Abstract
Background
Alcohol intake is a well-known lifestyle risk factor for CRC, and an increasing number of studies have revealed that alcohol intake is also tightly associated with CRC metastasis. However, the effect of alcohol on CRC metastasis and its underlying mechanism remain unclear.
Methods
A retrospective cohort study was performed to investigate the characteristics of patients with alcohol-related CRC. The effects of ethanol on the biological behaviours of CRC cells were assessed through in vivo and in vitro assays using the Lieber-DeCarli ethanol liquid diet and ethanol, respectively. The ethanol-mediated signalling pathway and downstream factors were screened through ELISA, western blot, immunofluorescence and co-immunoprecipitation.
Findings
Most patients with alcohol-related CRC, particularly those with tumour metastasis, were characterized by a notably higher circulating ethanol level and a lower systemic acetaldehyde level. Moreover, CRC cells accumulated in ethanol, but not acetaldehyde, to notably higher levels compared with adjacent normal cells. Alcohol intake significantly promoted CRC metastasis via the ethanol-mediated TGF-β/Smad/Snail axis, and ethanol induced the cytoplasmic mislocalization of RUNX3 and further promoted the aggressiveness of CRC by targeting Snail. Pirfenidone (PFD) significantly eliminated the effects of ethanol on CRC metastasis by specifically blocking TGF-β signalling.
Interpretation
Alcohol intake plays a vital role in CRC metastasis via the ethanol-mediated TGF-β/RUNX3/Snail axis, and PFD might be a novel therapeutic management strategy for CRC.
Keywords: Colorectal cancer, Ethanol, Tumour metastasis, Pirfenidone, RUNX3, TGF-β signalling
Research in context.
Evidence e prior to this study
An increasing numbers of studies have revealed that alcohol consumption is tightly associated with tumour metastasis of colorectal cancer (CRC), but its effect and underlying mechanism remain unclear. Recent studies on other diseases have demonstrated that ethanol significantly promotes the expression of TGF-β1, and the activation of this factor plays a vital role in the development and progression of multiple types of cancers. Although both ethanol and its metabolic product acetaldehyde are the major carcinogens in alcohol, the contribution and mechanical role of ethanol itself in alcohol-related CRC remain unknown.
Added value of this study
Alcohol intake significantly promotes tumour metastasis in CRC in vivo, and ethanol itself plays a vital role in promoting the metastasis of alcohol-related CRC via the TGF-β/RUNX3/Snail axis. In addition, ethanol activates this axis by inducing the upregulation of TGF-β1 and the cytoplasmic mislocalization of RUNX3. Pirfenidone (PFD) significantly eliminates the ethanol-induced promotion of the aggressiveness of CRC cells by blocking TGF-β signalling.
Implications of all available evidence
Alcohol intake is an important risk factor for CRC progression from early to advanced stages, and ethanol itself plays a vital role in the development and progression of alcohol-related CRC. PFD might be a novel therapeutic management strategy for CRC.
CRediT authorship contribution statement
Kehong Zheng: Conceptualization, Data curation. Jinlong Yu: Data curation. Zetao Chen: Data curation. Rui Zhou: Formal analysis. Chuang Lin: Data curation. Yuxuan Zhang: Writing - review & editing. Zonghai Huang: Funding acquisition, Formal analysis. Lina Yu: Funding acquisition, Visualization, Writing - review & editing. Liang Zhao: Funding acquisition, Writing - review & editing. Qian Wang: Conceptualization, Visualization.
Alt-text: Unlabelled box
1. Introduction
Colorectal cancer (CRC) is the third most common type of cancer and the second most common cause of cancer-related death worldwide [1]. Tumour metastasis is often the direct reason for the poor prognosis of most patients with CRC [2,3]. However, the mechanisms underlying CRC incidence and metastasis remain unclear because a multistep cascade of events is stimulated by various risk factors, including age, genetic mutation, inflammation, gut microflora composition, family history and harmful lifestyle habits [4,5]. Intriguingly, recent studies have shown a significantly higher incidence of CRC in highly developed countries [6,7]. Therefore, we hypothesized that environmental and lifestyle factors might exert a notably more crucial effect on CRC than previously suspected.
Alcohol consumption is very common in developed countries and has been confirmed to be an important risk factor for CRC, and particularly strong associations have been obtained with chronic and moderate to heavy alcohol intake [4]. Moreover, increasing numbers of epidemiological studies have revealed that alcohol consumption is also closely related to metastasis in patients with CRC, and this association was identified as the primary reason for the poor outcomes observed in these patients [8,9]. Due to the difficulty in detecting CRC at an early stage [10], it was conceivable to conclude that most CRC patients would not change their lifestyles until obvious symptoms and/or signs were detected. Thus, alcohol intake is also considered to play a vital role in accelerating the progression of early CRC.
The International Agency for Research on Cancer (IARC) has declared that ethanol and acetaldehyde are the major carcinogens associated with alcohol intake [11], [12], [13], but the mechanisms underlying the effects of ethanol itself on the development and progression of alcohol-related CRC remain unknown. Interestingly, recent studies on other diseases have revealed that ethanol significantly increases the expression of extracellular TGF-β1, which is a crucial trigger for the activation of TGF-β signalling and plays a vital role in the incidence and metastasis of CRC [14,15]. Therefore, ethanol itself might play an important role in the tumour metastasis of alcohol-related CRC via TGF-β signalling.
The present study aimed to investigate the effects of alcohol intake on CRC metastasis and the role of ethanol in alcohol-related tumour metastasis. Furthermore, our study might aid the development of novel therapeutic strategies for CRC.
2. Materials and methods
2.1. Clinical population and tumour tissue samples
The clinical data used in this study originated from 200 patients diagnosed with primary CRC between January 2012 and December 2017 by the Department of Pathology at Zhujiang Hospital of Southern Medical University in Guangdong, China. Two hundred healthy controls were randomly selected from the cohort of outpatients. The 45 pairs of CRC tissues with matched normal mucosa that were used for alcohol dehydrogenase 1C (ADH1C), alcohol dehydrogenase 1B (ADH1B), acetaldehyde dehydrogenase 2 (ALDH2), acetaldehyde dehydrogenase 1A1 (ALDH1A1) and acetaldehyde dehydrogenase 1B1 (ALDH1B1) detection were also diagnosed by the Department of Pathology at Zhujiang Hospital. We restricted our analysis to population-based cases that were not selected based on family history. This study was approved by the Ethics Committees of Zhujiang Hospital and Southern Medical University, and all aspects of the study comply with the criteria set by the Declaration of Helsinki. All the CRC patients provided informed consent to participate in this study. The ADH and ALDH expression profiles of the CRC samples, including any relevant clinical information, were identified through a search of the GEO: GSE87211 dataset (n = 160).
2.2. Measurement of alcohol intake
The alcohol intake was self-reported by participants during the baseline survey conducted in 2017 involving telephone follow-up. The drinking frequency (times/day) was calculated by asking the participants their weekly intake of alcohol during the 12 months before the follow-up (for healthy people) or the CRC diagnosis (for patients). The patients who did not usually drink some alcohol at least once a week were asked to provide their monthly or annual alcohol intake. The participants were further asked questions regarding the types of beverage (beer, grape wine, rice wine, weak spirits with an alcohol content < 40%, and strong spirits with an alcohol content ≥ 40%), the amount of alcohol consumed in a typical drinking day and the experience of flushing after drinking. The level of alcohol intake was calculated as grams (g) of pure alcohol per week based on the beverage type, amount consumed and frequency, and the following assumptions were used for the alcohol content by volume (v/v) in China: beer, 4%; grape wine, 12%; rice wine, 15%; weak spirits, 38%; and strong spirits, 53% [16]. According to the overall mean alcohol intake, we defined a patient who had not consumed any alcohol during the 12 months before the follow-up or CRC diagnosis as an “abstainer” and a participant who drank more than 5 g/week (regardless of his/her past drinking patterns) as a “drinker”.
2.3. Cell culture and treatment
The colorectal normal epithelial cell line FHC (Cat# CRL-1831, RRID: CVCL_3688) and the cancer cell lines HT29 (Cat# HCT-38, RRID: CVCL_0320), HCT116 (Cat# CCL-247, RRID: CVCL_0291), LS174T (Cat# CL-188, RRID: CVCL_1384), SW480 (Cat# CCL-228, RRID: CVCL_0546), SW620 (Cat# CCL-227, RRID: CVCL_0547), and RKO (Cat# CRL-2577, RRID: CVCL_0504) were obtained from American Type Culture Collection. The normal liver cell line L-O2 (Cat# CX0157) was obtained from BOSTER (Wuhan, China). All the cells were maintained as previously described and authenticated by short tandem repeat (STR) profiling after receipt. For use in this study, the cells were propagated for less than 6 months after resuscitation. These cells were grown in RPMI 1640 medium (Cat# A1049101, Thermo Fisher Life Technologies Corporation; Grand Island, NY, USA) supplemented with 10% foetal bovine serum (Cat# 10091130, Invitrogen, USA). Physiologically relevant concentrations of ethanol (0, 100, and 200 mg/dl, which correspond to 0, 22, and 44 mM, respectively; Cat# 459844, Sigma, USA) were accurately maintained for 48–72 h as previously indicated [17]. 4-Methylpyrazole (4-MP, 5 mM; Cat# HY-B0876, MedChemExpress, Shanghai, China) was used to inhibit the activity of alcohol dehydrogenase. The cells were also treated with pirfenidone (Cat# HY-B0673, MedChemExpress, Shanghai, China) and a TGF-β/Smad signalling inhibitor (Cat# SB-431542, Selleck, USA) at the recommended concentration (0.5 mg/ml) for 48–72 h. The RUNX3 vector with a nuclear localization sequence was purchased from GeneChem (Shanghai, China) and transfected using the lipofectamine® 3000 reagent (Cat# L3000015, Thermo Fisher Scientific, USA).
2.4. Analysis of the blood ethanol concentration
All Balb/c mice (aged 6 weeks) were treated with ethanol at 6 g/kg (by gavage, 32%, v/v solution). The blood alcohol concentrations were measured five times over 8 h (0.5, 1, 2, 4, and 8 h). For each sampling, 50 µl of whole blood was collected in a heparinized tube and centrifuged at 1800 × g for 5 min. The concentration of ethanol in serum was measured using an ethanol assay kit (Cat# KA4784, Abnova, USA). All experimental operations were performed in accordance with the instructions.
2.5. ADH activity assay
Total ADH activity was determined through the photometric method using an alcohol dehydrogenase assay kit purchased from Nanjing Jiancheng Bioengineering Institute (Cat# A083-1–1, Nanjing, China) according to the manufacturer's instructions. Briefly, the reaction mixture contained reagent 1 (0.65 ml), reagent 2 (0.05 ml), reagent 3 (0.75 ml) and the cell sample (0.05 ml). A mixture of the probe with ddH2o instead of the cell sample was used as a control. The absorbence at 340 nm was measured immediately after mixing (A1) and again after 10 min of incubation at 37 °C (A2). The activity of ADH was then computed using the following formula:
[One unit of ADH activity (U/mg prot) was defined as the amount of enzyme that catalyses the production of 1 nmol of products per minute per milligram of protein at 37 °C. RV: reaction volume. SV: sample volume. RT: reaction time. PC: protein concentration (mg/ml).]
2.6. Orthotopic xenograft colorectal cancer mouse model
The animals were fed the Lieber-DeCarli ethanol liquid diet (ethanol: 40 mg/ml) (Cat# TP4010A, TROPHIC Animal Feed High-Tech Co. LTD, China) for five days prior to cell injection. CT26 (Cat# CRL-2639, RRID: CVCL_7254, ATCC, USA) cells were suspended in fresh PBS at a concentration of 1 × 106 cells/50 μl and aspirated using a fine needle. Six-week-old Balb/c mice were anaesthetized and exposed to the caecum by laparotomy. In brief, a 0.5∼1-cm-long small nick was made in the skin, and the abdominal wall musculature was lifted. The abdominal cavity was opened, and the caecum was isolated and covered by warm saline and sterile gauze to keep the caecum moist. The cells (50 µl) were slowly injected into the caecal wall. The needle was carefully removed, and the injection site was inspected to ensure no leakage. The caecum was then returned to the abdominal cavity, and the abdominal wall and skin were closed. Twenty days later, the mouse was sacrificed, and the orthotopic xenograft colorectal cancer masses were measured and harvested for further study. If the tumour mass was invisible, the full intestine was embedded to calculate the maximum tumour size under microscopy.
2.7. Statistical analysis
The data were analysed using SPSS version 19.0 software (SPSS, Chicago, IL, USA). The clinical data were analysed using nonparametric tests (Wilcoxon and Mann-Whitney). Pearson's chi-squared (χ2) test, unpaired Student's t-test and paired t-test were used to evaluate the significance of the differences among different groups. All statistical tests were two-sided. The data are presented as the means ± SEMs.
2.8. Additional methods and material
All other methods are described in the online supplementary materials and methods.
3. Results
3.1. Most patients with alcohol-related CRC are characterized by a high circulating ethanol level
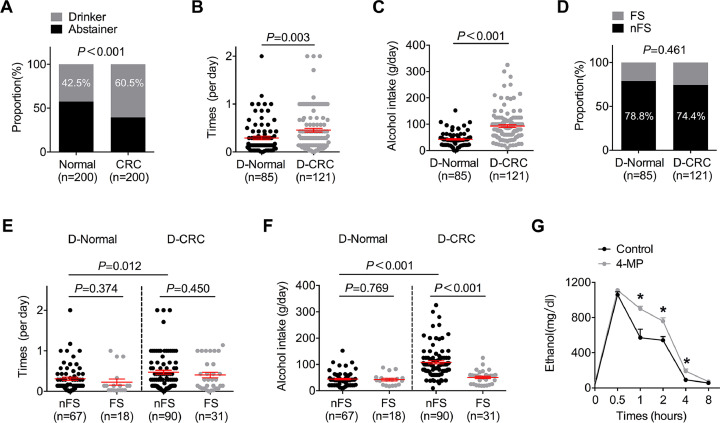
Our retrospective cohort study (n = 400, 200 healthy controls and 200 CRC patients) revealed that the proportion of CRC patients who consumed alcohol was significantly higher than that of healthy controls (control vs CRC = 42.5% vs 60.5%, Fig. 1A). Moreover, the frequency (control vs CRC = 0.294 ± 0.337 per day vs 0.455 ± 0.438 per day, Fig. 1B) and quantity (control vs CRC = 43.847 g ± 24.370 g vs 93.289 g ± 58.640 g) of alcohol intake (Fig. 1C) in patients with CRC were notably higher than those in the control subjects, which indicated that frequent and heavy alcohol intake is tightly associated with the incidence of CRC. Our study also found that subjects without flushing syndrome after alcohol intake in both the healthy and CRC groups were more likely to drink and had a higher risk of alcoholism (Fig. 1D). It is well known that a lack of flushing syndrome after drinking indicates strong acetaldehyde metabolic ability in the body and low acetaldehyde levels in tissues and the circulation [18]. Surprisingly, we found that the frequency and quantity of alcohol intake in alcohol-related CRC patients without flushing syndrome were substantially higher than those in the control subjects and patients with flushing syndrome (Fig. 1E and F). Accordingly, even with a high alcohol intake, most patients with alcohol-related CRC had low circulating levels of acetaldehyde, and thus, the guts of these patients might have only been exposed to these low acetaldehyde levels. A further in vivo study using mice revealed that the extent of circulating ethanol was primarily determined by the amount of alcohol consumed and that its duration was largely determined by the activity of alcohol dehydrogenase (ADH, Fig. 1G). Therefore, the exposure of the gut to high circulating levels of ethanol, but not acetaldehyde, after alcohol consumption might contribute to alcohol-related CRC in most patients.
Fig. 1.
The gut of most patients with alcohol-related CRC is exposed to a high circulating level of ethanol. (A) Pearson's chi-squared (χ2) test was used to analyse the relationship between alcohol intake and risk for CRC. (B) Unpaired Student's t-test was used to analyse the difference in drinking frequency between drinkers with and without CRC. “D-Normal” indicates drinkers in the normal group, “D-CRC” indicates drinkers with CRC. (C) Unpaired Student's t-test was used to analyse the difference in the average alcohol intake between drinkers with and without CRC. (D) Pearson's chi-squared (χ2) test was used to analyse the relationship between drinking and flushing syndrome in the normal and CRC groups. “FS” indicates subjects with flushing, and “nFS” indicates the absence of flushing syndrome. (E) Unpaired Student's t-test was used to analyse the difference in drinking frequency between drinkers with and without flushing syndrome. (F) Unpaired Student's t-test was used to analyse the difference in the average alcohol intake between drinkers with and without flushing syndrome. (G) A microplate reader was used to detect the ethanol concentration in mouse blood. *P < 0.05 and n.sP > 0.05 vs the control.
3.2. Alcohol intake promotes tumour metastasis in CRC by increasing the circulating ethanol level
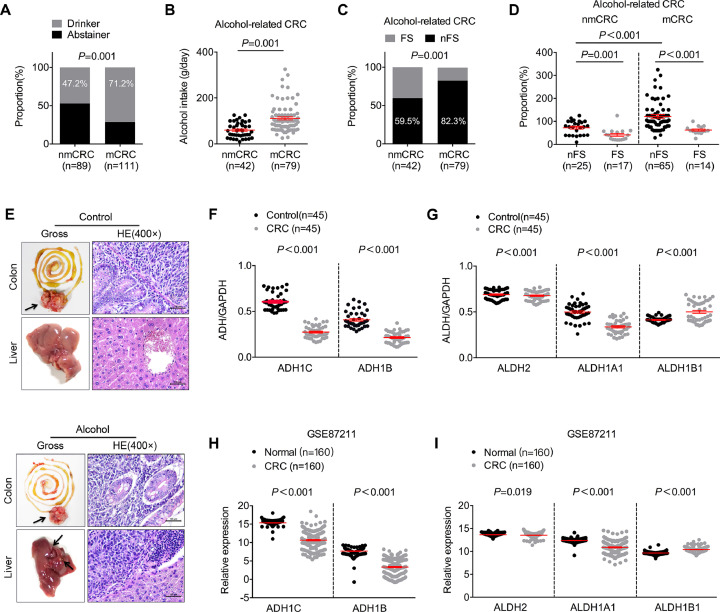
Further study revealed that alcohol intake was tightly associated with tumour metastasis in CRC (Fig. 2A). The average amount of alcohol intake in patients with tumour metastasis was significantly higher than that in patients without metastasis (nmCRC vs mCRC = 59.845 g ± 32.547 g vs 111.070 g ± 61.733 g, Fig. 2B), which indicated that a higher alcohol intake promoted CRC metastasis. In addition, a higher proportion of patients without flushing syndrome was obtained in the subgroup of subjects with tumour metastasis compared with the subgroup of patients without metastasis (nmCRC vs mCRC = 82.3% vs 59.5%, Fig. 2C). Moreover, we also found that patients with tumour metastasis and no flushing syndrome consumed a substantially higher amount of alcohol than the other patients (Fig. 2D). Thus, most alcohol-related CRC patients with tumour metastasis exhibited a high circulating level of ethanol, but not acetaldehyde, in the gut.
Fig. 2.
Ethanol itself might play a vital role in the incidence and metastasis of most alcohol-related CRC. (A) Pearson's chi-squared (χ2) test was used to analyse the relationship between alcohol intake and risk of CRC metastasis. “nmCRC” indicates non-metastatic CRC, and “mCRC” indicates metastatic CRC. (B) Unpaired Student's t-test was used to analyse the difference in the average alcohol intake between patients with and without tumour metastasis. (C) Pearson's chi-squared (χ2) test was used to analyse the relationship between flushing syndrome and risk of CRC metastasis. “FS” indicates patients with flushing syndrome, and “nFS” indicates the absence of flushing syndrome. (D) Unpaired Student's t-test was used to analyse the difference in the average alcohol intake between CRC patients with and without flushing syndrome. (E) Orthotopic xenograft colorectal cancer mouse model. (F) ADH1C and ADH1B expression in CRC and normal-matched tissues from our laboratory. The data were normalized to GAPDH and are expressed as the means ± SEMs. (G) ALDH2, ALDH1A1 and ALDH1B1 expression in CRC and normal-matched tissues from our laboratory. The data were normalized to GAPDH and are expressed as the means ± SEMs. (H) ADH1C and ADH1B expression in CRC and normal-matched tissues from the GEO GSE87211 datasets. The data were normalized to GAPDH and are expressed as the means ± SEMs. (I) ALDH2, ALDH1A1 and ALDH1B1 expression in CRC and normal-matched tissues from the GEO GSE87211 datasets. The data were normalized to GAPDH and are expressed as the means ± SEMs.
To confirm the effect of alcohol intake on tumour metastasis in CRC, an in vivo study was performed using the Lieber-DeCarli ethanol liquid diet. The results revealed that orthotopic xenograft tumours formed in all the mice, but none of the mice in the control group developed liver metastasis (Fig. 2A) compared with four out of six mice in the alcohol-treated group (Fig. 2B), which indicated that alcohol intake significantly promoted CRC metastasis. Taken together, the results indicated that a high circulating level of ethanol in the gut resulting from alcohol intake significantly promoted tumour metastasis in CRC.
In addition, our study also revealed that the incidence of alcohol-related CRC was associated with sex, which might be due to the prevalence of alcoholism in the male population. Moreover, we found that alcohol intake only significantly increased the risk of right colon cancer. We also found that most patients with alcohol-related CRC had smoking habits, which might also be an important risk factor for CRC [5] (Supplementary Table 1).
3.3. Ethanol, but not acetaldehyde, might be the crucial factor accounting for the incidence and metastasis of most alcohol-related CRC
Previous studies have confirmed that the metabolic rates of ethanol and acetaldehyde are primarily determined by the expression of key alcohol dehydrogenases (ADHs) and acetaldehyde dehydrogenases (ALDHs) [19]. Thus, we further investigated the alterations in the metabolism of alcohol in CRC by detecting the expression of key ADHs (ADH1C and ADH1B) and ALDHs (ALDH2, ALDH1B1 and ALDH1B1) in CRC and matched normal tissues. This analysis revealed that the transcriptional levels of ADH1C and ADH1B were substantially decreased in CRC compared with adjacent normal tissues (Fig. 1F). Further study showed significant decreases in the transcriptional levels of ALDH2 and ALDH1A1 and an increased transcriptional level of ALDH1B1 in CRC tissues. However, the absolute difference in the transcriptional level of ALDH2 between CRC and adjacent normal tissues was low (normal vs CRC = 0.6893 ± 0.0548 vs 0.6768 ± 0.0540, Fig. 1H). Interestingly, ALDH2 is the major enzyme that breaks down most ethanol-derived acetaldehyde, whereas ALDH1A1, as well as ALDH1B1, contributes to acetaldehyde metabolism only if ALDH2 is inactivated [19]. Thus, no significant changes in acetaldehyde metabolism might be found between CRC and normal tissues. Taken together, our results demonstrate that ethanol, but not acetaldehyde, accumulates more easily in CRC cells than in adjacent normal epithelial cells.
3.4. Ethanol enhances the migration/invasion, motility and homing capacity of CRC cells
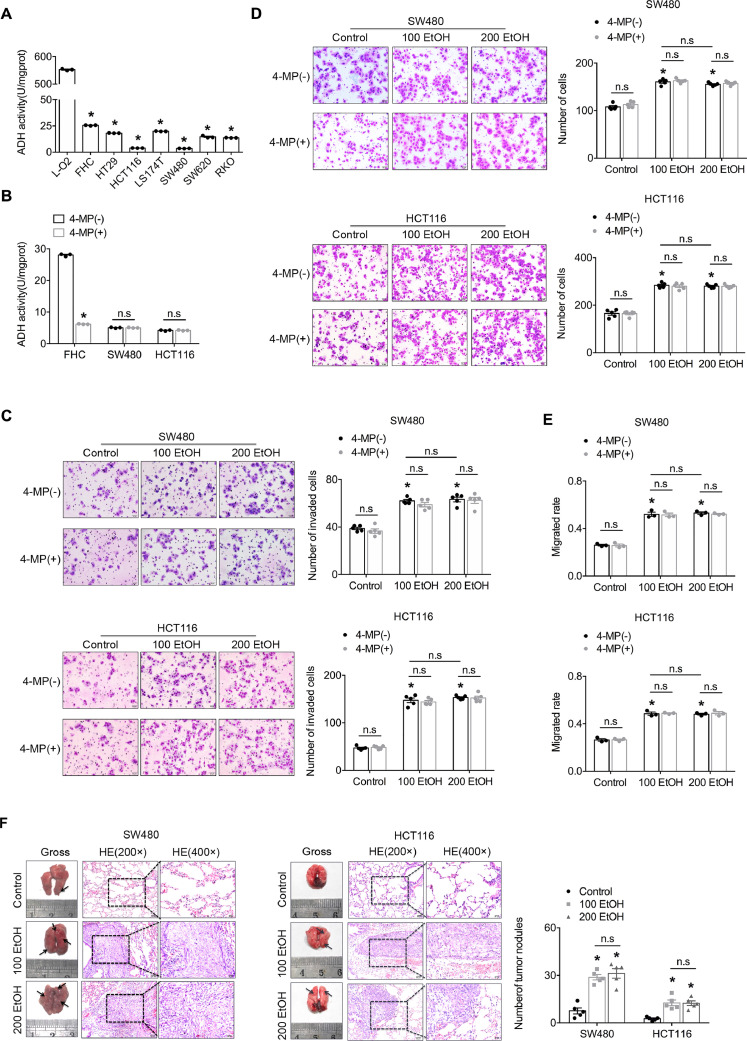
Our in vitro study revealed that the activity of ADH in all six CRC cell lines was notably weaker than that in the normal hepatic cell line L-O2, and particularly weak activity was detected in the SW480 and HCT116 cell lines, which presented up to a 157-fold decrease in activity (Fig. 3A). Consistent with the activity of ADH found in the above-mentioned cell lines, we also found that the transcriptional levels of ADH1C and ADH1B were significantly lower in the SW480 and HCT116 cell lines (Supplementary Fig. S1). In addition, 4-MP, a specific inhibitor of the activity of ADH, could not further inhibit the activity of ADH in both the SW480 and HCT116 cell lines, which indicated that both cells lines exhibited extremely weak ADH activity (Fig. 3B). We further investigated the biological effect of physiologically relevant concentrations (22 mM and 44 mM) of ethanol on CRC cells. Our study found that ethanol dose-independently promoted the migration/invasion and motility of SW480 and HCT116 cells, and this induction could not be further enhanced by 4-MP due to the already extremely weak ethanol metabolism in both cell lines (Fig. 3C–E, Supplementary Fig. S2). The in vivo study revealed that ethanol dose-independently promoted the formation of tumour nodules in the lungs compared with the controls, which indicated that ethanol promotes the homing capacity of cancer cells (Fig. 3F). Taken together, our findings demonstrate that ethanol plays an important role in promoting the migration/invasion of CRC cells.
Fig. 3.
Ethanol itself directly promotes the aggressiveness of CRC cells. (A) Ultraviolet spectrophotometer assay of the activity of ADH in normal hepatic and colorectal cell lines as well as six CRC cell lines. (B) Ultraviolet spectrophotometer assay of the activity of ADH in normal epithelial and CRC cell lines. “4-MP” indicates 4-methylpyrazole. (C) A Transwell assay with Matrigel was used to investigate the invasion ability of CRC cells. The bars in the right panel reflect the means ± SEMs. (D) A Transwell assay without Matrigel was used to investigate the migration ability of CRC cells. The bars in the right panel reflect the means ± SEMs. (E) A wound-healing assay was used to investigate the motility of CRC cells. (E) Representative images from the mouse tail-vein assay. The bars in the right panel reflect the means ± SEMs; *P < 0.05 and n.sP > 0.05 vs the control.
3.5. Ethanol, but not acetaldehyde, promotes epithelial-mesenchymal transition of CRC cells via the TGF-β/Smad/Snail axis
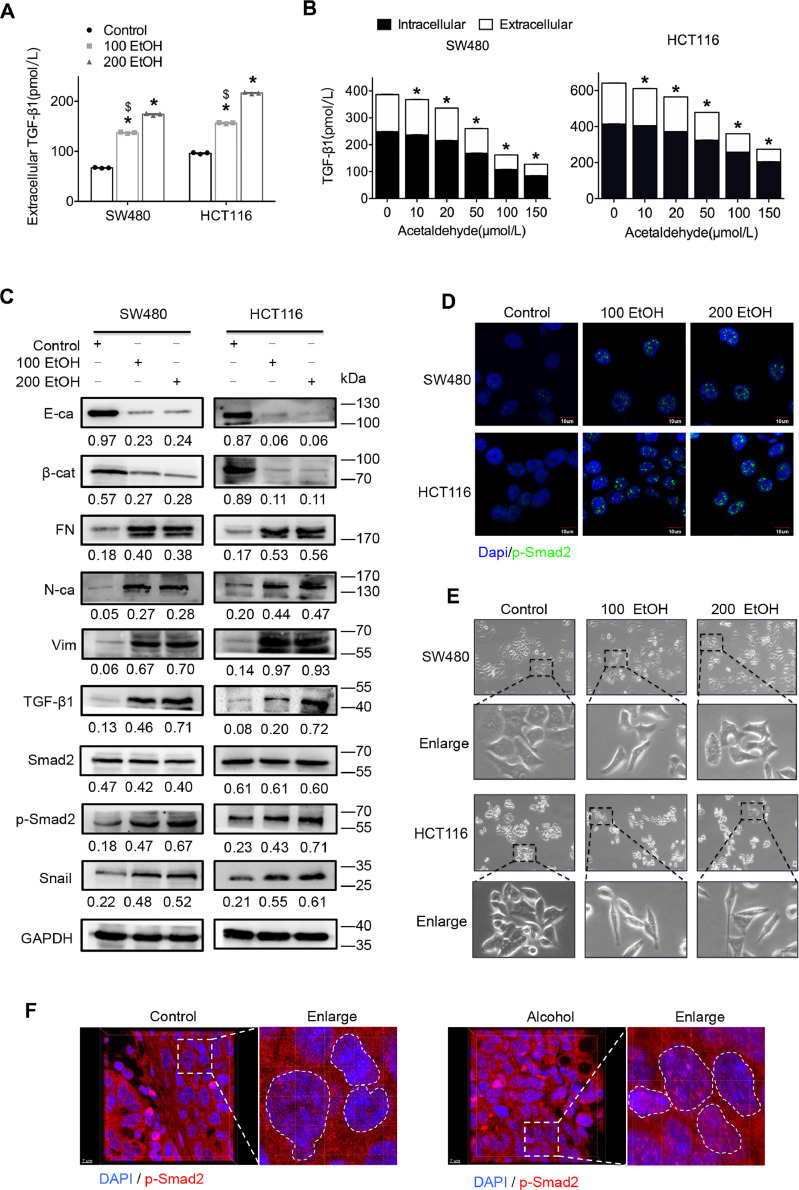
Consistent with the results from studies of other diseases [20], [21], [22], [23], we found that ethanol dose-dependently promoted intracellular and extracellular TGF-β1 expression (Fig. 4A and C). However, acetaldehyde intriguingly had the opposite effect on the expression of TGF-β1 (Fig. 4B). Further study showed that ethanol significantly promoted the phosphorylation and nuclear localization of Smad2 (Fig. 4C and D, Supplementary Fig. S3), which indicated the activation of TGF-β1/Smad signalling [24]. Interestingly, we found that SW480 and HCT116 cell lines exhibited significant higher levels of epithelial-mesenchymal transition (EMT) after treatment with ethanol at two different concentrations (Fig. 4E). Moreover, consistent alteration of the EMT signature (E-cadherin, β-catenin, fibronectin, N-cadherin and vimentin) was also found in both cell lines (Fig. 4C), which indicated that ethanol might enhance the aggressiveness of cancer cells by promoting EMT of CRC cells. Further study found that ethanol significantly promoted the expression of Snail (Fig. 4C), an important downstream effector of TGF-β signalling and a trigger of EMT [25,26]. In addition, the orthotopic xenograft CRC mouse model demonstrated that alcohol intake significantly increased the expression and nuclear location of phosphorylated-Smad2 in vivo (Fig. 4F, Supplementary Videos 1 and 2). Taken together, the results indicated that the accumulation of ethanol, but not acetaldehyde, might be the primary reason accounting for the development and progression of alcohol-related CRC through the activation of TGF-β signalling.
Fig. 4.
Ethanol itself promotes the epithelial-mesenchymal transition of CRC cells via the TGF-β/Smad/Snail axis. (A) An ELISA was used to detect the expression of extracellular TGF-β1 in cells treated with ethanol. (B) An ELISA was used to detect the expression of intracellular and extracellular TGF-β1 in cells treated with acetaldehyde. (C) Representative western blots for TGF-β signalling-associated proteins. The values under the membrane represent the expression of genes normalized to the expression of the reference gene GAPDH. “E-ca”, “β-cat”, “FN”, “N-ca”, and “Vim” indicate E-cadherin, β-catenin, fibronectin, N-cadherin and vimentin, respectively. (D) Immunofluorescence assay of p-Smad2 protein in ethanol-treated cells. Representative images are shown. The scale bars represent 7 μm. (E) Morphological observation of CRC cells treated with ethanol. (F) Immunofluorescence assay of p-Smad2 protein in ethanol-treated cells or samples from the orthotopic xenograft CRC mouse model. Representative images are shown. The scale bars represent 7 μm. *P < 0.05 vs the control; $P < 0.05 vs 200 EtOH.
3.6. TGF-β signalling plays a vital role in the ethanol-promoted aggressiveness of CRC cells
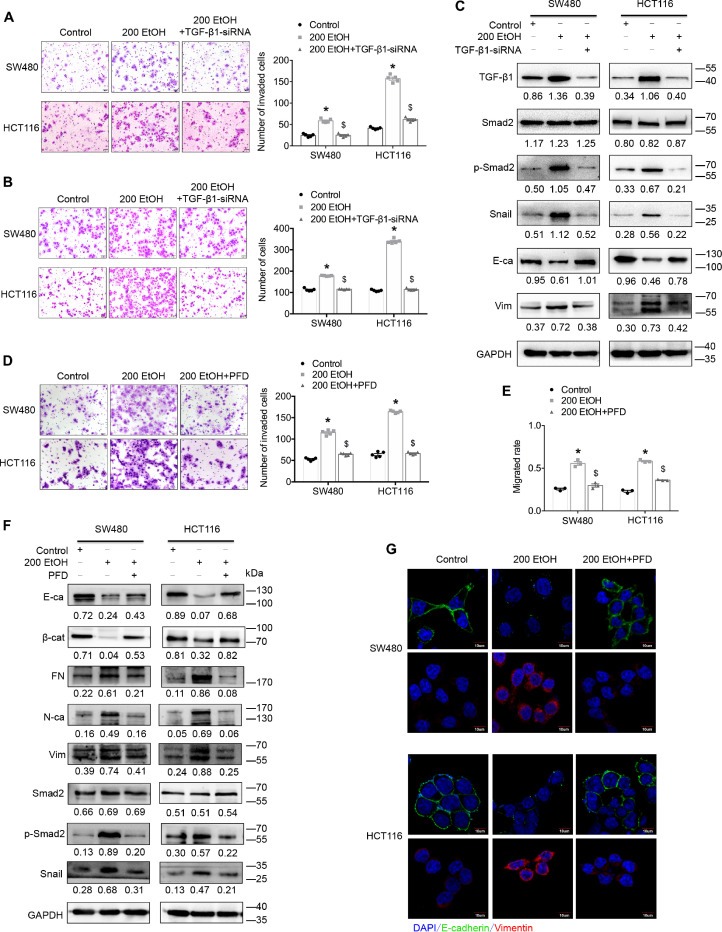
In this study, we used TGF-β1 small interfering RNA (siRNA) and a TGF-β/Smad signalling-specific inhibitor to confirm the role of TGF-β1 and TGF-β/Smad signalling in the ethanol-enhanced aggressiveness of CRC cells. The inhibition experiments revealed that decreasing the expression of TGF-β1 or blocking TGF-β/Smad signalling significantly eliminated the ethanol-promoted migration/invasion of CRC cells (Fig. 5A and B, Supplementary Figs. S4A and S4B). Consistent changes were also obtained in the analyses of the alteration of the EMT signature (E-cadherin and vimentin) and TGF-β (phosphorylated-Smad2 and Snail) signalling (Fig. 5C, Supplementary Fig. S4C). These findings strongly suggest that TGF-β1 plays a crucial role in the ethanol-enhanced aggressiveness of CRC cells via TGF-β/Smad signalling.
Fig. 5.
TGF-β signalling plays a vital role in the ethanol-promoted aggressiveness of CRC cells. (A) A Transwell assay with Matrigel was used to investigate the invasive ability of CRC cells. (B) A Transwell assay without Matrigel was used to investigate the migrated ability of CRC cells. (C) Representative western blots for TGF-β signalling-associated proteins. The values under the membrane represent the expression of genes normalized to the expression of the reference gene GAPDH. “E-ca” and “Vim” indicate E-cadherin and vimentin, respectively. (D) A Transwell assay with Matrigel was used to investigate the invasive ability of CRC cells. (E) A wound-healing assay was used to investigate the motility of CRC cells. (F) Representative western blots for TGF-β signalling-associated proteins. The values under the membrane represent the expression of genes normalized to the expression of the reference gene GAPDH. “E-ca”, “β-cat”, “FN”, “N-ca”, and “Vim” indicate E-cadherin, β-catenin, fibronectin, N-cadherin and vimentin, respectively. (G) Immunofluorescence assay showing the expression of EMT-associated proteins in cells. Representative merged images are shown. The scale bars represent 10 μm. “PFD” indicates pirfenidone. *P < 0.05 vs the control; $P < 0.05 vs 200 EtOH.
Intriguingly, pirfenidone (PFD) is a newly developed clinical anti-fibrotic drug that targets TGF-β signalling [27], but recent studies revealed that this drug can significantly inhibit the metastasis of human lung carcinoma by reverting the EMT of cancer cells [28,29]. However, its effects on CRC remain unknown. Thus, PFD was used in this study to determine its effect on CRC cells and the crucial role of ethanol-mediated TGF-β signalling in the aggressiveness of alcohol-related CRC. The results showed that PFD significantly decreased the phosphorylation and nuclear location of Smad2 in CRC cells (Supplementary Fig. S5A) and significantly inhibited the proliferation, migration/invasion and motility of CRC cells in vitro (Supplementary Figs. S5B–S5E). Further study revealed that PFD significantly eliminated the ethanol-promoted migration/invasion and motility of CRC cells in vitro (Fig. 5D and E, Supplementary Figs. S6–S8). Consistent with these biological phenotypes, PFD also reversed the ethanol-mediated alteration of the EMT signature (E-cadherin, β-catenin, fibronectin, N-cadherin and vimentin) and TGF-β (phosphorylated-Smad2 and Snail) signalling (Fig. 5F, Supplementary Fig. S9). Additional immunofluorescence assays revealed consistent alterations in the EMT signatures (E-cadherin and vimentin, Fig. 5G, Supplementary Figs. S10 and S11). These findings not only further confirmed the important role of TGF-β signalling in the ethanol-enhanced aggressiveness of CRC cells but also revealed that PFD might be a novel therapeutic strategy for the management of CRC patients.
3.7. The ethanol-induced cytoplasmic mislocalization of RUNX3 further promotes the aggressiveness of CRC cells via EMT by targeting Snail
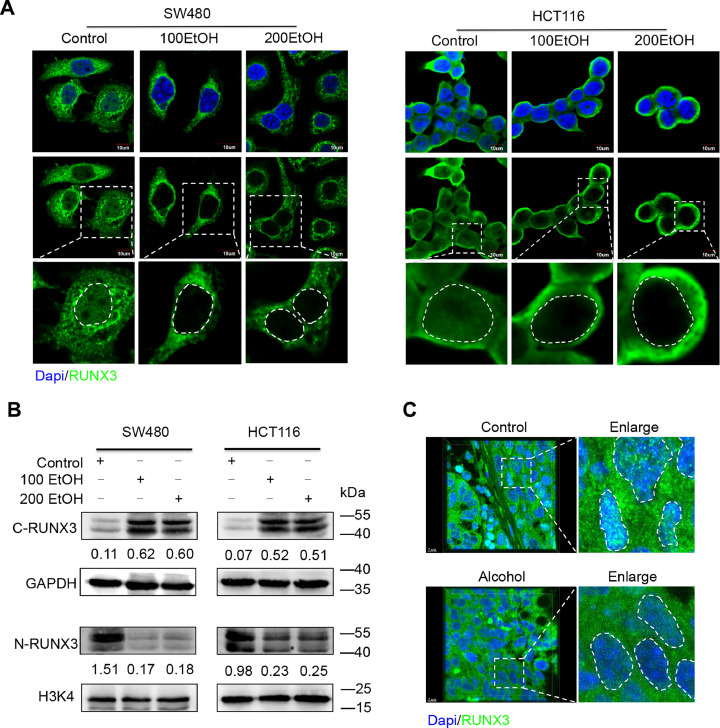
RUNX3 is a well-known cooperative factor for TGF-β signalling but acts as a tumour suppressor in the development and progression of multiple types of cancers (30, 31). Interestingly, our study found a significant absence of nuclear RUNX3 after treatment with different concentrations of ethanol (Fig. 6A). Further immunoblot assays confirmed that ethanol significantly decreased the expression of RUNX3 in the nucleus but promoted its expression in the cytoplasm. In addition, an orthotopic xenograft CRC mouse model demonstrated that alcohol intake significantly induced the cytoplasmic mislocalization of RUNX3 in CRC cells in vivo (Fig. 6C, Supplementary Videos 3 and 4). It is well known that the cytoplasmic mislocalization of RUNX3 could eliminate the RUNX3-mediated inhibition of tumour progression [32]. Therefore, the ethanol-mediated cytoplasmic mislocalization of RUNX3 might be an important mechanism accounting for the promotion of CRC metastasis.
Fig. 6.
Alcohol induces the cytoplasmic mislocalization of RUNX3 in vitro and in vivo. (A) Immunofluorescence assay of RUNX3 protein in cells treated with ethanol. (B) Representative western blots for the cytoplasmic and nuclear expression of RUNX3. The values under the membrane represent the expression of genes normalized to the expression of the reference gene GAPDH or H3K4. “C-RUNX3” and “N-RUNX3” indicate cytoplasmic and nuclear RUNX3, respectively. (C) Immunofluorescence assay of RUNX3 protein in samples from the orthotopic xenograft CRC mouse model.
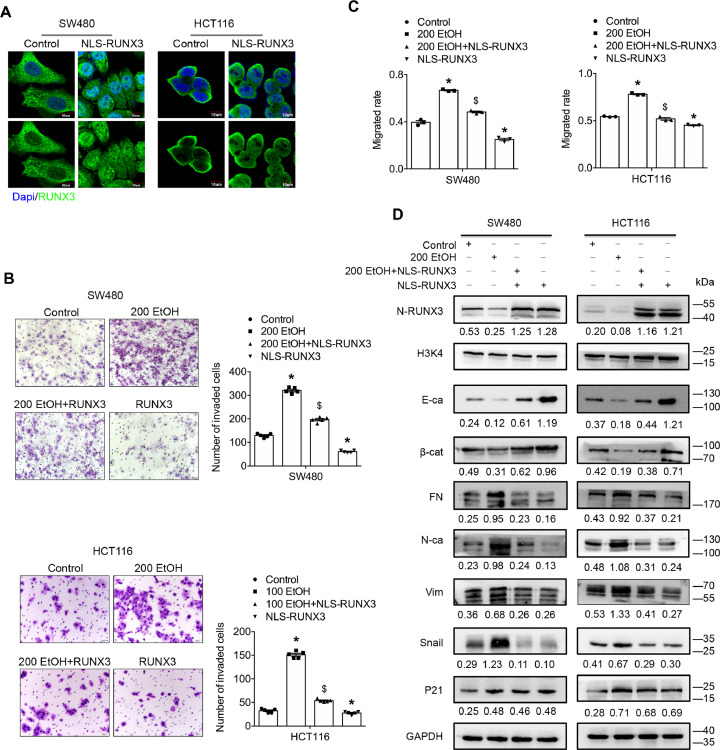
To clarify the role of RUNX3 in the metastasis of alcohol-related CRC, we recovered the nuclear expression of RUNX3 by constructing a RUNX3 vector with a nuclear localization sequence (NLS). Immunofluorescence and immunoblot assays were performed to confirm the transfection efficiency and nuclear localization of exogenous RUNX3 (Fig. 7A and D). Further studies found that nuclear RUNX3 overexpression significantly eliminated the ethanol-induced promotion of the migration/invasion abilities of both cell lines (Fig. 7B and C, Supplementary Figs. S13–S15). In addition, restoration of the nuclear expression of RUNX3 substantially reversed the ethanol-mediated alteration of Snail expression and the EMT signature. However, no significant difference in the expression of P21 was obtained after the restoration of nuclear RUNX3 expression (Fig. 7D, Supplementary S16). P21 is another downstream effector of TGF-β signalling that is tightly associated with cancer cell proliferation and anticancer drug resistance by inducing cell-cycle arrest [33]. Taken together, the findings indicate that the ethanol-induced cytoplasmic mislocalization of RUNX3 further promotes the aggressiveness of CRC cells by targeting Snail.
Fig. 7.
The ethanol-induced cytoplasmic mislocalization of RUNX3 further promotes the aggressiveness of CRC cells via EMT by targeting Snail. (A) Immunofluorescence assay of RUNX3 protein in cells transfected with the NLS-RUNX3 vector. (B) A Transwell assay with Matrigel was used to investigate the invasive ability of CRC cells treated with ethanol and/or NLS-RUNX3. (C) A wound-healing assay was used to investigate the motility of CRC cells treated with ethanol and/or NLS-RUNX3. (D) Representative western blots for TGF-β signalling-associated proteins. The values under the membrane represent the expression of genes normalized to the expression of the reference gene GAPDH or H3K4. “N-RUNX3”, “E-ca”, “βcat”, “FN”, “N-ca”, and “Vim” indicate nuclear-RUNX3, E-cadherin, β-catenin, fibronectin, N-cadherin and vimentin, respectively. *P < 0.05 vs the control; $P < 0.05 vs 200 EtOH.
3.8. Ethanol induces the cytoplasmic mislocalization of RUNX3 by enhancing the dissociation of the p-Smad2/3-RUNX3 complex
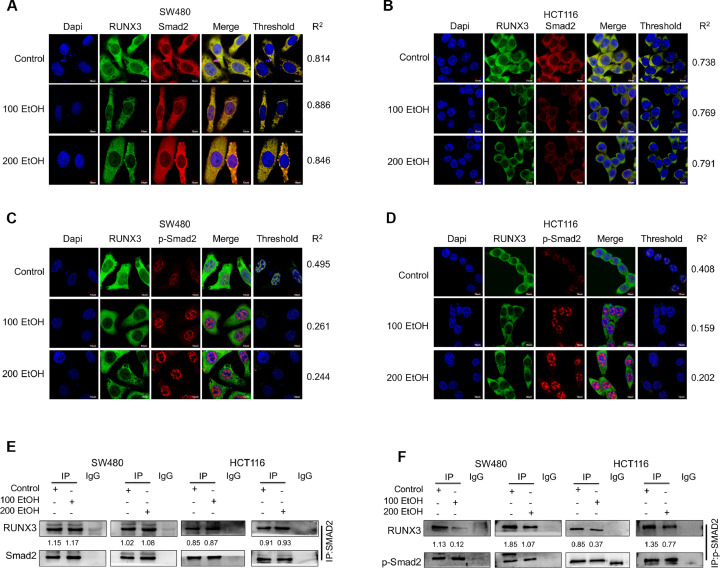
This study revealed that RUNX3 combines with the p-Smad2/3 complex to enter the nucleus and subsequently regulate the transcription of target genes [30]. Thus, we hypothesized that ethanol might induce the cytoplasmic localization of RUNX3 by promoting the dissociation of the phosphorylated-Smad2/3-RUNX3 and/or Smad2/3-RUNX3 complex. Immunofluorescence assays revealed that ethanol treatment significantly reduced the colocalization of RUNX3 and phosphorylated-Smad2 but not that of RUNX3 and Smad2 (Figs. 8A and B). Additional coimmunoprecipitation (co-IP) assays also determined that ethanol significantly reduced the binding efficiency of RUNX3 and phosphorylated Smad2 without change that of RUNX3 and Smad2 (Fig. 8C). These findings strongly suggest that ethanol induces the cytoplasmic localization of RUNX3 by promoting the dissociation of the phosphorylated-Smad2/3/RUNX3 complex.
Fig. 8.
Ethanol induces RUNX3 cytoplasmic mislocalization by dissociating the p-Smad2/3/RUNX3 complex. (A, B) An immunofluorescence assay was performed to investigate the co-localization of RUNX3 and Smad2 or p-Smad2. Representative images are shown. “R2” indicates the Pearson correlation coefficient. The scale bars represent 10 μm. (C) Co-immunoprecipitation assay of RUNX3 and Smad2 or p-Smad2 in CRC cells treated with ethanol. Representative images are shown (left panel). The bars in the right panel indicate the expression of the genes normalized to the expression of Smad2 or p-Smad2 and the control groups. (D) Hypothetical model mechanism. *P < 0.05 and NSP > 0.05 vs the control.
4. Discussion
Alcohol intake has generally been accepted as a common and legal lifestyle factor in many countries, particularly developed countries [34]. However, in 2012, based on a systematic review of evidence obtained from countries all over the world, the IARC drew the reliable conclusion that chronic and moderate to heavy alcohol intake is an important cause of cancer, including cancers of the oral cavity, pharynx, larynx, oesophagus, liver, colorectum and female breast [35]. Our study revealed that alcohol intake not only significantly increases the risk of CRC but also is tightly associated with tumour metastasis, which is the primary cause of the poor outcomes observed in most CRC patients [36,37]. Although new effective indicators and detecting strategies for CRC have been developed over the past decade, the detection and diagnosis of CRC at an early stage remain difficult [10,38]. Thus, it is possible that most CRC patients with chronic alcohol intake might continue drinking until significant symptoms and/or signs are detected. Further study confirmed that alcohol intake significantly promoted the migration/invasion of CRC cells via the ethanol-mediated TGF-β/RUNX3/Snail axis. Therefore, we hypothesized that alcohol intake not only increases the risk of CRC but also plays a vital role in accelerating its progression from early to advanced stages.
Ethanol and acetaldehyde are two major carcinogens associated with alcohol intake, and the effects of alcohol on the development and progression of CRC are primarily determined by the duration and extent of exposure in the colorectum [11], [12], [13]. Our study showed that most patients with alcohol-related CRC do not experience flushing syndrome after heavy alcohol intake, which indicates that their systemic level of acetaldehyde in the circulation and tissues was not substantially increased even after the consumption of a great amount of alcohol [19]. Because alcohol is absorbed rapidly from the gastrointestinal tract to the circulation system during ingestion [34], a patient with a heavy alcohol intake must have a notably high level of ethanol in his/her circulation after drinking, but the duration of this high level is determined by the capacity of the patient's body to break down alcohol [19]. In addition, most alcohol is absorbed into the circulation in the small intestine, and only some reaches the colorectum after ingestion [34]; thus, the ethanol level in colorectal tissue should be determined largely by the concentration of ethanol in the circulation. Taken together, the results indicated that an increased circulating level of ethanol in the colorectum is the major cause of the increased CRC incidence and metastasis detected in most patients with alcohol-related CRC.
The effects of ethanol on colorectal cells were determined mainly by the metabolic rates of ethanol and its byproduct acetaldehyde. In humans, ADHs and ALDHs are the major enzymes that break down ethanol and acetaldehyde, respectively [19]. Humans have seven ADHs, namely, ADH1A, ADH1B, ADH1C, ADH4, ADH5, ADH6 and ADH7. It has been well demonstrated that ADH1B, ADH1C and ADH4 are the key ADHs responsible for most of the breakdown of ethanol [19]. Because ADH4 is almost exclusively expressed in the liver [19], the ethanol metabolism in colorectal cells was primarily determined by the expression of ADH1B and ADH1C. Because the expression levels of both ADH1B and ADH1C were notably decreased in CRC compared with adjacent normal tissues, we hypothesized that almost all CRC patients exhibit an impaired ethanol metabolism. In addition, 18 enzymes in the ALDH superfamily are responsible for the maintenance of a non-toxic level of acetaldehyde in cells, and among these, ALDH2, ALDH1A1 and ALDH1B1 are the most relevant to acetaldehyde oxidation [39]. However, the rate of ethanol-derived acetaldehyde breakdown is mainly determined by the alteration of ALDH2 because both ALDH1A1 and ALDH1B1 contribute to acetaldehyde metabolism only in the absence of ALDH2 [19]. Interestingly, our study found a statistical difference in the expression of ALDH2 between CRC and adjacent normal tissues, but this difference was extremely low in absolute value. Therefore, we hypothesized the existence of a functional difference in the ethanol-derived acetaldehyde metabolism between CRC and adjacent normal tissues. Collectively, the results indicate that ethanol, but not acetaldehyde, might accumulate much easier in CRC cells compared with normal epithelial cells. Ethanol plays a vital role in the development and progression of CRC.
To investigate the mechanistic role of ethanol in the migration/invasion of CRC cells, we selected the SW480 and HCT116 cell lines, which have the lowest metabolic rate of ethanol, and used 4-MP to further slow the conversion of ethanol to acetaldehyde. However, no significant difference in the metabolic rate of ethanol and no biological changes were detected in either cell line. These findings suggest that the basal metabolic rate of ethanol might be extremely low in SW480 and HCT116 cells and that ethanol is the cause of the observed changes in CRC cells. Moreover, we surprisingly found that ethanol could significantly promote the expression and secretion of TGF-β1, whereas acetaldehyde exerted the opposite effects. It is well known that TGF-β1 is a vital ligand for the activation of TGF-β signalling [20], which plays an important role in the development and progression of cancers [40,41]. This finding also suggested that ethanol might play a crucial role in CRC metastasis by activating TGF-β signalling. In addition, although the duration of the exposure of CRC cells to ethanol was notably shorter compared with that used in studies of breast cancer [17,42], significant changes were also found in our study, which indicated that CRC cells might be highly sensitive to ethanol. Additional studies on CRC or other types of cancer are necessary to further investigate the role of ethanol itself in the development and progression of cancer.
Although previous studies have demonstrated that ethanol promotes the expression of TGF-β1, the underlying mechanism remain unclear [20], [21], [22], [23]. A recent study revealed that ethanol directly enhances the activation of TGF-β signalling by increasing the cell-surface and non-lipid raft microdomain localization of the type II TGF-β receptor [21]. Thus, ethanol might activate TGF-β signalling by enhancing the sensitivity of the TGF-β receptor to TGF-β1, which is already present in the tumour microenvironment and subsequently auto-induced in CRC cells. In contrast, it is well known that ethanol accumulation can significantly enhance oxidative stress in cells and thereby promote the expression of bioactive TGF-β1 [43,44]. Thus, the ethanol/P450s/ROS axis might be another mechanism underlying the ethanol-induced increase in the expression of TGF-β1.
Previous studies have demonstrated that RUNX3 is an independent regulator of TGF-β signalling and acts as a tumour suppressor in a variety of cancers [31,[45], [46], [47], [48], [49]]. Our investigation of the effects of ethanol on TGF-β signalling surprisingly revealed that ethanol dose-independently induced the cytoplasmic mislocalization of RUNX3, which is an important reason for the elimination of its suppressive effect on tumours [32]. Intriguingly, we found that the recruitment of nuclear RUNX3 significantly reversed the effects of ethanol on EMT and the migration/invasion of CRC cells by targeting Snail. However, no significant change was found in the expression of P21, another important gene downstream of TGF-β signalling [50]. These results suggested that the RUNX3-mediated regulation of the genes downstream of TGF-β signalling might be selective, and further studies are needed to investigate the underlying mechanism. In addition, because nuclear-localized RUNX3 plays a vital role in tumour suppression [51,52], we hypothesized that the cytoplasmic mislocalization of RUNX3 is another important mechanism underlying the ethanol-induced development and progression of CRC.
It has been well established that RUNX3 combines with the phosphorylated-Smad2/3 (p-Smad2/3) complex and subsequently enters the nucleus to regulate the transcription of downstream genes [30]. Our study demonstrated that RUNX3 could combine with the Smad2/3 complex before its phosphorylation and that exposure to ethanol did not change the binding of RUNX3 with the Smad2/3/RUNX3 complex. However, the combination of RUNX3 with the p-Smad2/3 complex was significantly dissociated after ethanol exposure. Thus, it is possible that ethanol induces the cytoplasmic mislocalization of RUNX3 by dissociating the p-Smad2/3/RUNX3 complex, which might result from chemical reactions with ethanol or ROS. Further investigations are needed to clarify the mechanism underlying the ethanol-induced dissociation of the p-Smad2/3/RUNX3 complex.
PFD is an orally available anti-fibrotic drug that exerts significant anti-inflammatory and anti-fibrotic effects in many organs, including the lung, renal system, liver, heart, muscle and eye [53]. PFD regulates fibroblast proliferation and collagen synthesis mainly by inhibiting TGF-β signalling [54]. Intriguingly, recent studies have revealed that PFD has important effects on the treatment of cancers by blocking TGF-β signalling [27,28]. Our studies also demonstrated that PFD significantly inhibited the aggressiveness of CRC cells. Because ethanol exerts crucial effects on tumour metastasis in CRC by activating TGF-β signalling, we consider PFD to be a novel therapeutic strategy for alcohol-related CRC.
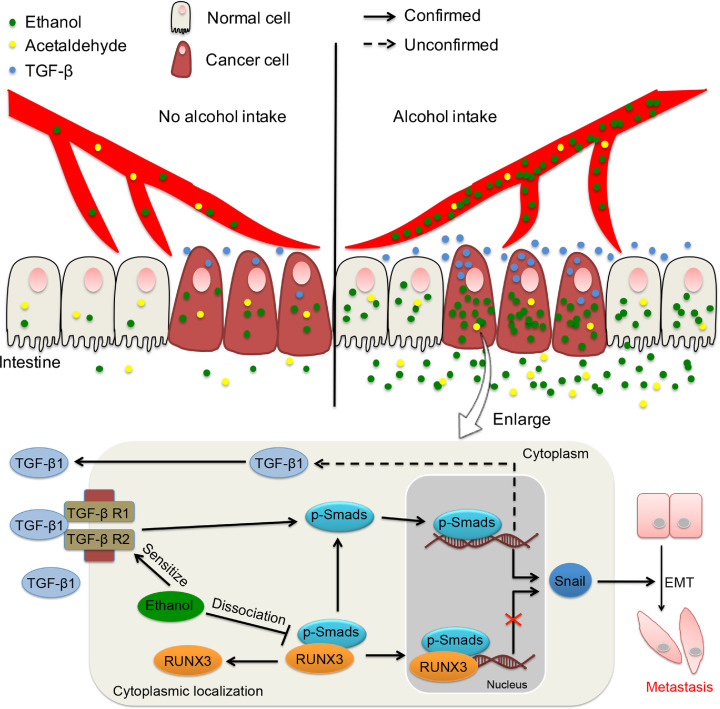
In conclusion, our study highlights the effects of alcohol intake on promoting CRC metastasis via the ethanol-mediated TGF-β/RUNX3/Snail axis (Fig. 9). Moreover, PFD might be a novel anticancer adjuvant therapy for CRC.
Fig. 9.
Graphical abstract of this study. The colorectum of most patients with alcohol-related CRC is exposed to a high circulating level of ethanol after drinking, and CRC cells exhibit an extremely impaired ethanol metabolism. Ethanol itself enhances the migration/invasion of CRC by promoting EMT via the TGF-β/RUNX3/Snail axis. PFD might be a novel therapeutic strategy for the management of CRC by targeting TGF-β signalling.
Declaration of Competing Interest
The authors have declared that no conflicts of interest exist.
Acknowledgments
Acknowledgements
Not applicable
Funding
This work was supported by the Foundation of High School Doctor Course of the National Department of Education (20134433110006) and the National Natural Science Foundation of China (Nos. 81572813, 81773082, and 81472319).
Ethics statement
This study was approved by the Ethics Committees of Zhujiang Hospital, Southern Medical University, and all aspects of the study comply with the criteria established by the Declaration of Helsinki. The CRC patients provided informed consent to participate in this study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.011.
Contributor Information
Kehong Zheng, Email: drzhengkh@163.com.
Lina Yu, Email: nana1800@smu.edu.cn.
Liang Zhao, Email: liangsmu@foxmail.com.
Qian Wang, Email: Wangqian@smu.edu.cn.
Appendix. Supplementary materials
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Sonoshita M., Itatani Y., Kakizaki F., Sakimura K., Terashima T., Katsuyama Y. Promotion of colorectal cancer invasion and metastasis through activation of notch-dab1-abl-rhogef protein trio. Cancer Discov. 2015;5(2):198–211. doi: 10.1158/2159-8290.CD-14-0595. [DOI] [PubMed] [Google Scholar]
- 3.He G.Y., Hu J.L., Zhou L., Zhu X.H., Xin S.N., Zhang D. The FOXD3/miR-214/MED19 axis suppresses tumour growth and metastasis in human colorectal cancer. Br J Cancer. 2016;115(11):1367–1378. doi: 10.1038/bjc.2016.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cernigliaro C., D'Anneo A., Carlisi D., Giuliano M., Marino Gammazza A., Barone R. Ethanol-Mediated stress promotes autophagic survival and aggressiveness of colon cancer cells via activation of nrf2/ho-1 pathway. Cancers. 2019;11(4) doi: 10.3390/cancers11040505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson A.S., Caswell S., Mowat C., Strachan J.A., Steele R.J.C. Lifestyle in patients at increased risk of colorectal cancer. J Hum Nutr Diet. 2019;32(5):570–577. doi: 10.1111/jhn.12663. [DOI] [PubMed] [Google Scholar]
- 6.Tuan J., Chen Y.X. Dietary and lifestyle factors associated with colorectal cancer risk and interactions with microbiota: fiber, red or processed meat and alcoholic drinks. Gastrointest Tumors. 2016;3(1):17–24. doi: 10.1159/000442831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNabb S., Harrison T.A., Albanes D., Berndt S.I., Brenner H., Caan B.J. Meta-analysis of 16 studies of the association of alcohol with colorectal cancer. Int J Cancer. 2019 doi: 10.1002/ijc.32377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandal P. Molecular signature of nitric oxide on major cancer hallmarks of colorectal carcinoma. Inflammopharmacology. 2018;26(2):331–336. doi: 10.1007/s10787-017-0435-z. [DOI] [PubMed] [Google Scholar]
- 9.McVicker B., Tuma D.J., Lazure K.E., Thomas P., Casey C.A. Alcohol, carcinoembryonic antigen processing and colorectal liver metastases. Adv Exp Med Biol. 2015;815:295–311. doi: 10.1007/978-3-319-09614-8_17. [DOI] [PubMed] [Google Scholar]
- 10.Raskov H., Pommergaard H.C., Burcharth J., Rosenberg J. Colorectal carcinogenesis–update and perspectives. World J Gastroenterol. 2014;20(48):18151–18164. doi: 10.3748/wjg.v20.i48.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter V., Jansen L., Ulrich A., Roth W., Blaker H., Chang-Claude J. Alcohol consumption and survival of colorectal cancer patients: a population-based study from germany. Am J Clin Nutr. 2016;103(6):1497–1506. doi: 10.3945/ajcn.115.127092. [DOI] [PubMed] [Google Scholar]
- 12.Svensson T., Yamaji T., Budhathoki S., Hidaka A., Iwasaki M., Sawada N. Alcohol consumption, genetic variants in the alcohol- and folate metabolic pathways and colorectal cancer risk: the JPHC study. Sci Rep. 2016;6:36607. doi: 10.1038/srep36607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pflaum T., Hausler T., Baumung C., Ackermann S., Kuballa T., Rehm J. Carcinogenic compounds in alcoholic beverages: an update. Arch Toxicol. 2016;90(10):2349–2367. doi: 10.1007/s00204-016-1770-3. [DOI] [PubMed] [Google Scholar]
- 14.Niu Y., Shao Z., Wang H., Yang J., Zhang F., Luo Y. LASP1-S100A11 axis promotes colorectal cancer aggressiveness by modulating TGFbeta/Smad signaling. Sci Rep. 2016;6:26112. doi: 10.1038/srep26112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H., Shi J., Luo Y., Liao Q., Niu Y., Zhang F. LIM and SH3 protein 1 induces TGFbeta-mediated epithelial-mesenchymal transition in human colorectal cancer by regulating S100A4 expression. Clin Cancer Res. 2014;20(22):5835–5847. doi: 10.1158/1078-0432.CCR-14-0485. An Official Journal of the American Association for Cancer Research. [DOI] [PubMed] [Google Scholar]
- 16.Millwood I.Y., Walters R.G., Mei X.W., Guo Y., Yang L., Bian Z. Conventional and genetic evidence on alcohol and vascular disease aetiology: a prospective study of 500 000 men and women in china. Lancet. 2019;393(10183):1831–1842. doi: 10.1016/S0140-6736(18)31772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu M., Wang S., Ren Z., Frank J.A., Yang X.H., Zhang Z. Chronic ethanol exposure enhances the aggressiveness of breast cancer: the role of p38gamma. Oncotarget. 2016;7(3):3489–3505. doi: 10.18632/oncotarget.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Offermans N.S.M., Ketcham S.M., van den Brandt P.A., Weijenberg M.P., Simons C. Alcohol intake, adh1b and adh1c genotypes, and the risk of colorectal cancer by sex and subsite in the Netherlands cohort study. Carcinogenesis. 2018;39(3):375–388. doi: 10.1093/carcin/bgy011. [DOI] [PubMed] [Google Scholar]
- 19.Hurley T.D., Edenberg H.J. Genes encoding enzymes involved in ethanol metabolism. Alcohol Res Curr Rev. 2012;34(3):339–344. [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnasamy Y., Ramshesh V.K., Gooz M., Schnellmann R.G., Lemasters J.J., Zhong Z. Ethanol and high cholesterol diet causes severe steatohepatitis and early liver fibrosis in mice. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0163342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang S.S., Chen C.L., Huang F.W., Johnson F.E., Huang J.S. Ethanol enhances TGF-beta activity by recruiting TGF-beta receptors from intracellular vesicles/lipid rafts/caveolae to non-lipid raft microdomains. J Cell Biochem. 2016;117(4):860–871. doi: 10.1002/jcb.25389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Z., Hou T., Shi W., Liu W., He H. Inhibition of hepatocyte apoptosis: An important mechanism of corn peptides attenuating liver injury induced by ethanol. Int J Mol Sci. 2015;16(9):22062–22080. doi: 10.3390/ijms160922062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown S.D., Brown L.A. Ethanol (EtOH)-induced TGF-beta1 and reactive oxygen species production are necessary for EtOH-induced alveolar macrophage dysfunction and induction of alternative activation. Alcohol Clin Exp Res. 2012;36(11):1952–1962. doi: 10.1111/j.1530-0277.2012.01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F., Luo Y., Shao Z., Xu L., Liu X., Niu Y. MicroRNA-187, a downstream effector of TGFbeta pathway, suppresses SMAD-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 2016;373(2):203–213. doi: 10.1016/j.canlet.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 25.Zheng H., Li W., Wang Y., Liu Z., Cai Y., Xie T. Glycogen synthase kinase-3 beta regulates snail and beta-catenin expression during Fas-induced epithelial-mesenchymal transition in gastrointestinal cancer. Eur J Cancer. 2013;49(12):2734–2746. doi: 10.1016/j.ejca.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Chen X.H., Liu Z.C., Zhang G., Wei W., Wang X.X., Wang H. TGF-beta and EGF induced HLA-I downregulation is associated with epithelial-mesenchymal transition (EMT) through upregulation of snail in prostate cancer cells. Mol Immunol. 2015;65(1):34–42. doi: 10.1016/j.molimm.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Li C., Rezov V., Joensuu E., Vartiainen V., Ronty M., Yin M. Pirfenidone decreases mesothelioma cell proliferation and migration via inhibition of ERK and AKT and regulates mesothelioma tumor microenvironment in vivo. Sci Rep. 2018;8(1):10070. doi: 10.1038/s41598-018-28297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurimoto R., Ebata T., Iwasawa S., Ishiwata T., Tada Y., Tatsumi K. Pirfenidone may revert the epithelial-to-mesenchymal transition in human lung adenocarcinoma. Oncol Lett. 2017;14(1):944–950. doi: 10.3892/ol.2017.6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujiwara A., Shintani Y., Funaki S., Kawamura T., Kimura T., Minami M. Pirfenidone plays a biphasic role in inhibition of epithelial-mesenchymal transition in non-small cell lung cancer. Lung Cancer. 2017;106:8–16. doi: 10.1016/j.lungcan.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Chen F., Liu X., Bai J., Pei D., Zheng J. The emerging role of RUNX3 in cancer metastasis (Review) Oncol Rep. 2016;35(3):1227–1236. doi: 10.3892/or.2015.4515. [DOI] [PubMed] [Google Scholar]
- 31.Tong D.D., Jiang Y., Li M., Kong D., Meng X.N., Zhao Y.Z. RUNX3 inhibits cell proliferation and induces apoptosis by TGF-beta-dependent and -independent mechanisms in human colon carcinoma cells. Pathobiol J Immunopathol Mol Cell Biol. 2009;76(4):163–169. doi: 10.1159/000218332. [DOI] [PubMed] [Google Scholar]
- 32.Kang K.A., Piao M.J., Ryu Y.S., Maeng Y.H., Hyun J.W. Cytoplasmic localization of RUNX3 via histone deacetylase-mediated src expression in oxidative-stressed colon cancer cells. J Cell Physiol. 2016;232(7):1914–1921. doi: 10.1002/jcp.25746. [DOI] [PubMed] [Google Scholar]
- 33.El-Deiry W.S. p21(WAF1) mediates cell-cycle inhibition, relevant to cancer suppression and therapy. Cancer Res. 2016;76(18):5189–5191. doi: 10.1158/0008-5472.CAN-16-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowan D.M., Maskrey J.R., Fung E.S., Woods T.A., Stabryla L.M., Scott P.K. Best-practices approach to determination of blood alcohol concentration (BAC) at specific time points: Combination of ante-mortem alcohol pharmacokinetic modeling and post-mortem alcohol generation and transport considerations. Regul Toxicol Pharmacol. 2016;78:24–36. doi: 10.1016/j.yrtph.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 35.Humans IWGotEoCRt Personal habits and indoor combustions. volume 100 E. a review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 2012;100(Pt E):1–538. [PMC free article] [PubMed] [Google Scholar]
- 36.Siegel R.L., Miller K.D., Fedewa S.A., Ahnen D.J., Meester R.G.S., Barzi A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 37.Zhou R., Shao Z., Liu J., Zhan W., Gao Q., Pan Z. COPS5 and LASP1 synergistically interact to downregulate 14-3-3sigma expression and promote colorectal cancer progression via activating PI3K/AKT pathway. Int J Cancer. 2018;142(9):1853–1864. doi: 10.1002/ijc.31206. [DOI] [PubMed] [Google Scholar]
- 38.Zarour L.R., Anand S., Billingsley K.G., Bisson W.H., Cercek A., Clarke M.F. Colorectal cancer liver metastasis: evolving paradigms and future directions. Cell Mol Gastroenterol Hepatol. 2017;3(2):163–173. doi: 10.1016/j.jcmgh.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson B., Brocker C., Thompson D.C., Black W., Vasiliou K., Nebert D.W. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genom. 2011;5(4):283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang S., Holzel M., Knijnenburg T., Schlicker A., Roepman P., McDermott U. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151(5):937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H., Verhaak R.G. Transcriptional mimicry by tumor-associated stroma. Nat Genet. 2015;47(4):307–309. doi: 10.1038/ng.3255. [DOI] [PubMed] [Google Scholar]
- 42.Xu M., Ren Z., Wang X., Comer A., Frank J.A., Ke Z.J. ErbB2 and p38gamma mapk mediate alcohol-induced increase in breast cancer stem cells and metastasis. Mol Cancer. 2016;15(1):52. doi: 10.1186/s12943-016-0532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Millonig G., Nair J., Patsenker E., Stickel F., Mueller S. Ethanol-induced cytochrome P4502E1 causes carcinogenic etheno-DNA lesions in alcoholic liver disease. Hepatology. 2009;50(2):453–461. doi: 10.1002/hep.22978. [DOI] [PubMed] [Google Scholar]
- 44.Hinge A., Xu J., Javier J., Mose E., Kumar S., Kapur R. p190-B rhogap and intracellular cytokine signals balance hematopoietic stem and progenitor cell self-renewal and differentiation. Nat Commun. 2017;8:14382. doi: 10.1038/ncomms14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Wang X., Cheng J., Wang Z., Jiang T., Hou N. MicroRNA-20a-5p targets RUNX3 to regulate proliferation and migration of human hepatocellular cancer cells. Oncol Rep. 2016;36(6):3379–3386. doi: 10.3892/or.2016.5144. [DOI] [PubMed] [Google Scholar]
- 46.Wang X.Z., Cheng Y., Wang K.L., Liu R., Yang X.L., Wen H.M. Peperomin e reactivates silenced tumor suppressor genes in lung cancer cells by inhibition of dna methyltransferase. Cancer Sci. 2016;107(10):1506–1519. doi: 10.1111/cas.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen F., Liu X., Cheng Q., Zhu S., Bai J., Zheng J. RUNX3 regulates renal cell carcinoma metastasis via targeting miR-6780a-5p/E-cadherin/EMT signaling axis. Oncotarget. 2016;8(60):101042–101056. doi: 10.18632/oncotarget.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song X.Y., Li B.Y., Zhou E.X., Wu F.X. The clinicopathological significance of RUNX3 hypermethylation and mRNA expression in human breast cancer, a meta-analysis. OncoTargets Ther. 2016;9:5339–5347. doi: 10.2147/OTT.S77828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong Y., Zou S., Yang F., Xu X., Bu W., Jia J. RUNX3-mediated up-regulation of miR-29b suppresses the proliferation and migration of gastric cancer cells by targeting kdm2a. Cancer Lett. 2016;381(1):138–148. doi: 10.1016/j.canlet.2016.07.038. [DOI] [PubMed] [Google Scholar]
- 50.Oshimori N., Oristian D., Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell. 2015;160(5):963–976. doi: 10.1016/j.cell.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Q.L., Ito K., Sakakura C., Fukamachi H., Inoue K., Chi X.Z. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109(1):113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 52.Voon D.C., Wang H., Koo J.K., Nguyen T.A., Hor Y.T., Chu Y.S. Runx3 protects gastric epithelial cells against epithelial-mesenchymal transition-induced cellular plasticity and tumorigenicity. Stem Cells. 2012;30(10):2088–2099. doi: 10.1002/stem.1183. [DOI] [PubMed] [Google Scholar]
- 53.Zou W.J., Huang Z., Jiang T.P., Shen Y.P., Zhao A.S., Zhou S. Pirfenidone inhibits proliferation and promotes apoptosis of hepatocellular carcinoma cells by inhibiting the wnt/beta-catenin signaling pathway. Med Sci Monit Int Med J Exp Clin Res. 2017;23:6107–6113. doi: 10.12659/MSM.907891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji T., Li S., Zhang Y., Lang J., Ding Y., Zhao X. An MMP-2 responsive liposome integrating antifibrosis and chemotherapeutic drugs for enhanced drug perfusion and efficacy in pancreatic cancer. ACS Appl Mater Interfaces. 2016;8(5):3438–3445. doi: 10.1021/acsami.5b11619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.