cancerimagingarchive.net/display/Public/TCGA-LUAD). All TCGA images were captured at X20 or X40 magnification and included both frozen and Formalin-Fixed, Paraffin-Embedded (FFPE) slides. The NLST data, including 345 tumor images from 201 patients, were acquired from the National Lung Screening Trial, which was performed by the National Cancer Institute. All NLST images were FFPE slides and captured at 40X magnification. The CHCAMS data, including 102 images from 102 stage I ADC patients, were obtained from the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (CHCAMS), China. All CHCAMS images were FFPE slides and captured at 20X magnification. The SPORE data, including 130 images from 112 patients, were acquired from the UT Lung SPORE tissue bank. All SPORE images were FFPE slides and captured at 20X magnification. The characteristics of the four datasets used in this study are summarized in Supplemental Table 1.
1.2. Extraction of image patches centering at nuclei centroids
A pathologist, Dr. Lin Yang, reviewed the H&E-stained pathology image slides and manually labeled Region of Interest (ROI) boundaries using the annotation tool of ImageScope (Leica Biosystem, Fig. 2a). ROIs were defined by the main malignant area within the pathology images. ConvPath randomly selected 10 sampling regions from each selected ROI. The sampling regions were sized 5000×5000 or 3000×3000 pixels in 40X or 20X magnification images, respectively. In each sampling region, ConvPath further extracted 80×80 pixel image patches (for 40X magnification images, 160×160 pixel image patches were extracted first and resized as 80×80 pixel) centering at nuclei centroids (Fig. 2b, Supplemental Figure 1).
Fig. 2.
Image preprocessing step of the ConvPath software. (a) Selection of regions of interest (ROIs) in whole pathological imaging slides. (b) Image segmentation pipeline to extract cell-centered image patches from selected ROIs.
In order to extract the image patches, RGB color space was first converted to H&E color space with the deconvolution matrix set as [0.550 0.758 0.351; 0.398 0.634 0.600; 0.754 0.077 0.652] [8]. Morphological operations consisting of opening and closing were adopted to process the hematoxylin channel image [9]. Then, ConvPath detected nuclei boundaries using a level set segmentation technique [10,11]. In this segmentation method, the initial contour was randomly given, the value of sigma in Gaussian filter was 1, the number of iterations was 30, and the velocity term was 60. Next, nuclei centroids were detected as the moments of centroids of connected targets in a binary image, where the foreground was the regional maximum locations in a distance map of the segmented image. Here, Euclidean distance was utilized for the distance transform and regional maximums were searched within 8-connected neighborhoods. Finally, image patches using the detected nuclei centroids as centers were extracted from the original pathological RGB image (Fig. 2b).
1.3. Deep learning algorithm in the convpath software
ConvPath incorporates a CNN [12], [13], [14] to recognize the major cell types, including tumor cells, stromal cells and lymphocytes, in the center of pathology image patches (Fig. 3a, Supplemental Table 2). The input to the CNN was an 80×80 image patch normalized to the range [−0.5, 0.5] with 3 channels corresponding to the red (R), green (G), and blue (B) channels. The output layer for the CNN was a softmax layer with 3 categories: tumor cell, stromal cell, and lymphocyte. For one image patch, a probability for each of the 3 categories was predicted by the CNN; the category with the highest probability was assigned as the predicted class for the image patch. The CNN was trained using a batch size of 10, a momentum of 0.9, a weight decay of 0.0001, an initial learning rate of 0.01, which shrinks by 0.99995 in each step, and training steps of 20,000. The image patches were rotated and flipped to augment sample size. A drop connect probability of 0.5 was used in all convolutional layer parameters. The NLST and TCGA datasets were combined and used as the training set for the CNN (Fig. 3b&c, Supplemental Table 3), and the SPORE dataset was used as the external validation set. The image patches in training and validation sets were labeled by the pathologist as ground truth.
Fig. 3.
Cell type recognition step of the ConvPath software. (a) Schema and structure of the convolutional neural network (CNN) to recognize the types of cells in the centers of image patches. (b) Confusion matrix of internal testing results of CNN on the NLST and TCGA training image slides. Prediction accuracies are calculated based on 3996 image patches for each cell type. (c) Confusion matrix of independent testing results of CNN on image patches of the SPORE dataset. Prediction accuracies are calculated based on 8245 lymphocyte, 2211 stroma, and 6836 tumor patches.
1.4. Tumor micro-environment feature extraction
Based on the prediction results of the CNN, ConvPath converted the pathology image into a “spatial map” of tumor cells, stromal cells and lymphocytes. From this spatial map, we can define the tumor cell regions, stromal cell regions and lymphocyte regions within each ROI, and characterize the distribution and interactions among these regions. For example, a stromal cell region is a small area with tumor tissue that consists of mostly stromal cells. Specifically, ConvPath used kernel smoothers to define regions of tumor cells, stromal cells and lymphocytes separately within the ROI (Fig. 4b). For instance, to define the tumor cell region, ConvPath extracted coordinates of the centers of all image patches and labeled them as 1 if they had been recognized as tumor cells from the previous step, 0 if not. For each point on the image, ConvPath then calculated the probability of being a tumor cell region by weighting all its neighbors with standard normal density kernel K (z/h), where z was defined as the distance between the point and center of each image patch, and h, the bandwidth, was defined as 2 times the estimated cell diameter. A region with probability larger than 0.5 was defined as a tumor cell region. The same approach was used to define stromal cell region and lymphocyte cell region. Next, ConvPath calculated 2 features for each region (Supplemental Table 4), which were the perimeter divided by the square root of region area and size divided by image size for the 3 kinds of cell regions separately.
Fig. 4.
Feature extraction step of the ConvPath software. (a) A zoomed-in part of a sampling region (Supplemental Figure 3) in which cell nuclei centroids are labeled with predicted cell types. Green, stroma; cyan, lymphocyte; yellow, tumor. (b) Cell type region detection using a kernel smoothing algorithm for the sampling region shown in Supplemental Figure 3. Area and perimeters are evaluated for regions of tumor, stroma, and lymphocyte.
1.5. Statistical analysis
R (version 3.2.4) [15] and R packages survival (version 2.38–3), glmnet (version 2.0–5), and clinfun (version 1.0.13) were used for statistical analysis. Survival time was defined as the period from diagnosis to death or last contact for the NLST and TCGA datasets, and from diagnosis to recurrence or last contact in the CHCAMS dataset. The prognostic model was trained on the NLST patients using a Cox regression model with elastic penalty, to predict a risk score for each sampling region. The final risk score of each patient was determined by averaging risk scores across 10 sampling regions of this patient. The performance of this prognostic model was evaluated on the TCGA and CHCAMS datasets by dichotomizing the patients by the median predicted risk score of each dataset. In the validation study, the maximum follow-up time was set to six years, since patient survival after six years may not directly relate to cancer-specific events. Kaplan-Meier (K-M) plots and log-rank tests were used to compare survival outcomes. In addition, a multivariate Cox proportional hazard model was used to test whether the prognostic risk scores were statistically significant after adjusting for clinical variables, including age, gender, tobacco history, and stage. A Jonckheere-Terpstra (J-T) k-sample test [16] was used to test whether higher risk scores were correlated with theoretically more severe ADC subtypes. The results were considered significant if the two-sided test (except for the J-T test, which is a one-sided test for trend) p value ≤ 0.05.
1.6. Data availability
Pathology images and clinical data in the NLST and TCGA datasets that support the findings of this study are available online in the NLST (https://biometry.nci.nih.gov/cdas/nlst/) and The Cancer Genome Atlas Lung Adenocarcinoma (TCGA-LUAD, https://wiki.cancerimagingarchive.net/display/Public/TCGA-LUAD). Data in the SPORE and CHCAMS datasets that support the findings of this study are available from the UT Lung SPORE Tissue bank and the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (CHCAMS), China, separately, but restrictions apply to the availability of these data.
2. Results
2.1. ConvPath classifies lung adenocarcinoma cell types with high accuracy
11,988 tumor, stromal, and lymphocyte image patches centered at cell nuclei centroids were extracted from 29 slides in the TCGA and NLST datasets (Fig. 2, Supplemental Table 3) and used to train the CNN model (Fig. 3a). Example image patches are shown in Supplemental Figure 1. The overall classification accuracies of the CNN model on training images were 99.3% for lymphocytes, 87.9% for stromal cells, and 91.6% for tumor cells, respectively (Fig. 3b). The independent cross-study classification rates in the SPORE dataset were 97.8% for lymphocytes, 86.5% for stromal cells, and 85.9% for tumor cells (Fig. 3c).
2.2. Tumor micro-environment features from predicted sampling regions correlate with overall survival
ConvPath was then used to generate cell type predictions for 10 random sampling regions within the ROI on each slide. Based on nuclei centroid locations together with accurate cell type predictions (Fig. 4a, Supplemental Figure 2), we investigated whether the spatial distributions of tumor cells, stromal cells, and lymphocytes correlated with the survival outcome of lung ADC patients. In each predicted sampling region, tumor, stromal, and lymphocyte cell regions were detected using a kernel smoothing algorithm (Fig. 4b, Method section). For regions of each cell type, simple parameters such as perimeter and size were measured. To ensure comparability across image slides captured at different magnitudes, the parameters were normalized by area of sampling region. In univariate Cox analysis, 4 of the 6 extracted features significantly correlated with survival outcome in the NLST dataset (Supplemental Table 4). Interestingly, both perimeter and area of stroma region were good prognostic factors, suggesting a protective effect of stromal cells in lung ADC patients (Supplemental Figure 3&4).
2.3. Development and validation of an image feature-based prognostic model
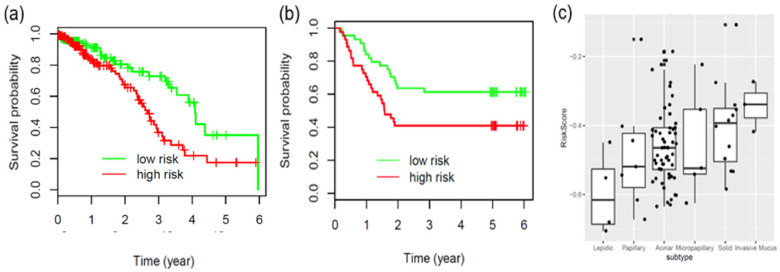
Utilizing the region features of each cell type extracted from the pathology images in the NLST dataset, we developed a prognostic model to predict patient survival outcome (coefficients of this model are shown in Supplemental Table 4). The model was then independently validated in the TCGA and CHCAMS datasets. The TCGA and CHCAMS patients were dichotomized according to the median predicted risk scores in each dataset. In both datasets, the patients in the predicted high-risk group had significantly worse survival outcomes than those in the predicted low-risk group (Fig. 5a&b, log rank test, p = 0.0047 for the TCGA dataset, p = 0.030 for the CHCAMS dataset). To evaluate whether the image features extracted by ConvPath were independent of clinical variables, multivariate Cox proportional hazard models were used to adjust the predicted risk scores with available clinical variables, including gender, age, stage and smoking status (Table 1). After adjustment, the still significant hazard ratios between high- and low-risk groups (p = 0.0021 for the TCGA dataset, p = 0.016 for the CHCAMS dataset) indicated that risk group as defined by ConvPath-extracted image features was an independent prognostic factor, in addition to other clinical variables.
Fig. 5.
Application of the prognostic model to independent datasets. (a, b) Validation of the prognostic model in the TCGA overall survival data (a, log rank test, p = 0.0047) and the CHCAMS recurrence data (b, log rank test, p = 0.030). (c) Boxplot for the distribution of predicted risk scores in the 5 histological subtypes of lung adenocarcinoma for the CHCAMS dataset patients. Jonckheere-Terpstra k-sample test, p = 0.0039. The boxes and whiskers show the lower (Q1) and upper (Q3) quartiles and the median for each histological subtype.
Table 1.
Multivariate analysis of the predicted risk scores in the CHCAMS and TCGA datasets adjusted by clinical variables.
| TCGA dataset (n = 346) | HR | 95% CI | p value |
|---|---|---|---|
| High risk vs. low risk | 2.19 | 1.33–3.60 | 0.0021 |
| Age (per year) | 1.03 | 1.01–1.06 | 0.014 |
| Male vs. female | 0.69 | 1.45–1.16 | 0.16 |
| Smoker vs. non-smoker | 0.88 | 0.53–1.47 | 0.62 |
| Stage | |||
| Stage I | ref | – | |
| Stage II | 2.69 | 1.45–5.00 | 0.0017 |
| Stage III | 5.04 | 2.69–9.43 | <0.001 |
| Stage IV | 6.06 | 2.49–14.73 | <0.001 |
| CHCAMS dataset (n = 88) | HR | 95% CI | p value |
| High risk vs. low risk | 2.21 | 1.16–4.21 | 0.016 |
| Age (per year) | 1.02 | 0.99–1.06 | 0.202 |
| Male vs. female | 1.85 | 0.69–4.91 | 0.22 |
| Smoker vs. non-smoker | 0.76 | 0.28–2.04 | 0.585 |
CHCAMS, National Cancer Center/Cancer Hospital of Chinese Academy of Medical Sciences, China;.
CI, confidence interval;.
HR, hazard ratio;.
TCGA, The Cancer Genome Atlas.
2.4. Predicted risk scores correlate with severity of adc subtypes
The 2015 WHO classification of lung cancer further divides invasive lung ADC into several subtypes, including acinar, lepidic, micropapillary, papillary, solid, and mucinous ADC [1]. The correlation of the predicted risk scores with the predominant histology subtypes identified by our pathologist for the CHCAMS dataset, according to the 2015 WHO classification guidelines, was tested (Fig. 5c). Higher risk scores correlated with more aggressive ADC subtypes, such as solid predominant ADC and invasive mucinous ADC (p = 0.0039). Noticeably, despite such correlation, image-derived risk score was independent of the ADC subtypes in multivariate survival analysis (Supplemental Table 5).
2.5. The convpath software and web server
To facilitate practical application of this pathological image analysis pipeline by pathologists and bioinformaticians, the image segmentation, deep learning, and feature extraction algorithms were incorporated into the ConvPath software. The ConvPath software is publicly accessible from the web server created for this study, which is at https://qbrc.swmed.edu/projects/cnn/ (Supplemental Figure 6).
3. Discussion
Since 2011, computer algorithms have been developed to analyze tissue pathology images for cancer diagnosis [17], [18], [19], [20], [21], grading [22], [23], [24], [25], [26] and prognosis [27], [28], [29], [30], [31], [32]. Recently, deep learning-based algorithms have made remarkable achievements in pathology image analysis [33], [34], [35], [36]. Several deep learning models for lung cancer pathology image analysis have been proposed for lung cancer H&E-stained pathology images. For example, a CNN model was developed to classify image patches of 300×300 pixel size as malignant or non-malignant in lung cancer pathology images, and has achieved an overall classification accuracy of 89.8% in an independent testing set [35]. This model could facilitate pathologists to quickly detect and locate tumor region from tissue pathology images. In addition to detecting tumor regions, Coudray et al. have developed a CNN model to distinguish different lung cancer subtypes [37].
To classify different cell types, several classic machine learning-based models and CNN models have also been developed. QuPath enabled semi-automatic detection of different types of objects (e.g., cell nuclei) through classic machine learning methods [38]. Sirinukunwattana et al. utilized CNN to classify nuclei into epithelial, inflammatory, fibroblast, and miscellaneous nuclei in colon cancer histology images [39].
Furthermore, several deep learning methods have been developed to characterize the tumor micro-environment, since the tumor micro-environment plays an important role in tumor progression and response to treatment. For example, a CNN model has been developed to classify lymphocytes from necrosis or other tissues in multiple cancer types [40]. In another study, Yi et al. developed a Fully Convolutional Neural Network (FCN) [41] to segment micro blood-vessels from lung ADC pathology images. An image segmentation CNN model was developed to classify each pixel in lung ADC pathology images as nucleus centroid, nucleus boundary, or non-nuclei [42]. Based on the results of this model, morphological, textural, and graphical features of cell nucleus were extracted and used to develop a prediction model for tumor recurrence in lung ADC patients.
The major cell types in a malignant tissue of lung include tumor cells, stromal cells, and lymphocytes. Stromal cells are connective tissue cells such as fibroblasts and pericytes, and their interaction with tumor cells plays an important role in cancer progression [43], [44], [45] and metastasis inhibition [46]. Tumor-infiltrating lymphocytes (TILs) are white blood cells that have migrated into a tumor. They are a mix of different types of cells, with T cells being the most abundant population. Tumor-infiltrating lymphocytes have been associated with patient prognosis in multiple tumor types [47], [48], [49], [50]. The spatial distributions of different types of cells could reveal a cancer cell's growth pattern, its relationships with the tumor microenvironment and the immune response of the body, all of which represent key “hallmarks of cancer”. For example, the crosstalk between cancer cells and stromal cells is needed for invasive growth and metastasis [3,4]. Spatial heterogeneity of TILs is associated with the tumor molecular profile and patient prognosis [40,51]. However, as there are more than 10,000 cells in each sampling region (Supplemental Figure 2), it is extremely labor-intensive and error-prone for a pathologist to manually recognize and localize every individual cell in a pathology slide. How to automatically classify different types of cells is a major technical challenge in studying the tumor microenvironment.
In this study, we developed a pathological image analysis and cell classification pipeline, which can perform nuclei segmentation, CNN-based cell type prediction, and feature extraction (Fig. 1). This pipeline successfully visualizes the spatial distributions of tumor, stromal, and lymphocyte cells in the ROI of lung ADC pathology images. It can potentially serve as a prognostic method independent of other clinical variables. The patient prognostic model based on extracted image features was trained in the NLST dataset and independently validated in the TCGA and CHCAMS datasets, which indicates the generalizability of this analysis pipeline to other lung ADC patients.
The accurate classification of cell types in pathology images was validated in an independent data cohort. While the qualities of H&E staining vary across different cohorts and there are inherent inter-patient differences, ConvPath still has 90.1% overall accuracy in the SPORE dataset (Fig. 3c). The ConvPath pipeline developed in ADC can be directly applied to Squamous Cell Carcinoma, another subtype of NSCLC; satisfactory results were shown in Supplemental Figure 7. The robustness of ConvPath benefits from the level set-based segmentation algorithm in the nuclei segmentation step. This segmentation algorithm is invariant to the location of the initial contour and can handle high variability across different H&E pathology images. Moreover, nuclei centroid extraction based on distance transform can separate most of the connected nuclei that are not properly processed by the commonly used CellProfiler software [30,52]. The robustness of prediction also benefits from the powerful structure of CNN [53].
The relationships between the extracted tumor micro-environment-related image features and patient prognosis were evaluated in this study (Supplemental Table 4). In univariate analysis, higher stromal cell abundance correlated with better prognosis (Supplemental Figure 4), which is consistent with a recent report on lung ADC patients [46]. However, disparate roles of stromal cells in tumor progression have been reported, including stimulation of tumor proliferation through growth signals and limitation of tumor cells metastatic spreading [44,45,54]. Combinatory analysis of cell spatial distribution detected in this study and the functionality of stromal cells, which could not be evaluated through H&E staining, will help elucidate whether these disparate roles arise from the different activation status of crosstalk between tumor and stroma. In contrast, higher lymphocyte abundance, reflected by region size rather than perimeter, correlated with worse prognosis (Supplemental Table 4, Supplemental Figure 5). Although the presence of both tumor- and stroma-infiltrating lymphocytes has been reported to correlate with tumor cell apoptosis and better patient survival in non-small cell lung cancer [47,50,55], the tumor-suppressive or tumor-promoting properties of lymphocytes depend on spatial distribution of the lymphocytes in the tumor microenvironment [56]. On the other hand, in this study, the “size of lymphocyte cell region/image size” (in Supplement Table 4) refers to regions mainly consisting of lymphocyte cells, which are aggregated lymphocytes. So the size of the lymphocyte cell region may not directly correlate or even negatively correlate with tumor- and stroma-infiltrating lymphocytes, which are individual lymphocytes that are in the tumor and stromal cell-enriched regions. As reported in other studies, the spatial organization of lymphocytes, as well as their interactions with cancer cells, may play a more important role in patient prognosis.
Quantifying distribution and interaction with tumor or stromal cells of lymphocytes can potentially provide a way to evaluate immune response status and serve as a biomarker for immunotherapy response. The analysis pipeline developed in this study could convert the pathology image into a “spatial map” of tumor cells, stromal cells and lymphocytes. This could greatly facilitate and empower comprehensive analysis of the spatial organization of cells [57], [58], [59], as well as their roles in tumor progression and metastasis.
In this study, we developed a computational tool to automatically segment and classify different types of cell nuclei. This tool could potentially assist pathologists in clinical practice: First, it can assist pathologists to quickly pinpoint the tumor cells. It is time consuming and difficult for pathologists to locate very small tumor regions in tissue images, so this could greatly reduce the time that pathologists need to spend on each image. Second, this tool could help pathologists and clinicians to predict the patient prognosis, and therefore to tailor the treatment plan of individual patients using readily available tissue images. Furthermore, this tool could be used to quantify cell-cell interactions and distributions of different types of cells, especially the spatial distribution of lymphocytes and their interaction with the tumor region, which could potentially provide information for patient response to immunotherapy.
The computation time of the Convpath could be reduced in several ways: 1) By applying our model only to the tumor Region of Interest (ROI), which could be either annotated by a pathologist or detected by our tumor detection algorithm. Depending on the tissue resected, this step will reduce the processing time by tenfold. 2) By using parallel processing by creating multiple threads. In summary, by leveraging other existing computational methods and hardware infrastructures, the whole-slide processing time can be reduced to less than 1 h.
There are several limitations of the ConvPath pathology image analysis pipeline. First, the sampling region selection and subsequent steps rely on ROI labeling, which is currently done by pathologists. The fully automated tumor region detection model [35] could potentially be used to locate the tumor region first and then apply the ConvPath pipeline only in the detected tumor region, so that we can largely reduce the computation time for the ConvPath pipeline to run across a whole slide image by ignoring the non-malignant regions. Second, only three major cell types are considered in the ConvPath CNN algorithm; thus, this CNN model is sensitive to out-of-focus cell types such as macrophages and epithelial cells. Also, different subtypes of lymphocytes, such as CD4+ and CD8+ T cells, are not distinguishable using our algorithm [47,60]. More comprehensive labeling and immune-histochemical staining will help solve this problem. Third, more comprehensive analysis of spatial distribution of cells is not included in this research [61,62]. Analyzing the spatial patterns, such as cell clustering and inter-cell interactions, will help us understand the mechanism of tumor progression and immune response to tumor cells.
Funding sources
This work was supported by the National Institutes of Health [1R01GM115473, 5R01CA152301, 5P30CA142543, 5P50CA070907, 5P30CA016672 and 1R01CA172211]; and the Cancer Prevention and Research Institute of Texas [RP120732].
The funders had no role in study design, data collection, data analysis, interpretation, writing of the manuscript.
Author contributions
G.X. and T.W. supervised the project. S.W., T.W., L.Y. and G.X. conceived the method. S.W., T.W., L.Y. and G.X. designed and performed the analyses and interpreted the results. L.Y., Y.Y, J.F. I.W., Y.M. and Y.X. collected and provided the data. S.W., T.W., L.Y., F.Y, X.L., Y.Y. and A, G. curated the data. C.L., S.W., S.L., and B.Y developed the web application with advice from G.X., T.W. and Y.X.
L.Y., A.G., J.F., and I.W. provided critical input. S.W. and T.W. drafted the article. All co-authors have read and edited the manuscript.
Declaration of Competing Interest
The other authors declare that they have no competing interests.
Acknowledgements
Jessie Norris for helping us to edit this manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.10.033.
Appendix. Supplementary materials
References
- 1.Travis W.D., Brambilla E., Nicholson A.G. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10(9):1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Egeblad M., Nakasone E.S., Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev. Cell. 2010;18(6):884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian B.-.Z., Pollard J.W. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Commun Acm. 2017;60(6):84–90. [Google Scholar]
- 6.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 7.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Ruifrok A.C., Johnston D.A. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
- 9.Veta M., van Diest P.J., Kornegoor R., Huisman A., Viergever M.A., Pluim J.P. Automatic nuclei segmentation in h&e stained breast cancer histopathology images. PLoS ONE. 2013;8(7):e70221. doi: 10.1371/journal.pone.0070221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K., Zhang L., Song H., Zhou W. Active contours with selective local or global segmentation: a new formulation and level set method. Image Vis Comput. 2010;28(4):668–676. [Google Scholar]
- 11.Yi F., Huang J., Yang L., Xie Y., Xiao G. Automatic extraction of cell nuclei from H&E-stained histopathological images. J Med Imaging (Bellingham) 2017;4(2) doi: 10.1117/1.JMI.4.2.027502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin H.C., Roth H.R., Gao M. Deep convolutional neural networks for computer-aided detection: cnn architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dosovitskiy A., Fischer P., Springenberg J.T., Riedmiller M., Brox T. Discriminative unsupervised feature learning with exemplar convolutional neural networks. IEEE Trans Pattern Anal Mach Intell. 2016;38(9):1734–1747. doi: 10.1109/TPAMI.2015.2496141. [DOI] [PubMed] [Google Scholar]
- 14.Neftci E.O., Pedroni B.U., Joshi S., Al-Shedivat M., Cauwenberghs G. Stochastic synapses enable efficient brain-inspired learning machines. Front Neurosci. 2016;10:241. doi: 10.3389/fnins.2016.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Team R.C. R Foundation for Statistical Computing; Vienna, Austria: 2016. R: a language and environment for statistical computing. [Google Scholar]
- 16.Jonckheere A.R. A distribution-free k-sample test against ordered alternatives. Biometrika. 1954;41(1/2):133–145. [Google Scholar]
- 17.Williams B.J., Hanby A., Millican-Slater R., Nijhawan A., Verghese E., Treanor D. Digital pathology for the primary diagnosis of breast histopathological specimens: an innovative validation and concordance study on digital pathology validation and training. Histopathology. 2018;72(4):662–671. doi: 10.1111/his.13403. [DOI] [PubMed] [Google Scholar]
- 18.Bauer T.W., Slaw R.J., McKenney J.K., Patil D.T. Validation of whole slide imaging for frozen section diagnosis in surgical pathology. J Pathol Inform. 2015;6:49. doi: 10.4103/2153-3539.163988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snead D.R., Tsang Y.W., Meskiri A. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology. 2016;68(7):1063–1072. doi: 10.1111/his.12879. [DOI] [PubMed] [Google Scholar]
- 20.Buck T.P., Dilorio R., Havrilla L., O'Neill D.G. Validation of a whole slide imaging system for primary diagnosis in surgical pathology: a community hospital experience. J Pathol Inform. 2014;5(1):43. doi: 10.4103/2153-3539.145731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ordi J., Castillo P., Saco A. Validation of whole slide imaging in the primary diagnosis of gynaecological pathology in a university hospital. J. Clin. Pathol. 2015;68(1):33–39. doi: 10.1136/jclinpath-2014-202524. [DOI] [PubMed] [Google Scholar]
- 22.Paul A., Mukherjee D.P. Mitosis detection for invasive breast cancer grading in histopathological images. IEEE Trans Image Process. 2015;24(11):4041–4054. doi: 10.1109/TIP.2015.2460455. [DOI] [PubMed] [Google Scholar]
- 23.Rathore S., Hussain M., Aksam Iftikhar M., Jalil A. Novel structural descriptors for automated colon cancer detection and grading. Comput Methods Programs Biomed. 2015;121(2):92–108. doi: 10.1016/j.cmpb.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen K., Sarkar A., Jain A.K. Prostate cancer grading: use of graph cut and spatial arrangement of nuclei. IEEE Trans Med Imaging. 2014;33(12):2254–2270. doi: 10.1109/TMI.2014.2336883. [DOI] [PubMed] [Google Scholar]
- 25.Waliszewski P., Wagenlehner F., Gattenlohner S., Weidner W. [Fractal geometry in the objective grading of prostate carcinoma] Der Urologe Ausg A. 2014;53(8):1186–1194. doi: 10.1007/s00120-014-3472-x. [DOI] [PubMed] [Google Scholar]
- 26.Atupelage C., Nagahashi H., Yamaguchi M., Abe T., Hashiguchi A., Sakamoto M. Computational grading of hepatocellular carcinoma using multifractal feature description. Comput Med Imaging Graph. 2013;37(1):61–71. doi: 10.1016/j.compmedimag.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Beck A.H., Sangoi A.R., Leung S. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3(108):108ra13. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 28.Yuan Y., Failmezger H., Rueda O.M. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med. 2012;4(157):157ra43. doi: 10.1126/scitranslmed.3004330. [DOI] [PubMed] [Google Scholar]
- 29.Luo X., Zang X., Yang L. Comprehensive computational pathological image analysis predicts lung cancer prognosis. J Thorac Oncol. 2017;12(3):501–509. doi: 10.1016/j.jtho.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu K.H., Zhang C., Berry G.J. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coudray N., Ocampo P.S., Sakellaropoulos T. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24(10):1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kather J.N., Pearson A.T., Halama N. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019 doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Gadepalli K, Norouzi M, et al. Detecting cancer metastases on gigapixel pathology images. arXiv preprint arXiv:1703024422017.
- 34.Wang D, Khosla A, Gargeya R, Irshad H, Beck A.H. Deep learning for identifying metastatic breast cancer. arXiv preprint arXiv:1606057182016.
- 35.Wang S.D., Chen A., Yang L. Comprehensive analysis of lung cancer pathology images to discover tumor shape and boundary features that predict survival outcome. Sci Rep-Uk. 2018;8 doi: 10.1038/s41598-018-27707-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehteshami Bejnordi B., Veta M., Johannes van Diest P. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318(22):2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coudray N., Ocampo P.S., Sakellaropoulos T. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bankhead P., Loughrey M.B., Fernandez J.A. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sirinukunwattana K., Ahmed Raza S.E., Yee-Wah T., Snead D.R., Cree I.A., Rajpoot N.M. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging. 2016;35(5):1196–1206. doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- 40.Saltz J., Gupta R., Hou L. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23(1):181–193. doi: 10.1016/j.celrep.2018.03.086. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi F.L., Yang L., Wang S.D. Microvessel prediction in h&e stained pathology images using fully convolutional neural networks. BMC Bioinformatics. 2018;19 doi: 10.1186/s12859-018-2055-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X.X., Janowczyk A., Zhou Y. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital h&e images. Sci Rep-Uk. 2017;7 doi: 10.1038/s41598-017-13773-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura H., Ichikawa T., Nakasone S. Abundant tumor promoting stromal cells in lung adenocarcinoma with hypoxic regions. Lung Cancer. 2018;115:56–63. doi: 10.1016/j.lungcan.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Bremnes R.M., Donnem T., Al-Saad S. The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J Thorac Oncol. 2011;6(1):209–217. doi: 10.1097/JTO.0b013e3181f8a1bd. [DOI] [PubMed] [Google Scholar]
- 45.Pietras K., Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316(8):1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 46.Ichikawa T., Aokage K., Sugano M. The ratio of cancer cells to stroma within the invasive area is a histologic prognostic parameter of lung adenocarcinoma. Lung Cancer. 2018 doi: 10.1016/j.lungcan.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 47.Gooden M.J., de Bock G.H., Leffers N., Daemen T., Nijman H.W. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer. 2011;105(1):93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyashita M., Sasano H., Tamaki K. Prognostic significance of tumor-infiltrating CD8+ and FOXP3+ lymphocytes in residual tumors and alterations in these parameters after neoadjuvant chemotherapy in triple-negative breast cancer: a retrospective multicenter study. Breast Cancer Res. 2015;17:124. doi: 10.1186/s13058-015-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huh J.W., Lee J.H., Kim H.R. Prognostic significance of tumor-infiltrating lymphocytes for patients with colorectal cancer. Arch Surg. 2012;147(4):366–372. doi: 10.1001/archsurg.2012.35. [DOI] [PubMed] [Google Scholar]
- 50.Brambilla E., Le Teuff G., Marguet S. Prognostic effect of tumor lymphocytic infiltration in resectable non-small-cell lung cancer. J Clin Oncol. 2016;34(11):1223–1230. doi: 10.1200/JCO.2015.63.0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mani N.L., Schalper K.A., Hatzis C. Quantitative assessment of the spatial heterogeneity of tumor-infiltrating lymphocytes in breast cancer. Breast Cancer Res. 2016;18(1):78. doi: 10.1186/s13058-016-0737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carpenter A.E., Jones T.R., Lamprecht M.R. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S., Yang D.M., Rong R., Zhan X., Xiao G. Pathology image analysis using segmentation deep learning algorithms. Am. J. Pathol. 2019 doi: 10.1016/j.ajpath.2019.05.007. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xian X., Hakansson J., Stahlberg A. Pericytes limit tumor cell metastasis. J Clin Invest. 2006;116(3):642–651. doi: 10.1172/JCI25705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Shibli K.I., Donnem T., Al-Saad S., Persson M., Bremnes R.M., Busund Ll-T. Prognostic effect of epithelial and stromal lymphocyte infiltration in non–small cell lung cancer. Clinical cancer research. 2008;14(16):5220–5227. doi: 10.1158/1078-0432.CCR-08-0133. [DOI] [PubMed] [Google Scholar]
- 56.Bremnes R.M., Al-Shibli K., Donnem T. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non-small cell lung cancer. J Thorac Oncol. 2011;6(4):824–833. doi: 10.1097/JTO.0b013e3182037b76. [DOI] [PubMed] [Google Scholar]
- 57.Li Q., Wang X., Liang F., Xiao G. A bayesian mark interaction model for analysis of tumor pathology images. Annals of Applied Statistics. 2019 doi: 10.1214/19-aoas1254. https://ui.adsabs.harvard.edu/abs/2018arXiv180208308L accessed 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Q., Wang X., Liang F. A bayesian hidden potts mixture model for analyzing lung cancer pathology images. Biostatistics. 2018 doi: 10.1093/biostatistics/kxy019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Q., Yi F., Wang T., Xiao G., Liang F. Lung cancer pathological image analysis using a hidden potts model. Cancer Inform. 2017;16 doi: 10.1177/1176935117711910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wakabayashi O., Yamazaki K., Oizumi S. CD4+ t cells in cancer stroma, not CD8+ t cells in cancer cell nests, are associated with favorable prognosis in human non-small cell lung cancers. Cancer Sci. 2003;94(11):1003–1009. doi: 10.1111/j.1349-7006.2003.tb01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rampias T., Favicchio R., Stebbing J., Giamas G. Targeting tumor-stroma crosstalk: the example of the NT157 inhibitor. Oncogene. 2016;35(20):2562–2564. doi: 10.1038/onc.2015.392. [DOI] [PubMed] [Google Scholar]
- 62.Castino G.F., Cortese N., Capretti G. Spatial distribution of b cells predicts prognosis in human pancreatic adenocarcinoma. Oncoimmunology. 2016;5(4) doi: 10.1080/2162402X.2015.1085147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pathology images and clinical data in the NLST and TCGA datasets that support the findings of this study are available online in the NLST (https://biometry.nci.nih.gov/cdas/nlst/) and The Cancer Genome Atlas Lung Adenocarcinoma (TCGA-LUAD, https://wiki.cancerimagingarchive.net/display/Public/TCGA-LUAD). Data in the SPORE and CHCAMS datasets that support the findings of this study are available from the UT Lung SPORE Tissue bank and the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (CHCAMS), China, separately, but restrictions apply to the availability of these data.