Abstract
Background
Rasal2 has diametric effects on progression of oestrogen receptor-positive (ER+) and -negative (ER–) breast cancers. The relevant causes are unknown. It is also unknown whether the effects of Rasal2 are mediated by an exosome-transport process.
Methods
Exosomes were purified from breast cancer cells and identified by transmission electron microscopy and flow cytometry analysis. In vivo and in vitro experiments were conducted to investigate the role of Rasal2 in exosome-mediated breast cancer progression. Western blot analysis was performed to detect Rasal2 and p-Rasal2 (phosphorylated Rasal2) expression in ER+/ER− breast cancer cells and in exosomes, cancer tissues and blood of patients with ER+ or ER− breast cancer.
Findings
Phosphorylation of Rasal2 at Serine 237 promoted tumour growth in both ER+ and ER− tumour cells and tissues. The functions of both p-Rasal2 and non-p-Rasal2 (non-phosphorylated-Rasal2) in the modulation of breast cancer progression are exosome-mediated. p-Rasal2 expression in ER+ breast cancer cells and exosomes, cancer tissues and blood was significantly lower than in ER− tumour cells and patients.
Interpretation
p-Rasal2 facilitates tumour progression in both ER+ and ER− breast cancers. The ratio of p-Rasal2/non-p-Rasal2 in ER+ and ER− breast cancers is one of the factors deciding the role of Rasal2 (or total Rasal2) as a suppressor in ER+ breast cancers or as a promoter in ER− breast cancers. Targeting the phosphorylation of Rasal2 machinery may therefore be useful as a therapy to restrain breast cancer progression by reducing p-Rasal2/non-p-Rasal2 ratio, especially in ER− breast cancers.
Fund
NSFC and Hong Kong Research Grants Council.
Keywords: ER+ and ER– breast cancer, Phosphorylation, Phosphorylated Rasal2 (p-Rasal2) and Rasal2, Tumour progression, Exosomal transport
Abbreviations: CM, Conditional medium; DLS, Dynamic Light Scattering; EMT, Epithelial-mesenchymal-transition; ERK, Extracellular signal-regulated kinase; ER+, Estrogen receptor-positive; ER–, Estrogen receptor-negative; EVs, Extracellular vesicles; MEK, Mitogen-activated extracellular signal-regulated kinase; non-p-Rasal2, Non-phosphorylated Rasal2; PP2Cβ, Protein phosphatase 2C beta (= PPM1B, metal-dependent protein phosphatase 1B; p-Rasal2, Phosphorylated Rasal2; p-Rasal2 (S237), Phosphorylation of Rasal2 at Serine 237; TEM, Transmission Electron Microscopy; TNBC, Triple-negative breast cancer
Research in context.
Evidence before this study
Rasal2, which encodes a RAS-GTPase-activating protein (RAS-GAP), functions as a tumour suppressor in luminal breast cancers which are typically oestrogen receptor-positive (ER+), or as a promoter in triple-negative or oestrogen receptor-negative (ER–) breast cancers (TNBC) which have a high incidence of early relapse and metastasis. The relevant causes of why Rasal2 plays diametrical effects in ER+ and ER–breast cancers are unknown. It is also unknown whether the effects of Rasal2 are mediated by an exosome-transport process.
Added value of this study
In the in vitro, in vivo and in patient experiments here, we demonstrate for the first time that the phosphorylation of Rasal2 (p-Rasal2) at S237 in PH domain facilitates tumour progression in both ER+ and ER– breast cancers. The ratio of p-Rasal2/non-p-Rasal2 in ER+ and ER– breast cancers is one of the factors deciding the role of Rasal2 (or total Rasal2) as a suppressor in ER+ breast cancers and as a promoter in ER– breast cancers. We also provide evidence that the functions of both p-Rasal2 and non-p-Rasal2 in the modulation of breast cancer progression are exosome-mediated.
Implications of all the available evidence
Targeting the phosphorylation of Rasal2 machinery may therefore be useful as a therapy to restrain breast cancer progression by reducing p-Rasal2/non-p-Rasal2 ratio, especially in ER–breast cancers.
CRediT authorship contribution statement
Wang Xuan: Conceptualization, Formal analysis, Investigation, Methodology. Qian Christopher: Formal analysis, Writing - review & editing. Yang Yinlong: Data curation, Formal analysis. Liu Meng-Yue: Investigation, Methodology. Ke Ya: Conceptualization, Funding acquisition, Supervision, Validation, Writing - original draft, Writing - review & editing. Qian Zhong-Ming: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Validation, Writing - original draft, Writing - review & editing.
Alt-text: Unlabelled box
1. Introduction
Breast cancer is one of the most common malignancies in women worldwide and remains the top cause of cancer death in females [1,2]. Transcriptional profiling studies demonstrate that breast cancer is an extremely heterogeneous disease, comprising a number of different subtypes [3], [4], [5]. However, the molecular basis of different types of breast cancers remain poorly understood. A better mechanistic understanding of the signals that drive the progression of breast cancer would not only help identify individuals who could benefit from additional up-front adjuvant treatment, but might also provide insight into new therapeutic strategies [6].
Rasal2, which encodes a RAS-GTPase-activating protein (RAS-GAP), has been demonstrated to function as a tumour and metastasis suppressor in luminal breast cancers [6,7] which are typically oestrogen receptor-positive (ER+) and represent the majority of breast cancers [6,8]. Additional findings reveal that Rasal2 plays the same role ininhibiting bladder cancer [9], renal cell carcinoma [10] and ovarian cancer progression [11]. As opposed to its role in luminal breast cancers (ER+), however, Rasal2 is found to be oncogenic in triple-negative or oestrogen receptor-negative (ER–) breast cancers (TNBC) which have a high incidence of early relapse and metastasis [12]. The oncogenic role of Rasal2 has also been found in colorectal cancer [13]. The relevant causes of why Rasal2 plays diametrical effects in ER+ and ER–breast cancers are unknown. We hypothesized that the phosphorylated Rasal2 might play a completely different role from Rasal2 or non-phosphorylated Rasal2 in breast cancer progression because reversible phosphorylation has been demonstrated to be a widespread molecular mechanism to regulate the function of cellular proteins [14,15]. Studies on the role of phosphorylated Rasal2 in breast cancer progression should be able to provide key insights into answering to the question of why Rasal2 plays diametrical effects in ER+ and ER– breast cancers.
It is also unknown whether the effects of Rasal2 on tumour growth, progression, and metastasis are mediated by an exosome-transport process. Growing evidence points to exosomes as key mediators of cell-cell communication, transferring their specific cargo (e.g., proteins, lipids, DNA and RNA molecules) from producing to receiving cells [16], [17], [18]. A number studies have found that the cancer-derived exosomes are important mediators of metastasis, angiogenesis and the tumour macro-/micro-environment [19], [20], [21]. These findings led us to speculate that Rasal2 affects breast cancer progression via exosome-transport.
The in vitro, in vivo and in patient experiments of the present study were designed to test these two hypotheses. We demonstrated for the first time that the phosphorylation of Rasal2 at S237 in PH domain facilitates tumour progression in both ER+ and ER– breast cancers. Our findings support the hypothesis that the difference in the ratio of p-Rasal2/non-p-Rasal2 (non-phosphorylated-Rasal2) in ER+ (< 1, p-Rasal2 expression is less than non-p-Rasal2 expression) and ER– (> 1, p-Rasal2 expression is more than non-p-Rasal2 expression) breast cancers is one of the factors deciding the role of Rasal2 (or total Rasal2) as a suppressor in ER+ breast cancers and a promoter in ER– breast cancers. We also provide evidence that the functions of both p-Rasal2 and non-p-Rasal2 in the modulation of breast cancer progression are exosome-mediated.
2. Materials and methods
2.1. Cell lines, animals and specimens
MDA-MB-231 (ATCC Cat# CRM-HTB-26, RRID:CVCL_0062) and MCF-7 (ATCC Cat# CRL-12584, RRID:CVCL_0031) cells, both obtained from the American Type Culture Collection, were maintained in RPMI-1640 and DMEM containing 10% foetal bovine serum (FBS) respectively. Female Balb/c nude mice (4–6-week-old, Sino-British SIPPR/BK Lab, Animal Ltd, China) were used for xenografts experiment. The use and care of experimental animals was approved by the Institutional Animal Care and Use Committee (IACUC), Roche R & D Centre, Shanghai, China. Cancer tissues and adjacent normal tissues were obtained from 20 pairs of patients with ER+ or ER– breast cancers. Whole blood (1–1.5 mL) was collected from 10 patients with ER+ or ER– breast cancers and 10 healthy controls, and then centrifuged at 3000 g for 10 min. The supernatant was stored at −80 °C. Information on all the patients was provided by the Fudan University Affiliated Tumour Hospital.
2.2. Reagents
Unless otherwise stated, all chemicals were obtained from the Sigma Chemical Company, St. Louis, MO, USA. Exosome-Human CD63 Isolation/Detection Reagent was purchased from Invitrogen, Carlsbad, CA, USA; anti-E-cadherin (BD Biosciences Cat# 560061, RRID:AB_1645347) from BD Biosciences, Franklin Lakes, NJ, USA; anti-Rasal2 (Novus Cat# NBP1-19149, RRID:AB_2284853) from NOVUS Biologicals, Colorado, USA; anti-Myc from Santa Cruz Biotechnology, Delaware, USA; anti-CD63 (Abcam Cat# ab59479, RRID:AB_940915) and pp2cβ (Abcam Cat# ab58146, RRID:AB_944952) from Abcam, Cambridge, MA, USA; the antibodies against vimentin (Cell Signalling Technology Cat# 3295, RRID:AB_2216129), against ERK (extracellular signal-regulated kinase)(Cell Signalling Technology Cat# 9107, RRID:AB_10695739), phospho-ERK (p-ERK)(Cell Signalling Technology Cat# 9101, RRID:AB_331646), snail (Cell Signalling Technology Cat# 3895, RRID:AB_2191759), MEK (Mitogen-activated extracellular signal-regulated kinase) (Cell Signalling Technology Cat# 9122, RRID:AB_823567), phospho-MEK (p-MEK)(Cell Signalling Technology Cat# 9120, RRID:AB_330745) from Cell Signalling Technology, Beverly, MA, USA. Rasal2 S237 phospho-specific antibody was produced by Youke Biosciences, Shanghai, China. All of the secondary antibodies were purchased from Jackson Immuno Research, West Grove, PA, USA, except PE Goat anti-mouse secondary antibody which was obtained from Biolegend, California, USA.
2.3. Construction of stable Rasal2 knockout (KO) cell lines by crispr/cas9 technique and establishment of Rasal2 S237D and S237A mutant by transfection
Rasal2 KO cell lines were generated by using a lentiCRISPRv2-sgRNA-RASAL2 plasmid (5′-CCTGAGATACTCCGTCTACG-3′) purchased from Addgene, Cambridge, MA, USA. Empty vector was transfected into cells as control (CT). Stable cell lines were established by selection with puromycin. Myc-Rasal2 (WT, S237D, S237A) plasmids were transfected into Rasal2 KO cells. Stable cell lines were established by choice with geneticin (G418).
2.4. Conditional medium (CM)
Conditional medium (CM) or exosome-depleted medium was prepared as described by Takasugi et al. [22]. In brief, the serum was centrifuged at 100,000 g for 16-h. The supernatant was filtered through a 0.22-μm filter (Merck Millipore, Billerica, USA), and then was diluted with a medium supplemented with all the nutrients and antibiotics to reach about 10% FBS concentration. The ultracentrifugation was done in polycarbonate tubes with a P70AT fixed-anglerotor.
2.5. Exosome purification
Exosomes were purified from the cell culture supernatant as described by Mo et al. [23]. In brief, CM was first treated at 300 g for 10-min to remove cells, and then centrifuged at 2000 g for 20-min to remove dead cells. The debris in the medium were removed by centrifugation at 10,000 g for 30-min, the supernatant was then subjected to ultracentrifugation at 100,000 g for 70-min. The pelleted EVs were re-suspended in PBS and centrifuged again at 100,000 g for 70-min. All centrifugations were carried out at 4 °C.
2.6. Transmissionelectron microscopy
For transmission electron microscopy (TEM), purified exosomes from cells were re-suspended in PBS and imaged as detailed by Jia et al. [24]. Briefly, EV samples were added onto formvar coated copper grids for 2-min, then washed in ultrapure water and negatively stained with 2% phosphotungstic acid (PTA). The samples were visualised using a Phillip CM120 transmission electron microscope (Eindhoven, The Netherlands) and photographed by a Gatan CCD camera (Gatan, Pleasanton, CA).
2.7. Dynamic light scattering
The size distribution of exosomes was determined by dynamic light scattering (DLS) on a Zetasizer Nano ZS90 Malvern Instrument as described by Lason et al. [25]. In brief, EV samples were diluted in PBS to generate suitable scattering intensity. The samples were automatically introduced into the sample chamber. Each sample was analysed in triplicate, and each replicate was measured six times.
2.8. Flow cytometry analysis of exosomes-bound dynabeads
Dynabeads® magnetic beads were incubated with 25 μg of EVs overnight, then washed by isolation buffer and incubated with PE Goat anti-mouse secondary antibody (Biolegend, 3 μl of antibody in 20 μl of 2% BSA). Secondary antibody incubation alone was used as control. The percentage of positive beads was calculated relative to the total number of beads analysed per sample [26].
2.9. Western blot analysis
The identification of exosomes was also confirmed by measuring expression of exosome-specific markers CD63 by western blot analysis.EVs in PBS were added 1:1 to 2× RIPA lysis buffer supplemented with protease and phosphatase inhibitor cocktail and PMSF. Western blot samples were then processed as described by Du et al. [27] and Sheller et al. [28]. The following anti-human antibody was used for western blot: exosome marker CD63 (Abcam, Cambridge, MA, USA) diluted 1:500.
2.10. Exosome labelling and uptake assay
Exosomes were labelled with PKH26 fluorescent dye using the PKH26 fluorescent cell linker kit [29]. Briefly, exosome pellets and 4 µl PKH26 were suspended in 1 ml diluent C respectively. The exosome suspension was then mixed with the stain solution and incubated for 4-min. The labelling reaction was stopped by adding an equal volume of 1% BSA. Labelled exosomes were ultra-centrifuged at 100,000 g for 70-min, washed with PBS, ultra-centrifuged again and then filtrated through 0.22 μm filter. MCF-7 and MDA-MB-231 cells were then co-cultured with their autologous exosomes, individually. The uptake was observed by a Leica TCS SP8 X confocal microscope (Leica, Germany) as described byMuller et al. [30] and Kim et al. [31].
2.11. Cell counting kit-8 (CCK-8) assay
Cell proliferation was determined by Cell Counting Kit-8 (CCK-8, Dojindo Laboratories), as described by Wang et al. [32]. Briefly, tumour cells were seeded in 96-well plates in triplicate prior to treat with autologous exosomes, respectively. After 24-h treatment, 10 μl of CCK-8 solution was added to each well in the plates, followed by incubation for 2-h. The absorption value was measured at 450 nm and the cell proliferation rate [(%) = [(ODExo group − ODblank group) − (ODPBS group − ODblank group)]/(ODPBS group − ODblank group) × 100%] was calculated using the data obtained from the wells [33].
2.12. Transwell migration assay
Migration assay was conducted in 24-well transwell plate as described by Shao et al. [34]. In brief, a total of 0.5 ml DMEM containing 10% FBS was added to the lower chamber at 37 °C and 0.2 ml (1 × 106 cells/ml) of cell solution containing 0.05 μg/μl exosomes was seeded into the upper chamber in serum-free culture medium. Cells were incubated for 24-h at 37 °C, subsequently fixed with paraformaldehyde, stained using 0.1% crystal violet for 15-min and photographed by Leica DM500 microscope (Leica, Germany). All experiments were performed independently in triplicate.
2.13. Matrigel invasion assay
Invasion assay was performed by using the Cultrex Cell Invasion Assay kit (RandD Systems) as described by Yang et al. [35]. Briefly, serum-starved cells in 50 μL serum-free medium with exosomes were placed in the top chamber, the lower chamber was filled with 10% FBS medium. After 24-h incubation, invaded cells on the upper chamber of the membrane were dissociated with cell dissociation solution containing Calcein AM at 37 °C for 1-h and then read the bottom plate at 485 nm excitation and 520 nm emission.
2.14. Wound-healing assay
For wound-healing assay, cells were plated into six-well plates as described by Jia et al. [36]. Briefly, the cell monolayers were wounded with a pipette tip to draw a gap on the plates. After being treated with tumour-derived exosomes for 24-h, cells migrated into the cleared section were observed by an inverted microscope (Leica, Germany). Image-J software was used to measure the area of scratches before and after incubation. The wound closure percent was calculated: (the scratch area before incubation – the scratch area after incubation)/(the scratch area before incubation) × 100%.
2.15. Immunohistochemical analysis
Cancer tissues and adjacent normal tissues from the patients with ER+ and ER– breast cancer were used to detect the expression of Rasal2 by immunohistochemical analysis as described by Du et al. [37] and Yan et al. [38]. Briefly, the slides were incubated with primary antibody referred above (1:200), followed by incubation with horseradish peroxidase-conjugated secondary antibody. Finally, the staining processes were performed with diaminobenzidine colorimetric reagent solution and hematoxylin (Sigma Chemical Co, USA). Images were captured with Aperio Scan-Scope AT Turbo (Aperio, USA). For evaluation of the staining score, intensity was graded from 0-iii (0: no staining; i: weak staining; ii: moderate staining; and iii: strong staining) and the abundance of positive cells was graded from 0 to iv (0: <5% positive cells; i: 5–25% positive cells; ii: 26–50% positive cells; iii: 51–75% positive cells; and iv: >75% positive cells) [39]. The staining score was obtained by multiplying the two values.
2.16. Human xenograft assay
To assess the effects of exosomes on tumour growth in vivo, female Balb/c nude mice underwent breast pad injections with 1.5 × 106 MDA-MB-231 cells and MCF-7 cells in 100 μL PBS, respectively [40]. Exosomes (5 μg) were injected into mice in tail vein for 2 times a week continued for two months. Mice were observed for increases in tumour volume to assess tumour growth. Tumour diameters were measured at regular intervals and tumour volumes were calculated as volume = (width) 2 × length/2.
2.17. Ethics statement
Informed consent was acquired from all patients with approval from the Institute Research Ethics Committee of the Fudan University Affiliated Tumour Hospital. All animal care and experimental protocols were performed according to the Animal Management Rules of the Ministry of Health of China, and approved by the Animal Ethics Committees of Fudan University and The Chinese University of Hong Kong.
2.18. Statistical analysis
Data were presented as the mean ± S.D. and analysed using the Graphpad software (La Jolla, CA, USA). Differences between the means from the groups were determined using Student t-test. P < 0.05 was considered to indicate a statistically significant difference.
3. Results
3.1. The expression of Rasal2 in ER+ and ER–breast cancer tissues
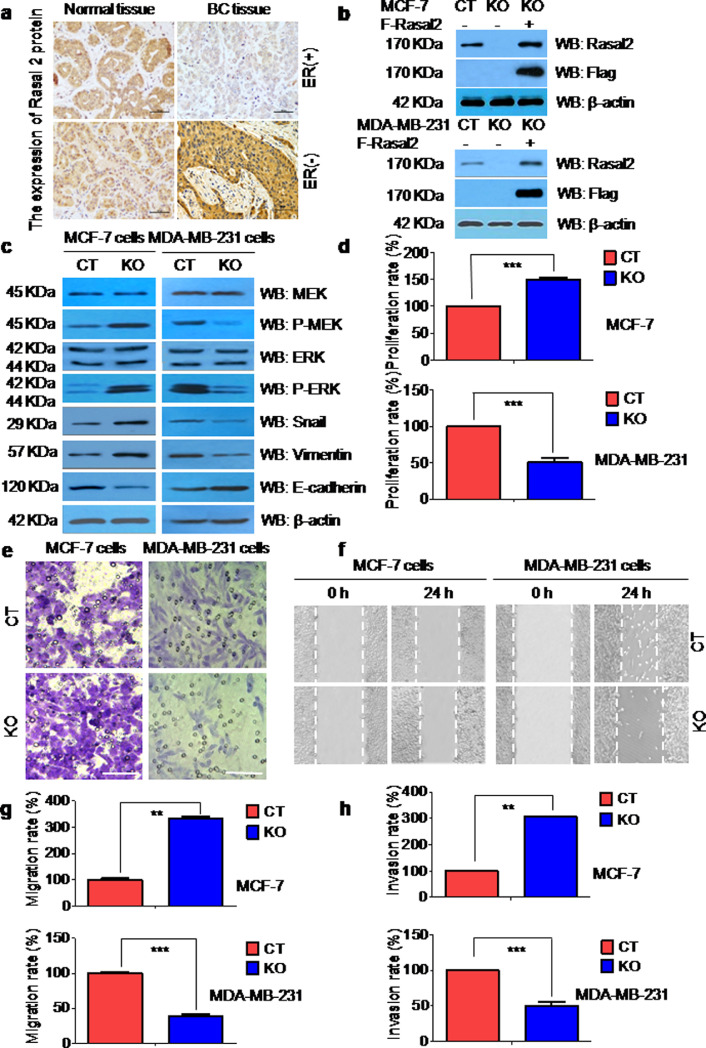
We first investigated the expression of Rasal2 in the cancer and adjacent normal tissues of the patients with breast cancers by an immunohistochemical analysis. Rasal2 expression was significantly lower in ER+breast cancer tissues, and higher in ER–breast cancer tissues when compared to adjacent normal tissues (Fig. 1(a)) and also correlated with ER status in breast cancer specimens (Supplementary Figure 1). The findings were consistent with those reported by others [6,12].
Fig. 1.
(a) The expression of Rasal2 in ER+ and ER–breast cancer tissues. Representative images of immunohistochemistry (IHC) analysis of Rasal2 expression in the tumour tissues and adjacent normal tissues of ER+ and ER–human breast cancer (scale bar: 100 μm). (b)–(h) The expression and roles of Rasal2 in ER+MCF-7 and ER–MDA-MB-231 breast cancer cells. (b) Construction of stable Rasal2 knockout (KO) MCF-7 cells and MDA-MB-231 cells using Crispr/cas9 technique. Immunoblot analysis for Rasal2 and Flag was used to detect the KO efficiency in MCF-7 cells and MDA-MB-231 cells; (c) Rasal2 affects diametrically Epithelial-Mesenchymal Transition (EMT)-related proteins expression in MCF-7 and MDA-MB-231 cells. EMT-related proteins in the lysates of control (CT) and KO MCF-7 and MDA-MB-231 cells were analysed by immunoblot analysis; (d)–(h) Rasal2 regulates diametrically tumour cell proliferation, migration and invasion in MCF-7 cells and MDA-MB-231 cells. The proliferation rate in CT and KO MCF-7 and MDA-MB-231 cells was examined by CCK-8 assay, the migration rate by wound healing assay and transwell migration assay, and the invasion rate by matrigel invasion assay (Scale bar: 100 μm). Data are presented as the mean ± S.D. (n = 3) ** p < 0.01, *** p < 0.001 vs. CT (t-test, two-tailed).
3.2. The expression and roles of Rasal2 in ER+MCF-7 and ER–MDA-MB-231 breast cancer cells
To find out the effects of Rasal2 KO on the progression of breast cancer in vitro, we first constructed stable Rasal2 KO cell lines in ER+MCF-7 and ER–MDA-MB-231 cells using a CRISPR Cas9 technique. Western blot analysis showed that there was no expression of Rasal2 in KO cells, however, the expression was recovered again in KO cells transfected with Flag-Rasal2 plasmid, demonstrating the establishment of the constructs (Fig. 1(b)). We then examined the expression of tumour cell progression-related proteins p-MEK, p-ERK, snail, vimentin and E-cadherin in these two KO cell lines by immunoblot assay. It was found that the expression of p-MEK, p-ERK, vimentin, snail was significantly higher and E-cadherin lower in ER+MCF-7 cells compared to CT cells, while in ER–MDA-MB-231 cells, the expression of p-MEK, p-ERK, vimentin, snail was significantly lower and E-cadherin higher compared to CT cells (Fig. 1(c)). Also, we conducted CCK-8 (Fig. 1(d)), transwell migration (Fig. 1(e)), wound-healing (Fig. 1(f)–(g)), and invasion assays (Fig. 1(h)) and demonstrated that proliferation (Fig. 1(d)), migration (Fig. 1(e)–(g)) and invasion rates (Fig. 1(h)) in ER+MCF-7 cells were significantly higher, and in ER–MDA-MB-231 cells lower than those in the corresponding CT cells.
3.3. Breast cancer cells secreted extracellular vesicles (EVs)
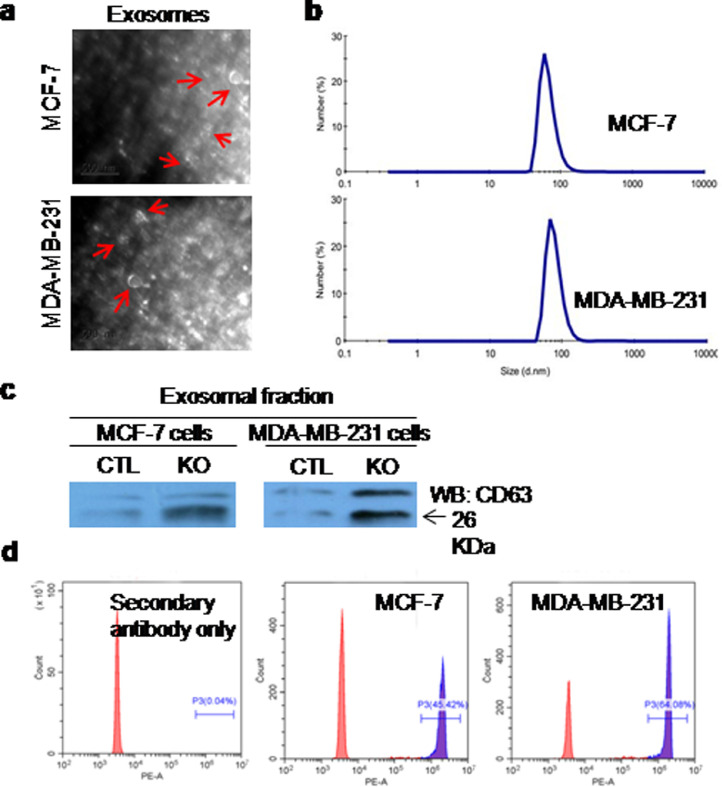
In order to know whether exosomes could be released by breast cancer cells, we cultured ER+MCF-7 cells and ER–MDA-MB-231 cells in CM for 24-h, and then isolated EVs (or exosomes) from CM by ultracentrifugation. Transmission electron microscope (TEM) analysis showed that the shape of EVs were spherical (Fig. 2(a)), and size were approximately 30–150 nm. Dynamic light scattering identified that the diameters of EVs were about 30–100 nm (Fig. 2(b)), which coincided with the results of TEM. The immunoblot analysis demonstrated the positive expression of exosome-specific surface protein marker CD63 in EVs (Fig. 2(c)). Flow cytometry analysis of exosomes-bound dynabeads also detected CD63+ exosomes (Fig. 2(d)).
Fig. 2.
Identification of the extracellular vesicles (EVs) or exosomes derived from breast cancer cells. (a) The shape of EVs was observed by transmission electron microscopy (TEM). Scale bar: 500 nm; (b) The size distribution of EVs was determined by dynamic light scattering (DLS) with Malvern Zetasizer Nano ZS90; (c) the expression of exosomal surface protein marker CD63 was detected by immunoblot analysis; (d) exosome-specific biomarker CD63 was analysed by flow cytometry. Negative control: secondary antibody alone.
3.4. Rasal2 regulated tumour progression via exosome in vitro
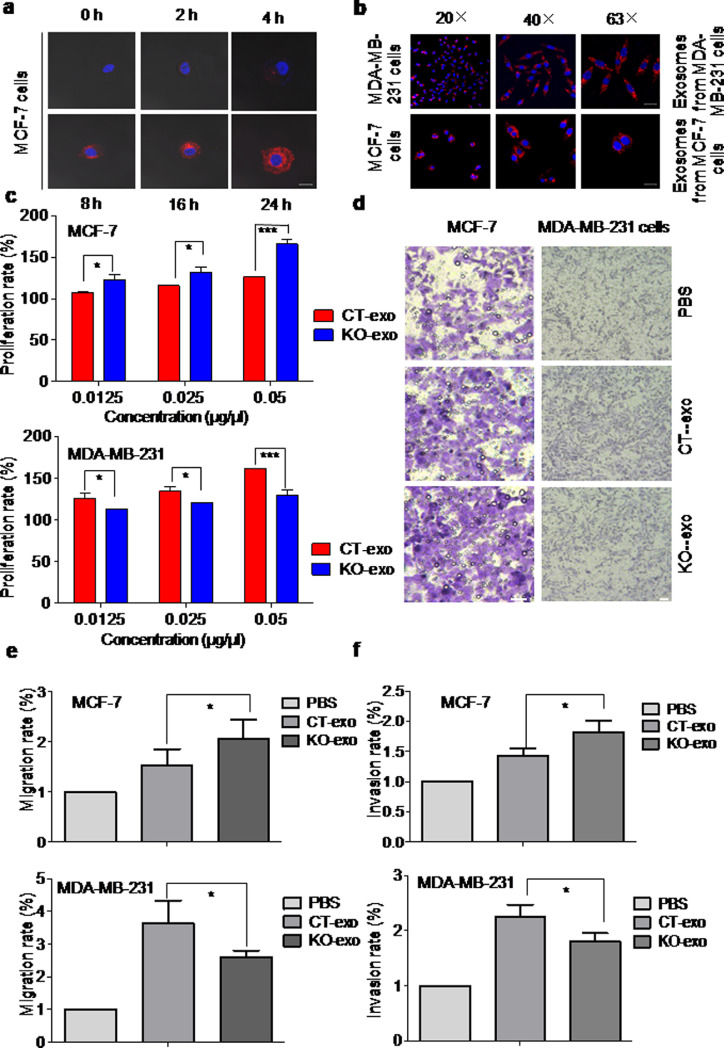
In order to evaluate the role of exosomes in Rasal2-regulated tumour progression, we investigated whether ER+MCF-7 cells can take up exosomes by incubation of the cells with their autologous PKH26-labelled exosomes for 0, 2, 4, 8, 16, 24-h. Observation by confocal microscopy demonstrated that MCF-7 cells can take up exosomes in a time-dependent manner (Fig. 3(a)). Also, incubation of ER–MDA-MB-231 and ER+MCF-7 cells with their autologous PKH26-labelled exosomes for 8-h under different magnifications (20×, 40×, 63×) showed that both ER+MCF-7 and ER–MDA-MB-231 cells have the ability to take up their autologous exosomes (Fig. 3(b)).
Fig. 3.
Rasal2 regulates tumour progression via an exosomal pathway in vitro. (a) MCF-7 cells were cultured with 10 μg of PKH26-labelled exosomes secreted from MCF-7 cells for different times (0, 2, 4, 8, 16, 24-h) and then the cells were observed by confocal microscopy. MCF-7 cells were found to take up autologous exosomes in a time-dependent manner (Scale bar: 20 μm); (b) the internalization of autologous exosomes was also visualized in MDA-MB-231 cells. After 8-h treatment with 10 μg of PKH26-labelled autologous exosomes, the internalization of exosomes was confirmed by confocal microscopy under different magnifications (20×, 40×, 63×) in MCF-7 and MDA-MB-231 cells(Scale bar: 20 μm); (c)–(e) the cell proliferation rate was measured by CCK-8 assay after incubation with different concentrations (0.0125, 0.025, 0.05 μg/μl) of autologous exosomes for 24-h, and the migration and invasion rates were determined by transwell migration assay and matrigel invasion assay after 0.05 μg/μl of CT-exo and KO-exo were co-cultured with autologous cells for 24-h. The findings showed that autologous exosomes regulate diametrically progression of MCF-7 cells and MDA-MA-231 cells (Scale bar: 100 μm); data are presented as the mean ± S.D. (n = 3) *p < 0.05,*** p < 0.001 vs. CT-exo (t-test, two-tailed).
Subsequently, we investigated the effects of exosomes isolated from CT (CT-exo) or KO (KO-exo) ER+MCF-7 and ER–MDA-MB-231 cells on the proliferation, migration and invasion rates of cancer cells. After 24-h incubation of the cells with their autologous CT-exo and KO-exo in different concentrations (0.0125, 0.025, 0.05 μg/μL), CCK-8 assay showed that proliferation rate in all concentrations in KO-exo group was significantly higher than that in CT-exo group in ER+MCF-7 cells and lower than that in CT-exo group in ER–MDA-MB-231 cells, indicating Rasal2 affected tumour cell proliferation via exosomal secretion in the opposing ways (Fig. 3(c)). Transwell migration assay and matrigel invasion assay, by incubation of the cells with their autologous CT-exo and KO-exo (0.05 μg/μL), showed that tumour-derived exosomes induced a significant increase in migration (Fig. 3(d)–(e)) and invasion rates (Fig. 3(f)) in ER+MCF-7 cells, but a reduction in ER–MDA-MB-231 cells as compared with the corresponding CT-exo group.
3.5. Autologous exosomes regulated the growth of MCF-7 and MDA-MB-231 xenograft tumour in vivo
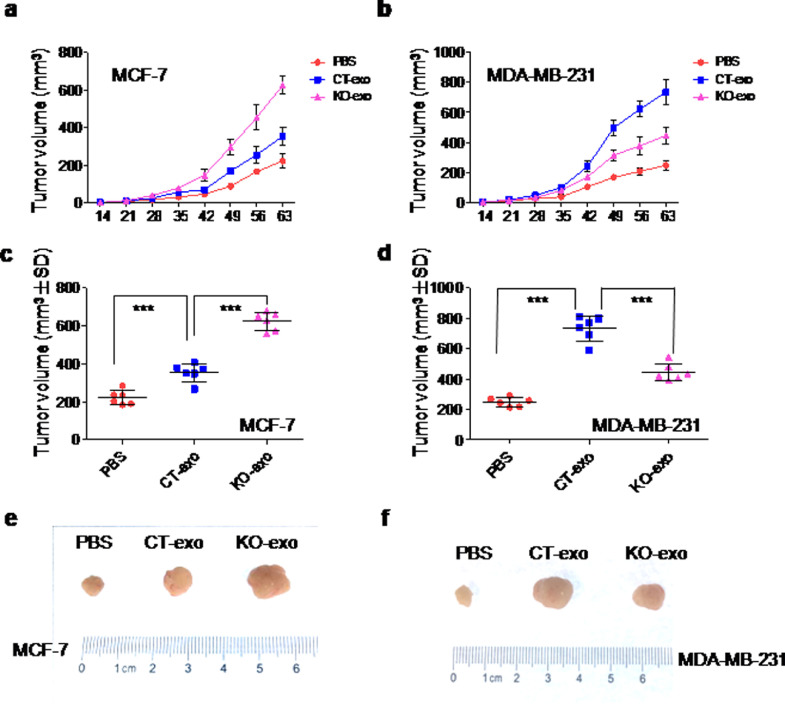
To test whether the effects of exosomes on tumour cells in vitro could be replicated in vivo, the effects of tumour-derived exosomes on tumour growth were assessed in mice xenograft models by injecting 5 μg of exosomal protein derived from ER+MCF-7 or ER–MDA-MB-231 cells (in a total volume of 100 μL PBS) into nude mice via the tail vein and twice a week for seven weeks. As expected, treatment of ER+MCF-7 xenograft mice with KO-exo significantly enhanced the tumour growth rate at day 35 and after as compared to those treated with CT-exo (Fig. 4(a)). On the contrary, treatment of ER–MDA-MB-231 xenograft mice with KO-exo induced a significant reduction in the tumour growth rate as compared to those treated with CT-exo (Fig. 4(b)). Consistently, volume and size of the tumour harvested from ER+MCF-7 xenograft mice treated with KO-exo were significantly larger than those treated with CT-exo (Fig. 4(c)–(e)), while those from ER–MDA-MB-231 xenograft mice were smaller than those treated with CT-exo (Fig. 4(d)–(f)).
Fig. 4.
Autologous exosomes regulate the growth of MCF-7 and MDA-MB-231 xenograft tumour in vivo. (a) and (b). MCF-7 cells and MDA-MB-231 cells (1.5 × 106) were injected into nude mice (6 mice/group), respectively. After two weeks’ inoculation, 5 μg of autologous CT-exo and KO-exo were respectively injected into mice in tail vein two times a week continued for seven weeks. The control group was injected with PBS only. The growth rate of tumours was assessed and tumour volumes (mm³) were calculated at 14, 21, 28, 35, 42, 49, 56 or 63 days after inoculation and each point represents the mean calculated volume of six tumours; (c)–(f) the tumour volume (c and d) and size (e and f) were measured after nine weeks’ inoculation. Data are presented as the mean ± S.D. (n = 6). **p < 0.01, ***p < 0.001 vs. PBS or CT-exo (t-test, two-tailed).
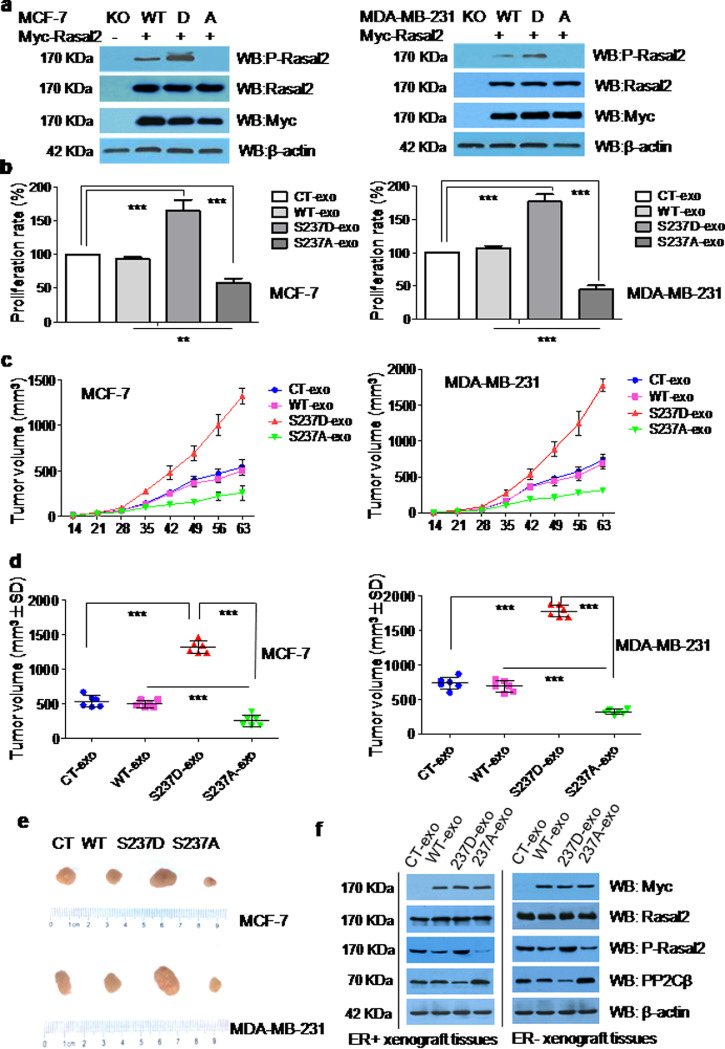
3.6. The phosphorylation of Rasal2 at S237 in PH domain facilitated tumour progression via exosomal secretion
To find out the effects of the phosphorylation of Rasal2 at S237 in PH domain on tumour progression, we constructed A and D mutant to simulate the non-phosphorylation and complete phosphorylation at S237 of Rasal2 in ER+MCF-7 and ER–MDA-MB-231 cells (Fig. 5(a)) and then detected the proliferation rate of these cells by co-culturing the cells with exosomes extracted from CT (CT-exo), WT (WT-exo), S237D (S237D-exo) or S237A (S237A-exo) tumour cells. The S237 domain was selected because in our preliminary study, we found that AMPK-mediated phosphorylation of Rasal2 S237 in PH domain enhances the interaction between Rasal2 and Vps34, and promotes the activation of autophagy, thereby affecting the development of breast cancer. It was found that exosomes from S237D mutant significantly increased, whereas exosomes from S237A mutant decreased the proliferation ratein both ER+ and ER–breast cancer cells as compared withexosomes from CT and WT tumour cells (Fig. 5(b)). The xenograft experiment was then carried out to test whether similar results could also be obtained in vivo. As shown in Fig. 5(c)–(e), tumour volume and size were significantly larger in both ER+MCF-7 and ER–MDA-MB-231 xenograft models injected with S237D-exo and smaller in both models injected with S237A-exo than that in both models injected by CT-exo or WT-exo. Correspondingly, the expression of p-Rasal2 (S237D) was found to be significantly higher in tumour tissues lysates of both ER+ and ER– xenograft injected with S237D-exo and lower in both models injected by S237A-exo,when compared to both models injected by CT-exo or WT-exo (Fig. 5(f)).
Fig. 5.
The phosphorylation of Rasal2 at S237 in PH domain facilitates tumourprogression via exosomal secretion. (a) Construction of A and D mutant of Rasal2 at S237. Myc-Rasal2 (WT, S237D, S237A) plasmids were transfected into Rasal2 KO cells and transfection efficiency was analysed with anti-Rasal2 and anti-myc antibodies by immunoblot analysis in the lysates of MCF-7 and MDA-MB-231 cells; (b) proliferation rate. 0.05 μg/μl of autologous CT-exo, WT-exo, S237D-exo and S237A-exo were respectively co-incubated with tumour cells for 24-h, the proliferation rate was then evaluated by CCK-8 assay. Data are presented as the mean ± S.D. (n = =3). ***p < 0.001 vs. CT-exo or WT-exo (t-test, two-tailed); (c) tumour volumes. MCF-7 cells and MDA-MB-231 cells were injected into nude mice (6 mice/group). After two weeks’ inoculation, 5 μg of autologous CT-exo, WT-exo, S237D-exo and S237A-exo were respectively injected into mice in tail vein for two times a week continued for seven weeks and the growth rate of tumours were measured and tumour volumes (mm³) calculated on each of the indicated days after inoculation, each point representing the mean calculated volume of six tumours; (d) tumour volumes were measured after nine weeks inoculation. Data are presented as the mean ± S.D. (n = =6). **p < 0.01,***p < 0.001; vs. CT-exo, WT-exo or S237D-exo(t-test, two-tailed); (e) the size of tumours; (f) the expression of Rasal2, P-Rasal2 (S237) and PP2Cβ by immunoblot analysis in the lysates of mice xenograft tumour tissues.
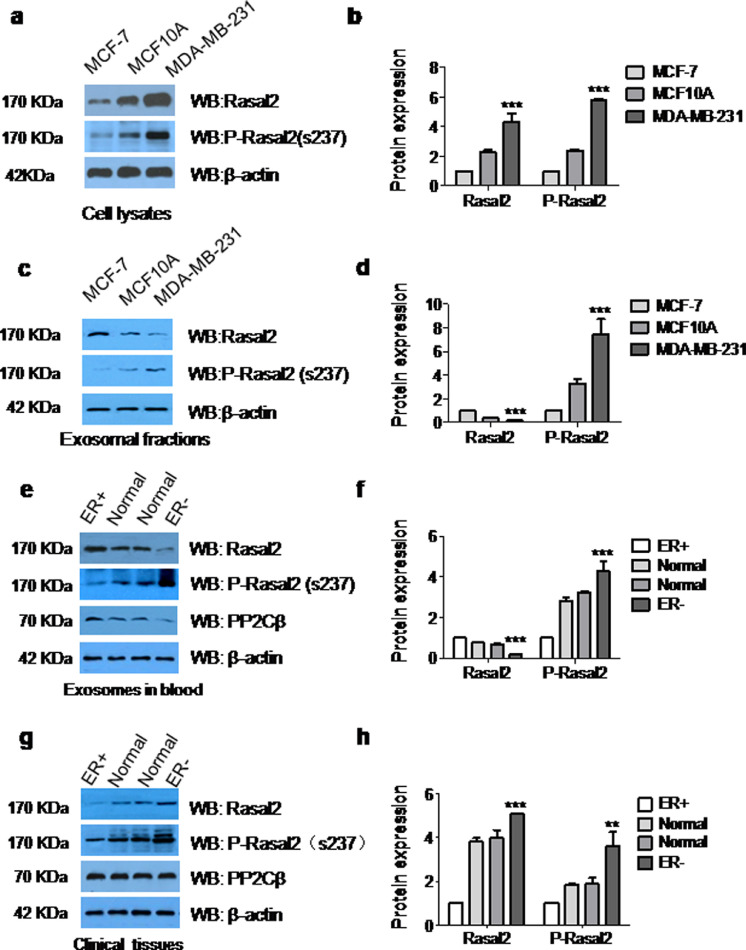
3.7. The expression of p-Rasal2 and PP2Cβin the exosomes derived from ER+ or ER–breast cancer cells and cancer tissues, and the exosomes derived from blood of patients with ER+ or ER– breast cancer
To further confirm the role of the phosphorylation of Rasal2 at S237 in tumour progression, we measured the expression of Rasal2 and p-Rasal2 (S237) in the lysates of ER+ and ER– breast cancer cell and the exosomes isolated from these cells by immunoblot analysis. The results showed that levels of both Rasal2 and p-Rasal2 (S237) in the lysates of ER+MCF-7 cell were much lower than those in the lysates of ER–MDA-MB-231 cells (Fig. 6(a) and (b)). However, the expression of Rasal2 was higher and p-Rasal2 (S237) lower in exosomes derived from ER+MCF7 cells, when compared to exosomes derived from ER–MDA-MB-231 cells (Fig. 6(c) and (d)). We also purified exosomes from the blood of patients with ER+ and ER– breast cancer and examined the expression of Rasal2 and p-Rasal2 (S237). As presented in Fig. 6(e) and (f), the expression of p-Rasal2 (S237) was lower in the exosomes derived from blood of patients with ER+ breast cancer, and higher in those of patients with ER– breast cancer ascompared with normal people. We meanwhile also measured the expression of p-Rasal2 (S237) in tumour tissues and adjacent normal tissues from 20 pairs of patients with ER+ and ER– breast cancer by Western blot analysis. Similar results were observed in tumour tissues of patients, showing that ER– cancer tissues were more enriched with p-Rasal2 (S237), while less expression was found in ER+ tumour tissuescompared with their adjacent normal tissues (Fig. 6(g) and (h)).
Fig. 6.
The expression of Rasal2, p-Rasal2 and PP2Cβin the exosomesderived from ER+ or ER–breast cancer cells and cancer tissues, and the exosomes derived from blood of patients with ER+ or ER–breast cancer. (a)–(d) The expression of Rasal2 and p-Rasal2 were detected by immunoblot analysis in the lysates of cells (a and b) and the exosomes (c and d) derived from MCF-7, MCF10A and MDA-MB-231 cells. Data are presented as the mean ± S.D. (n = =3). ***p < 0.001 vs. MCF-7 (t-test, two-tailed); (e) and (f) immunoblot analysis for Rasal2, p-Rasal2 and PP2Cβ in the exosomes purified from the blood of patients with ER+ and ER− breast cancers.Data are presented as the mean ± S.D. (n = =10). ***p < 0.001 vs. ER+ (t-test, two-tailed); (g) and (h) immunoblot analysis for Rasal2 and, p-Rasal2 and PP2Cβ in cancer tissues and adjacent normal tissues of patients with ER+ and ER− breast cancers. Data are presented as the mean ± S.D. (n = =20). **p < 0.01,***p < 0.001 vs. ER+ (t-test, two-tailed).
In addition, we examined the expression of Type 2 C protein phosphatase β (PP2Cβ) in tumour tissues of ER+ and ER– xenograft mice with injected 237D-exo and 237A-exo (Fig. 5(f)), in the exosomes from blood (Fig. 6(e) and (f)) and in the cancer tissues (Fig. 6(g) and (h)) of patients with ER+ or ER– breast cancer. In contrast to the expression of p-Rasal2, the expression of PP2Cβ was lower in tumour tissues of ER+ and ER–xenograft mice with injected 237D-exo and higher in those with injected 237A-exo (Fig. 5(f)), and higher in the exosomes from blood (Fig. 6(e) and (f)) and cancer tissues (Fig. 6(g) and (h)) of patients with ER+ breast cancer, and lower in those of patients with ER– breast cancer.
4. Discussion
In the present study, we demonstrated that phosphorylated Rasal2 (p-Rasal2) facilitates tumour growth, progression, and metastasis in both ER+ and ER– breast cancers. This conclusion is evidenced by our findings from in vitro and in vivo studies. In our in vitro experiments, treatment of the cells with the exosomes from S237D mutant (complete-phosphorylation) was found to significantly increase, whereas treatment with the exosomes from S237A mutant (non-phosphorylation) was found to decrease the proliferation rate in both ER+ and ER– breast cancer cells. In our in vivo investigation, injection with S237D-exo increases, whereas injection with S237A-exo reduces the tumour volume and sizeas well as the expression of p-Rasal2 in the lysates of tumour tissues in both ER+MCF-7 and ER–MDA-MB-231 xenograft mice.
Our investigations in vitro and in patients also demonstrated that the expression of p-Rasal2 is lower in ER+ and higher in ER–breast cancer. Firstly,the levels of p-Rasal2 in both the lysates of ER+MCF-7 cells and the exosomes derived from ER+MCF7 cells were found to be much lower than those in both the lysates of ER–MDA-MB-231 cells and the exosomes derived from ER–MDA-MB-231 cells in vitro. Secondly, the expression of p-Rasal2 was shown to be lower in the cancer tissues of patients withER+breast cancer and higher in the cancer tissues of patients with ER–breast cancer as compared with their adjacent normal tissues. Finally, the expression p-Rasal2 (S237) was also demonstrated to be lower in the exosomes purified from blood of patients with ER+ breast cancer, and higher in those of patients with ER–breast cancer as compared with those in exosomes purified from normal people.
Based on the existence of p-Rasal2 not only in the lysates of ER+MCF-7 and ER–MDA-MB-231 cells in vitro, in tumour tissues of ER+ and ER–xenograft mice injected with 237D-exo or 237A-exo in vivo, but also in the exosomes from blood and the cancer tissues of patients with ER+ or ER– breast cancer, we concluded that Rasal2 or total Rasal2 includes two different types, p-Rasal2 and non-phosphorylated-Rasal2 (non-p-Rasal2), in both ER+ and ER–breast cancers. The former functions as a promoter, while the latter might work as a suppressor in the progression of both ER+ and ER–breast cancer.
This indicated that the function of Rasal2 (or total Rasal2) as a promoter or a suppressor in breast cancer progression depends on the ratio of p-Rasal2/non-p-Rasal2, rather than the amount of total Rasal2. In other words, the ratio of p-Rasal2/non-p-Rasal2 is the major factor determining the function of Rasal2 (or total Rasal2) in different types of breast cancers. In the present study, we demonstrated that the ratio of p-Rasal2/non-p-Rasal2 in ER+ (<1) is much lower than that in ER− (>1) breast cancers. This likely is one of the causes for why Rasal2 (or total Rasal2) has diametrical effects, functioning as a suppressor in ER+ breast cancers and a promoter in ER–breast cancers. Therefore, the ratio of p-Rasal2/non-p-Rasal2 may be able to be considered as an indicator or predictor of cancer development and disease outcomes in both ER+ and ER− breast cancers.
To find out the potential mechanisms involved in the control of p-Rasal2 expression, we examined the expression of Type 2 C protein phosphatase β (PP2Cβ) as well as p-Rasal2 in tumour tissues of ER+ and ER−xenograft mice injected 237D-exo or 237A-exo, and in the exosomes from blood and cancer tissues of patients with ER+ or ER−breast cancer. PP2Cβ is a dephosphorylase and is also known as PPM1B [PP (protein phosphatase), Mg(2+)/Mn(2+) dependent, 1B] [15,41,42]. We demonstrated that the expression of PP2Cβ is negatively correlated with that of p-Rasal2 (S237) not only in tumour tissues of ER+ and ER–xenograft mice injected 237D-exo or 237A-exo, but also in the exosomes from blood and the cancer tissues of patients with ER+ or ER– breast cancer. This indicated that PP2Cβ might be one of the factors controlling the phosphorylation of Rasal2 in breast cancer tissues. Increasing the activity of dephosphorylase by promoting the expression of PP2Cβ as a target may be useful as a therapy to restrain breast cancer progression.
In the present study, we also show that both ER+MCF-7 cells and ER–MDA-MB-231 cells have the ability to release extracellular vesicles or exosomes by transmission electron microscope (TEM) and flow cytometry analysis, and to take up their autologous exosomes in a time-dependent manner by using fluorescent lipid membrane dyes PKH26. Furthermore, we discovered that the exosomes isolated from tumour cells with KO-exo can up-regulate proliferation, migration and invasion rates in ER+MCF-7 cells and down-regulate these rates in ER–MDA-MB-231 cells, inCCK-8, transwell migration, matrigel and invasion assays in vitro, as well as enhance tumour growth rate, tumour volume and size in ER+MCF-7 xenograft mice with KO-exo and reduce these indexes in ER–MDA-MB-231 xenograft mice with KO-exo in vivo. These findings imply that the effects of Rasal2 (or total Rasal2) on tumour growth, progression, and metastasis are mediated by an exosome-transport process.
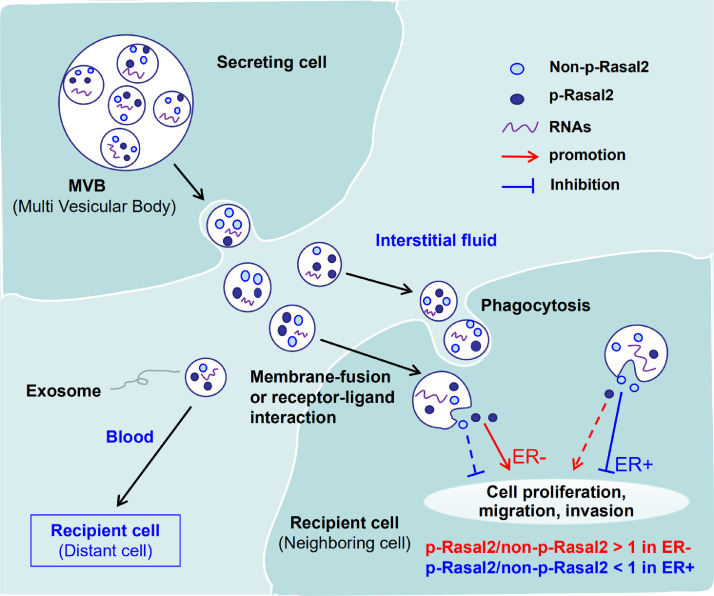
Many different cell types including cancer cells release exosomes not only into the cancer microenvironment but also into the circulation [43,44]. Cancer-derived exosomes can mediate directional tumour metastasis [45]. Exosomes are involved in intercellular signalling, both by membrane fusion to recipient cells with deposition of exosomal contents into the cytoplasm and by the binding of recipient cell membrane receptors [46]. Therefore, our findings imply that in ER+ and ER–breast cancers,p-Rasal2 and non-p-Rasal2 are both encapsulated into exosomes in the cancer cells and then secreted into interstitial fluid (or the blood) by these cells. The exosomes were taken up by the target or recipient cells (neighbouring or distant cells) via the membrane-fusion, phagocytosis or receptor-ligand interactions, where afterp-Rasal2 and non-p-Rasal2 are released from the exosomes and play their functions to facilitate or inhibit the development of proliferation, migration and invasion in target or recipient cells (Fig. 7).
Fig. 7.
The schematic diagram of the phosphorylated Rasal2 facilitates breast cancer progression via exosomal pathway. The exosomes carrying Rasal2 and p-Rasal2 released into the interstitial fluid (or the blood) by breast cancer cells and then taken up by the target or recipient cells via the membrane-fusion, phagocytosis or receptor-ligand interactions, whereafterp-Rasal2 and Rasal2 were released from the exosomes and played their functions to facilitate (ratio of p-Rasal2 and non-p-Rasal2 > 1 in ER− breast cancer) or inhibit (ratio of p-Rasal2 and non-p-Rasal2 < 1 in ER+ breast cancer) the development of proliferation, migration and invasion in target or recipient cells.
In summary, we demonstrated for the first time that the phosphorylation of Rasal2 at S237 in PH domain facilitates tumour progression. Our findings also support that the ratio of p-Rasal2/non-p-Rasal2 is the major factor determining the function of Rasal2 (or total Rasal2) as a promoter or suppressor in different types of breast cancers and that PP2Cβ might be one of the factors controlling the phosphorylation of Rasal2 in breast cancer tissues. We also provide evidence that the functions of Rasal2 (or total Rasal2) in the modulation of breast cancer progression are exosome-mediated. Targeting the phosphorylation of Rasal2 machinery may therefore be useful as a therapy to restrain breast cancer progression by reducing the ratio of p-Rasal2/non-p-Rasal2, especially in ER–breast cancers.
Funding source
This work was supported by the National Natural Science Foundation of China (Nos. 81572721,81272391 and 31571195) and the Hong Kong Research Grants Council (General Research Fund 14110418, 14167817, 141076160). The funders above did not have any involvement in the study design, data collection, data analysis, interpretation, and manuscript writing.
Declaration of competing interest
The authors declare no competing interests with relevance to this study.
Data sharing
The data, analytical methods, and study materials for the purposes of reproducing the results or replicating procedures can be made available on request to the corresponding author who manages the information.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.019.
Appendix. Supplementary materials
References
- 1.Autier P., Boniol M., La Vecchia C. Disparities in breast cancer mortality trends between 30 European countries: retrospective trend analysis of who mortality database. Brit Med J. 2010;341:c3620. doi: 10.1136/bmj.c3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Perou C.M., Sørlie T., Eisen M.B. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Sørlie T., Perou C.M., Tibshirani R. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. ProcNatlAcadSci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sotiriou C., Neo S.-.Y., McShane L.M. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsen S.N., Wronski A., Castaño Z. Loss of rasgap tumor suppressors underlies the aggressive nature of luminal b breast cancers. Cancer Discov. 2017;7:202–217. doi: 10.1158/2159-8290.CD-16-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLaughlin S.K., Olsen S.N., Dake B. The rasgap gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell. 2013;24:365–378. doi: 10.1016/j.ccr.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sears R., Gray J.W. Epigenomic inactivation of rasgaps activates ras signaling in a subset of luminal b breast cancers. Cancer Discov. 2017;7:131–133. doi: 10.1158/2159-8290.CD-16-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hui K., Wu S., Yue Y. RASAL2 inhibits tumor angiogenesis via p-AKT/ETS1 signaling in bladder cancer. Cell Signal. 2018;48:38–44. doi: 10.1016/j.cellsig.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Hui K., Yue Y., Wu S. The expression and function of RASAL2 in renal cell carcinoma angiogenesis. Cell Death Dis. 2018;9(9):881. doi: 10.1038/s41419-018-0898-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y., Zhao M., Xu H. RASAL2 down-regulation in ovarian cancer promotes epithelial-mesenchymal transition and metastasis. Oncotarget. 2014;5:6734–6745. doi: 10.18632/oncotarget.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng M., Bao Y., Li Z. RASAL2 activates RAC1 to promote triple-negative breast cancer progression. J Clin Invest. 2014;124:5291–5304. doi: 10.1172/JCI76711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y., Tong J.H.M., Lung R.W.M. RASAL2 promotes tumor progression through LATS2/YAP1 axis of hippo signaling pathway in colorectal cancer. Mol Cancer. 2018;17:102. doi: 10.1186/s12943-018-0853-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moorhead G.B.G., Trinkle-Mulcahy L., Ulke-Lemee A. Emerging roles of nuclear protein phosphatases. Nat Rev Mol Cell Biol. 2007;8:234–244. doi: 10.1038/nrm2126. [DOI] [PubMed] [Google Scholar]
- 15.Tasdelen I., van Beekum O., Gorbenko O. The serine/threonine phosphatase PPM1B (PP2Cβ) selectively modulates PPARγ activity. Biochem J. 2013;451:45–53. doi: 10.1042/BJ20121113. [DOI] [PubMed] [Google Scholar]
- 16.Katzmann D.J., Babst M., Emr S.D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 17.Bobrie A., Krumeich S., Reyal F. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920–4930. doi: 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- 18.Conigliaro A., Cicchini C. Exosome-mediated signaling in epithelial to mesenchymal transition and tumor progression. J Clin Med. 2018;8(1) doi: 10.3390/jcm8010026. pii: E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thery C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 20.Ostrowski M., Carmo N.B., Krumeich S. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. sup pp 1-13. [DOI] [PubMed] [Google Scholar]
- 21.Javeed N., Mukhopadhyay D. Exosomes and their role in the micro-/macro-environment: a comprehensive review. J Biomed Res. 2017;31:386–394. doi: 10.7555/JBR.30.20150162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takasugi M., Okada R., Takahashi A. Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through Epha2. Nat Commun. 2017;8:15729. doi: 10.1038/ncomms15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo L.J., Song M., Huang Q.H. Exosome-packaged miR-1246 contributes to bystander DNA damage by targeting LIG4. Br J Cancer. 2018;119(4):492–502. doi: 10.1038/s41416-018-0192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia X., Chen J., Megger D.A. Label-free proteomic analysis of exosomes derived from inducible hepatitis b virus-replicating Hepad38 cell line. Mol Cell Proteomics. 2017;16:S144–SS60. doi: 10.1074/mcp.M116.063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lason E., Sikora E., Ogonowski J. Influence of process parameters on properties of nanostructured lipid carriers (NLC) formulation. ActaBiochim Pol. 2013;60:773–777. [PubMed] [Google Scholar]
- 26.Welch J.L., Madison M.N., Margolick J.B. Effect of prolonged freezing of semen on exosome recovery and biologic activity. Sci Rep. 2017;7:45034. doi: 10.1038/srep45034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du F., Qian Z.M., Luo Q., Yung W.H., Ke Y. Hepcidin suppresses brain iron accumulation by downregulating iron transport proteins in iron-overloaded rats. MolNeurobiol. 2015;52:101–114. doi: 10.1007/s12035-014-8847-x. [DOI] [PubMed] [Google Scholar]
- 28.Sheller S., Papaconstantinou J., Urrabaz-Garza R. Amnion-epithelial-cell-derived exosomes demonstrate physiologic state of cell under oxidative stress. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0157614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura R., Uemoto S., Tabata Y. Immunosuppressive effect of mesenchymal stem cell-derived exosomes on a concanavalin A-induced liver injury model. InflammRegen. 2016;36:26. doi: 10.1186/s41232-016-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller L., Simms P., Hong C.S. Human tumor-derived exosomes (TEX) regulateTreg functions via cell surface signaling rather than uptake mechanisms. Oncoimmunology. 2017;6(8) doi: 10.1080/2162402X.2016.1261243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H., Kang J.Y., Mun D. Calcium chloride enhances the delivery of exosomes. PLoS ONE. 2019;14(7) doi: 10.1371/journal.pone.0220036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Wang H., Cao J. Exosomes from adipose-derived stem cells promotes VEGF-C-Dependent lymphangiogenesis by regulating miRNA-132/TGF-beta pathway. Cell Physiol Biochem. 2018;49:160–171. doi: 10.1159/000492851. [DOI] [PubMed] [Google Scholar]
- 33.Huang J., Ding Z., Luo Q., Xu W. Cancer cell-derived exosomes promote cell proliferation and inhibit cell apoptosis of both normal lung fibroblasts and non-small cell lung cancer cell through delivering alpha-smooth muscle actin. Am J Transl Res. 2019;11(3):1711–1723. [PMC free article] [PubMed] [Google Scholar]
- 34.Shao N., Xue L., Wang R. MiR-454-3p is an exosomal biomarker and functions as a tumor suppressor in glioma. Mol Cancer Ther. 2019;18(2):459–469. doi: 10.1158/1535-7163.MCT-18-0725. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y., Rao R., Shen J. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008;68:4833–4842. doi: 10.1158/0008-5472.CAN-08-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia Y., Zhu Y., Qiu S. Exosomes secreted by endothelial progenitor cells accelerate bone regeneration during distraction osteogenesis by stimulating angiogenesis. Stem Cell Res Ther. 2019;10:12. doi: 10.1186/s13287-018-1115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du F., Zhu L., Qian Z.M., Wu X.M., Yung W.H., Ke Y. Hyperthermic preconditioning protects astrocytes from ischemia/reperfusion injury by up-regulation of HIF-1 alpha expression and binding activity. Biochim Biophys Acta. 2010;1802:1048–1053. doi: 10.1016/j.bbadis.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 38.Yan M., Li X., Tong D. MiR-136 suppresses tumor invasion and metastasis by targeting RASAL2 in triple-negative breast cancer. Oncol Rep. 2016;36:65–71. doi: 10.3892/or.2016.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang J.F., Zhao H.P., Wang Z.F., Zheng S.S. Upregulation of RASAL2 promotes proliferation and metastasis, and is targeted by miR-203 in hepatocellular carcinoma. Mol Med Rep. 2017;15(5):2720–2726. doi: 10.3892/mmr.2017.6320. [DOI] [PubMed] [Google Scholar]
- 40.Yoon J.H., Ham I.H., Kim O. Gastrokine 1 protein is a potential theragnostic target for gastric cancer. Gastric Cancer. 2018;21:956–967. doi: 10.1007/s10120-018-0828-8. [DOI] [PubMed] [Google Scholar]
- 41.Yien Y.Y., Bieker J.J. Functional interactions between erythroid Krüppel-like factor (EKLF/KLF1) and protein phosphatase PPM1B/PP2Cβ. J BiolChem. 2012;287:15193–15204. doi: 10.1074/jbc.M112.350496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salminen A., Kaarniranta K., Kauppinen A. Age-related changes in AMPK activation: role for AMPK phosphatases and inhibitory phosphorylation by upstream signaling pathways. Ageing Res Rev. 2016;28:15–26. doi: 10.1016/j.arr.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Ge R., Tan E., Sharghi-Namini S., Asada H.H. Exosomes in cancer microenvironment and beyond: have we overlooked these extracellular messengers? Cancer Microenviron. 2012;5:323–332. doi: 10.1007/s12307-012-0110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kahlert C., Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng G., Qu J., Zhang Y. Gastric cancer-derived exosomes promote peritoneal metastasis by destroying the mesothelial barrier. FEBS Lett. 2017;591:2167–2179. doi: 10.1002/1873-3468.12722. [DOI] [PubMed] [Google Scholar]
- 46.Blackwell R.H., Foreman K.E., Gupta G.N. The role of cancer-derived exosomes in tumorigenicity& epithelial-to-mesenchymal transition. Cancers (Basel) 2017;9(8) doi: 10.3390/cancers9080105. pii: E105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.