Abstract
Background
Long non-coding RNAs (lncRNAs) have drawn increasing attention because they play a pivotal role in various types of autoimmune diseases, including rheumatoid arthritis (RA). Fibroblast-like synoviocytes (FLSs), a prominent component of hyperplastic synovial pannus tissue, are the primary effector cells in RA synovial hyperplasia and invasion which can lead to joint destruction. In this study, we investigated whether lncRNAs could act as competing endogenous RNAs to regulate the pathological behaviors of RA-FLSs.
Methods
LncRNA microarray was conducted to establish lncRNA expression profiles in FLSs isolated from RA patients and healthy controls (HCs). Differentially expressed lncRNAs were verified by quantitative real-time PCR (qRT-PCR) on RA-FLSs and synovial fluid. The functional role of lncRNA PICSAR downregulation was evaluated in RA-FLSs. We conducted molecular biological analysis to predict miRNAs which have a potential binding site for PICSAR and further refined the results by qRT-PCR. Luciferase reporter assay was adopted to validate the interaction of lncRNA PICSAR and miR-4701-5p. Western Blot and qPCR were used to identify the target gene and protein. The functional role of miR-4701-5p upregulation was examined in RA-FLSs.
Findings
We identified a long intergenic non-protein-coding RNA162 (LINC00162), also known as lncRNA PICSAR (p38 inhibited cutaneous squamous cell carcinoma associated lincRNA), has significantly higher expression in RA-FLSs and RA synovial fluid. The cell proliferation, migration, invasion and proinflammatory cytokines production of RA-FLSs showed significant alterations after the lncRNA PICSAR suppression. Mechanistically, lncRNA PICSAR functioned through sponging miR-4701-5p in RA-FLSs.
Interpretation
Our results reveal PICSAR may exert an essential role in promoting synovial invasion and joint destruction by sponging miR-4701-5p in RA and that lncRNA PICSAR may act as a biomarker of RA.
Keywords: Rheumatoid arthritis, Fibroblast-like synoviocytes, Long non-coding RNAs, PICSAR
Research in Context.
Evidence before this study
Long non-coding RNAs (lncRNAs) play essential roles in many diseases, including rheumatoid arthritis (RA). The transition behavior of fibroblast-like synoviocytes (FLSs) stimulates the progression of RA and promotes joint destruction. Although the important biological functions of lncRNAs have been gradually explored, their underlying mechanism in RA-FLSs remains unclear.
Added value of this study
In the present study, we found that the lncRNA expression profile of RA-FLSs was different from that of control group. LncRNA PICSAR is significantly overexpressed in RA-FLSs and synovial fluid and may contribute to the occurrence and development of RA by sponging miR-4701-5p.
Implications of all the available evidence
This study demonstrated the crucial role of lncRNA PICSAR in the development of RA and lncRNA PICSAR could be an essential biomarker of RA.
Alt-text: Unlabelled box
1. Introduction
Rheumatoid arthritis (RA) is a common chronic inflammatory joint disease characterized by persistent synovial hyperplasia and progressive destruction of joint cartilage and bone [1,2]. It is well recognized that RA can lead to decreased functional status, disability, and increased mortality [3,4]. The main pathological feature of established RA is the proliferative synovial lining tissue, which contains a greater amount of activated fibroblast-like synoviocytes (FLSs). Through the interaction with T cells, B cells, macrophages and many pro-inflammatory cytokines, FLSs play an essential role in the pathogenesis of the disease and promote the continued joint inflammation [5], [6], [7], [8]. As a major source of inflammatory cytokines and catabolic enzymes that promote joint degeneration, RA-FLSs also exhibit some tumor-like behavior when activated in the chronic inflammatory environment, such as contact inhibition escape, anchorage-independent growth and increased ability of migration and invasion [9], [10], [11]. This transition behavior stimulates the progression of RA and eventually gives rise to joint destruction [12]. Therefore, elucidating the underlying molecular mechanism whereby FLSs serve as pathological factors of RA may address the urgent medical need in the therapy of RA and related diseases.
Long non-coding RNAs (lncRNAs), which are defined as RNAs longer than 200 nucleotides in length and without protein-coding capacity, were initially dismissed as merely transcriptional “noise” [13]. However, with the development of RNA microarray and RNA sequencing techniques, accumulating evidence has revealed that lncRNAs play important roles in the regulation of a series of cellular processes, such as gene transcription, cell differentiation, cell proliferation and cell cycle [14], [15], [16], [17]. On account of these multiple mechanisms, numerous autoimmune diseases including RA were demonstrated to be connected with lncRNAs [18], [19], [20]. Although in-depth studies of lncRNAs have been conducted, their functional and pathological significance is still largely unknown. In this report, we identified lncRNA PICSAR (LINC00162) and systemically investigated its regulatory and functional role in RA-FLSs.
2. Materials and methods
2.1. Patients and tissue samples
The synovial tissues of RA groups were taken from active patients with RA (five females and three males, 45–67 years of age) who underwent synovectomy of the knee joint or knee replacement surgery. RA patients were diagnosed according to the 2010 American College of Rheumatology /European League Against Rheumatism (ACR/EULAR) classification criteria [21]. The synovial tissues of healthy control groups were taken from four patients (one female and three males, 19–38 years of age) who underwent arthroscopic surgery for knee meniscus injuries or cruciate ligament rupture and who had no medical history of acute or chronic joint abnormalities or systemic disease. The synovial fluid from RA patients and non-RA control groups were taken from additional six RA patients and four traumatic arthritis patients according to the standard procedure as described above. The basic clinical parameters for RA patients were provided in the Table 3 of Supplementary Material. All samples were obtained from discarded tissues at the Third Affiliated Hospital of Sun Yat-sen University in Guangzhou, Guangdong, China, during June 2017 to August 2018. The research was approved by the ethics committee of the Third Affiliated Hospital of Sun Yat-sen University and all subjects provided the written informed consent in this study according to the Declaration of Helsinki principles.
2.2. Culture and characterization of FLSs
The FLSs were isolated form synovial tissues by means of a tissue block culture method. Synovial tissues were dissected free of fat, blood vessels and fibrous tissues, rinsed with phosphate-buffered solution (PBS), and transferred to culture flasks after shredding into the approximate size of 1 mm3. The culture flasks, which contained an appropriate amount of Dulbecco's modified Eagle's medium (DMEM, Gibco Laboratories, Invitrogen, USA) culture medium supplemented with 10% fetal bovine serum (FBS, Gibco Laboratories), were placed upright for tissue adherence in an incubator with 5% CO2 at 37 °C. After 4–5 h, the culture flasks were carefully laid flat and cell cultures were followed by passaging the culture medium once every 2–3 days until the logarithmic growth phase. In general, FLSs migrated out from the tissue explant and were grown into a firmly adherent cell monolayer of 90%−95% confluency within 2 weeks. Then FLSs were trypsinized, collected, re-suspended, and implanted for cell subculture. The FLSs passaged 3–5 times were used for the following experiments.
The morphology characters of FLSs were observed under the light microscope. As for the characterization of FLSs, expression levels of specified surface markers, CD68 (a macrophage marker) and CD90 (a fibroblast marker), were detected by flow cytometry. FLSs at passages 3 were trypsinized, centrifuged and stained with monoclonal antibodies CD68 FITC and CD90 PE (Biolegend, USA) in the dark. Isotype matched control antibodies were used as methodologic controls. The flow cytometric analysis was performed by FACSCalibur Flow Cytometer (Becton Dickinson Biosciences, Franklin Lakes, USA) and the data analysis were performed by the software FlowJo 7.6 (BD Biosciences, USA).
2.3. RNA extraction and quality control
Total cellular RNA was isolated using the TRIzol® reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The quantity and quality of RNA samples were measured by NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA). RNA integrity was assessed by standard denaturing agarose gel electrophoresis and the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
2.4. Microarray and data analysis
The expression of lncRNAs and mRNAs in FLSs was analyzed using the Arraystar Human LncRNA Microarray v4.0 (Arraystar Inc, Rockville, MD), which is designed for the global profiling of 40,173 human lncRNAs and 20,730 protein-coding transcripts. Sample labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology) with minor modifications. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were conducted using the GeneSpring GX v12.1 software package (Agilent Technologies).
LncRNAs and mRNAs were chosen for further data analysis after quantile normalization of the raw data. Differentially expressed lncRNAs and mRNAs with statistical significance between the two groups were identified through P-value filtering using the Student's t-test. Differentially expressed lncRNAs and mRNAs between the two samples were identified through Fold Change filtering. Finally, Hierarchical Clustering and combined analysis was performed to show the distinguishable lncRNAs expression patterns between the two groups using in-house scripts. The microarray data were deposited to the NCBI's Gene Expression Omnibus (GEO) database and are provided at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128813. The accession number is GSE128813.
2.5. GO and KEGG pathway analysis
Gene ontology (GO) analysis (http://geneontology.org/), a common useful method for high‐throughput genome or transcriptome data, was performed to identify characteristic biological functions of the differently expressed mRNAs. The ontology covers three domains: Biological Process, Cellular Component and Molecular Function [22]. KEGG pathway analysis (Kyoto Encyclopedia of Genes and Genomes, https://www.genome.jp/kegg/) is a knowledge base for systematic analysis of gene function, linking genomic information with high‐order functional information [23] and was applied to determine the roles of the differentially expressed mRNAs played in biological pathways. We performed the GO and KEGG pathway analysis using the clusterProfiler packages in R language (version 3.4.3) (https://www.r-project.org/).
2.6. Quantitative real-time reverse transcription PCR
To measure the expression of five lncRNAs with the most significant up-regulation fold change, cDNAs were synthesized from equal amounts of RNA of different samples using the PrimeScript™RT reagent Kit with gDNA Eraser (Takara Biotechnology, Tokyo, Japan) and applied to PCR reaction using TB Green™ Premix Ex Taq™ II (Takara Biotechnology). GAPDH was used as an internal control. Sequences of primer sets are listed in Supplementary Table 1. The expressions of miRNAs were detected by stem-loop real-time qPCR. MicroRNAs from total RNA were reverse transcribed and subjected to subsequent amplification using special Hairpin-it™ MicroRNAs Quantitation kits (GenePharma, Shanghai, China), which contained both the specific primers of miRNAs and internal control U6. All qPCR reactions were performed on the ABI 7500 Fast real-time PCR amplification equipment (Applied Biosystems, Foster City, CA). In regard to normalization of the data, relative expression levels of target genes to control gene GAPDH/U6 were calculated by using the 2−ΔΔCt method.
2.7. Transfection and RNA interference
The chemically synthesized small interference RNAs (siRNAs) specifically targeting PICSAR (PICSAR-siRNA) and negative control (NC-siRNA) were designed and commercially constructed by GenePharma. The miR-4701-5p mimics and negative controls (NC-mimics) were purchased from RiboBio (Guangzhou, China). All of the sequences are mentioned in Supplementary Table S2. RA-FLSs were cultured in 6-well plates (7.5 × 105 cells/well) or in 96-well plates (3 × 104 cells/well) at 24 h before transfection in accordance with the different experiment needs. When cells confluence reached 70%–90%, transient transfections of RA-FLSs were performed using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's instructions. The cultural supernatants of transfected cells were replaced with fresh culture medium at 6 h after the transfection. Transfection efficiency was observed by qPCR at 24 h after transfection to determine whether subsequent experiments could be performed. Then, the cells were collected at the appropriate time points based upon experimental requirements of the further experiments.
2.8. Cell Counting Kit-8 assay
Cell proliferation was detected with the Cell Counting Kit-8 (Dojindo Laboratories, Japan) assay according to the manufacturer's protocol. Initially, 3 × 104 RA-FLSs per well were seeded into 96-well plates and incubated to 70%–90% confluence. Cells in logarithmic growth phase were then transfected according to the above method. Three triplicate wells were applied to cell culture for another 24, 48, 72, and 96 h in each group. At each experimental point, 10 μL CCK-8 solution was added to each well and cells were incubated at 37 °C for 1.5 h. Cell viability was evaluated by absorbance at 450 nm measured with a microplate absorbance reader (Bio-Rad Laboratories, Hercules, CA).
2.9. Cell cycle analysis
Cell cycle phase analysis was detected by flow cytometry with the Cell Cycle Detection Kit (KeyGEN BioTECH, Nanjing, China) assay according to the manufacturer's protocol. Briefly, 7.5 × 105 RA-FLSs per well were seeded into 6-well plates and incubated to 70%–90% confluence. Cells in logarithmic growth phase were then transfected according to the above method. After following the transient transfection steps above, the cells were then harvested and washed twice with cold with PBS. Next, the pellet was resuspended, fixed in 70% prechilled ethanol, and stored overnight at 4 °C. Fixed cells were then washed again with PBS and treated with 500 μl propidium iodide (PI) / RNase A staining buffer at 37 °C in the dark for 30 min. Stained cells were subsequently analyzed with a flow cytometer (FACSCalibur, BD Bioscience Influx, Franklin Lakes, NJ, USA). Red fluorescence emitted from the PI–DNA complex was estimated at 488 nm and 10,000 events in each sample were analyzed by using Modfit software (Verity Software House, Topsham, ME, USA).
2.10. Western blot analysis
The total proteins were extracted using Total Protein Extraction Kit (Keygen, Nanjing, China) following the manufacturer's instructions and deactivated at 100 °C for 5 min. Protein lysates (35 mg) were fractionated on Tris-glycine-buffered 10% SDS-PAGE by electrophoresis and blotted onto a polyvinylidene fluoride membrane (Millipore). Membranes were blocked at room temperature for 2 h, then incubated overnight at 4 °C with primary antibodies. The primary antibodies included the phosphorylation extracellular signal-regulated kinases (p-ERK1/2) (1:2000, Cell Signaling Technology, Beverly, MA, USA) and ST3Gal I, (1:1000, Abcam, Cambridge, UK). Subsequently, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. The protein expression was normalized to GAPDH or ERK1/2 expression. Protein signals were detected using Enhanced chemiluminescence (ECL, Keygen) and the bands were analyzed with ImageJ2x software.
2.11. Cell migration assay
Cell migration and invasion assays of FLSs was performed in the Boyden chamber method using a filter with pore size of 8.0 μm (Transwell, Corning Labware Products, USA). RA-FLSs transfected for 48 h were resuspended in serum-free medium at a final concentration of 8 × 104 cells/ml and then seeded in the upper well. Additional 600 μl DMEM containing 10% FBS were placed as a chemoattractant in the lower well. After incubation at 37 °C in 5% CO2 for 12 h, the nonmigrating cells were removed from the filter's upper surface using a cotton swab. The cells that had migrated through the membrane were fixed in 4% paraformaldehyde (Boster, China) for 20 min and stained with crystal violet (KeyGEN BioTECH) for another 20 min. Chemotaxis was quantified using an optical microscope to count the stained cells that had migrated to the lower side of the filter. The mean number of migrated cells was recorded in five randomly selected fields at 50 × magnification for each assay.
2.12. Cell invasion assay
For the cell invasion assay, similar experiments were conducted using the same Transwell chamber methodology described previously. The upper chamber wells were coated individually with 30μl diluted BD Matrigel Basement Membrane Matrix (BD Biosciences, USA). The matrigel was diluted 1:10 with DMEM. After air-drying and concretion of matrigel, transfected RA-FLSs resuspended in serum-free medium at a final concentration of 105 cells/ml were seeded in the upper well. Similarly, the lower well contained 600 μl DMEM with 10% FBS. After 24 h of culture at 37 °C in 5% CO2, microscopic visualization and cells counting were conducted as described with the migration assays.
2.13. Cytokines secretion by ELISA
RA-FLSs transfected for 48 h were collected and analyzed for proinflammatory cytokines (IL-6, IL-8) and matrix metalloproteinases-3 (MMP-3) with ELISA kits (R&D Systems, USA) as recommended by the manufacturer.
2.14. Prediction of PICSAR combined miRNAs
MiRNAs that could bind to lncRNA PICSAR via complementary base pairing were predicted jointly by using DIANA-LncBase v2 (http://www.microrna.gr/LncBase/) and starBase v3.0 (http://starbase.sysu.edu.cn/), which both present an extensive collection of microRNA and lncRNA interactions using the data from high throughput AGO CLIP-Seq libraries [24,25]. The microRNA intersections of the databases are presented by Venn graph.
2.15. Dual luciferase reporter assays
The wild type and a mutant 3′UTR sequence of PICSAR which contained the potential miR-4701-5p binding sites were inserted into pMIR-REPORT™ Luciferase vector (Ambion, Austin, USA) using the Spe I and Hind III restriction sites. The plasmids were named PICSAR-WT and PICSAR-MUT. RA-FLSs were harvested after co-transfection of PICSAR-WT, PICSAR-MUT and pMIR Vector with miR-4701-5p mimic or negative control respectively using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's protocol. Luciferase activity was measured after 48 h using the Dual- Glo luciferase reporter assay kit (Promega, Madison, WI, USA) according to manufacturer's instructions. The relative fluorescence intensity ratio is then presented as the ratio of RLU1/RLU2(firefly luciferase activity/Renilla luciferase activity). Normalized firefly luciferase activity for each construct was compared to that of the pMIR Vector no-insert control. For each transfection, luciferase activity was averaged from six replicates.
2.16. Statistical analysis
SPSS statistical software v20.0 (Chicago, IL, USA) was used for statistical analyses. The experimental data were presented as the mean ± standard deviation (S.D.) or normalized to the fold change over the mean of the control. Student's t-test was used for data comparison between two groups and statistical differences among groups were tested by one-way ANOVA or the Kruskal–Wallis test. Differences were considered statistically significant when P-values were less than 0.05.
3. Results
3.1. Identification of FLSs
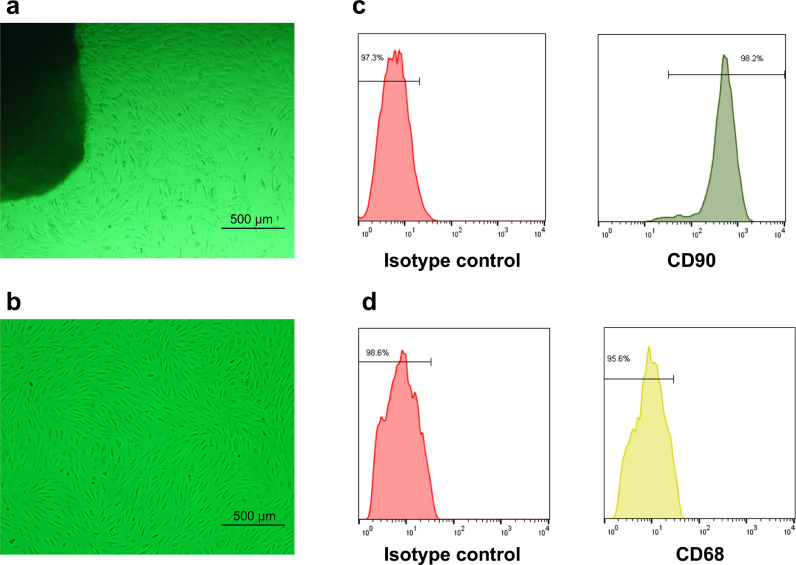
The morphology of RA-FLSs was observed under the light microscope. Primary fibroblast-like synoviocytes crawled out from the edge of the synovial tissues from RA patients after adherent culture for 3–5 days (Fig. 1a). RA-FLSs at passages 3 have unique morphology and are spindle-shaped and whirlpool-like in growth (Fig. 1b). FLSs are characterized by the surface markers include positive staining for VCAM-1, CD44, CD55, CD90 (Thy-1) and the absence of macrophage markers such as CD14 or CD68 [26]. We detected the expression level of surface markers (CD90, CD68) on FLSs for characterization. The flow cytometry analysis showed that the expression rate of CD90 of FLSs was up to 98% compared with isotype control (Fig. 1c). On the contrary, the expression of CD68 was negative (Fig. 1d).
Fig. 1.
The cell morphological characters and surface markers expression of rheumatoid arthritis (RA) fibroblast-like synoviocytes (FLSs). Observed with a light microscope, (a) the primary cell crawled out from synovial tissues and (b) the cells at passages 3 had unique morphology. The surface markers (CD90 and CD68) of RA-FLSs at passages 3 was detected by Flow cytometry. Cells were identified by (c) positive staining for CD90 and (d) CD68 negative staining, compared with isotype control respectively.
3.2. Expression profile of lncRNAs and mRNAs in RA-FLSs
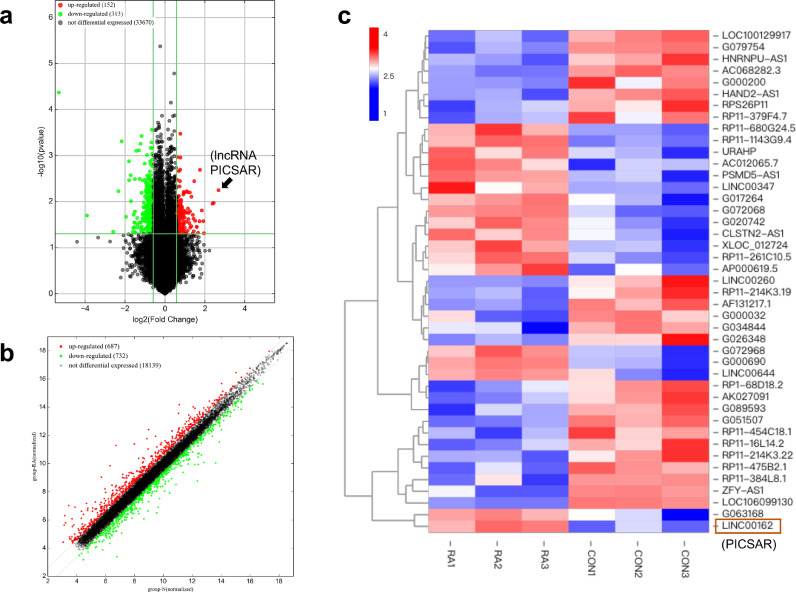
The microarray data (lncRNAs and mRNAs) were acquired from the two groups between RA-FLS and non-RA control FLSs. We analyzed statistical significance of differently expressed lncRNAs and mRNAs with the threshold of fold changes > 1.5 and P-values < 0.05. Differently expressed lncRNAs in the samples were shown using Volcano plot (Fig. 2a) and a Scatter Plot showed differently expressed mRNAs (Fig. 2b). These results showed 465 differently expressed lncRNAs (152 upregulation and 313 downregulation) and 1410 mRNAs (678 upregulation and 732 downregulation) in the RA group compared with the control group, respectively. As the Heat Map displayed in Fig. 2c, we increased the lncRNA screening conditions to the difference multiple of more than 2.5 and the most upregulated lncRNA was LINC00162 (lncRNA PICSAR) with 6.4-fold change. These data suggested that these differently expressed lncRNAs and mRNAs may play significant and complex roles in the RA-FLSs transition behavior and contribute to synovial aggression and joint destruction in RA.
Fig. 2.
Expression Profiles of differently expressed lncRNAs and mRNAs screened by microarray analysis of fibroblast-like synoviocytes (FLSs) samples from rheumatoid arthritis (RA) group (n = 3) and control group (n = 3). With the threshold of fold changes > 1.5 and P < 0.05, (a) the Volcano Plot showed differentially expressed lncRNAs between RA-FLSs and HC-FLSs and (b) the Scatter Plot showed differentially expressed mRNAs. (c) The Heat Map showing distinguishable lncRNA expression profiles with fold changes > 2.5 and P < 0.05.
3.3. GO and KEGG pathway analysis
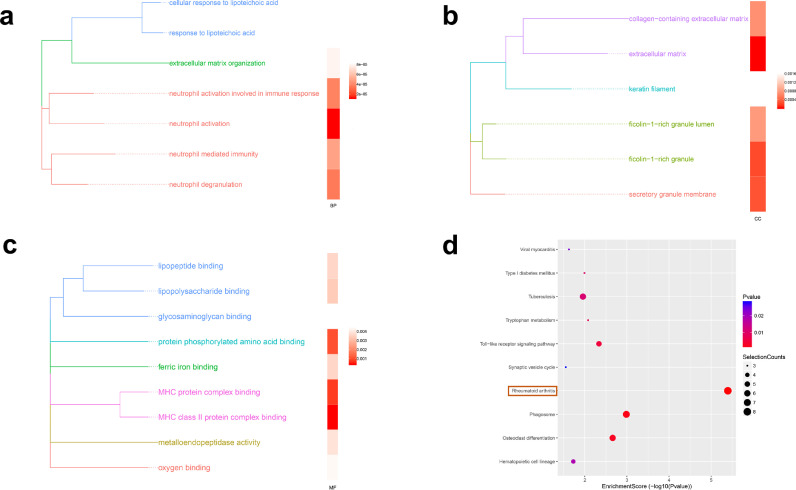
In the pathway analysis of the results, we focused our attention on the GO and KEGG pathway of up-regulated mRNAs with the threshold of fold changes > 1.5 and P < 0.05. GO terms are grouped according to functional theme and shown with the enrichment P-values in each category. The result showed that these mRNAs are mainly related to neutrophil activation (Biological Process) (Fig. 3a), extracellular matrix (Cellular Component) (Fig. 3b) and MHC class II protein complex binding (Molecular Function) (Fig. 3c). KEGG pathway analysis revealed that there were 16 pathways enriched in these mRNAs. Of these, rheumatoid arthritis, phagosome, osteoclast differentiation and Toll-like receptor signaling pathway were the most enriched (Fig. 3d).
Fig. 3.
GO and KEGG pathway analysis for the up-regulated mRNAs with the threshold of fold changes > 1.5 and P < 0.05. GO terms are grouped according to functional theme and shown with the enrichment P-values in each category: (a) Biological Process, (b) Cellular Component and (c) Molecular Function. (d) The Bubble Chart showed the top 10 of mRNAs KEGG pathway enrichment results.
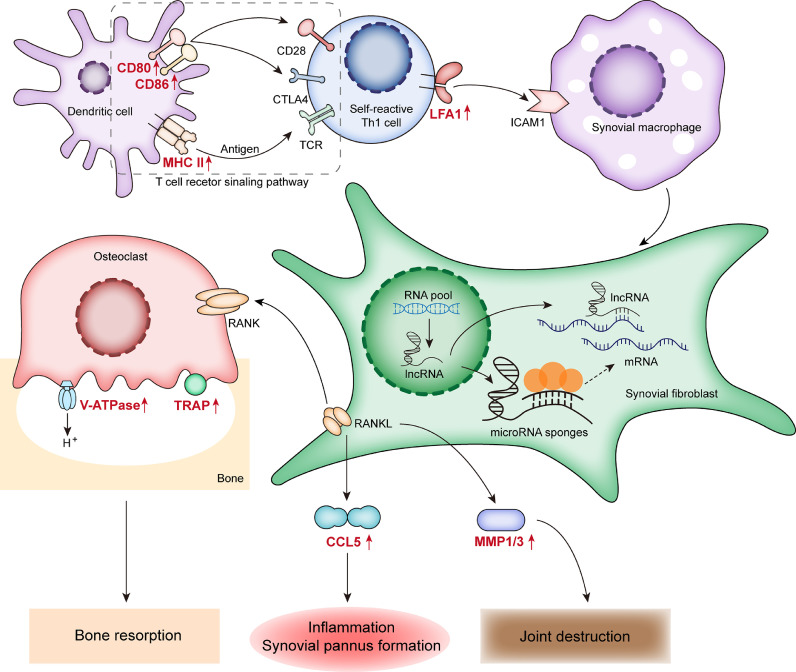
KEGG Pathway Map displays the genes (proteins) and small molecules in each pathway according to the function of molecular participation. In the map of rheumatoid arthritis pathway (Fig. 4), we inferred the specific biological roles in which these up-regulated mRNAs may participate during the process of osteoclast activity promotion, FLS cell secretion and Th1 cell self-reaction (P < 0.05). Overall, these analyses illustrate the close relationship between epigenetic regulation in RA-FLSs and processes related to RA progression.
Fig. 4.
The rheumatoid arthritis pathway map. The red labels represent the enriched genes according to the KEGG pathway result (P < 0.05).
3.4. Identification of PICSAR as a candidate lncRNA
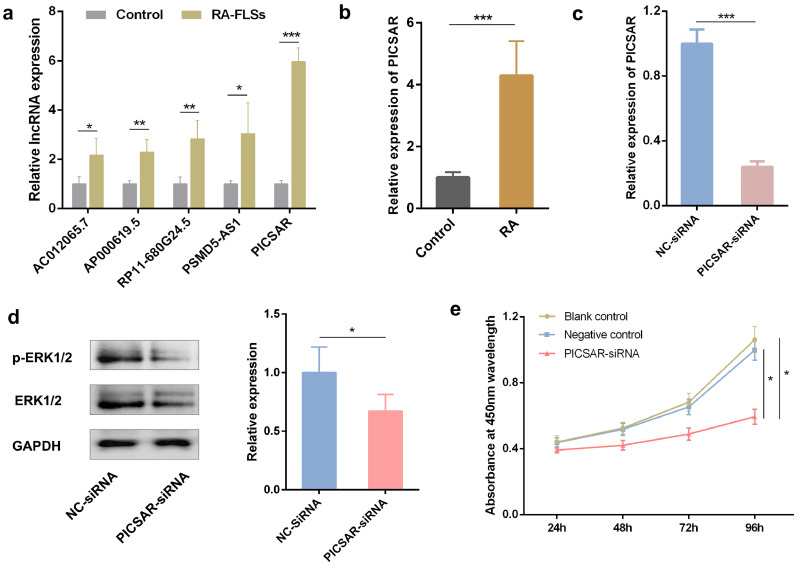
Based on fold changes > 3.7 in lncRNA expression between RA-FLSs and control group and P-values < 0.05, we found the 5 most significantly upregulated lncRNAs and these were selected for further verification. In RA-FLSs, we analyzed the expression of these 5 lncRNAs by qRT-PCR and found that LINC00162 (lncRNA PICSRA) was the one that was most significantly over-expressed compared to control (P < 0.001, Fig. 5a). We also noted that lncRNA PICSRA was significantly increased in RA synovial fluid than in control group (P < 0.001, Fig. 5b). Hence, PICSAR was selected for the subsequent functional and molecular mechanism experiments.
Fig. 5.
Verification of lncRNAs and effect of lncRNA PICSAR suppression on the proliferation of rheumatoid arthritis (RA) fibroblast-like synoviocytes (FLSs). (a) Quantitative real-time reverse transcription PCR (qRT-PCR) verified 5 selected lncRNAs with the highest up-regulated differential expression according to the microarray analysis. LncRNA PICSAR is the most significantly up-regulated in RA-FLSs. (b) LncRNA PICSAR was also over-expressed in the synovial fluid of RA patients compared with the control group by qPCR. (c) The inhibition efficiency of specific PICSAR small interference RNA (siRNA) in RA-FLSs was detected by qRT-PCR. (d) Compared with negative control, the expression of p-ERK1/2 in the total ERK1/2 protein was significantly decreased in the RA-FLSs after transfection. (e) PICSAR knockdown in RA-FLSs decreases cell proliferation assessed by a Cell Counting Kit-8 assay. The absorbance at 450 nm wavelength of PICSAR-siRNA group were significantly reduced at the indicated time points compared to the other two control groups. All the results were presented as the mean ± standard deviation (S.D.) based on ≥ 3 replicates involving ≥ 3 samples. *P < 0.05, **P < 0.01, and ***P < 0.001 versus control groups.
3.5. Suppression of PICSAR inhibits the proliferation of RA-FLSs
Since PICSAR expression was found to be upregulated in RA-FLSs, we further performed PICSAR- siRNA silencing to determine whether the elevated lncRNA PICSAR expression on RA-FLSs was functional. PICSAR-siRNA, the PICSAR targeted RNA interference, remarkably decreased the expression level of PICSAR compared with the negative control (NC-siRNA) and the inhibition efficiency was verified by qRT-PCR (P < 0.001, Fig. 5c).
The expression of p-ERK1/2 and total ERK1/2 which related to the proliferation of RA-FLSs was detected by the Western Blot. As shown in Fig. 5d, compared with the control group, the expression of p-ERK1/2 in the total ERK1/2 protein was significantly decreased in the RA-FLSs after PICSAR-siRNA transfection (P < 0.01).
The CCK-8 assay was used to assess RA-FLSs cell viability. As shown in Fig. 5e, growth curves of RA-FLSs demonstrated an obvious difference in the proliferation levels between the PICSAR knockdown and control groups. After PICSAR-siRNA transfection of RA-FLSs, the decline trend of cell proliferation was first observed at 24 h, then the difference gradually increased, and finally the inhibition rate in growth was significantly maintained (40.48 ± 3.56%, P < 0.05) at 96 h compared to the NC-siRNA group. The absorbance at 450 nm wavelength of the negative control group was close to the blank control group at each time point.
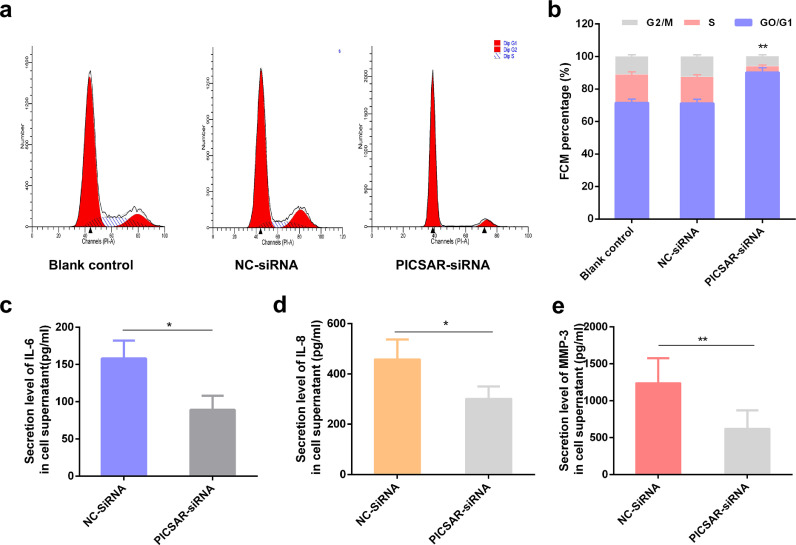
Flow cytometry was carried out to determine if PICSAR reduction also affects the cell-cycle changes in RA-FLSs. Similarly, downregulation of PICSAR produced cell-cycle arrest at G0/G1 phase (Fig. 6a). The percentages of RA-FLSs that stayed in G2/M + S phase were markedly decreased after PICSAR-siRNA transfection compared with negative control (10.03 ± 3.80% vs 28.46 ± 4.21%, P < 0.01, Fig. 6b) and no significant difference was found between blank control and negative control. Thus, we concluded that PICSAR had significant regulatory effects on the proliferation of RA-FLSs.
Fig. 6.
The effect of lncRNA PICSAR suppression on cell cycle and MMPs, proinflammatory cytokines production or secretion in FLSs. (a and b) Knocking-down PICSAR in RA-FLSs influences cell cycle. The flow cytometry analysis was performed to analyze the effects of PICSAR on cell-cycle distribution. (c–e) Compared to the NC-siRNA group, the concentrations of proinflammatory cytokines (IL-6, IL-8) and matrix metalloproteinases-3 (MMP-3) in the supernatant of RA-FLS cells were significantly reduced after PICSAR-siRNA treatment. All the results were presented as the mean ± standard deviation (S.D.) based on ≥ 3 replicates involving ≥ 3 samples. *P < 0.05, **P < 0.01, and ***P < 0.001 versus control groups.
3.6. Suppression of PICSAR decreases IL-6, IL-8 and MMP-3 production of RA-FLSs
In addition to the tumor-like behaviors, RA-FLSs also showed pathological destructions through production or secretion of a variety of proinflammatory cytokines or proteinases such as IL-6, IL-8, and MMPs. ELISA was conducted to assess the secretion levels of IL-6, IL-8, and MMPs in the supernatants of RA-FLS cells at 48 h after PICSAR-siRNA transfection. The results suggested that PICSAR knockdown significantly decreased IL-6 (Fig. 6c, P < 0.05), IL-8 (Fig. 6d, P < 0.05) and MMP-3 (Fig. 6e, P < 0.01) production of RA-FLSs compared to the NC-siRNA control group.
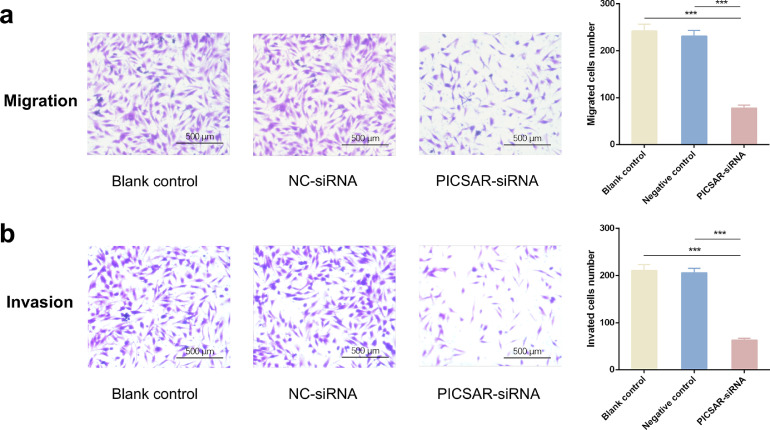
3.7. Suppression of PICSAR decreases the migration and invasion of RA-FLSs in vitro
Given that migration and invasion are important biological characteristics of RA-FLSs, we next determined the effect of PICSAR on these cellular properties. Transwell and Matrigel Transwell assays were performed to detect RA-FLSs migration and invasion ability respectively after PICSAR suppression. In the migration assay, the number of trans-membrane cells was dramatically suppressed in the treatment group with PICSAR-siRNA transfection compared to the NC-siRNA group and blank control groups (Fig. 7a). Simultaneously, Transwell chambers with Matrigel were used to detect the invasive capacity of RA-FLSs. The results showed a distinct decrease in the number of cells that penetrated the coated filter, suggesting impaired invasive ability of RA-FLSs after PICSAR silencing (Fig. 7b). There was no significant difference between blank control and negative control in both of the assays. Taken together, these findings demonstrated that the downregulation of PICSAR attenuated RA-FLSs migration and invasion.
Fig. 7.
lncRNA PICSAR knockdown suppressed the migration and invasion ability of rheumatoid arthritis (RA) fibroblast-like synoviocytes (FLSs). (a) The migration of RA-FLSs was examined by Transwell assay and (b) the invasion of RA-FLSs was detected by Matrigel Transwell assay. In both panels, the left was representative images of transmembrane cells and the right was quantification of different groups. All the results were presented as the mean ± standard deviation (S.D.) based on ≥ 3 replicates involving ≥ 3 samples. ***P < 0.001 versus control groups.
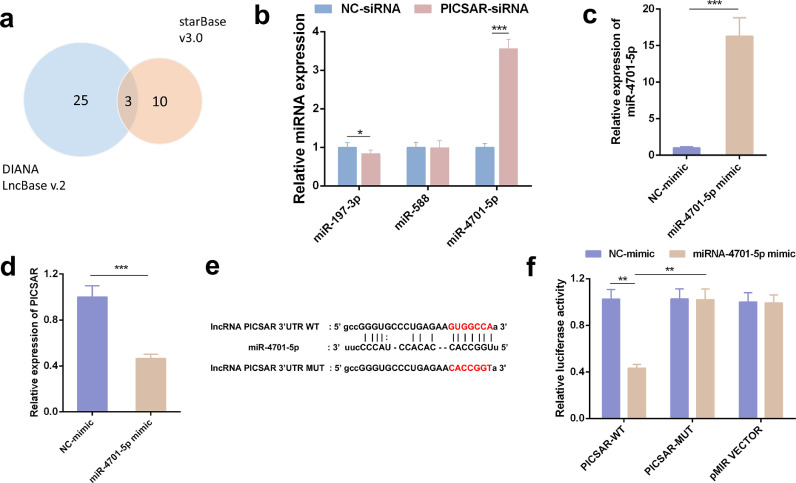
3.8. Screening and verification of miRNAs that can bind to PICSAR
Accumulating evidence has shown that lncRNAs could be competitively binding to miRNAs and function as a competing endogenous RNAs (ceRNAs) [27]. To verify whether PICSAR could act as a ceRNA for a certain miRNA, the bioinformatics programs starBase v3.0 and LncBase v2, the most used high throughput AGO CLIP-Seq libraries, were used to seek miRNAs which may have interacted with PICSAR (Fig. 8a). The microRNA intersections in both of the software programs suggested that 3 microRNAs (hsa-miR-588, hsa-miR-197-3p and hsa-miR-4701-5p) could be highly related to PICSAR, so we performed RT-qPCR for further screening after downregulation of PICSAR and among these miRNA candidates, the expression of miR-4701-5p was increased (Fig. 8b). The miRNA mimics are small, chemically modified double-stranded RNAs designed to mimic endogenous mature miRNA molecules and we used the specific mimic to increase miR-4701-5p expression efficaciously (Fig. 8c). Moreover, we demonstrated that overexpression of miR-4701-5p lowered the level of PICSAR in RA-FLSs (Fig. 8d). To confirm the targeting relationship between PICSAR and miR-4701-5p, a luciferase reporter assay was conducted according to the predicted binding site (Fig. 8e). As shown in Fig. 8f, the results revealed that overexpression of miR-4701-5p significantly inhibited luciferase reporter activities of PICSAR-WT, while no obvious impact on the activities of cells transfected with PICSAR-MUT was observed. Together, these data suggest that miR-4701-5p can directly interact with miR-4701-5p and serve as a negative upstream regulator of miR-4701-5p.
Fig. 8.
Screening and validation of miRNAs regulated by PICSAR in rheumatoid arthritis (RA) fibroblast-like synoviocytes (FLSs). (a) LncBase and starBase are used to predict miRNAs that can bind to PICSAR. A total of 3 miRNAs, common to both databases, can bind to PICSAR. (b) Quantitative real-time reverse transcription PCR (qRT-PCR) shows that miR-4701-5p expression was significantly up-regulated among 3 selected miRNAs after PICSAR knockdown in RA-FLSs. (c) Verification of interference efficiency by qRT-PCR. The specific mimic up-regulated miR-4701-5p expression in RA-FLSs. (d) The overexpression of miR-4701-5p lowered the level of PICSAR in RA-FLSs. (e) miR-4701-5p and PICSAR binding sequences and PICSAR mutant sequences. (f) Luciferase reporter assay was conducted to verify the targeting effect of miR-4701-5p on PICSAR sequence. All the results were presented as the mean ± standard deviation (S.D.) based on ≥ 3 replicates involving ≥ 3 samples. *P < 0.05, **P < 0.01, and ***P < 0.001 versus control groups.
3.9. The interaction between PICSAR and miRNA-4701-5p in RA-FLSs
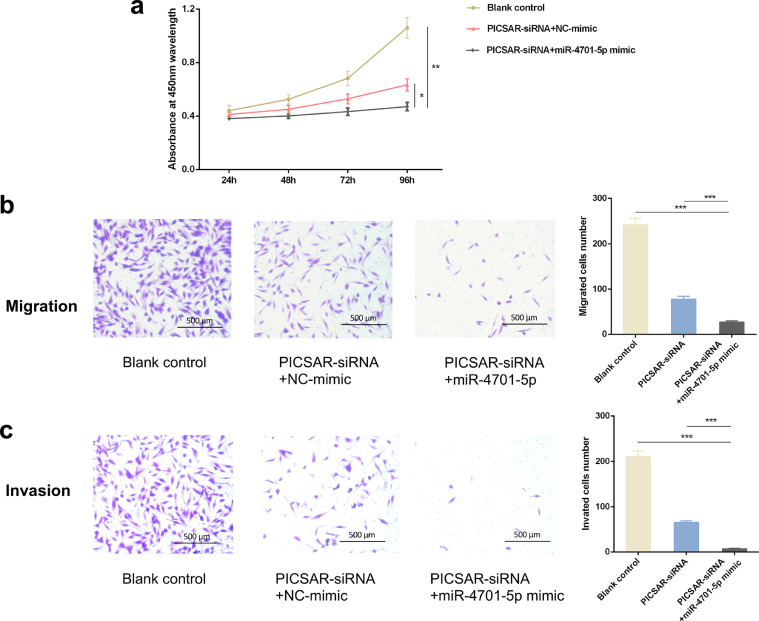
To investigate whether miR-4701-5p exerts biological effects via interacting with PICSAR, we performed CCK-8 experiments by upregulating the expression of miR-4701-5p in PICSAR silencing RA-FLSs. The results showed that the proliferation was further suppressed. The inhibition in growth rate was notably maintained (25.71 ± 2.48%, P < 0.05) at 96 h compared to the NC-mimic group and (55.61 ± 3.92%, P < 0.01) to the blank control (Fig. 9a). In addition, miR-4701-5p mimic could also further strengthen the effect of PICSAR knockdown on cell migration and invasion in RA-FLSs (Fig. 9b and c). These data intimate that the deceased expression of PICSAR caused by sponging miR-4701-5p could further modulate cell growth and mobility negatively.
Fig. 9.
PICSAR regulated cell proliferation, migration and invasion by suppressing miR-4701-5p in rheumatoid arthritis (RA) fibroblast-like synoviocytes (FLSs). (a) Overexpression of miR-4701-5p further suppressed the proliferation of RA-FLSs after PICSAR knockdown. (b and c) Evaluated by the quantification of transmembrane cells in Transwell assay and Matrigel Transwell assay respectively, suppressed migratory and invasive capacities of the PICSAR-depleted RA-FLSs were depressed further with miR-4701-5p up-regulation. All the results were presented as the mean ± standard deviation (S.D.) based on ≥ 3 replicates involving ≥3 samples. *P < 0.05, **P < 0.01, and ***P < 0.001 versus control groups.
We further conducted the Western Blot to test whether the target protein of miR-4701-5p such as ST3Gal I expression could be affected by the differential expression of lncRNA PICSAR. However, the protein ST3Gal I expression showed no significant difference after the knockdown of lncRNA PICSAR (Supplemental Fig. 1). Then, the expression of ST3GAL1 mRNA was detected in RA-FLSs by qPCR at 48 h after transfection of PICSAR-siRNA. Consistent with the result of protein expression, we observed no significant difference in the ST3GAL1 mRNA expression compared with negative control (Supplemental Fig. 2).
4. Discussion
RA, one of the most common rheumatic diseases, involves complex traits in which multiple genetic and environmental factors interact. As genetic factors have clear causal relationship to RA, it is important to explore the underlying mechanisms and pathways that lead to disease from a genomic standpoint [28]. Recently, there are increasing studies of complex traits of diseases and these have shown that many disease susceptibility variants regulate the expression levels of genes which act in a cell-specific manner and the epigenome is thought to play a significant role in this phenomenon [29]. With the continuous development of high-through put genomic technologies and epigenetic studies during the last decade, the study of lncRNAs has stimulated vigorous debate over the question of whether noncoding RNAs are truly functional biomolecules. It is obvious that there is no unifying answer because meaningful understanding of lncRNA function can only be achieved from associated in-depth studies [30].
However, there is growing evidence suggesting that a number of lncRNAs contribute to the pathogenesis of disease, especially cancer, at the levels of epigenetic modification, transcription, post-transcription, translation and post-translation [31]. To select lncRNAs related to rheumatic diseases, strategies were adopted as follows: using microarray and whole transcriptome sequencing to identify differentially expressed lncRNAs; investigating functional lncRNAs that already have an identified role in the immune system or disease-causing inflammatory pathways by qPCR; and identifying lncRNA genes containing disease-associated single nucleotide polymorphisms (SNPs) by genome-wide association studies (GWAS) [32]. In the field of RA research, recent studies have detected abnormal expression patterns of some lncRNAs in RA [33], [34], [35]. However, the overall pathophysiological contributions of lncRNAs to RA remain largely unknown. Therefore, further studies are urgently required to clarify the functions and underlying mechanisms of more disease-related lncRNAs in RA.
The use of FLSs is wildly applied in RA-related research because they permit evaluation of a homogeneous cell lineage of primary cells from the site of disease and play a critical role in rheumatoid joint damage [36]. In our present study, we conducted a lncRNA microarray between RA-FLSs and healthy control, then revealed a large amount of differentially expressed lncRNAs and mRNAs between two different groups. Moreover, we first identified a long intergenic non-protein-coding RNA 162 (LINC00162), also named as lncRNA PICSAR (p38 inhibited cutaneous squamous cell carcinoma associated lincRNA) or NLC1-C, which was found to have the most significant higher-than-normal expression levels in RA-FLSs and synovial fluid. PICSAR has also been reported to have expression and plays crucial roles in initiation and development of multiple human cancers. High-throughput sequencing of RNA was performed to identify LINC00162 differential expression between esophageal squamous cell carcinoma tissue samples and paired non-tumor tissues [37]. PICSAR was also discovered to be over-expressed in MDA-MB-231 cells which are the most common cell lines used in breast cancer research by RNA-seq [38]. Downregulation NLC1-C promotes cell proliferation by repressing miR-320a and miR-383 in testicular embryonal cell carcinoma [39]. Knockdown of LINC00162 inhibited the proliferation and migration of cutaneous squamous cells by suppressing ERK1/2 activity [40]. Furthermore, PICSAR downregulated the expression of α2β1 and α5β1 integrins to decrease adhesion and promoted the migration of cutaneous squamous cells [41]. Taken into consideration collectively, these observations raise the possibility that overexpressed of PICSAR in RA-FLSs could play a regulatory role in promoting cell pathological behaviors.
Given the compact size of the RNA effectors, RNAi experiments have been successfully scaled up for high throughput and pooled genetic screens for lncRNA function [42]. To investigate the role of PCSAR in the progression of RA, we performed the loss-of-function approach using specific chemically synthesized siRNA. Joint swelling and synovial hyperplasia, the significant features of RA, are caused by the inflammatory hyperplasia of synovial cells, especially the FLSs. The results of Western Blot, CCK-8 cell viability assay and cell cycle analysis showed PICSAR knockdown could markedly suppress the proliferation of RA-FLSs, indicating that PICSAR may play a significant regulatory role in the RA-FLSs growth. Additionally, RA-FLSs contribute to RA pathological process through the production and secretion of a variety of proinflammatory cytokines or proteinases such as IL-6, IL-8, and MMPs. A series of inflammatory reactions are stimulated by multiple proinflammatory cytokines in synovial fluid and MMPs promote the development of cartilage and joint damage [43,44]. The results showed that PICSAR suppression reduced IL-6, IL-8, and MMP-3 production of RA-FLSs, indicating another significant function of RA-FLSs affected by PICSAR. Cartilage erosion and bone destruction are the other main pathohistological properties of RA. Early inflammatory environments stimulate FLSs to migrate and invade intra-articular structures, leading to the damage of cartilage and subchondral bone eventually [45]. This suggests that the enhanced migratory and invasive ability of RA-FLSs may be major alterations during the pathogenetic process of RA. As in this study, the reduced numbers of transmembrane cells in Transwell chambers showed PICSAR knockdown could markedly suppress the migration and invasive capacity of RA-FLSs, suggesting that PICSAR may be involved in the regulation of RA-FLSs migration and invasion. These data imply that PICSAR positively regulates the proliferation, secretion and motility of FLSs in vitro and that the increased expression of PICSAR in RA-FLSs might contribute to increasing rheumatoid synovial hyperplasia and aggression, leading ultimately to joint destruction.
As our knowledge of the transcriptome space has expanded, it has become increasingly clear that lncRNAs harbor multiple recognition binding sites for microRNAs, functioning as competing endogenous RNAs (ceRNAs) or natural microRNA sponges to abolish the endogenous suppressive effect of these miRNAs on their targeted transcripts. Interactions between lncRNAs and miRNAs have been reported recently and have received extensive attention in various cancers and human development [46]. In the RA research during the past two years, a few studies have conducted analysis of ceRNA crosstalk about some interesting lncRNAs, such as HOTAIR, ZFAS1, GAPLINC [47], [48], [49]. Nevertheless, none of them used microarray to identify the most significant differentially expressed lncRNA. Therefore, in this study, we explored the underlying molecular mechanisms by which PICSAR regulates the proliferation and motility of RA-FLSs based upon this hypothesis.
The miRNAs that targeted PICSAR were predicted by molecular biological analysis. Among these candidate miRNAs, miR-4701-5p was selected as a superior target of PICSAR according to significant up-regulation after PICSAR knockdown. One research group has proved that miR-4701-5p directly targeted ST3GAL1 to regulate the susceptibility of CML cells to multiple chemotherapeutics in vitro [50]. Another study conducted miRNA microarray of peripheral blood CD14+ monocytes among Sjögren's syndrome, systemic lupus erythematosus, rheumatoid arthritis patients and healthy controls. Unlike in the SjS and SLE groups, miR-4701-5p expression level was downregulated in the RA group compared with healthy controls [51]. Based on these observations, miR-4701-5p could be highly related to cell pathologic behaviors in RA.
To verify the hypothesis, subsequent analyses showed that the overexpression of miR-4701-5p reduced the expression level of PICSAR in RA-FLSs and the targeting relationship between miR-4701-5p and PICSAR was confirmed through luciferase reporter assay. Moreover, miR-4701-5p overexpression can reinforce the effect of PICSAR knockdown on cell proliferation, migration and invasion in RA-FLSs. However, the expression of protein ST3Gal I and ST3GAL1 mRNA bears little relation to the suppression of PICSAR. These results indicate that PICSAR could sponge to miR-4701-5p, but does not affect ST3GAL1 in RA-FLSs that seems different from CML cells [50]. It is likely that miR-4701-5p targets on different gene(s) in RA-FLSs. It deserves a deep study in the future to search the exact target gene(s) of miR-4701-5p in RA-FLSs. Therefore, we propose herein a mechanism reported for the first time which PICSAR acts as a miR-4701-5p sponge to promote cell pathologic behaviors of RA-FLSs, thereby expediting the progression of RA.
In conclusion, we demonstrated that PICSAR could promote cell proliferation, migration, and invasion through sponging interaction with miR-4701-5p in RA-FLSs. Therefore, lncRNA PICSAR may play a valuable role as biomarker in RA and provide a new insight into the pathogenesis of RA.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
Acknowledgments
This work was supported by grants provided from the National Natural Science Foundation of China (81771750; 81671611 to YFP); Program from Guangdong Introducing Innovative and Entrepreneurial Teams (2016ZT06S252 to YFP); S.Z is supported from the NIH R01 AR059103 and R61 AR073409.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.024.
Contributor Information
Yun Feng Pan, Email: panyunf@mail.sysu.edu.cn.
Song Guo Zheng, Email: SongGuo.Zheng@osumc.edu.
Appendix. Supplementary materials
References
- 1.Smolen J.S., Aletaha D., Barton A., Burmester G.R., Emery P., Firestein G.S. Rheumatoid arthritis. Nat Rev Dis Primers. 2018;4:18001. doi: 10.1038/nrdp.2018.1. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Wang J., Xu Z., Huang F., Qian W., Ma J. Apremilast ameliorates experimental arthritis via suppression of Th1 and Th17 cells and enhancement of CD4+Foxp3+ regulatory T cells differentiation. Front Immunol. 2018;9:1662. doi: 10.3389/fimmu.2018.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aletaha D., Smolen J.S. Diagnosis and management of rheumatoid arthritis: a. review. JAMA. 2018;320(13):1360–1372. doi: 10.1001/jama.2018.13103. [DOI] [PubMed] [Google Scholar]
- 4.Kong N., Lan Q., Su W., Chen M., Wang J., Yang Z. Induced T regulatory cells suppress osteoclastogenesis and bone erosion in collagen-induced arthritis better than natural T regulatory cells. Ann Rheum Dis. 2012;71(9):1567–1572. doi: 10.1136/annrheumdis-2011-201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottini N., Firestein G.S. Duality of fibroblast-like synoviocytes in RA: passive responders and imprinted aggressors. Nat Rev Rheumatol. 2013;9(1):24–33. doi: 10.1038/nrrheum.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z., Bozec A., Ramming A., Schett G. Anti-inflammatory and immune-regulatory cytokines in rheumatoid arthritis. Nat Rev Rheumatol. 2019;15(1):9–17. doi: 10.1038/s41584-018-0109-2. [DOI] [PubMed] [Google Scholar]
- 7.Smolen J.S., Aletaha D., McInnes I.B. Rheumatoid arthritis. Lancet. 2016;388(10055):2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 8.Bartok B., Firestein G.S. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233(1):233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattaram P., Jones K. Regulation of fibroblast-like synoviocyte transformation by transcription factors in arthritic diseases. Biochem Pharmacol. 2019 doi: 10.1016/j.bcp.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu X.X., Wu Y.J., Zhang J., Wei W. T-cells interact with B cells, dendritic cells, and fibroblast-like synoviocytes as hub-like key cells in rheumatoid arthritis. Int Immunopharmacol. 2019;70:428–434. doi: 10.1016/j.intimp.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y., Pan Y.F., Xue Y.Q., Fang L.K., Guo X.H., Guo X. uPAR promotes tumor-like biologic behaviors of fibroblast-like synoviocytes through PI3K/Akt signaling pathway in patients with rheumatoid arthritis. Cell Mol Immunol. 2018;15(2):171–181. doi: 10.1038/cmi.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber L.C., Distler O., Tarner I., Gay R.E., Gay S., Pap T. Synovial fibroblasts: key players in rheumatoid arthritis. Rheumatology. 2006;45(6):669–675. doi: 10.1093/rheumatology/kel065. [DOI] [PubMed] [Google Scholar]
- 13.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 15.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jandura A., Krause H.M. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet. 2017;33(10):665–676. doi: 10.1016/j.tig.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Hung T., Wang Y., Lin M.F., Koegel A.K., Kotake Y., Grant G.D. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu G.C., Pan H.F., Leng R.X., Wang D.G., Li X.P., Li X.M. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015;14(9):798–805. doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Li Z., Li X., Jiang C., Qian W., Tse G., Chan M.T.V. Long non-coding RNAs in rheumatoid arthritis. Cell Prolif. 2018;51(1) doi: 10.1111/cpr.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson M.J., Jones S.W. Review: long noncoding RNAs in the regulation of inflammatory pathways in rheumatoid arthritis and osteoarthritis. Arthritis Rheumatol. 2016;68(11):2575–2583. doi: 10.1002/art.39759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aletaha D., Neogi T., Silman A.J., Funovits J., Felson D.T., Bingham C.O. Rheumatoid arthritis classification criteria: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann Rheum Dis. 2010;69(9):1580–1588. doi: 10.1136/ard.2010.138461. [DOI] [PubMed] [Google Scholar]
- 22.Falcon S., Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 23.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paraskevopoulou M.D., Vlachos I.S., Karagkouni D., Georgakilas G., Kanellos I., Vergoulis T. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosengren S., Boyle D.L., Firestein G.S. Acquisition, culture, and phenotyping of synovial fibroblasts. Methods Mol Med. 2007;135:365–375. doi: 10.1007/978-1-59745-401-8_24. [DOI] [PubMed] [Google Scholar]
- 27.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okada Y., Eyre S., Suzuki A., Kochi Y., Yamamoto K. Genetics of rheumatoid arthritis: 2018 status. Ann Rheum Dis. 2019;78(4):446–453. doi: 10.1136/annrheumdis-2018-213678. [DOI] [PubMed] [Google Scholar]
- 29.Consortium G. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang G., Lu X., Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Tang Y., Zhou T., Yu X., Xue Z., Shen N. The role of long non-coding RNAs in rheumatic diseases. Nat Rev Rheumatol. 2017;13(11):657–669. doi: 10.1038/nrrheum.2017.162. [DOI] [PubMed] [Google Scholar]
- 33.Song J., Kim D., Han J., Kim Y., Lee M., Jin E.J. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med. 2015;15(1):121–126. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y., Xu Y.Z., Sun N., Liu J.H., Chen F.F., Guan X.L. Long noncoding RNA expression profile in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Res Ther. 2016;18(1):227. doi: 10.1186/s13075-016-1129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou Y., Xu S., Xiao Y., Qiu Q., Shi M., Wang J. Long noncoding RNA LERFS negatively regulates rheumatoid synovial aggression and proliferation. J Clin Invest. 2018;128(10):4510–4524. doi: 10.1172/JCI97965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ai R., Laragione T., Hammaker D., Boyle D.L., Wildberg A., Maeshima K. Comprehensive epigenetic landscape of rheumatoid arthritis fibroblast-like synoviocytes. Nat Commun. 2018;9(1):1921. doi: 10.1038/s41467-018-04310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C.Q., Huang G.W., Wu Z.Y., Xu Y.J., Li X.C., Xue Y.J. Integrative analyses of transcriptome sequencing identify novel functional lncRNAs in esophageal squamous cell carcinoma. Oncogenesis. 2017;6(2):e297. doi: 10.1038/oncsis.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y., Ye P., Long X. Differential expression profiles of the transcriptome in breast cancer cell lines revealed by next generation sequencing. Cell Physiol Biochem. 2017;44(2):804–816. doi: 10.1159/000485344. [DOI] [PubMed] [Google Scholar]
- 39.Lu M., Tian H., Cao Y.X., He X., Chen L., Song X. Downregulation of miR-320a/383-sponge-like long non-coding RNA NLC1-C (narcolepsy candidate-regi-on 1 genes) is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Cell Death Dis. 2015;6:e1960. doi: 10.1038/cddis.2015.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piipponen M., Nissinen L., Farshchian M., Riihila P., Kivisaari A., Kallajoki M. Long noncoding RNA PICSAR promotes growth of cutaneous squamous cell carcinoma by regulating ERK1/2 activity. J Invest Dermatol. 2016;136(8):1701–1710. doi: 10.1016/j.jid.2016.03.028. [DOI] [PubMed] [Google Scholar]
- 41.Piipponen M., Heino J., Kahari V.M., Nissinen L. Long non-coding RNA PICSAR decreases adhesion and promotes migration of squamous carcinoma cells by downregulating α2β1 and α5β;1 integrin expression. Biol Open. 2018;7(11) doi: 10.1242/bio.037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S.J., Lim D.A. Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep. 2018;19(12) doi: 10.15252/embr.201846955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alam J., Jantan I., Bukhari S.N.A. Rheumatoid arthritis: recent advances on its etiology, role of cytokines and pharmacotherapy. Biomed Pharmacother. 2017;92:615–633. doi: 10.1016/j.biopha.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 44.Tolboom T.C., Pieterman E., van der Laan W.H., Toes R.E., Huidekoper A.L., Nelissen R.G. Invasive properties of fibroblast-like synoviocytes: correlation with growth characteristics and expression of MMP-1, MMP-3, and MMP-10. Ann Rheum Dis. 2002;61(11):975–980. doi: 10.1136/ard.61.11.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harre U., Schett G. Cellular and molecular pathways of structural damage in rheumatoid arthritis. Semin Immunopathol. 2017;39(4):355–363. doi: 10.1007/s00281-017-0634-0. [DOI] [PubMed] [Google Scholar]
- 46.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H.J., Wei Q.F., Wang S.J., Zhang H.J., Zhang X.Y., Geng Q. LncRNA HOTAIR alleviates rheumatoid arthritis by targeting miR-138 and inactivating NF-κB pathway. Int Immunopharmacol. 2017;50:283–290. doi: 10.1016/j.intimp.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 48.Ye Y., Gao X., Yang N. LncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Human Cell. 2018;31(1):14–21. doi: 10.1007/s13577-017-0179-5. [DOI] [PubMed] [Google Scholar]
- 49.Mo B.Y., Guo X.H., Yang M.R., Liu F., Bi X., Liu Y. Long non-coding RNA GAPLINC promotes tumor-like biologic behaviors of fibroblast-like synoviocytes as microRNA sponging in rheumatoid arthritis patients. Front Immunol. 2018;9:702. doi: 10.3389/fimmu.2018.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y., Luo S., Dong W., Song X., Zhou H., Zhao L. Alpha-2, 3-sialyltransferases regulate the multidrug resistance of chronic myeloid leukemia through miR-4701-5p targeting ST3GAL1. Lab Invest. 2016;96(7):731–740. doi: 10.1038/labinvest.2016.50. [DOI] [PubMed] [Google Scholar]
- 51.Williams A.E., Choi K., Chan A.L., Lee Y.J., Reeves W.H., Bubb M.R. Sjögren’s syndrome-associated microRNAs in CD14(+) monocytes unveils targeted TGFβ signaling. Arthritis Res Ther. 2016;18(1):95. doi: 10.1186/s13075-016-0987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.