Abstract
Background
The mechanisms underlying autoimmune thyroid disease (AITD) remain elusive. Identification of such mechanisms would reveal novel and/or better therapeutic targets. Here, we use integrated analysis of miRNAs and mRNAs expression profiling to identify potential therapeutic targets involved in the mechanisms underlying AITD.
Methods
miRNA and mRNA from twenty fresh-frozen thyroid tissues (15 from AITD patients and 5 from healthy controls) were subjected to next-generation sequencing. An anti-correlated method revealed potential pathways and disease targets, including proteins involved in the formation of primary cilia. Thus, we examined the distribution and length of primary cilia in thyroid tissues from AITD and controls using immunofluorescence and scanning electron microscopy, and parsed cilia formation in thyroid cell lines in response to inflammatory stimuli in the presence of miRNA mimics.
Findings
We found that the expression of miR-21-5p, miR-146b-3p, miR-5571-3p and miR-6503-3p was anti-correlated with Enolase 4 (ENO4), in-turned planar cell polarity protein (INTU), kinesin family member 27 (KIF27), parkin co-regulated (PACRG) and serine/threonine kinase 36 (STK36) genes. Functional classification of these miRNA/mRNAs revealed that their differential expression was associated with cilia organization. We demonstrated that the number and length of primary cilia in thyroid tissues was significantly lower in AITD than in control (frequency of follicular ciliated cells in controls = 67.54% vs a mean of 22.74% and 21.61% in HT and GD respectively p = 0.0001, by one-way ANOVA test). In addition, pro-inflammatory cytokines (IFNγ and TNFα) and specific miRNA mimics for the newly identified target genes affected cilia appearance in thyroid cell lines.
Interpretation
Integrated miRNA/gene expression analysis has identified abnormal ciliogenesis as a novel susceptibility pathway that is involved in the pathogenesis of AITD. These results reflect that ciliogenesis plays a relevant role in AITD, and opens research pathways to design therapeutic targets in AITD.
Funding
Instituto de Salud Carlos III, Comunidad de Madrid, Grupo Español de Tumores Neuroendocrinos y Endocrinos, Ministerio de Economía y Empresa and FEDER.
Keywords: microRNAs, mRNA, Next generation sequencing, Autoimmune thyroid disease, Graves’ disease, Hashimoto's thyroiditis, Cilia
Research in context
Evidence before this study
The mechanisms underlying autoimmune thyroid disease (AITD) remain elusive. The synergy between susceptibility genes and environmental stimulation, together with epigenetic modulation, denote the multifactorial interplay involved in the pathogenesis of AITD. MicroRNAs (miRNA), which are small noncoding RNA that regulate gene expression at a posttranscriptional level, have also been implicated in the pathogenesis of AITD. Understanding the regulatory role of these miRNAs could led to identify potential susceptibility pathways, diagnostic biomarkers and therapeutic targets of the disease.
The use of integrative analysis of omics data such as miRNAs and mRNAs expression profiling could help to identify novel pathways and biomarkers, and reveal novel and/or better therapeutic targets to treat AITD.
Added value of this study
The major findings of our study include the following: (i) Expression of miR-21-5p, miR-146b-3p, miR-5571-3p and miR-6503-3p are up-regulated in AITD and anti-correlated with Enolase 4 (ENO4), in-turned planar cell polarity protein (INTU), kinesin family member 27 (KIF27), parkin co-regulated (PACRG) and serine/threonine kinase 36 (STK36) genes. (ii) The functional classification of these miRNA/mRNAs are associated with cilia organization. (iii) The percentage and length of primary cilia in thyroid tissues is lower in AITD compared to controls. (iv) Pro-inflammatory cytokines (IFNγ and TNFα) affect cilia appearance in thyroid cell lines. (v) Treatment of thyrocytes with cytokines and specific miRNA mimics repress the expression of cilia-related genes for the newly identified target genes, and display less and shorter cilia.
Implication of all available evidence
We have identified a molecular signature that regulates primary cilia as a novel susceptibility pathway that controls the pathogenesis of AITD. These results reflect that ciliogenesis plays a relevant role in AITD, and opens novel research pathways to protect against the disease, including therapeutic targeting of miRNAs.
Introduction
Autoimmune thyroid disorders (AITD) – Graves’ disease (GD) and Hashimoto's thyroiditis (HT) – result from a dysregulation of the immune system directed against the thyroid gland. Both diseases feature circulating thyroid antibodies and infiltration of autoreactive lymphocytes in the thyroid gland, and sometimes the orbit. HT is a predominantly cellular autoimmune disease with prominent inflammatory infiltration, leading to destruction of the thyroid gland. Conversely, GD emerges as an essentially humoral autoimmune response, featuring autoantibodies directed against the thyrotropin receptor (TSHR-Ab). These antibodies stimulate the growth and function of thyroid follicular cells and lead to the development of goiter and hyperthyroidism [1], [2], [3]. A major occurrence in GD disease is Graves’ ophthalmopathy (GO), which shares cellular and humoral traits and is arguably the most common extra-thyroid manifestation of AITD [4]. The specific mechanisms underlying the pathogenesis of AITD have remained elusive, but seem to include the combination of specific susceptibility genes together with environmental stimulation, causing a breakdown of CD4+ T cell-dependent self-tolerance, and the subsequent development of autoimmune diseases. Different studies have associated the presence of other autoimmune disorders in patients with AITD, which underlines the importance of a common immunopathogenesis of these disorders [5]. In this context, the synergy between susceptibility genes and environmental stimulation is also accompanied by epigenetic modulation, e.g. DNA demethylation, histone modification and abnormalities in the expression of RNA interference, including microRNAs [6,7]. MicroRNAs (miRNAs) are small non-protein-coding RNA molecules of ≈22 nucleotides that interact with specific RNA target(s) in a sequence-dependent manner, regulating gene expression at a post-transcriptional level [8]. As such, they control a plethora of biological phenomena, including immune activation, apoptosis, differentiation and development, proliferation and metabolism. This role makes miRNA attractive candidates as disease biomarkers or even therapeutic devices.
In the specific setting of AITD, transcriptomic data from different sources, including miRNA and mRNA, have improved our understanding on the complex gene regulatory networks involved in the pathophysiology of disease [9,10]. Specifically, abnormal miRNA expression underlies the differentiation and activation of immune cells (reviewed in [7]). In line with these findings, we have recently identified a collection of deregulated miRNAs in thyroid tissue and serum of AITD patients using Next Generation Sequencing (NGS), which constitutes the backbone of a novel 5-miRNA signature useful as a AITD risk biomarker [11]. Thus, modeling context-specific miRNA–mRNA networks has the potential to identify mechanisms that can cause AITD. In this regard, an integrative analysis of miRNA and mRNA in GD identified a clear association between the retinoic acid pathway and the development of GD [12]. Herein, we used NGS techniques to develop an integrative analysis of the gene expression profiling of miRNAs and mRNAs, from thyroid tissue to identify and characterize novel molecular pathways of the disease. The negative correlation analysis of the differentially expressed miRNAs and mRNAs led to the identification of key signaling pathways altered in AITD. Interestingly, most of these pathways control the formation of primary cilia. Indeed, primary cilia in the thyroid gland emerge from the apex of follicular cells of different mammals [13]. In humans, cilia are usually located in a central position on the thyrocyte surface [14]. Although the role of primary cilia in thyroid tissue remains obscure, some authors have suggested that they could be involved in thyroglobulin secretion and/or control mechanosensory events [13], [14], [15]. Recently, defective ciliogenesis have been associated with thyoid pathology, however no molecular mechanisms were identified [16].
In this study, we identify a molecular mechanism that could control primary cilia formation as an important mechanism underlying AITD. Furthermore, the effect of pro-inflammatory cytokines and miRNA targeting in primary cilia formation in thyroid cells suggest the potential usefulness of this pathway as a novel therapeutic avenue to treat these diseases.
Materials and methods
Human samples
Thyroid tissue samples were collected from patients with AITD who underwent thyroidectomy at the Surgical Department of the Hospital Universitario de la Princesa. As controls, normal thyroid parenchyma samples adjacent to other thyroid lesions were collected. All thyroid tissues were reviewed by an experienced pathologist, and diagnosis was reconfirmed by an endocrinologist, based on well-established clinical, laboratory and histological criteria [2]. Serum free thyroxine 4 (FT4), thyroid stimulating hormone (TSH), and levels of antibodies against thyroglobulin (Tg), thyroperoxidase (TPO) and thyrotropin receptor (TR) were measured as previously described at the time of thyroid surgery [17]. Thyroid glands were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned (4–5 µm thick slices), and mounted on FLEX IHC Microscope Slides (Dako) for immunofluorescence. Sections were also stained with hematoxylin-eosin for histological diagnosis. Clinical data are summarized in Tables 1 and 2.
Table 1.
Clinical features of patients with autoimmune thyroid disease (AITD) from whom NGS was performed.
| AITD | HT | GD | GO | p-value |
|---|---|---|---|---|
| n | 5 | 4 | 5 | |
| Gender (F/M) | 5/0 | 4/0 | 5/0 | |
| Age (year) | 54.2 ± 12.3 | 47.2 ± 15.4 | 40.6 ± 11.6 | 0.3024 (one way ANOVA) |
| TSH mU/mL | 1.7 (1.04–15.22) | 2.8 (0.48–7.6) | 2.5 (0.5–21.72) | 0.9463 (Kruskal–Wallis test) |
| T4 ng/dL | 1.1 (1.1–1.12) | 0.86 (0.81–1.5) | 1.19 (0.67–1.27) | 0.9452 (Kruskal–Wallis test) |
| Tg-Ab UI/mL | 342 (133.8–446.8) | – | – | – |
| TPO-Ab UI/mL | 228 (52.5–1577) | – | – | – |
| TSHR-Ab U/L | – | 3.655 (1.46–23.05) | 4.37 (1.55–16.63) | > 0.9999 (Mann–Whitney test) |
Values show number for categorical values and median (interquartile intervals 25–75) for continuous variables. HT = Hashimoto's thyroiditis; GD = Graves’ disease. F/M = female/male; TSH, thyrotropin (Normal range = 0.27–4.20); T4, thyroxine (Normal range = 0.93–1.7); Tg-Ab, anti-thyroglobulin antibody (Negative <344); TPO-Ab, anti-thyroid peroxidase antibody (Negative <100); TSHR-Ab, anti-thyrotropin receptor antibody (Negative <0.7).
Table 2.
Clinical features of patients with autoimmune thyroid disease (AITD) from whom RT-PCR was performed.
| AITD | HT | GD | GO | p-value |
|---|---|---|---|---|
| n | 13 | 13 | 13 | |
| Gender (F/M) | 12/1 | 12/1 | 12/1 | |
| Age (year) | 49.7 ± 15.9 | 43.2 ± 16.4 | 38.9 ± 9.9 | 0.1704 (one way ANOVA) |
| TSH mU/mL | 1.97 (0.84–2.72) | 0.3 (0.01–1.38) | 0.4 (0.01–1.76) | 0.0527 (Kruskal–Wallis test) |
| T4 ng/dL | 1.22 (1.04–1.41) | 1.49 (1.02–2.27) | 1.12 (0.96–1.23) | 0.3692 (Kruskal–Wallis test) |
| Tg-Ab UI/mL | 334 (180–412) | 360 (344–1134) | 643.5 (20–1483) | 0.4569 (Kruskal–Wallis test) |
| TPO-Ab UI/mL | 306 (150–724.5) | 125.5 (61.6–2235) | 354 (101–812) | 0.6506 (Kruskal–Wallis test) |
| TSHR-Ab U/L | – | 3.8 (1.82–28.9) | 2.4 (1.4–5.63) | 0.4313 (Mann–Whitney test) |
Values show number for categorical values and median (interquartile intervals 25–75) for continuous variables. HT = Hashimoto's thyroiditis; GD = Graves’ disease; GO=Graves'Ophthalmopathy. F/M = female/male; TSH, thyrotropin (Normal range = 0.27–4.20); T4, thyroxine (Normal range = 0.93–1.7); Tg-Ab, anti-thyroglobulin antibody (Negative <344); TPO-Ab, anti-thyroid peroxidase antibody (Negative <100); TSHR-Ab, anti-thyrotropin receptor antibody (Negative <0.7).
This study was carried out in accordance with the principles set out in the Declaration of Helsinki under a protocol approved by the Internal Ethical Review Committee of the Hospital de la Princesa (Committee Register Number: 2796). All subjects gave written informed consent in accordance with the Declaration of Helsinki.
Tissue miRNA and mRNA isolation
Thyroid tissue samples were immediately snap-frozen in liquid nitrogen cooled isopentane and transferred to a − 80 °C freezer. RNA was isolated from 50 fresh-frozen thyroid tissues (13 with HT, 13 with GD, 13 with GO and 11 controls): 19 samples were used for miRNA and mRNA NGS, 36 samples to validate miRNAs and all 50 for qRT-PCR mRNA validation test. Total RNA was isolated using miRNeasy Mini Kit (Qiagen). The quality and quantity of RNA and microRNA were assessed in a 2100 Bioanalyzer using a RNA 6000 Nano kit and Small RNA kit (Agilent Technologies). Only samples with RIN>7 were used.
Next generation sequencing of mRNA and microRNA
NGS procedure was performed as previously described [11]. Briefly, 1 µg of RNA was used for NGS. Integrity and quality of the ribosomal depleted RNA was checked with Agilent Bioanalyzer 2100 (Agilent Technologies). RNA sequencing was carried out by Sistemas Genómicos (https://www.sistemasgenomicos.com/web_sg/) with the Next Generation Sequence (NGS) platform using the Illumina HiSeq 2000 sequencing instrument (Illumina). Ribosomal-depleted samples were used to generate whole transcriptome libraries. Amplified cDNA quality was analyzed with the Bioanalyzer 2100 DNA 1000 kit (Agilent Technologies) and quantified using the Qubit 2.0 Fluorometer (Invitrogen). The bioinformatic analysis of NGS and raw data availability is described in Supplementary Materials and Methods.
Quantitative real-time RT-PCR for miRNAs
First-strand cDNA was generated using cDNA synthesis kit and subsequent quantitative Real Time PCR (q-RT-PCR) was performed by triplicate using miRCURY LNA™ Universal RT microRNA PCR and SYBR Green (both from Qiagen) with the CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Expression of miRNAs was performed using microRNA LNA™ PCR primer sets for hsa-miR-21-5p, hsa-miR-146b-3p, hsa-miR-5571-3p, and hsa-miR-6503-3p. Simultaneously, 5S, U6, SNORD48 and RNU1A1 were used as candidate reference genes (Qiagen). Data were normalized using the geometric mean Ct of the best gene combination generated by the NormFinder algorithm [18] (RNU1A1 and SNORD48) and as previously reported [11]. Relative expression was determined using the log base 2 values of the difference Cts between miRNAs and the geometric mean of the selected housekeeping genes.
Quantitative real-time RT-PCR for gene expression
To validate the different expression levels of the mRNA genes determined by NGS, qPCR primers were selectively designed for 5 mRNA genes (STK36, INTU, ENO4, PACRG and KIF27 – Supplementary Table 1a for the sequence of primers) that were found to be differentially expressed in the discovery screen. β-Actin, GAPDH and HPRT were used as reference genes. RNA from each sample was reverse transcribed for cDNA synthesis using high-capacity cDNA reverse transcription kit with a ribonuclease inhibitor (ThermoFisher), following the manufacturer's instructions. cDNAs were amplified using Power SYBR Green PCR master mix (ThermoFisher Scientific). Relative expression was determined using the log base 2 values of the difference Cts between mRNAs and the geometric mean of β-Actin and GAPDH (selected reference genes by NormFinder [18], Supplementary Table 1b).
Immunofluorescence staining
Serial sections from 20 paraffin embedded thyroid tissues (5 controls, 5 nodular hyperplasia (NH), 5 HT and 5 GD) were placed in an oven at 65 °C for 30 min. Thyroid-sections were then deparaffinised in xylene and rehydrated through graded alcohols. Antigen retrieval was performed in a PTLink instrument using EnVision Flex target retrieval solution high pH (Dako). Non-specific binding was blocked using 5% bovine serum albumin and 10% normal goat serum for 30 min.
Cells were cultured on round coverslips in 6-well plates. After incubation under the indicated conditions, cells were washed with 1 × PBS and fixed with 4% paraformaldehyde in PBS for 15 min, and then permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. Permeabilized cells were blocked with 5% bovine serum albumin and 10% normal goat serum in PBS for 15 min at room temperature. Thereafter, tissue sections and cultured cells were incubated with primary antibodies. Primary antibodies were incubated over night at 4 ᵒC. On the following day, slides were washed three times with PBS and incubated 1 h with secondary antibody. Primary antibodies against acetylated alpha-tubulin (Sigma-Aldrich) and ADP ribosylation factor like GTPase 13B (Arl13B) (Proteintech) were used. Alexa 488 labeled goat anti-mouse and Alexa 568 labeled goat anti-rabbit (Invitrogen) were used as secondary antibodies. 4′,6-diamidino-2-phenylindole (DAPI) was used for nuclei counterstained. Stacks of 5–7 μm-thick were observed using a Leica Sp5 for cilia quantification. For high resolution images, sections were acquired in STED mode in a Leica SP8 confocal microscope.
Morphometric analysis
The frequency of cilia in a total of 20 thyroid sections was estimated manually by analyzing Z-stacked images captured in confocal microscope. Low and higher magnification images were analyzed by two independent observers in a blinded way. The frequency of follicular ciliated cells was estimated by analyzing the relative number of cilia protruding from the apical surface of the epithelium vs. the number of nuclei in adequately oriented sections of those thyroid follicles as previously reported [13]. A total of 5796 nuclei in control samples, 7902 in HT, 6492 in GD and 1123 in NH in 41, 70, 54 and 24 thyroid tissue fields was analyzed, respectively. Primary cilia length was measured using the ROI measurement tool of Image J Software (NIH) in a total of 3863 cilia in controls, 1886 in HT, 1299 in GD and 863 in NH.
Scanning electron microscopy
For scanning electron microscopy (SEM) studies, two control and 4 AITD (2 HT and 2 GD) thyroid tissue samples were fixed in 2% glutaraldehyde and 2% paraformaldehyde in phosphate buffer. All images were acquired with a high resolution FEI Verios 460 Field Emission SEM in high vacuum conditions (10E-6–10E-7 mbar) with 2 kV beam acceleration voltage and 1 kV beam deceleration (sample bias), resulting in 1 kV incidence voltage. The specimens were imaged directly on glass substrates without any metal coating after being dehydrated in a graded alcohol series and dried thoroughly by evaporation of hexamethyldisilazane (HMDS, Sigma-Aldrich). Other imaging parameters were: current, 13 pA; working distance, 3–4 mm; scanning mode, and integration of 100 frames with 100 nS dwell time.
Cell preparation
Human cell lines Nthy-ori 3-1 (kindly provided by Dr. Pilar Santisteban, Instituto de Investigaciones Biomédicas ‘Alberto Sols’, Spain) were cultured in RPMI 1640 with GlutaMAX medium supplemented with 10% FBS (Hyclone), 1% of penicillin/streptomycin (Gibco) and 10 mU/ml of thyroid stimulated hormone (TSH, Sigma-Aldrich). For experiments, cells were seeded at confluence and were grown in media without serum for 48 h to induce cell cycle arrest and promote primary cilia assembly as previously described [13,19]. These cells were incubated at the same time with or without proinflammatory cytokines, including 1000 U/ml IFN-γ (R&D systems) and 800 mU/ml TNF-α (R&D systems), as previously described [20]. All experiments were performed in triplicate wells for each condition, and three independent experiments were carried out.
miRNA mimics transfection
Nthy-ori 3-1 cell lines were seeded in 6-well plates 18–24 h prior to transfection. Following manufacturer's instructions, media were replaced with antibiotics-free media 6–12 h before transfection. Negative control miRCURY LNA miRNA mimic 5′FAM (339173 YM00479902-ADB, Qiagen) and a pool of hsa-miR-146b-3p, hsa-miR-21-5p and hsa-miR-6503-3p miRNA mimics (339176 YM00473138-ADB, YM00473093-ADB and YM00471117-ADB respectively, Qiagen) were transfected using Lipofectamine® RNAiMAX (Invitrogen), according to the manufacturer's protocol. Twenty-four hours later, media were replaced with the same conditions, as previously described in cell preparation methods.
Western blot analysis
Cells and tissues were washed twice with PBS and lysed in RIPA lysis buffer (Sigma-Aldrich) supplemented with Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo-Fisher Scientific). Lysates were sonicated and protein concentrations were measured using a Pierce™ BCA Protein Assay Kit (Thermo-Fisher Scientific). Samples were resolved in a 8% SDS-PAGE and transferred to nitrocellulose membranes, blocked with 10% skimmed milk and incubated with the following primary antibodies overnight at 4 °C: α-tubulin, acetylated α-tubulin, γ-tubulin, GAPDH (Sigma-Aldrich) and β-actin (Santa Cruz Biotechnology). Membranes were washed, incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody, and visualized using a chemiluminescent detection reagent kit (Thermo-Fisher Scientific).
Statistics
Descriptive results were expressed as mean ± standard deviation (SD) or mean ± error of the mean (SEM), as appropriate. Spearman bivariate correlations were performed for all quantitative variables and differences between groups were compared using analysis of variance (Mann–Whitney U or Kruskal–Wallis analysis of variance, as appropriate).The analysis for all quantitative variables and differences between groups were compared using analysis of variance (U-Mann Whitney or Kruskal Wallis ANOVA, as appropriate). Samples from all groups within an experiment were processed at the same time. The p-values were two sided and statistical significance was considered when p < 0.05. Data is presented with the specific p-values: p < 0.05, p < 0.01, p < 0.005 and p < 0.001. All statistical analyses were performed using STATA 12.0 and GraphPad Prism 4 software.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information files, or are available in a persistent repository or upon reasonable requests to the authors.
Next generation sequencing data supporting the findings of this study are deposited in Bioproject: PRJNA396505 and PRJNA530166 and Biosamples: SAMN07427434, SAMN07427435, SAMN07427436, SAMN07427437, SAMN07427438, SAMN07427439, SAMN07427440, SAMN07427441, SAMN07427442, SAMN07427443, SAMN07427444, SAMN07427445, SAMN07427446, SAMN07427447, SAMN07427448, SAMN07427449, SAMN07427450, SAMN07427451 and SAMN07427452.
Results
Integrated miRNA and mRNA profiling reveals cilia related genes in AITD pathophysiology
Analysis of miRNA and mRNA samples of AITD vs. control samples identified 19 deregulated miRNAs and 3690 deregulated genes. As reported previously [11], control samples formed a separate sub-cluster from AITD samples. However, the analysis revealed a slightly but not substantial separation between AITD groups. We, therefore, decided to assess the differential miRNA/mRNA expression of all AITD samples together in the same group, in comparison with the control group (Supplementary Fig. 1).
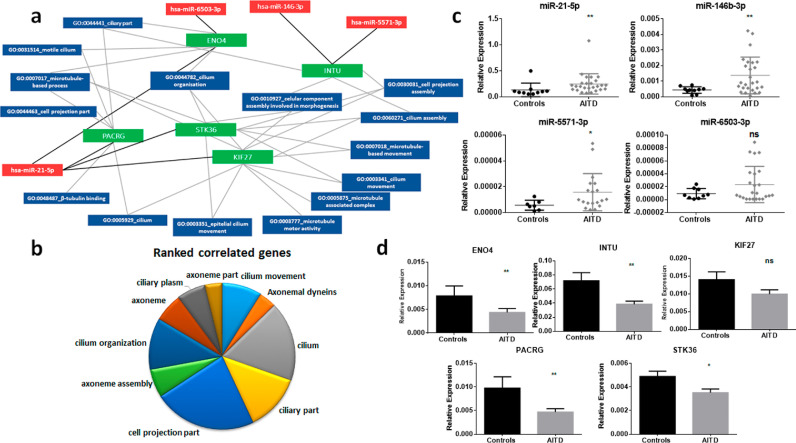
We then examined the expression of miRNAs that negatively correlated with the expression levels of their target genes. We found a specific anti-correlation of 17 miRNAs and 160 genes. The interactions were visualized as a network using Cytoscape (Fig. 1a), which revealed a high number of genes related with ciliary organization (Fig. 1b). The most represented anti-correlated miRNAs and genes were: miR-21-5p, miR-146b-3p, miR-5571-3p and miR-6503-3p and ENO4 (Enolase 4), INTU (in-turned planar cell polarity protein), KIF27 (kinesin family member 27), PACRG (parkin co-regulated) and STK36 (serine/threonine kinase 36). These findings were validated by qPCR, and revealed an upregulation of miR-21-5p, miR-146b-3p, miR-5571-3p (p < 0.01; p < 0.001 and p < 0.05 respectively, by Mann–Whitney test), and miR-6503-3p, which did not reach significance (Fig. 1c). Conversely, their target genes were significantly downregulated: ENO4 (p < 0.01, by Mann-Whitney test), INTU (p < 0.01, by Mann-Whitney test), KIF27 (it did not reach significance), PACRG (p < 0.01, by Mann-Whitney test) and STK36 (p < 0.05, by two sided unpaired t-test) (Fig. 1d).
Fig. 1.
(a) Visualization of a subnetwork of the miRNA–mRNA anti-correlation and of the involved Gene Ontology (GO) biological processes. MiRNAs are represented in red, genes in green and biological processes in blue. (b) Pie representation of the top ten biological processes generated in the miRNA–mRNA analysis. (c) Comparison of the relative expression of the miRNAs involved in the subnetwork between 10 controls and 26 AITD thyroid tissues: miR-21-5p, miR146b-3p, miR-5571-3p and miR-6503-3p were upregulated in AITD compared to controls. (d) Validation by qPCR of the expression of the downregulated genes ENO4, INTU, KIF27, PACRG and STK36 in 39 AITD and 11 controls thyroid tissues. Data are presented as mean +/- standard deviation relative expression: *p < 0.05; **p < 0.01; ns=not significant (by Mann–Whitney test or by two sided unpaired t-test as appropriate).
To assess the existence of any possible associations between miRNA and mRNA levels and clinical features we investigated the correlations of all the analyzed miRNAs and genes with patients’ corresponding serum levels of free thyroxine (T4), thyrotropin (TSH), antithyroid peroxidase antibody (TPO-Ab), antithyroglobulin antibody (Tg-Ab), and TSHR-Ab (correlation heatmaps in Supplementary Fig. 2). Spearman r analyses revealed that miR-146b-3p was positively associated with levels of Tg-Ab (r = 0.52 and p = 0.048), miR-6503-3p positively correlated with levels of TSHR-Ab (r = 0.56 and p = 0.02), however was negatively associated with T4 levels (r = −0.62 and p = 0.0086). We therefore associated clinical data with mRNA levels, however, any of the genes analyzed correlated with clinical features (Supplementary Fig. 2).
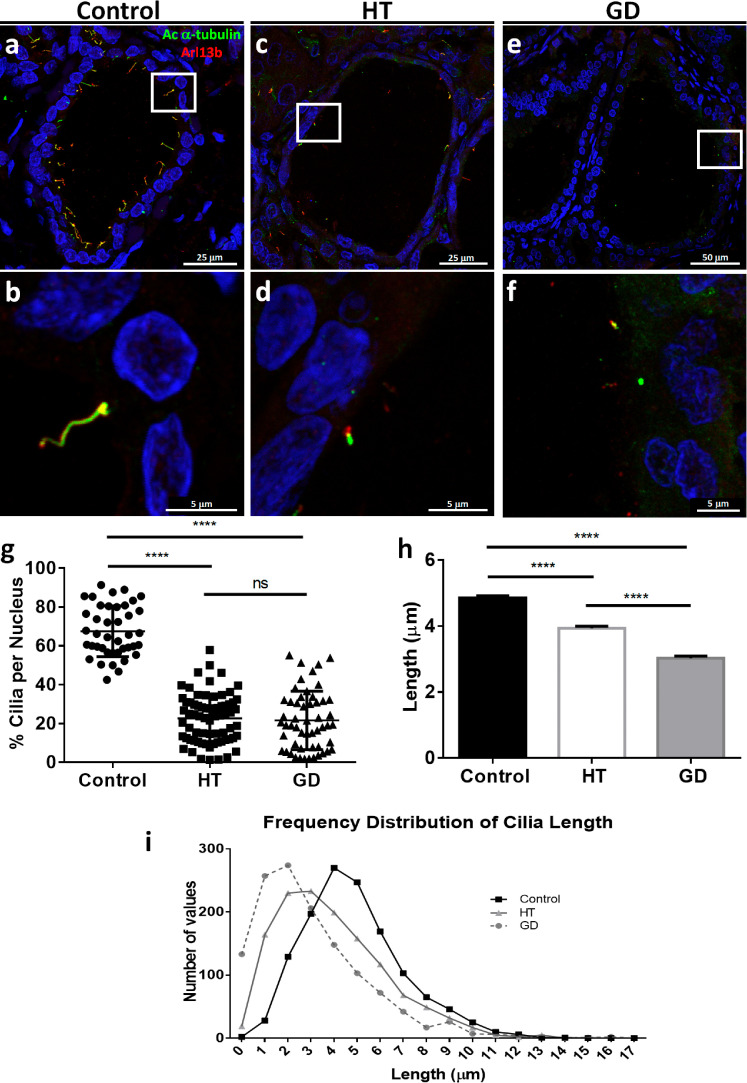
Decreased number and length of cilia in AITD tissues
To identify primary cilia, we used acetylated α-tubulin at K40 and ADP ribosylation factor like GTPase 13B (Arl13B), which localize to the cilia [21]. Fig. 2a and b show control thyroid glands comprised by follicular cells with a primary cilium that extend from the apical surface of each thyrocyte towards the colloid-rich follicular lumen. Five cases with nodular hyperplasia were also quantified and no remarkable changes in cilia frequency with control thyroids were found (Supplementary Fig. 3). The frequency of follicular ciliated cells in control thyroid tissue was of 67.54% (of a total count of 5796 nuclei). In thyroid tissue from HT and GD patients (Fig. 2c–f), however, the presence of primary cilia was significantly reduced, with a mean of 22.74% ciliated cells in HT and 21.61% in GD (of a total count of 7902 and 6492 nuclei, respectively) (p < 0.0001, by one-way ANOVA test) (Fig. 2g). We also examined ciliary length and observed significantly shorter primary cilia in both HT and GD, compared to controls (average length 4.85 ± 2.13; 3.93 ± 2.3 and 3.02 ± 2.3, respectively; p < 0.0001, by Kruskal–Wallis test) (Fig. 2h and i). To determine if ciliary defects were associated with acetylation defects in thyroid tissue, we measured the acetylation of α-tubulin at K40 in tissue extracts and found no statistically significant differences between tissue samples (Supplementary Fig. 4).
Fig. 2.
(a–f) Representative images captured by confocal microscopy characterizing the primary cilia of controls (a and b), HT (c and d) and GD (e and f) thyroid follicles stained with acetylated α-tubulin (in green) and Arl13b (in red). In controls, primary cilia usually appear as one cilium per thyrocyte; however in HT and GD thyroid tissues, primary cilia exhibited a shorter length and some cells even lacked this structure (g). Quantification of primary cilia in a total of 5796 nuclei of 41 confocal images in control tissue, 7902 nucluei in 70 images captured in HT tissue and 6492 nuclei in 54 images of GD thyroid samples. Control samples had higher ciliated cells compared to HT and GD (****p < 0. 0001; by one-way ANOVA test). (h) Length quantification of a total of 3900 cilia (1300 cilia per group), as observed in the column graph, HT and GD presented shorter cilia compared to controls (****p < 0.0001, by Kruskal–Wallis test). (i) Histogram representing the frequency distribution of cilia length in the three groups. In HT and GD the distribution of cilia length overlaps around 0–3 μm, with the GD group having the shorter cilia length values. Control samples portrayed higher length values, around 4–5 μm.
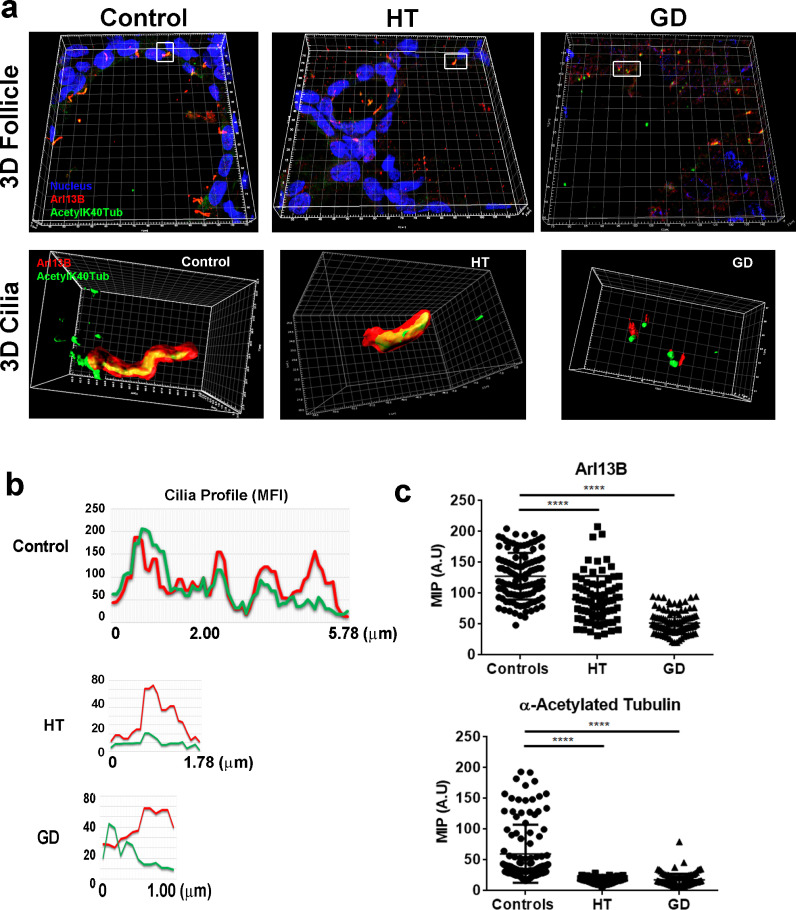
To further characterize cilia structures and elucidate differences in their architecture, we used stimulated emission depletion (STED) microscopy. As observed in the 3D reconstruction of follicles, the number of cilia was reduced in HT and GD (Fig. 3a). Quantification of the mean intensity of the cilia along its length (ROI; region of interest) allowed the projection of its mean intensity of fluorescence per pixel (MFI) profile. The distribution and expression of the pixel count in controls was significantly different from that observed in AITD (Fig. 3b and c). Cilia profiles from control samples showed that both fluorochromes, Arl13b and acetylated K40 α-tubulin, overlapped towards the ROI (maximum intensity of ≈200 AU, length ≈6 μm), whereas acetyl-K40 α-tubulin appeared severely reduced (maximum intensity below 80 AU and a length <2 μm) in HT and GD samples. Interestingly, the profile distribution was different in HT and GD; whilst HT fluorescence spectrum of both markers coincides along the cilium, the GD profile showed a cluster of acetyl-K40 α-tubulin at the proximal part of the cilium, which decreased as Arl13b fluorescence intensified until the distal tip.
Fig. 3.
(a) Representative 3D reconstructions from Confocal stacks of thyroid follicles from control, HT and GD patients. Magnifications from insets (white squares) correspond to 3D projections of volume segmentations from cilia structures imaged with STED confocal (xy optimized). Imaris was used to generate the 3D volumes and segmentation (surfaces). Surfaces represent the intensity of fluorescence detected for the analyzed fluorochromes (Red, anti-Arl13b plus alexa-568-conjugated anti-rabbit antibody; Green, anti-Acetylated-K40 α-Tubulin plus alexa-488-conjugated anti-mouse antibody). (b) Profiles of the mean intensity of fluorescence per pixel (MFI) of representative cilia from samples of healthy controls, GD and HT patients. Red, MFI of Arl13b; Green, Acetylated-K40 α-tubulin. (c) Quantification of the MFI showed that Arl13b and acetylated α-tubulin are significantly reduced in HT and GD samples (****p < 0.0001; by Kruskal–Wallis test).
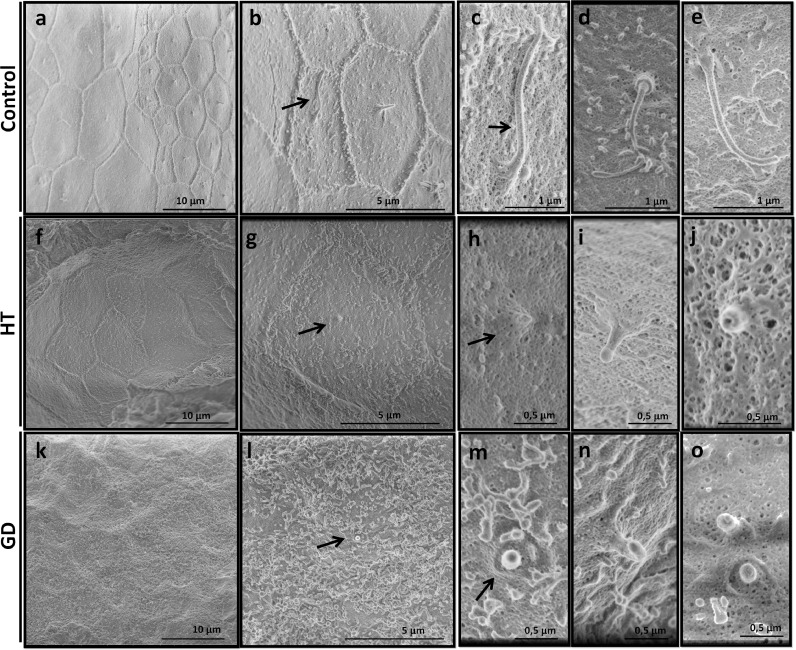
High resolution Scanning Electron Microscopy (SEM) revealed further morphological details of cilia defects in both HT and GD samples (Fig. 4). In control samples follicular cells displayed a polyhedral outline averaging 6 sides, and one or two cilia were present in the center of the cell's follicular surface. In addition, the cell surface had unevenly distributed microvilli, which tended to concentrate along cell borders (Fig. 4a–e). Cilia protruded towards the follicular lumen, without ciliary pocket, and the morphology of the ciliary base resembled the arrangement described for the appendages of the basal body (Fig. 4d) [22]. AITD samples contained thyroid cells of different sizes, with irregularly outlined polyhedral sides, from which cilia emerged as vestigial structures from the center. In HT, follicular cells had different shapes with irregular margins, which sometimes even jeopardized the distinction between cells (Fig. 4f–j). Hypomorphic cilia emerged from the center of each cell, sometimes appearing as “volcano-like” structures (Fig. 4j). In GD, follicular cells displayed a convex surface filled with microvilli; and short, microvilli-like cilia were also occasionally observed at the center of the cell (Fig. 4k–o).
Fig. 4.
High resolution SEM images from control (a–e), HT (f–j) and GD (k–o) thyroid gland. (a) At lower magnification, thyrocytes in normal thyroid appeared as polyhedral cells with, normally, 5 or 6 sides. In the center of the cell, a long primary cilium is usually observed. (b–e) At higher magnification, the distributed microvilli can be observed in the surface of the cell, which concentrates at the cell border, shaping epithelial cells. In HT (f–j), follicular cells presented different shapes and with few microvilli in their surface. Very short cilia with different shapes and sizes were present in the center of each cell reminding stumped-cilia. (k–o) In GD, follicular cells showed a convex surface with abundant microvilli. Shortened cilia were also seen at the center of the cell. An example of two small cilia is shown in o.
Inflammatory cytokines decreased primary cilia appearance in thyroid cell cultures
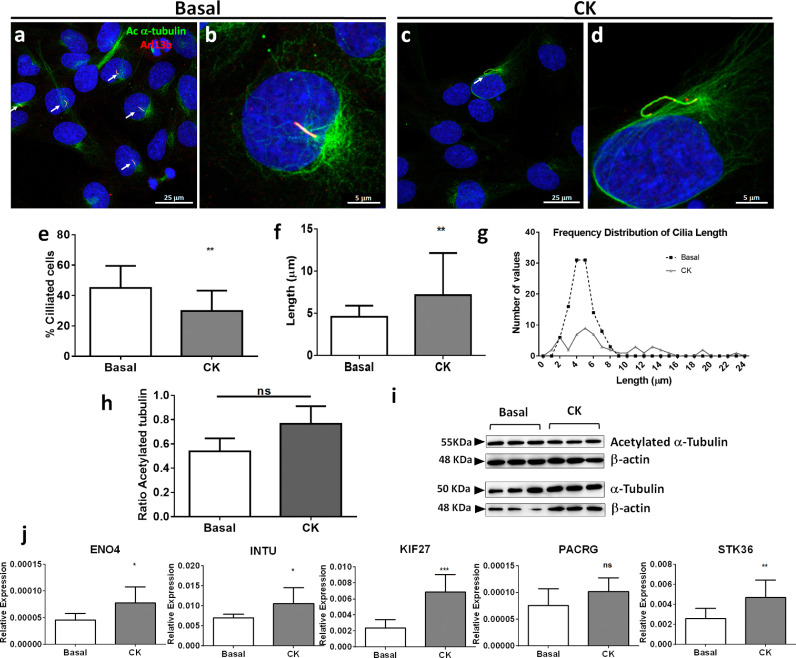
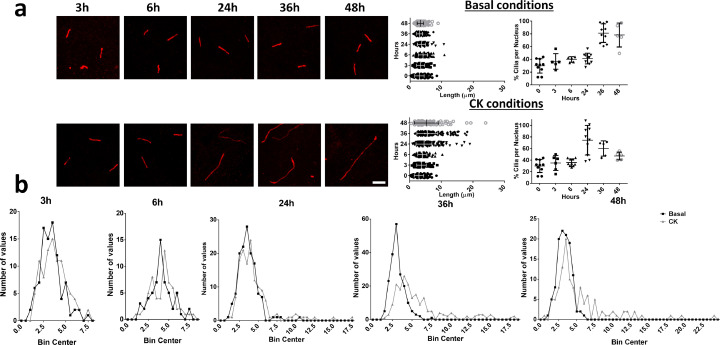
Combination of IFN-γ with TNF-α in thyroid follicular cell cultures mimic the high inflammatory scenario found in AITD [20]. Stimulation of the untransformed human thyroid follicular cell line N-Thy Ori with pro-inflammatory cytokines in serum-starved medium significantly reduced ciliogenesis (Fig. 5a–e; p < 0.005, by two sided unpaired t-test). Interestingly, the length of the scarce cilia in treated thyroid cells was increased in comparison to those in the normal (Basal) condition (mean of 7.2 μm vs mean of 4.5 μm, respectively, Fig. 5f and g; p < 0.005, by Mann–Whitney test). Performing a timing of ciliogenesis at basal and proinflammatory conditions, we observed at basal conditions a moderate increase of the percentage of cilia as time goes by. However after CK treatment, the highest percentage of ciliogenesis was reached at 24 h and then decreased, suggesting a fragile behavior of cilia in proinflammatory cultures (Fig. 6). Notably, acetyl-K40 α-tubulin was similar in total extracts from stimulated vs. non-stimulated cells (Fig. 5h and i). To assess expression differences of the ciliary-related genes observed in the miRNA–mRNA correlation study, we determined the expression of ENO4, INTU, KIF27, PACRG and STK36 in both conditions. Quantitative analysis indicated a marked increase of all ciliary genes in the presence of inflammatory cytokines (Fig. 5j: ENO p < 0.05, by two-sided unpaired t-test; INTU p < 0.05, by two-sided unpaired t-test; KIF27 p < 0.001, by Mann-Whitney test; PACRG n.s and STK36 p < 0.01 by two-sided unpaired t-test). This finding indicates that stimulation of thyrocytes with TNFα and IFNγ affects ciliary formation, suggesting that cilia assembly is affected by the pro-inflammatory environment.
Fig. 5.
Primary cilia in NThyOri cell line cultured at basal (without cytokines) and at proinflammatory (CK: with TNF-α and IFN-γ) serum starved conditions. (a and b) Immunofluorescent staining of acetylated α-tubulin (green) and Arl13b (red) at basal conditions showed a higher percentage of ciliated cells (arrows) compared with those presented in CK (c and d). (e) The frequency of ciliated cells were statistically significantly between both conditions. (f and g) Average and frequency distribution of the length in the medium with and without cytokines. (h and i) Quantification and western blot of acetylated α-tubulin in protein cell extractions. (j) Relative expression of ciliary genes at basal and CK conditions: ENO4, INTU, KIF27, PACRG and STK36. Data are represented as mean ± SD (*p < 0.05; **p < 0.01, ***p < 0.005 and ns=not significant; by two sided unpaired t-test or by Mann–Whitney test as appropriate).
Fig. 6.
Timing of ciliogenesis (at 3, 6, 24, 36 and 48 h) in arrested basal conditions and after TNFα and IFNγ treatment (CK conditions). (a) Representative confocal images of cilia at both conditions (Arl13b in red, Scale bar = 5 μm) and quantification of length and percentage of cilia. (b) Frequency distribution histograms of the length of cilia during the timing experiment.
miRNAs transfection affects ciliary growth in thyrocytes
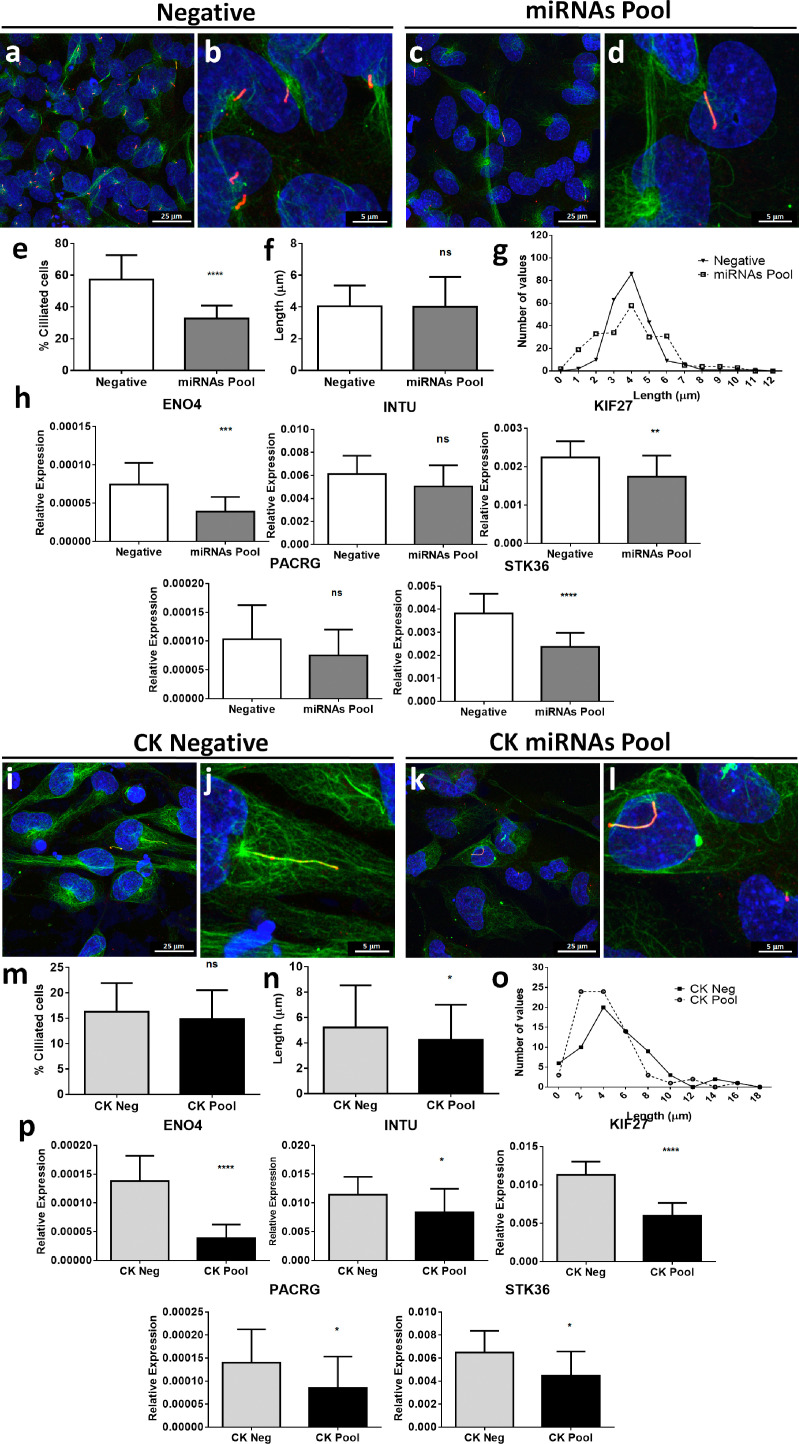
To analyze the role of miRNAs in thyroid cultures we transfected N Thy Ori cells with a negative control miRNA mimic (Negative) and a pool of miR-146b-3p, miR-21-5p and miR-6503-3p miRNA mimics (miRNAs Pool) (Supplementary Fig. 5). As observed in Fig. 7, in basal conditions, miRNAs transfection (miRNAs Pool) triggered a reduction from 57.33% to 32.78% ciliated cells (p < 0.0001, by two-sided unpaired t-test) (Fig. 7a–e). However, cilia elongation was not affected in these conditions (Fig. 7f and g). In any case, comparison of the expression levels of ciliary implicated genes between negative and transfected cells demonstrated a significantly reduction of ENO4 (p < 0.001, by two-sided unpaired t-test), KIF27 (p < 0.005, by two-sided unpaired t-test), STK36 (p < 0.0001, by two-sided unpaired t-test); INTU and PACRG had a moderate reduction of expression, although it did not reach significant differences (Fig. 7h). Thus, miRNA mimics dysregulated the expression of these target genes at basal conditions, producing an imbalance in ciliary growth in culture thyrocytes.
Fig. 7.
Primary cilia in NThyOri cell line cultured at basal (a-h) or with CK (i–o) -serum starved conditions transfected with a pool of miR-21-5p, miR-146b-3p and miR-6503-3p mimics. (a–d and i–l) Immunofluorescence staining of acetylated α-tubulin (green) and Arl13b (red) in negative and in pool transfected (miRNAs pool) cells. (e–g and m–o) Percentage, average length and frequency distribution of the length of ciliated cells in negative and transfected cells. (h and p) Quantification of ciliary related genes presented as relative expression. Data are represented as mean ± SD (**p < 0.01, ***p < 0.005, ****p < 0.001 and ns = not significant; by two sided unpaired t-test or by Mann–Whitney test as appropriate).
When we stimulated thyrocytes with TNFα and IFNγ (CK) concomitantly with miRNA mimics (Fig. 7i–l), the percentage of ciliated cells remained similar, but with a reduction in length from 5.2 μm to 4.2 μm (p < 0.05, by Mann–Whitney test) (Fig. 7m–o). Furthermore, expression of ENO4, KIF27, INTU and PACRG decreased significantly in a higher proportion than in basal conditions (Fig. 7p), suggesting a synergistic effect of cytokines and miRNAs in the ciliary process. Quantification of acetylated α-tubulin protein in cell extracts between conditions did not reveal significant differences (Supplementary Fig. 4).
Discussion
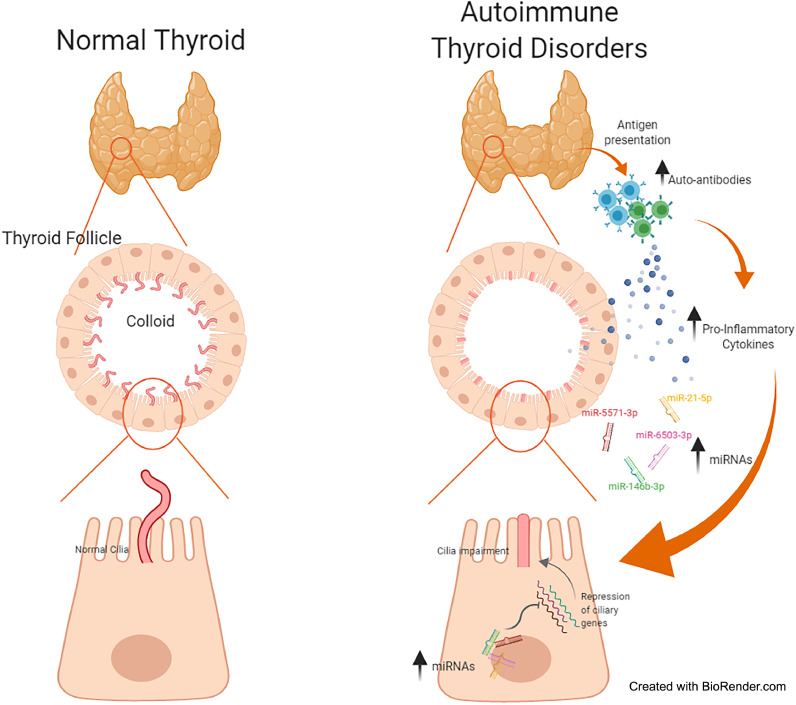
In this study, we have identified novel molecular AITD susceptibility pathways using an integrated analysis of miRNAs and mRNAs expression profiling of thyroid tissues from AITD patients and controls. Our results revealed that mechanisms involved in ciliary development are significantly deregulated in AITD. In addition, we have demonstrated that genes involved in ciliary pathways (ENO4, INTU, KIF27, PACRG and STK36) are downregulated in thyroid tissue of AITD patients, and that the percentage of follicular ciliated cells and cilia length are markedly reduced in HT and GD patients (Fig. 8). Furthermore, we have shown that thyrocytes cultured in pro-inflammatory conditions and transfected with miRNA mimics, exhibit impaired ciliary phenotypes, indicating that ciliogenesis plays an important role in the development of AITD.
Fig. 8.
Proposed model of the mechanisms underlying the ciliary defects in AITD. Abnormal antigen presentation in thyroid and breakdown of tolerance to self-antigens lead to complex and interrelated intrathyroidal immune processes. For instance, thyroid-specific B and T cells produce inflammatory cytokines such as IFNγ and TNFα. This pro-inflammatory environment may increase the expression of miRNAs (miR-21-5p, miR-146b-3p and miR-6503-3p). The combination of both the pro-inflammatory environment and increased miRNA expression may repress the expression of ciliary related genes, leading to an impairment of ciliogenesis.
Primary cilia typically appear alone as a single cilium in human thyroid follicular cells. The relationship between these structures and the Golgi apparatus suggests that cilium may act as sensory regulators in the thyroid to modulate hormone secretion [14]. Indeed, trace amine-associated receptor 1 (Taar1), a G-protein-coupled receptor, have been described to be present in primary cilium of thyroid rodent cells and has been attributed to function in thyroid hormones liberation, thyroid autoregulation and homeostatic thyroid stimulating hormone receptor maintenance [23,24]. Alterations of primary cilia have also been associated in the regulation of cancer development towards the control of metabolic phenotypes [25,26]. Lee et al. have described defective ciliogenesis in thyroid Hürthle cell lesions associated with mitophagy or autophagy [19]. Their findings circumscribe ciliary defects to Hürthle cells found in HT, although we observed a generalized effect throughout the tissue. Our findings in GD however, are consistent with a recent report in the literature which also described decreased ciliary frequencies and cilia length in GD [16]. However, in the contrary with these authors, we did not find any differences in cilia frequency in nodular hyperplasia, as has been previously reported by others [19]. Although these variable findings could be attributed to thyroid tissue heterogeneity [16], they could also be related to some degree of inflammation and/or autoimmunity that could have been overlooked in the pathological diagnosis.
MiRNAs identified herein (miR-21-5p, miR-146b-3p, miR-5571-3p and miR-6503-3p) target genes involved in the different ciliary processes. One such gene is INTU, which is part of the CPLANE complex (Ciliogenesis and Planar Polarity Effector), which is an important physical and functional unit governing the recruitment of microtubule-based intraflagellar transport proteins to the base of cilia. Indeed, mutation of CPLANE genes elicits specific ciliopathy phenotypes in mouse models and it is associated with novel ciliopathies in human patients [27]. The parkin-co-regulated gene (PACRG) and Enolase 4 (ENO4) appear in the flagella of various eukaryotes. PACRG directly associates with microtubules and tubulin dimers and is involved in doublet microtubule assembly, being crucial for the stability and function of primary cilia [28], [29], [30]. It is therefore conceivable that downregulation of these proteins in AITD patients affects the stability of cilia in thyroid cells, and contributes to the impairment of ciliary growth in thyroid. Finally, STK36 encodes a serine/threonine kinase that enhances the activity of Gli transcription factors, targeting the Hedgehog signaling pathway, which is involved in cilia formation [31,32]. Interestingly, STK36 has been associated with kinesin Kif27 (kinesin family member 27), which was also downregulated in AITD samples. Both proteins are involved in the recruitment and/or activation of central pair proteins along the cilium [33], thus abnormalities in these proteins could be associated with a defective cilia elongation in AITD. Using high resolution microscopy techniques we have also found that the expression profile of α-acetylated tubulin and Arl13b distribution between HT and GD are somewhat different. In SEM images, stumped-cilia appeared at the center of thyroid cells in both diseases. However in HT, several “volcano-like” cilia were observed with a hole in the center which, as far as we know, has never been described before.
The AITD microenvironment is clearly pro-inflammatory, indeed, several studies have shown the importance of cytokines and chemokines in its pathogenesis [34]. In this context, it was pertinent to prove if inflammatory cytokines affected cilia formation. We found that pro-inflammatory cytokines IFN-γ and TNF-α decreased the number of cilia despite increasing the expression of the aforementioned genes. Importantly, addition of miRNA to the system further decreased the number of cilia and repressed the expression of the genes identified in this study. This apparent discrepancy can be reconciled by the fragile behavior of cilia and the possible effect of pro-inflammatory cytokines on the expression of cilia-related genes at a transcriptional level. In short, those cilia that overcome the constriction imposed by the pro-inflammatory conditions would be longer as a consequence of the increased expression of the pro-cilia genes triggered by cytokines. However, the fragile phenotype of these elongated cilia would, in time, curtail or break their axoneme which is consistent with an overall decrease in the number of cilia in these cultures. Indeed, previous reports have described the elongation of cilia in response of IL-1 via a protein kinase A dependent mechanism, suggesting a fundamental importance of inflammation in the regulation of cilia structure and function [35]. It is also important that the combination of cytokines with miRNA, which is likely a better representation of the actual microenvironment of thyroid during the onset of the disease, displayed less and shorter cilia, consistent with a miRNA-dependent repression of the cilia-related genes that is not compensated by the pro-inflammatory cytokines. Several studies have described the linkage between miRNA expression and cytokine activities in two complementary ways: from a regulation of the expression and secretion of miRNAs by cytokines and vice versa, a regulation of cytokine expression by miRNAs [36,37]. Interestingly, miR-21 controls the production of pro-inflammatory cytokines in TGF-β receptor II mutant mice, contributing in the development of autoimmune cholangitis and colitis [37].
Together, data herein strongly support that cilia deregulation in thyroid cells is part of the molecular mechanism that underlies AITD. This is likely related to the normal function of primary cilia in this tissue. Primary cilia activate a wide range of responses and numerous intracellular signal transduction cascades essential for cell development, proliferation, differentiation, survival and migration. Major signaling routes influenced by primary cilia include Hedgehog, platelet-derived growth factor (PDGF)-α signaling, and cell polarity Wnt pathways, as well as the insulin growth factor (IGF-1)-receptor signaling path. Some of these are associated to extra-thyroid manifestations of AITD, for example Graves’ Orbitopathy [22,[38], [39], [40], [41]]. In fact, the TSHR and IGF-1R pathways form a functional complex in thyroid and orbital tissue [41] that is involved in the recruitment of T cells to the affected organs during the onset of GD [40].
This research, however, is subject to some limitations. MiRNA and mRNA NGS were performed in whole thyroid tissue, thus, it was difficult to differentiate the roles of infiltrating lymphocytes and thyrocytes. In addition, although we did not observe any statistical correlations between thyroid hormone levels and gene expression, whether thyroid dysfunction could also had a role in ciliogenesis should be address in further studies.
To conclude, it is important to emphasize that novel molecular approaches, as the integration of heterogeneous types of omics data like the miRNA–mRNA network, have the potential to identify new molecular susceptibility pathways. This contributes to the development of new therapeutic approaches, e.g. targeting miRNAs, to treat inflammatory conditions such as AITD or other autoimmune diseases.
CRediT authorship contribution statement
Rebeca Martínez-Hernández: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Ana Serrano-Somavilla: Data curation, Formal analysis, Investigation, Methodology, Validation. Ana Ramos-Leví: Data curation, Writing - original draft, Writing - review & editing. Miguel Sampedro-Nuñez: Data curation, Formal analysis, Investigation, Methodology, Software. Alberto Lens-Pardo: Data curation, Formal analysis, Methodology, Validation. Muñoz De Nova: Data curation. Juan Carlos Triviño: Formal analysis, Methodology, Software. María Ujue González: Funding acquisition, Methodology, Visualization. Lorena Torné: Methodology, Visualization. Javier Casares-Arias: Methodology, Visualization. Noa B. Martín-Cófreces: Formal analysis, Investigation, Methodology, Software, Visualization, Writing - original draft. Francisco Sánchez-Madrid: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Writing - original draft, Writing - review & editing. Mónica Marazuela: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors have declared that no competing financial interests exist.
Acknowledgments
We thank Francisca Molina-Jimenez from the Instituto Universitario Princesa for technical assistance with confocal microscopy. We thank the Institut d'Investigació en Ciències de la Salut Germans Trias i Pujol (IGTP-HUGTIP) Biobank and Magdalena Adrados from the Department of Pathological Anatomy of HUP for her help classifying thyroid specimens. We also warmly thank Miguel Vicente Manzanares for English corrections and Miguel Angel Alonso for his technical support.
This work was supported by the following grants: Proyectos de Investigación en Salud (FIS) PIE13-0041 and PI16-02091 (funded by Instituto de Salud Carlos III), TIRONET2-CM, B2017/BMD-3724 (funded by Comunidad de Madrid), GETNE G1707 (funded by Grupo Español de Tumores Neuroendocrinos y Endocrinos) and cofinanced by FEDER funds to MM. We also acknowledge the service from the MiNa Laboratory at IMN, and funding from CM (project S2018/NMT-4291 TEC2SPACE), MINECO (project CSIC13-4E-1794) and EU (FEDER, FSE) to M.U.G. The funders had no role in study design, data collection, data analysis, interpretation or writing of the report.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.10.061.
Appendix. Supplementary materials
References
- 1.Ramos-Leví A.M., Marazuela M. Pathogenesis of thyroid autoimmune disease: the role of cellular mechanisms. Endocrinol Nutr. 2016;63:421–429. doi: 10.1016/j.endonu.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Weetman A.P. Autoimmune thyroid disease. Autoimmunity. 2004;37:337–340. doi: 10.1080/08916930410001705394. [DOI] [PubMed] [Google Scholar]
- 3.Weetman A.P. Cellular immune responses in autoimmune thyroid disease. Clin Endocrinol (Oxf) 2004;61:405–413. doi: 10.1111/j.1365-2265.2004.02085.x. [DOI] [PubMed] [Google Scholar]
- 4.Bartalena L., Fatourechi V. Extrathyroidal manifestations of Graves’ disease: a 2014 update. J Endocrinol Invest. 2014;37:691–700. doi: 10.1007/s40618-014-0097-2. [DOI] [PubMed] [Google Scholar]
- 5.Fallahi P., Ferrari S.M., Ruffilli I., Elia G., Biricotti M., Vita R. The association of other autoimmune diseases in patients with autoimmune thyroiditis: review of the literature and report of a large series of patients. Autoimmun Rev. 2016;15:1125–1128. doi: 10.1016/j.autrev.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Cañas C.A., Cañas F., Bonilla-Abadía F., Ospina F.E., Tobón G.J. Epigenetics changes associated to environmental triggers in autoimmunity. Autoimmunity. 2016;49:1–11. doi: 10.3109/08916934.2015.1086996. [DOI] [PubMed] [Google Scholar]
- 7.Wang B., Shao X., Song R., Xu D., Zhang J.-.A. The emerging role of epigenetics in autoimmune thyroid diseases. Front Immunol. 2017;8:396. doi: 10.3389/fimmu.2017.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Andrés-León E., Rojas A.M. miARma-Seq, a comprehensive pipeline for the simultaneous study and integration of miRNA and mRNA expression data. Methods. 2019;152:31–40. doi: 10.1016/j.ymeth.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Xu J., Shao T., Ding N., Li Y., Li X. miRNA–miRNA crosstalk: from genomics to phenomics. Brief Bioinform. 2017;18:1002–1011. doi: 10.1093/bib/bbw073. [DOI] [PubMed] [Google Scholar]
- 11.Martínez-Hernández R., Sampedro-Núñez M., Serrano-Somavilla A., Ramos-Leví A.M., de la Fuente H., Triviño J.C. A microrna signature for evaluation of risk and severity of autoimmune thyroid diseases. J Clin Endocrinol Metab. 2018;103:1139–1150. doi: 10.1210/jc.2017-02318. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z., Fan X., Zhang R., Lin Z., Lu T., Bai X. Integrative analysis of mRNA and miRNA array data reveals the suppression of retinoic acid pathway in regulatory T cells of Graves’ disease. J Clin Endocrinol Metab. 2014;99:E2620–E2627. doi: 10.1210/jc.2014-1883. [DOI] [PubMed] [Google Scholar]
- 13.Utrilla J.C., Gordillo-Martínez F., Gómez-Pascual A., Fernández-Santos J.M., Garnacho C., Vázquez-Román V. Comparative study of the primary cilia in thyrocytes of adult mammals. J Anat. 2015;227:550–560. doi: 10.1111/joa.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin A., Hedinger C., Häberlin-Jakob M., Walt H. Structure and motility of primary cilia in the follicular epithelium of the human thyroid. Virchows Arch B Cell Pathol. 1988;55:159–166. doi: 10.1007/BF02896572. [DOI] [PubMed] [Google Scholar]
- 15.Rupik W. Ultrastructural studies of cilia formation during thyroid gland differentiation in grass snake embryos. Micron. 2013;44:228–237. doi: 10.1016/j.micron.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Fernández-Santos J.M., Utrilla J.C., Vázquez-Román V., Villar-Rodríguez J.L., Gutiérrez-Avilés L., Martín-Lacave I. Primary cilium in the human thyrocyte: changes in frequency and length in relation to the functional pathology of the thyroid gland. Thyroid. 2019;29:595–606. doi: 10.1089/thy.2018.0401. [DOI] [PubMed] [Google Scholar]
- 17.Vitales-Noyola M., Serrano-Somavilla A., Martínez-Hernández R., Sampedro-Nuñez M., Ramos-Levi A.M., González-Amaro R. Patients with autoimmune thyroiditis show diminished levels and defective suppressive function of Tr1 regulatory lymphocytes. J Clin Endocrinol Metab. 2018;103:3359–3367. doi: 10.1210/jc.2018-00498. [DOI] [PubMed] [Google Scholar]
- 18.Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 19.Lee J., Yi S., Kang Y.E., Chang J.Y., Kim J.T., Sul H.J. Defective ciliogenesis in thyroid hürthle cell tumors is associated with increased autophagy. Oncotarget. 2016;7:79117–79130. doi: 10.18632/oncotarget.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-López M.A., Sancho D., Sánchez-Madrid F., Marazuela M. Thyrocytes from autoimmune thyroid disorders produce the chemokines IP-10 and Mig and attract CXCR3+ lymphocytes. J Clin Endocrinol Metab. 2001;86:5008–5016. doi: 10.1210/jcem.86.10.7953. [DOI] [PubMed] [Google Scholar]
- 21.Menzl I., Lebeau L., Pandey R., Hassounah N.B., Li F.W., Nagle R. Loss of primary cilia occurs early in breast cancer development. Cilia. 2014;3:7. doi: 10.1186/2046-2530-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernabé-Rubio M., Alonso M.A. Routes and machinery of primary cilium biogenesis. Cell Mol Life Sci. 2017;74:4077–4095. doi: 10.1007/s00018-017-2570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szumska J., Qatato M., Rehders M., Führer D., Biebermann H., Grandy D.K. Trace amine-associated receptor 1 localization at the apical plasma membrane domain of fisher rat thyroid epithelial cells is confined to cilia. Eur Thyroid J. 2015;4:30–41. doi: 10.1159/000434717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qatato M., Szumska J., Skripnik V., Rijntjes E., Köhrle J., Brix K. Canonical tsh regulation of cathepsin-mediated thyroglobulin processing in the thyroid gland of male mice requires taar1 expression. Front Pharmacol. 2018;9 doi: 10.3389/fphar.2018.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J., Yi S., Won M., Song Y.S., Yi H.-.S., Park Y.J. Loss-of-function of IFT88 determines metabolic phenotypes in thyroid cancer. Oncogene. 2018;37:4455–4474. doi: 10.1038/s41388-018-0211-6. [DOI] [PubMed] [Google Scholar]
- 26.Fabbri L., Bost F., Mazure N. Primary cilium in cancer hallmarks. IJMS. 2019;20:1336. doi: 10.3390/ijms20061336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toriyama M., Lee C., Taylor S.P., Duran I., Cohn D.H., Bruel A.-.L. The ciliopathy-associated CPLANE proteins direct basal body recruitment of intraflagellar transport machinery. Nat Genet. 2016;48:648–656. doi: 10.1038/ng.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda T. Parkin-co-regulated gene (PACRG) product interacts with tubulin and microtubules. FEBS Lett. 2008;582:1413–1418. doi: 10.1016/j.febslet.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 29.Dawe H.R., Farr H., Portman N., Shaw M.K., Gull K. The Parkin co-regulated gene product, PACRG, is an evolutionarily conserved axonemal protein that functions in outer-doublet microtubule morphogenesis. J Cell Sci. 2005;118:5421–5430. doi: 10.1242/jcs.02659. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura N., Dai Q., Williams J., Goulding E.H., Willis W.D., Brown P.R. Disruption of a spermatogenic cell-specific mouse enolase 4 (eno4) gene causes sperm structural defects and male infertility. Biol Reprod. 2013;88:90. doi: 10.1095/biolreprod.112.107128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nozawa Y.I., Yao E., Lin C., Yang J.-.H., Wilson C.W., Gacayan R. Fused (Stk36) is a ciliary protein required for central pair assembly and motile cilia orientation in the mammalian oviduct. Dev Dyn. 2013;242:1307–1319. doi: 10.1002/dvdy.24024. [DOI] [PubMed] [Google Scholar]
- 32.Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 33.Wilson C.W., Nguyen C.T., Chen M.-.H., Yang J.-.H., Gacayan R., Huang J. Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature. 2009;459:98–102. doi: 10.1038/nature07883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antonelli A., Ferrari S.M., Corrado A., Di Domenicantonio A., Fallahi P. Autoimmune thyroid disorders. Autoimmun Rev. 2015;14:174–180. doi: 10.1016/j.autrev.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Wann A.K.T., Knight M.M. Primary cilia elongation in response to interleukin-1 mediates the inflammatory response. Cell Mol Life Sci. 2012;69:2967–2977. doi: 10.1007/s00018-012-0980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benes V., Collier P., Kordes C., Stolte J., Rausch T., Muckentaler M.U. Identification of cytokine-induced modulation of microRNA expression and secretion as measured by a novel microRNA specific qPCR assay. Sci Rep. 2015;5 doi: 10.1038/srep11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ando Y., Yang G.-.X., Kenny T.P., Kawata K., Zhang W., Huang W. Overexpression of microRNA-21 is associated with elevated pro-inflammatory cytokines in dominant-negative TGF-β receptor type II mouse. J Autoimmun. 2013;41:111–119. doi: 10.1016/j.jaut.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wann A.K.T., Chapple J.P., Knight M.M. The primary cilium influences interleukin-1β-induced NFκB signalling by regulating IKK activity. Cell Signal. 2014;26:1735–1742. doi: 10.1016/j.cellsig.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu D., Shi S., Wang H., Liao K. Growth arrest induces primary-cilium formation and sensitizes IGF-1-receptor signaling during differentiation induction of 3T3-L1 preadipocytes. J Cell Sci. 2009;122:2760–2768. doi: 10.1242/jcs.046276. [DOI] [PubMed] [Google Scholar]
- 40.Pritchard J., Han R., Horst N., Cruikshank W.W., Smith T.J. Immunoglobulin activation of T cell chemoattractant expression in fibroblasts from patients with graves’ disease is mediated through the insulin-like growth factor i receptor pathway. J Immunol. 2003;170:6348–6354. doi: 10.4049/jimmunol.170.12.6348. [DOI] [PubMed] [Google Scholar]
- 41.Tsui S., Naik V., Hoa N., Hwang C.J., Afifyan N.F., Hikim A.S. Evidence for an association between TSH and IGF-1 receptors. J Immunol. 2008;181:4397–4405. doi: 10.4049/jimmunol.181.6.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information files, or are available in a persistent repository or upon reasonable requests to the authors.
Next generation sequencing data supporting the findings of this study are deposited in Bioproject: PRJNA396505 and PRJNA530166 and Biosamples: SAMN07427434, SAMN07427435, SAMN07427436, SAMN07427437, SAMN07427438, SAMN07427439, SAMN07427440, SAMN07427441, SAMN07427442, SAMN07427443, SAMN07427444, SAMN07427445, SAMN07427446, SAMN07427447, SAMN07427448, SAMN07427449, SAMN07427450, SAMN07427451 and SAMN07427452.