Abstract
Background: Head and neck squamous cell carcinoma (HNSCC) is a disastrous disease with substantial morbidity and mortality. This study aims to explore the effective diagnostic and prognostic biomarkers for HNSCC.
Methods: MiRNA expression data and corresponding clinical information of HNSCC from The Cancer Genome Atlas (TCGA) database were analyzed comprehensively to identify the miRNAs with diagnostic and prognostic power. The predictive ability of different classifications was analyzed for the three-miRNA combinations. Diagnostic and prognostic value were then evaluated and verified in clinical patients.
Findings: 128 differentially expressed miRNAs in HNSCC tissues were identified in the TCGA dataset, and 10 miRNAs were finally selected for further study. Classification analysis developed a three-miRNA signature of hsa-mir-383, hsa-mir-615, and hsa-mir-877 with the best diagnosis power, which was verified in validation patients. Survival analysis indicated that different expression levels of hsa-mir-383, rather than that of hsa-mir-615 or hsa-mir-877 led to significantly different survival rates in both cohorts. Furthermore, the multivariate Cox hazards analysis suggested that the microRNA signature yielded statistical significance to predict clinical outcome independently from other clinical variables in validation patients.
Interpretation: A three-miRNA signature of hsa-mir-383, hsa-mir-615, and hsa-mir-877 may serve as an excellent diagnostic biomarker for HNSCC, and potential prognostic significance for HNSCC patients.
Funding: This work was supported by the grants of the National Natural Science Foundation of China (81901021), Key Research and Development Program of Shandong (2019GSF108277), China postdoctoral Scinence Foundation Grant (2019M652380), Fundamental Research Funds of Shandong University (2018CJ047).
Keywords: Head and neck squamous cell carcinoma, Microrna, Biomarker, Diagnosis, Prognosis
Abbreviations: HNSCC, Head and neck squamous cell carcinoma; miRNAs, MicroRNAs; TCGA, The Cancer Genome Atlas; RPM, reads per million miRNAs mapped; SVM, Support Vector Machine; RBF, radial basis function; ROC, Receiver operating characteristic; AUC, Area Under the Curve; OSCC, oral squamous cell carcinoma; LSCC, laryngeal squamous cell carcinomas; NPC, nasopharyngeal carcinoma; AATK, apoptosis-associated tyrosine kinase; CRC, colorectal cancer
Research in context.
Evidence before this study
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer globally with significant mortality. Despite the recent advances in treatments, there is still no obvious improvement in the five-year survival rate of HNSCC patients. In approximately forty-two percent of the patients, tumors have already developed to the advanced stage when diagnosed, with extensive lymph vascular invasion or distant metastasis. Therapy is often unsuccessful with a high postoperative recurrence rate and a poor prognosis. It is therefore imperative to explore more effective diagnostic and prognostic biomarkers for optimizing the management of HNSCC. Since the specific expression profiles of circulating miRNAs have been linked with the ability to act as biomarkers for the diagnosis, classification, surveillance, and prognosis evaluation of the tumor, we aimed to identify a panel of miRNA markers for evaluating the diagnostic and prognostic value in HNSCC.
Added value of this study
In this study, we first elucidated the differentially expressed miRNAs. We developed 3 miRNAs as a signature for diagnosis and prognostic biomarkers. We further examined the diagnosis and prognostic power in validation patients. These results indicate that the miRNA signature has potential diagnosis and prognostic significance for HNSCC.
Implications of all the available evidence
The miRNA signature in the present study was completely an optimal diagnosis and prognostic model for HNSCC, the prediction of HNSCC mortality was successfully developed and carefully evaluated. It provides a theoretical basis for the development of miRNA diagnostic reagents and the improvement of mortality evaluation.
Alt-text: Unlabelled box
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) refers to a group of malignant neoplasms, primarily deriving from the oral and nasal cavity, oropharynx, hypopharynx, and larynx, and is the sixth most common cancer globally with significant mortality [1]. Statistically, more than 650,000 new cases emerge, and 350,000 cases die from HNSCC worldwide annually [1,2]. Despite the recent advances in treatments including surgical skills, radiochemotherapy, and systematic drug therapy, there is still no obvious improvement in the five-year survival rate of HNSCC patients [3]. In approximately forty-two percent of the patients, tumors have already developed to the advanced stage when diagnosed, with extensive lymph vascular invasion or distant metastasis. Therapy is often unsuccessful with a high postoperative recurrence rate and a poor prognosis [4,5]. It is therefore imperative to explore more effective diagnostic and prognostic biomarkers for optimizing the management of HNSCC.
MicroRNAs (miRNAs), a class of small non-coding RNAs with a length of ~21 nucleotides, have been proven to play vital roles in the biological processes of eukaryotic organisms by regulating gene expression at the posttranscriptional level [6]. To date, the dysregulation of miRNAs has been widely involved in the tumorigenesis of many cancers, such as breast cancer [7], ovarian cancer [8], pancreatic cancer [9], lung cancer [10,11], colorectal cancer [12], cholangiocarcinoma [13], and so on. Furthermore, the specific expression profiles of circulating miRNAs have been linked with the ability to indicate the types or subtypes of tumors, and when quantified can act as biomarkers for the diagnosis, classification, surveillance, and prognosis evaluation of the tumor [14,15]. Many miRNAs participating in the pathogenesis of HNSCC have now been revealed [16], some of which are currently being evaluated for their diagnostic or prognostic performance [17]. However, none of these have been broadly used as an efficient biomarker in a clinical setting.
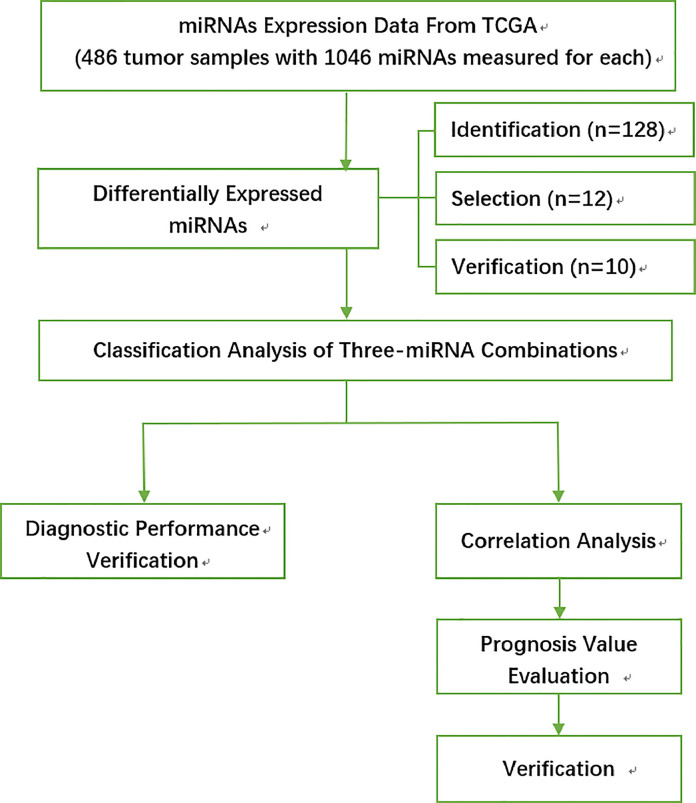
In the present study, to investigate potential miRNA biomarkers for HNSCC, we have comprehensively analyzed the miRNA expression data and corresponding clinical information HSNCC from The Cancer Genome Atlas (TCGA) database. Differentially expressed miRNAs were identified, and the diagnostic and prognostic value was evaluated. Results were further verified in clinical patients. A flowchart of this study is shown in Fig. 1. Finally, we determined a three-miRNA signature that may serve as a potential diagnostic and prognostic biomarker for HNSCC.
Fig. 1.
An overall flowchart of this work.
2. Materials and methods
2.1. Data acquisition from TCGA database
The miRNA expression data for HSNCC were downloaded from TCGA database. The HSNCC data contain 486 tumor samples and 44 normal samples, each measuring 1046 miRNAs. Specifically, we used the miRNA-seq data which record the RPM (reads per million miRNAs mapped) values. The data were then log2-transformed for the subsequent analysis. Clinical information for HSNCC was also obtained from the TCGA data portal.
2.2. Identification of differentially expressed miRNAs
Differential analysis was performed by the Wilcoxon test with the built-in R function “wilcox.test”. MiRNAs with a log-2 fold change ≥ 1 and adjusted p-value<0.05 were considered to be differentially expressed.
2.3. Classification prediction
Classification analysis was performed by Support Vector Machine (SVM) with R package e1071. For each classification task, the radial basis function (RBF) was chosen as the kernel function, and the best values of the two parameters cost (C) and gamma (γ) in the kernel function were obtained by a grid-search approach using cross-validation. Finally, the classification accuracy was evaluated by five-fold cross-validation.
2.4. Survival analysis
The survival analysis was carried out with the R package “survival”, in which the clinical information and the miRNA expression data were used. Specifically, in TCGA dataset, for each miRNA the tumor samples were divided into two groups according to their expression levels in individual samples compared with their mean expression level across all samples. In the validation patients, the cut-off was defined based on the ROC analysis at the highest Youden index when the log-rank test p-value was under 0.05. Otherwise, the median of the expression data was used for classifying HNSCC samples into patients with low or high miRNA expression.
2.5. Cox regression analysis
We evaluated the impact of miRNAs on survival time and clinical survival data by Cox proportional hazards regression analysis based on the R package “KMsurv”. Patients with a high-risk score were considered to have poor survival.
2.6. Functional analyses of miRNAs
The target genes of the miRNAs were obtained based on three databases, i.e. miRanda, miRDB, and TargetScan. Specifically, only the regulatory relationships recorded in all three databases were selected for further analysis. The Database for Annotation, Visualization, and Integrated Discovery (DAVID; https://david.ncifcrf.gov/) was applied to investigate the Gene Ontology (GO) functional analysis as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the identified target genes. p < 0.05 was chosen as the cut-off criterion.
2.7. Clinical patients and serum samples for validation
A total of 75 HNSCC patients and 92 normal controls were recruited in this study between July 1, 2016 and July 1, 2017 at the Shandong Provincial Hospital Affiliated to Shandong University for validation. The inclusion criteria for HNSCC patients were as follows: i) Patients with pathologically confirmed HNSCC; ii) patients who underwent curative surgical resection; and iii) patients >18 years old. The exclusion criteria were as follows: i) Patients who received preoperative chemotherapy or radiotherapy; and ii) patients with two or more primary tumors, asynchronously, or synchronously. The inclusion criteria for normal controls were as follows: i) without pathologically confirmed any type of cancer; ii) not underwent trauma or curative surgery, and iii) >18 years old. The exclusion criteria were as follows: i) pregnancy, and ii) with acute or chronic inflammation. For both patients and controls, 5 mL venous peripheral blood samples were collected in Vacutainer® serum separating tubes (BD Biosciences, Franklin Lakes, NJ, USA) before surgery. After coagulation by silence for 30 min, the blood samples were centrifugated at 2000 × g for 10 min at 4°C to obtain serum, which was preserved at −80 °C [18]. All patients and normal controls gave informed consent. This study was approved by the Medical Institutional Ethical Committee of the Provincial Hospital Affiliated to Shandong University.
2.8. miRNA extraction, reverse transcription, and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the prepared serum samples using the mercury RNA kit (Exiqon A/S, Vedbaek, Denmark). The SYBR II Prime Script miRNA RT-PCR Kit (Takara Bio, China) was used for reverse transcription and real-time PCR. RNU6B (U6) was used as an internal reference for normalization, relative to which the expression of miRNAs was calculated using the comparison 2-∆∆CT method [18]. Primers for miRNAs and U6 were purchased from Takara Biotechnology Co., Ltd. The manufacturer's instructions for each kit were followed.
2.9. Ethics statement
The study was approved by the Research Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong University. Informed consent was obtained from all patients and their families.
2.10. Statistical analysis
SPSS PASW Statistics v18.0(SPSS Inc., Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA) were used to analyzed and display the data. Processing procedures of data from TCGA database have been described above in the relevant sections. A receiver operating characteristic (ROC) curve analysis was performed, and the area under the ROC curve (AUC) was calculated to evaluate the diagnostic value. The optimal cut-off value, sensitivity, and specificity were determined by calculating the Youden index. The Kaplan–Meier method and log-rank test were used to evaluate the prognostic value. P < 0.05 was considered statistically significant.
3. Results
3.1. Clinicopathological characteristics
The clinicopathological characteristics of the patients in both the TCGA database and the validation patients were listed in Table 1.
Table 1.
Clinicopathological characteristics of HNSCC patients from two cohorts. A total of 75 HNSCC patients were recruited in this study between July 1, 2016 and July 1, 2017 at the Shandong Provincial Hospital Affiliated to Shandong University, while the TCGA samples were retrieved from the TCGA database.
| Characteristics | Validation patients (N = 75) |
TCGA patients (N = 486) |
||
|---|---|---|---|---|
| N | % | N | % | |
| Age(years) | ||||
| <60 | 32 | 42.7 | 215 | 44.3 |
| ≥60 | 43 | 57.3 | 270 | 55.7 |
| [Not Available] | 0 | 0.0 | 1 | 0.2 |
| Gender | ||||
| Male | 47 | 62.7 | 354 | 72.8 |
| Female | 28 | 37.3 | 132 | 27.2 |
| Lesion site | ||||
| Alveolar Ridge | 3 | 4.0 | 17 | 3.5 |
| Base of tongue | 8 | 10.7 | 24 | 4.9 |
| Buccal Mucosa | 13 | 17.3 | 19 | 3.9 |
| Floor of mouth | 4 | 5.3 | 58 | 11.9 |
| Hard Palate | 5 | 6.7 | 6 | 1.2 |
| Hypopharynx | 3 | 4.0 | 9 | 1.9 |
| Larynx | 5 | 6.7 | 106 | 21.8 |
| Lip | 10 | 13.3 | 3 | 0.6 |
| Oral Cavity | 3 | 4.0 | 70 | 14.4 |
| Oral Tongue | 13 | 17.3 | 125 | 25.7 |
| Oropharynx | 5 | 6.7 | 9 | 1.9 |
| Tonsil | 3 | 4.0 | 40 | 8.2 |
| HPV infection | ||||
| [Not Available] | 75 | 100 | 261 | 53.7 |
| [Not Evaluated] | 0 | 0.0 | 114 | 23.5 |
| [Unknown] | 0 | 0.0 | 8 | 1.6 |
| Positive | 0 | 0.0 | 64 | 13.2 |
| Negative | 0 | 0.0 | 39 | 8.0 |
| TNM staging | ||||
| [Discrepancy] | 0 | 0.0 | 4 | 0.8 |
| [Not Available | 0 | 0.0 | 65 | 13.4 |
| I | 22 | 29.3 | 26 | 5.3 |
| II | 41 | 54.7 | 71 | 14.6 |
| III | 9 | 12.0 | 76 | 15.6 |
| IV | 3 | 4.0 | 244 | 50.2 |
| Histological grade | ||||
| Good | 19 | 25.3 | [Not Available]: 4 | 0.8 |
| G1: 60 | 12.8 | |||
| Moderate | 47 | 62.7 | G2: 282 | 58.0 |
| G3: 117 | 24.1 | |||
| Poor | 9 | 12.0 | G4: 7 | 1.4 |
| GX: 16 | 3.3 | |||
| Tumor recurrence | ||||
| [Not Available] | 0 | 0.0 | 276 | 56.8 |
| [Unknown] | 0 | 0.0 | 14 | 2.9 |
| Yes | 19 | 25.3 | 49 | 10.1 |
| No | 56 | 74.7 | 147 | 30.2 |
| Death | ||||
| Yes | 28 | 37.3 | 155 | 31.9 |
| No | 47 | 62.7 | 331 | 68.1 |
| Smoke | ||||
| Yes | 20 | 26.7 | 357 | 73.5 |
| No | 55 | 73.3 | 116 | 23.9 |
| [Not Available] | 0 | 0.0 | 10 | 2.1 |
| [Unknown] | 0 | 0.0 | 3 | 0.6 |
| Alcohol | ||||
| Yes | 18 | 24.0 | 322 | 66.3 |
| No | 57 | 76.0 | 156 | 32.1 |
| [Not Available] | 0 | 0.0 | 8 | 1.6 |
3.2. Differentially expressed miRNAs in HNSCC patients from TCGA database
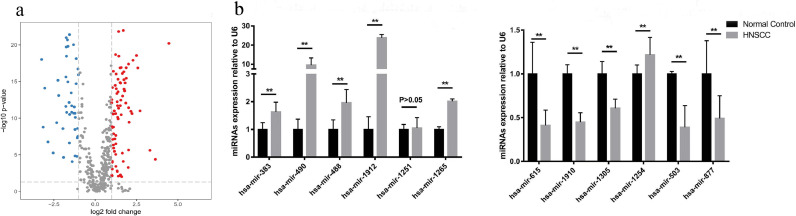
A total of 486 HNSCC samples and 44 normal samples were obtained from the TCGA database, with 1046 miRNAs measured for each. According to the differential analysis by the Wilcoxon test, we identified 128 miRNAs as significantly differentially expressed in HNSCC tissue compared with the normal samples, which is displayed in the volcano plot in Fig. 2a. By sorting the miRNAs according to their p-value, 30 miRNAs were then further selected from the pool of 128 as being more significantly differentially expressed (Table S1). Considering the remarkable expression changes of these 30 miRNAs in HNSCC patients, there is a strong likelihood that they participate in the pathogenesis of HNSCC to some extent. Based on our previous study and work by other authors, from these 30 more significantly differentially expressed miRNAs we selected 12 promising miRNAs for further study, including the up-regulated miRNAs, i.e. hsa-mir-383, hsa-mir-490, hsa-mir-488, hsa-mir-1912, hsa-mir-1251, hsa-mir-1265, and the down-regulated miRNAs, i.e. hsa-mir-615, hsa-mir-1910, hsa-mir-1305, hsa-mir-1254, hsa-mir-503, and hsa-mir-877.
Fig. 2.
(a). A volcano plot showing the log2 fold change of 128 significantly differentially expressed miRNAs in HNSCC patients from the TCGA database; (b) The expression level of the 12 miRNAs in the serum of 75 validation patients. Among the 6 up-regulated miRNAs, 5 miRNAs, i.e., hsa-mir-383, hsa-mir-490, hsa-mir-488, hsa-mir-1912, hsa-mir-1265, showed the same expression variation tendency as revealed by the bioinformatics analysis, while hsa-mir-1251 showed no significant expression difference (P>0.05). Among the 6 down-regulated miRNAs, 5 miRNAs, i.e., hsa-mir-615, hsa-mir-1910, hsa-mir-1305, hsa-mir-503, and hsa-mir-877, showed the same tendency, while hsa-mir-1254 showed the opposite.
3.3. Verification of the differentially expressed miRNAs in HNSCC patients from the clinic
For the purpose of verification, we collected 75 HNSCC patients and 92 normal controls from the clinic. We detected the expression levels of the 12 selected miRNAs in the serum of 75 validation patients and compared them with that of 92 normal controls. Results indicated that, among these 12 miRNAs, 10 miRNAs, i.e. hsa-mir-383, hsa-mir-490, hsa-mir-488, hsa-mir-1912, hsa-mir-1265, hsa-mir-615, hsa-mir-1910, hsa-mir-1305, hsa-mir-503, and hsa-mir-877, showed the same expression variation tendency as revealed by the bioinformatic analysis, while hsa-mir-1251 showed no significant expression difference (P > 0.05) and hsa-mir-1254 showed the opposite tendency (Fig. 2b). As a result, 10 differentially expressed miRNAs were verified in serum, not only further affirming their possible roles in HNSCC pathogenesis but also suggesting their potential capacity as biomarkers.
3.4. Classification analysis of the three-miRNA combinations in tissue from TCGA database
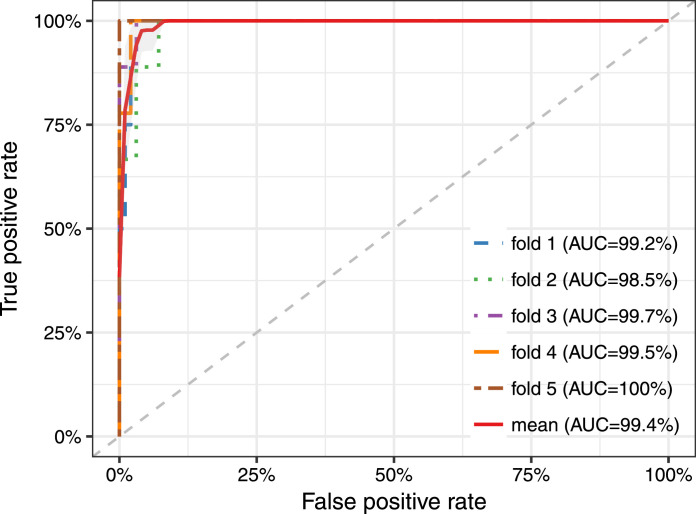
Before evaluating the diagnosis performance of these 10 miRNAs, the classification ability in differentiating tumor tissues from normal tissues was first analyzed using the samples from the TCGA database. To date, no single miRNA has been demonstrated to show a prominent diagnostic value, thus more attention was paid to their synergistic power. We picked three out of ten miRNAs each time and evaluated their classification ability by five-fold cross-validation. All of the 120 combinations were tested in turn (Table S2) and the top 30 combinations were listed in Table S3. Results suggested that the three-miRNA combination of hsa-mir-383, hsa-mir-615, and hsa-mir-877 demonstrated the best prediction accuracy with an average AUC (Area Under the Curve) of 99.4% (Fig. 3).
Fig. 3.
Classification accuracy of the combination of hsa-mir-383, hsa-mir-615 and hsa-mir-877 in TCGA dataset (486 cancer patients and 44 normal controls). The five-fold cross-validation result indicated that the combination of three miRNAs hsa-mir-383, hsa-mir-615, and hsa-mir-877 demonstrated the best prediction accuracy with an average AUC (Area Under the Curve) of 99.4%.
3.5. Diagnostic value of the combination of hsa-mir-383, hsa-mir-615, and hsa-mir-877 in serum
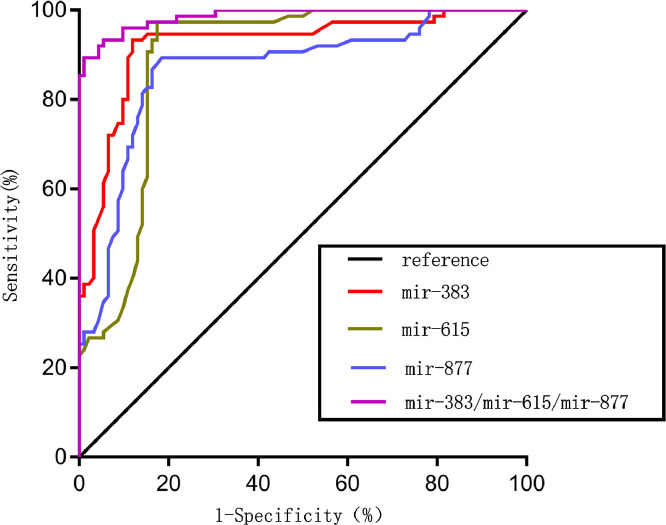
In order to explore the diagnostic value of the three-miRNA combination of hsa-mir-383, hsa-mir-615, and hsa-mir-877, we detected their expression level in the serum of 75 HNSCC patients and 92 normal controls and analyzed their synergistic diagnostic value. As shown in Fig. 4, the combination of the three miRNAs yielded an AUC value of 0.986 (95%CI, 0.973–0.999; P < 0.0001) with 89.3% sensitivity and 98.9% specificity in distinguishing patients with HSNCC from healthy subjects. For comparison, we also analyzed the diagnostic power of single miRNAs in isolation (Table 2). Specifically, each of the three miRNAs hsa-mir-383, hsa-mir-615, hsa-mir-877 yielded AUC values of 0.924 (95%CI, 0.880–0.967; P < 0.0001), 0.890 (95%CI, 0.837–0.942; P < 0.0001), and 0.868 (95%CI, 0.810–0.926; P < 0.0001) with 93.3%, 82.6%, 81.5% sensitivity and 88%, 97.3%, 89.3% specificity, respectively. We can see that the diagnostic power of the three miRNAs combined was superior to that of a single miRNA signature, and thus can serve as a potential effective diagnostic biomarker for HNSCC.
Fig. 4.
Diagnostic value of hsa-mir-383, hsa-mir-615, hsa-mir-877 and the combination of hsa-mir-383, hsa-mir-615, and hsa-mir-877 in serum. A receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic power of the miRNAs. Each of the three miRNAs hsa-mir-383, hsa-mir-615, hsa-mir-877 yielded AUC values of 0.924 (95%CI, 0.880–0.967; P < 0.0001), 0.890 (95%CI, 0.837–0.942; P < 0.0001), and 0.868 (95%CI, 0.810–0.926; P < 0.0001) with 93.3%, 82.6%, 81.5% sensitivity and 88%, 97.3%, 89.3% specificity, respectively, while the combination of the three miRNAs yielded an AUC value of 0.986 (95%CI, 0.973–0.999; P < 0.0001) with 89.3% sensitivity and 98.9% specificity in distinguishing patients with HSNCC from healthy subjects.
Table 2.
Diagnostic power of the single and three-miRNA signatures in validation patients. A receiver operating characteristic (ROC) curve analysis was performed, and the area under the ROC curve (AUC) was calculated to evaluate the diagnostic value. The optimal cut-off value, sensitivity, and specificity were determined by calculating the Youden index.
| miRNAs | Optimal cut-off value | Se (%) | Sp (%) | AUC | 95% CI | P |
|---|---|---|---|---|---|---|
| mir-383 | 1.175 | 93.3 | 88.0 | 0.924 | 0.880–0.967 | <0.0001 |
| mir-615 | 0.855 | 82.6 | 97.3 | 0.890 | 0.837–0.942 | <0.0001 |
| mir-877 | 0.715 | 81.5 | 89.3 | 0.868 | 0.810–0.926 | <0.0001 |
| mir-383/mir-615/mir-877 | 0.129 | 89.3 | 98.9 | 0.986 | 0.973–0.999 | <0.0001 |
Se, Sensitivity; Sp, Specificity; AUC, Area Under the Curve; CI, Confidence Interval.
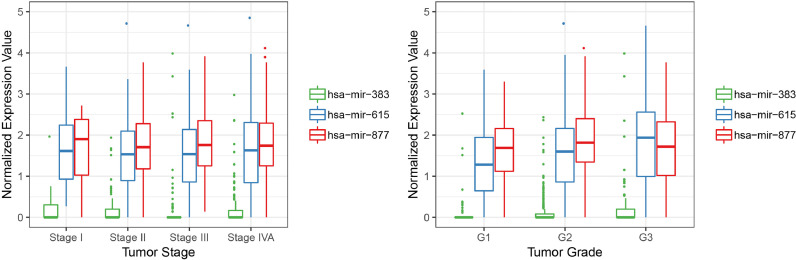
3.6. Correlations between hsa-mir-383, hsa-mir-615, or hsa-mir-877 and tumor stage or grade
The correlations between the hsa-mir-383, hsa-mir-615, or hsa-mir-877 expression levels and tumor stages or grades of HNSCC patients from the TCGA database were assessed. According to the one-way ANOVA test results (Fig. 5) for tumor stage, we obtained the p-values of 0.152, 0.982, and 0.762 for hsa-mir-383, hsa-mir-615, and hsa-mir-877, respectively, while for tumor grade, we obtained the p-values of 0.098, 0.003, and 0.082 for hsa-mir-383, hsa-mir-615, and hsa-mir-877, respectively. From the test results, we can see that the expression levels of hsa-mir-615 were significantly changed with different tumor grades, which indicated their potential functional role in tumor grade progression. We also analyzed the correlation between other clinical variables and miRNAs expression (such as HPV infection, clinical T, N, M, etiologic agents and so on), and the results were listed in Figure S1.
Fig. 5.
Correlations between hsa-mir-383, hsa-mir-615, or hsa-mir-877 and tumor stage or grade. One-way ANOVA test results suggested the expression levels of hsa-mir-615 was significantly changed with different tumor grades with the p-values of 0.003, while hsa-mir-383 and hsa-mir-877showed no significant correlation with tumor grade with the p-values of 0.098 and 0.082, respectively. No significant correlations were found between hsa-mir-383, hsa-mir-615, or hsa-mir-877 and tumor stage with the p-values of 0.152, 0.982, and 0.762, respectively.
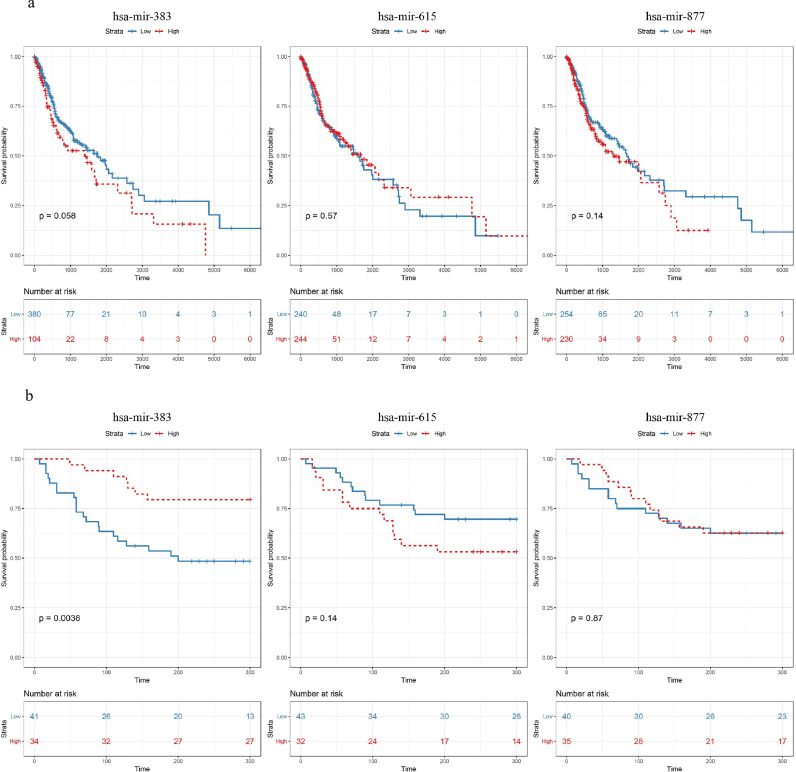
3.7. Prognostic value of hsa-mir-383, hsa-mir-615, or hsa-mir-877 in HNSCC
We performed the Kaplan–Meier survival analysis to examine the potential prognostic power of the three miRNAs (Fig. 6a). Notably, different expression levels of hsa-mir-383 have led to significantly different survival rates (log-rank test p-value = 0.058), while no significant survival differences were observed between the high and low expression set of hsa-mir-615 (log-rank test p-value = 0.57) or hsa-mir-877 (log-rank test p-value = 0.14). We then verified this result in validation patients. The 75 HNSCC patients recruited were also divided into a high expression group and a low expression group relative to the cut off expression level of the three miRNAs across all samples. Kaplan–Meier survival analysis and log-rank test results are shown in Fig. 6b. Similar to the results obtained from the TCGA database, low expression levels of hsa-mir-383 were significantly correlated with a poor prognosis (log-rank test p-value = 0.0038), while the expression levels of hsa-mir-615 or hsa-mir-877 showed no significant relationship to the prognosis (hsa-mir-615, log-rank test p-value = 0.14; hsa-mir-877, log-rank test p-value = 0.87).
Fig. 6.
Prognostic value of hsa-mir-383, hsa-mir-615, or hsa-mir-877 in both cohorts. (a) In TCGA patients, Kaplan–Meier Survival analysis result indicated that different expression levels of hsa-mir-383 led to significantly different survival rates (log-rank test p-value = 0.058), while no significant survival differences were observed between the high and low expression set of hsa-mir-615 (log-rank test p-value = 0.57) or hsa-mir-877 (log-rank test p-value = 0.14); (b). In validation patients, Kaplan–Meier survival analysis indicated that low expression level of hsa-mir-383 was significantly correlated with poor prognosis (log-rank test p-value = 0.0038), while the expression level of hsa-mir-615 or hsa-mir-877 showed no significant relationship with the prognosis (hsa-mir-615, log-rank test p-value = 0.14; hsa-mir-877, log-rank test p-value = 0.87).
As different expression levels of hsa-mir-383 have led to significantly different survival rates, we performed functional analysis on the target genes of miR-383. By selecting the regulatory relationships between miR-383 and its target genes that existed in three reliable miRNA-target interactions databases TargetScan, miRDB, and miRanda, a total of 252 genes were discovered to be regulated by miR-383 (Table S4). GO functional and KEGG pathway enrichment analysis of the target genes were shown in Table S5-S6.
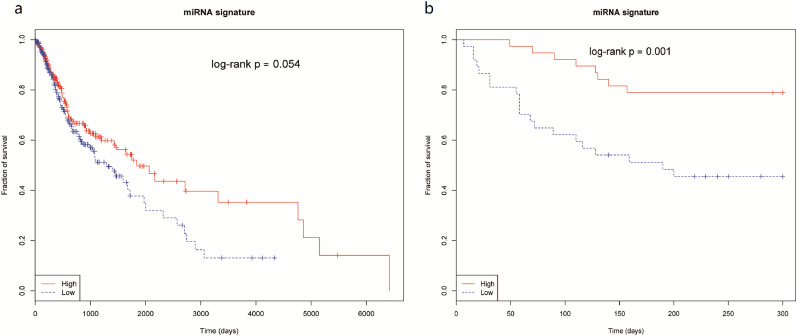
3.8. Prognostic value of hsa-mir-383, hsa-mir-615, and hsa-mir-877 as a signature in HNSCC
To evaluate the ability of these miRNAs as a signature to predict poor prognosis, we first carried out a univariate Cox proportional hazards regression analysis to evaluate the correlation between each of the three miRNAs and the survival outcome. The P values for outcome correlation were calculated using the Wald test. As a result, only mir-383 was significantly correlated with the overall survival in both HNSCC cohorts (Table 3). We then used the Z score from the Cox regression model as the coefficient for each miRNA and established a single prognostic model for the TCGA dataset and the validation patients respectively as follows:
Table 3.
Univariate Cox regression analysis to evaluate the prognostic value of each miRNAs. Beta and HR represent the regression coefficients and hazard ratio, respectively.
| miRNA | beta | waldScore | HR..95..CI.for.HR. | wald.test | p.value | |
|---|---|---|---|---|---|---|
| TCGA dataset | hsa-mir-383 | 0.15 | 1.9 | 1.2 (0.99–1.3) | 3.5 | 0.061 |
| hsa-mir-615 | −0.039 | −0.46 | 0.96 (0.81–1.1) | 0.21 | 0.65 | |
| hsa-mir-877 | 0.11 | 1 | 1.1 (0.91–1.4) | 1 | 0.31 | |
| validation patients | hsa-mir-383 | −2 | −3.4 | 0.14 (0.045–0.44) | 11 | 0.00079 |
| hsa-mir-615 | 1.2 | 1.2 | 3.2 (0.48–22) | 1.4 | 0.23 | |
| hsa-mir-877 | −0.38 | −0.48 | 0.68 (0.14–3.2) | 0.23 | 0.63 |
In the two prediction models above, a high-risk score predicts poor survival for patients. Moreover, to perform the Kaplan-Meier survival analysis and examine the potential prognostic power of three miRNAs as a signature in both datasets, we divided the patients from both cohorts into two subsets in terms of their median risk scores. Notably, in both the TCGA dataset and validation dataset, the miRNA signature showed significant survival differences between patients with high-risk score and those with low-risk score (log-rank test p-value = 0.054 for TCGA, and log-rank test p-value=0.001 for validation patients) (Fig. 7).
Fig. 7.
Prognostic value of hsa-mir-383, hsa-mir-615, and hsa-mir-877 as signature in both cohorts. Kaplan-Meier survival analysis was used to evalute the prognostic value of hsa-mir-383, hsa-mir-615 and hsa-mir-877 as a signature in (a) TCGA patients; (b) validation patients. The patients in each cohort were divided into either low-risk group or high-risk group based on the risk score. The p-values were obtained by log rank test.
Besides, a multivariate Cox regression analysis was performed to assess the independent prognostic power of the miRNA signature within the context of several common clinical parameters, including age at diagnosis, sex, smoking history, lesion site, alcohol history, stage and histological grade in both the TCGA dataset and validation patients. As expected, the miRNA signature was still found to be statistically significant in the multivariate survival analysis (Table 4, p-value = 0.019 for TCGA dataset and p-value= 0.002 for validation patients). Therefore, the miRNA signature could be considered as a potential prognostic indicator for the diagnosis of HNSCC.
Table 4.
Multivariate Cox regression analysis to evaluate the indepentent prognostic value of the miRNA signature and clinical parameters in both cohorts. HR represents the hazard ratio, and the p values were calculated using Wald test.
| Parameter | TCGA dataset |
Validation patients |
||
|---|---|---|---|---|
| HR | P-value | HR | P-value | |
| miRNA signature | 1.513445 | 0.019061 | 3.58578 | 0.00248 |
| Age | 1.347121 | 0.106108 | 1.17869 | 0.69827 |
| Sex | 1.224615 | 0.312622 | 0.78619 | 0.57673 |
| Stage(I/II/III vs IV) | 1.010694 | 0.95046 | 1.06384 | 0.81699 |
| Grade | 0.988297 | 0.904352 | 0.78029 | 0.48750 |
| Tobacco | 1.001398 | 0.985877 | 0.78070 | 0.57995 |
| Alcohol | 0.853304 | 0.394126 | 1.11181 | 0.81072 |
| Race | 0.797077 | 0.347236 | – | – |
| Lesion site | 0.990915 | 0.765786 | 0.98943 | 0.86104 |
4. Discussion
miRNAs can affect cell proliferation, differentiation, and apoptosis through restraining the post-transcriptional translation or causing the degradation of target mRNAs by base pairing to their 3′-untranslated region (3′-UTR), contributing to the pathogenesis of various diseases when their expression dysregulates, including cancers [19], [20], [21]. Studies revealed that circulating miRNAs secreted by tumor tissues or released from lytic cells due to apoptosis or necrosis are sufficiently stable in serum and display similar expression variation tendency as in tissue, which enables them to be used as tumor biomarkers [22]. In the present study, we identified 128 differentially expressed miRNAs in HNSCC tissues by analyzing the miRNA sequencing data downloaded from TCGA datasets, among which 10 miRNAs were finally selected for further study after a battery of comprehensive screening including p-value sorting, literature review, our previous findings and validation in serum. These miRNAs consisted of the up-regulated, i.e., hsa-mir-383, hsa-mir-490, hsa-mir-488, hsa-mir-1912, hsa-mir-1265, and the down-regulated, i.e., hsa-mir-615, hsa-mir-1910, hsa-mir-1305, hsa-mir-503, and hsa-mir-877. Considering the experiences from a previous study that showed that a combination of multiple miRNAs manifests more efficiently than a single miRNA as a diagnostic or prognostic biomarker [23], we developed a three-miRNA signature of hsa-mir-383, hsa-mir-615, and hsa-mir-877 with excellent diagnostic value in HNSCC patients. We also predicted the possible facilitating functions of the three miRNAs in the progression of tumor grades according to their correlativity and further demonstrated the potential prognostic significance of them for HNSCC patients.
Until now, several studies have focused on investigating differentially expressed miRNAs in HNSCC patients and attempted to evaluate their clinical significance for the diagnosis or prognosis of HNSCC [17]. Circulating miR-142-3p, miR-186-5p, miR-195-5p, miR-374b-5p, and miR-574-3p were found up-regulated in HNSCC plasma by the comparison of 18 patients and 12 healthy controls, of which miR-186-5p demonstrated the highest sensitivity (0.938) and specificity (0.917) to distinguish patients from healthy individuals, however, the diagnostic ROC curve and the AUC value were not reported. In addition, significant correlations were also found between the high expression of these five miRNAs and a poorer prognosis [24]. The other two up-regulated miRNAs, miR-21 and miR-26b, were also observed in HNSCC tissues and plasma. Furthermore, their decline after surgery was associated with a good prognosis, suggesting their potential prognostic roles [25]. There are also several miRNAs responsive to treatment, i.e., miR-99a, miR-21, miR-223, miR-425-5p, miR-21-5p, miR-106b-5p, miR-590-5p, miR-574-3p, and miR-885-3p, and can thus be considered as novel biomarkers for prognosis [26]. However, there were some limitations to these studies. Most of this research drew its conclusions based on limited samples, while some of them inferred the diagnostic or prognostic functions of miRNAs according to their expression variation compared to normal controls or pre-treatment without quantitative evaluation by performing a ROC curve or survival analysis. To overcome the limitation of sample size, we resorted to the TCGA database, which provided abundant resources of miRNA sequencing data and clinical information of HNSCC patients. Our TCGA-based results not only have strong reliability but are somewhat superior due to the increased sample size. What's more, our results were confirmed in a validation set from provided by a clinic. Therefore, we believe that the three-miRNA signature identified in the present study has enormous potential applications for diagnosing HNSCC and that hsa-mir-383, a component of this signature, maybe serve as a potential prognostic biomarker.
Previous studies have performed microRNA analysis in HNSCC based on TCGA datasets. Specifically, Wong et al. [27] screened the prognostic biomarkers of OSCC and OPCC in TCGA data and clinical samples respectively. And Hui et al. also carried out a research from the perspective of patients' prognosis, in which the most valuable microRNA was screened and its function was analyzed and studied accordingly [28]. Different from the previous two studies, although our research is also based on TCGA data, our focus is to find out the microRNAs that can distinguish cancer patients from normal samples. Moreover, in our study, we aimed to find a universal diagnostic or prognostic non-invasive biomarker for HNSCC instead of specific types of tumors. On this basis, we further studied and analyzed the relationship between the selected microRNAs and the development and prognosis of the tumor.
A major limitation of this study should not be ignored. HNSCC is a group of highly heterogeneous diseases originating from many sites. Several miRNAs differentially expressed in specific sites were identified. MiR-24, miR-181, miR-196a, miR-10b, and miR-187 were found at elevated levels in oral squamous cell carcinoma (OSCC) tissues and plasma, and miR-155 was up-regulated in laryngeal squamous cell carcinomas (LSCC) [29], [30], [31], [32], [33], [34]. Differentially expressed miRNAs, such as miR-17, miR-20a, mi-29cand miR-223 were reported in nasopharyngeal carcinoma (NPC) [35]. However, we left aside the difference of lesion site in this study, which may be one of the key answers to explain why a combination of miRNAs is superior to a single miRNA for the diagnosis of HNSCC, and we believe that further distinguishing the subtypes of HNSCC according to lesion sites can lead to a more accurate conclusion.
An understanding of the functions and mechanisms of miRNAs during the occurrence and progress of tumor growth can help make full use of their indicative function in diagnosis and prognosis. MiR-383, situated in the seventh intron of the apoptosis-associated tyrosine kinase (AATK) gene, has been identified as a tumor suppressor in several cancers. In colorectal cancer (CRC), miRNA-383 was demonstrated to inhibit tumor growth and metastasis by directly targeting Paired box 6 [36]. An inhibitory effect on cell invasion and metastasis of miR-383 was also illustrated in cervical cancer via suppression of the PI3K-AKT-mTOR signaling pathway by down-regulating PARP2 [37]. More mechanisms of the inhibitory roles of miR-383 were studied in other cancers, such as lung adenocarcinoma and human epithelial ovarian cancer [38,39]. However, the biological role of miR-383 in HNSCC remains unclear. In this study, we found that these miRNAs, which were up-regulated in HNSCC comparing with the control, were also linked with a good prognosis in HNSCC patients, and their high expression was correlated with a favorable prognosis, which indicated their potential role in strengthening the therapy sensitiveness of the HNSCC patients.
Yet our correlation analysis with tumor stage and grade failed to provide supporting information. MiR-615 and miR-877 were also demonstrated to act as tumor suppressors in several cancers via various mechanisms. For instance, miR-615 blocks the growth of tumor cells by targeting AKT2-related cell-cycle progression signaling pathways in breast cancer [40]. In non-small cell lung cancer, miR-615-3p restrains tumor growth and metastasis by inhibiting IGF2 [41]. MiR-877 inhibits the proliferation and migration abilities of renal cell carcinoma cells by modulating the eEF2K/eEF2 signaling cascade [42]. In the current study, the significant expression changes of hsa-mir-615 and hsa-mir-877 between HNSCC tissues with different tumor grades also suggested the potential function of them in HNSCC progression, which needs to be further studied in cellular experiments.
In summary, our study developed a three-miRNA signature for the diagnosis of HNSCC and revealed the prognostic value of hsa-mir-383 therein. We believe that all of the three miRNAs play roles in the pathogenesis of HNSCC and a preliminary correlation was observed between hsa-mir-383 and hsa-mir-383 with the tumor grade. However, there is still a long way to go before illuminating their concrete function and mechanisms in HNSCC.
Declarations of Competing Interests
The authors have no conflicts of interest to declare.
Acknowledgments
Acknowledgements
We thank all the members of the CSBL Lab at UGA for their help to Chao Liu on bioinformatics training.
Funding sources
This work is also supported by Qilu Young Scholar Program of Shandong University. https://doi.org/10.13039/501100001809. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.016.
Contributor Information
Chao Liu, Email: ibuliuchao@gmail.com.
Yanyan Jiang, Email: yanyan.jiang@sdu.edu.cn.
Dongsheng Zhang, Email: ds63zhang@sdu.edu.cn.
Appendix. Supplementary materials
References
- 1.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in globocan 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Marur S., Forastiere A.A. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91(3):386–396. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Matar N., Haddad A. New trends in the management of head and neck cancers. J Med Liban. 2011;59(4):220–226. [PubMed] [Google Scholar]
- 5.Cohan D.M., Popat S., Kaplan S.E., Rigual N., Loree T., Hicks W.L., Jr. Oropharyngeal cancer: current understanding and management. Curr Opin Otolaryngol Head Neck Surg. 2009;17(2):88–94. doi: 10.1097/moo.0b013e32832984c0. [DOI] [PubMed] [Google Scholar]
- 6.Krol J., Loedige I., Filipowicz W. The widespread regulation of microrna biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 7.Mulrane L., McGee S.F., Gallagher W.M., O'Connor D.P. Mirna dysregulation in breast cancer. Cancer Res. 2013;73(22):6554–6562. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 8.Cramer D.W., Elias K.M. A prognostically relevant mirna signature for epithelial ovarian cancer. Lancet Oncol. 2016;17(8):1032–1033. doi: 10.1016/S1470-2045(16)30149-8. [DOI] [PubMed] [Google Scholar]
- 9.Niu Y., Jin Y., Deng S.C., Deng S.J., Zhu S., Liu Y. Mirna-646-mediated reciprocal repression between hif-1alpha and miip contributes to tumorigenesis of pancreatic cancer. Oncogene. 2018;37(13):1743–1758. doi: 10.1038/s41388-017-0082-2. [DOI] [PubMed] [Google Scholar]
- 10.Zaporozhchenko I.A., Morozkin E.S., Ponomaryova A.A., Rykova E.Y., Cherdyntseva N.V., Zheravin A.A. Profiling of 179 mirna expression in blood plasma of lung cancer patients and cancer-free individuals. Sci Rep. 2018;8(1):6348. doi: 10.1038/s41598-018-24769-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Yang X., Xu Y., Cao J., Chen L. Genomic analysis of drug resistant small cell lung cancer cell lines by combining mrna and mirna expression profiling. Oncol Lett. 2017;13(6):4077–4084. doi: 10.3892/ol.2017.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostenfeld M.S., Jensen S.G., Jeppesen D.K., Christensen L.L., Thorsen S.B., Stenvang J. Mirna profiling of circulating epcam(+) extracellular vesicles: promising biomarkers of colorectal cancer. J Extracell Vesicles. 2016;5:31488. doi: 10.3402/jev.v5.31488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puik J.R., Meijer L.L., Le Large T.Y., Prado M.M., Frampton A.E., Kazemier G. Mirna profiling for diagnosis, prognosis and stratification of cancer treatment in cholangiocarcinoma. Pharmacogenomics. 2017;18(14):1343–1358. doi: 10.2217/pgs-2017-0010. [DOI] [PubMed] [Google Scholar]
- 14.Di Leva G., Croce C.M. Mirna profiling of cancer. Curr Opin Genet Dev. 2013;23(1):3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen J., Stass S.A., Jiang F. Micrornas as potential biomarkers in human solid tumors. Cancer Lett. 2013;329(2):125–136. doi: 10.1016/j.canlet.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen B., Schneider A., Victoria B., Nunez Lopez Y.O., Muller M., Szewczyk M. Blood serum from head and neck squamous cell carcinoma patients induces altered microrna and target gene expression profile in treated cells. Front Oncol. 2018;8:217. doi: 10.3389/fonc.2018.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arantes L., De Carvalho A.C., Melendez M.E., Lopes Carvalho A. Serum, plasma and saliva biomarkers for head and neck cancer. Expert Rev Mol Diagn. 2018;18(1):85–112. doi: 10.1080/14737159.2017.1404906. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Q., Sun X., Liu C., Li T., Cui J., Qin C. Expression of the microrna-143/145 cluster is decreased in hepatitis b virus-associated hepatocellular carcinoma and may serve as a biomarker for tumorigenesis in patients with chronic hepatitis b. Oncol Lett. 2018;15(5):6115–6122. doi: 10.3892/ol.2018.8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chekulaeva M., Filipowicz W. Mechanisms of mirna-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21(3):452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Micrornas Bartel DP. Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Carleton M., Cleary M.A., Linsley P.S. Micrornas and cell cycle regulation. Cell Cycle. 2007;6(17):2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 22.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K. Characterization of micrornas in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 23.Liang B., Zhao J., Wang X. A three-microrna signature as a diagnostic and prognostic marker in clear cell renal cancer: an in silico analysis. PLoS ONE. 2017;12(6) doi: 10.1371/journal.pone.0180660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Summerer I., Unger K., Braselmann H., Schuettrumpf L., Maihoefer C., Baumeister P. Circulating micrornas as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer. 2015;113(1):76–82. doi: 10.1038/bjc.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu C.M., Lin P.M., Wang Y.M., Chen Z.J., Lin S.F., Yang M.Y. Circulating mirna is a novel marker for head and neck squamous cell carcinoma. Tumour Biol. 2012;33(6):1933–1942. doi: 10.1007/s13277-012-0454-8. [DOI] [PubMed] [Google Scholar]
- 26.Hou B., Ishinaga H., Midorikawa K., Shah S.A., Nakamura S., Hiraku Y. Circulating micrornas as novel prognosis biomarkers for head and neck squamous cell carcinoma. Cancer Biol Ther. 2015;16(7):1042–1046. doi: 10.1080/15384047.2015.1045692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong N., Khwaja S.S., Baker C.M., Gay H.A., Thorstad W.L., Daly M.D. Prognostic microrna signatures derived from the cancer genome atlas for head and neck squamous cell carcinomas. Cancer Med. 2016;5(7):1619–1628. doi: 10.1002/cam4.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hui L., Wu H., Yang N., Guo X., Jang X. Identification of prognostic microrna candidates for head and neck squamous cell carcinoma. Oncol Rep. 2016;35(6):3321–3330. doi: 10.3892/or.2016.4698. [DOI] [PubMed] [Google Scholar]
- 29.Lin S.C., Liu C.J., Lin J.A., Chiang W.F., Hung P.S., Chang K.W. Mir-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol. 2010;46(3):204–208. doi: 10.1016/j.oraloncology.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Yang C.C., Hung P.S., Wang P.W., Liu C.J., Chu T.H., Cheng H.W. Mir-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J Oral Pathol Med. 2011;40(5):397–404. doi: 10.1111/j.1600-0714.2010.01003.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu C.J., Tsai M.M., Tu H.F., Lui M.T., Cheng H.W., Lin S.C. Mir-196a overexpression and mir-196a2 gene polymorphism are prognostic predictors of oral carcinomas. Ann Surg Oncol. 2013;20(3):S406–S414. doi: 10.1245/s10434-012-2618-6. Suppl. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y.C., Chen Y.J., Wang H.M., Tsai C.Y., Chen W.H., Huang Y.C. Oncogenic function and early detection potential of mirna-10b in oral cancer as identified by microrna profiling. Cancer Prev Res (Phila) 2012;5(4):665–674. doi: 10.1158/1940-6207.CAPR-11-0358. [DOI] [PubMed] [Google Scholar]
- 33.Liu C.J., Lin J.S., Cheng H.W., Hsu Y.H., Cheng C.Y., Lin S.C. Plasma mir-187* is a potential biomarker for oral carcinoma. Clin Oral Investig. 2017;21(4):1131–1138. doi: 10.1007/s00784-016-1887-z. [DOI] [PubMed] [Google Scholar]
- 34.Wang J.L., Wang X., Yang D., Shi W.J. The expression of microrna-155 in plasma and tissue is matched in human laryngeal squamous cell carcinoma. Yonsei Med J. 2016;57(2):298–305. doi: 10.3349/ymj.2016.57.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng X., Xiang J., Wu M., Xiong W., Tang H., Deng M. Circulating mir-17, mir-20a, mir-29c, and mir-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS ONE. 2012;7(10):e46367. doi: 10.1371/journal.pone.0046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan F., Tu Z., Duan L., Wang D., Lin F. Microrna-383 suppresses cell proliferation and invasion in colorectal cancer by directly targeting paired box 6. Mol Med Rep. 2018;17(5):6893–6901. doi: 10.3892/mmr.2018.8682. [DOI] [PubMed] [Google Scholar]
- 37.Teng P., Jiao Y., Hao M., Tang X. Microrna-383 suppresses the pi3k-akt-mtor signaling pathway to inhibit development of cervical cancer via down-regulating parp2. J Cell Biochem. 2018;119(7):5243–5252. doi: 10.1002/jcb.26585. [DOI] [PubMed] [Google Scholar]
- 38.Zhao S., Gao X., Zang S., Li Y., Feng X., Yuan X. Microrna-383-5p acts as a prognostic marker and inhibitor of cell proliferation in lung adenocarcinoma by cancerous inhibitor of protein phosphatase 2a. Oncol Lett. 2017;14(3):3573–3579. doi: 10.3892/ol.2017.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J., Dou Y., Sheng M. Inhibition of microrna-383 has tumor suppressive effect in human epithelial ovarian cancer through the action on caspase-2 gene. Biomed Pharmacother. 2016;83:1286–1294. doi: 10.1016/j.biopha.2016.07.038. [DOI] [PubMed] [Google Scholar]
- 40.Bai Y., Li J., Li J., Liu Y., Zhang B. Mir-615 inhibited cell proliferation and cell cycle of human breast cancer cells by suppressing of akt2 expression. Int J Clin Exp Med. 2015;8(3):3801–3808. [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J., Jia Y., Jia L., Li T., Yang L., Zhang G. Microrna-615-3p inhibits the tumor growth and metastasis of nsclc via inhibiting igf2. Oncol Res. 2018 doi: 10.3727/096504018X15215019227688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Q., Xu X., Liu Q., Luo F., Shi J., He X. Microrna-877 acts as a tumor suppressor by directly targeting eef2k in renal cell carcinoma. Oncol Lett. 2016;11(2):1474–1480. doi: 10.3892/ol.2015.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.