Abstract
Background
Sleep disordered breathing (SDB) is a common disorder that results in oxidative stress and inflammation and is associated with multiple age-related health outcomes. Epigenetic age acceleration is a DNA methylation (DNAm)-based marker of fast biological aging. We examined the associations of SDB traits with epigenetic age acceleration.
Methods
A sample of 622 participants from the Multi-Ethnic Study of Atherosclerosis (MESA) had blood DNAm measured and underwent Type 2 in-home polysomnography that assessed apnea-hypopnea index (AHI), percentage of sleep time with oxygen saturation lower than 90% (Per90), and arousal index. DNAm data provided measures of DNAm-Age acceleration and DNAm-PhenoAge acceleration. The association of each SDB trait with age acceleration was estimated using linear regression, controlling for covariates. In secondary analyses, we studied the associations of SDB traits with epigenetic age acceleration 2–10 years after sleep study in 530 individuals from the Framingham Heart Study (FHS).
Findings
In MESA, AHI was associated with greater DNAm-PhenoAge acceleration (β = 0.03; 95% CI [0.001, 0.06]). Arousal index was associated with greater DNAm-Age acceleration (β = 0.04; 95% CI [0.01, 0.07]). Both associations were stronger in women than men. In the secondary FHS analyses, Per90 was associated with greater DNAm-Age acceleration and this association was stronger in men.
Interpretation
More severe SDB was associated with epigenetic age acceleration in both cohorts. Future work should prospectively study short- and long-term effects of SDB, and whether treatment reduces epigenetic age acceleration among those individuals with SBD.
Funding
National Institutes of Health.
Keywords: Sleep disordered breathing, Apnea-hypopnea index, Hypoxemia, Arousal index, Epigenetic age acceleration, Aging
Research in context.
Evidence before this study
We searched PubMed up to May 1, 2019, with the terms “DNA methylation age”, “epigenetic age”, “epigenetic age acceleration”, “epigenetic aging”, “epigenetic clock”, “epigenetic biomarker of aging”, and “sleep” for studies investigating any association between epigenetic age acceleration and sleep. We also searched for relevant references in the identified articles and review articles on DNA methylation age. Prior studies have linked modifiable lifestyle factors (e.g., smoking) to accelerated epigenetic age. Sleep behaviors and disorders represent another set of behavioral and physiological mechanisms that may be modified. But their role in epigenetic age acceleration has been minimally explored. One initial study has reported that insomnia was associated with advanced epigenetic age. However, no studies to date have described the association between sleep disordered breathing and epigenetic age acceleration.
Added value of this study
To our knowledge, this is the first empirical study to investigate the associations between sleep disordered breathing traits and epigenetic age acceleration and to assess potential sex differences in the associations. In a cohort of racially/ethnically diverse adults, we found that more severe sleep disordered breathing was associated with epigenetic age acceleration. This finding was validated in another, predominantly White, cohort. A meta-analysis of the cohorts confirmed the finding.
Implications of all the available evidence
Our analyses suggest an association between sleep disordered breathing severity with epigenetic age acceleration. Sleep disordered breathing, one of the most common yet under diagnosed and under recognized sleep disorders, might carry unique prospects in terms of developing interventions to delay aging progression and preventing age-related disorders, which might help to extend the human health span and lifespan.
Alt-text: Unlabelled box
1. Introduction
Sleep disordered breathing (SDB) is a highly prevalent, chronic, and treatable condition characterized by recurrent episodes of complete (apnea) or partial (hypopnea) upper airway obstruction during sleep, which lead to fragmentation of sleep and decreases in oxyhemoglobin saturation [1]. The estimated prevalence of SDB was 9% for women and 24% for men in the 1990s [1], which has been increasing over the past two decades [2,3]. SDB has been associated with increased vulnerability for mortality [4], age-related cognitive and functional decline [2], and chronic diseases such as stroke, coronary heart disease, and heart failure [5,6].
While it is established that SDB prevalence and characteristics change with age [3], little is known on whether SDB is associated with age acceleration. Aging processes differentially progress between individuals, who, accordingly, may have different susceptibilities to age-related diseases and death [7]. Hence, a current area of research distinguishes between chronological and biological age: the first being years since birth, and the latter estimated using various biomarkers [7,8]. Identifying contributors to so-called biological aging may help understand aging processes as well as risks and mechanisms for chronic diseases and disability. In turn, this knowledge may result in proposed health behavior and lifestyle interventions to delay aging progression and preventing age-related disorders.
A novel measure of biological age that has gained a marked interest in recent years is epigenetic age [7,[9], [10], [11], [12]]. Epigenetic age estimators use DNA methylation (DNAm) levels in a set of CpG sites, coupled with a mathematical algorithm, to compute an estimate of age. This estimate is referred to as epigenetic age/clock or DNAm age. The epigenetic clock theory of aging views biological aging as unintended consequences of developmental and maintenance programs, whose molecular footprints give rise to epigenetic age estimators [11]. Epigenetic age measures have been shown as valid and reliable indicators of biological age. Accelerated epigenetic age (estimated epigenetic age being higher than expected on the basis of chronological age) has been associated with several age-related diseases such as cardiovascular disease (CVD) [12], cancer [13], dementia [12], Parkinson's disease [14], and mortality [15].
Previous studies have linked modifiable lifestyle risk factors such as diet, exercise, alcohol consumption, and smoking status to epigenetic aging [12,16]. Sleep behaviors and disorders represent another set of behavioral and physiological mechanisms that may be modified. However, their role in epigenetic age acceleration has been minimally explored. An initial investigation using data from the Women's Health Initiative Study found that insomnia symptoms were associated with advanced epigenetic age [17]. While prior literature has reported that SDB was associated with shorter telomere length (another measure of biological age) [18], there are no reports of the association of SDB with epigenetic age acceleration, or no reports of sex dimorphism in the association of SDB with age acceleration.
The goal of this paper was to examine the relationships between polysomnography-measured SDB traits and epigenetic age acceleration among adults from the Multi-Ethnic Study of Atherosclerosis (MESA). We hypothesized that indices of SDB severity, as measured by a greater frequency of breathing disturbances, hypoxemia, and sleep fragmentation, would be associated with greater epigenetic age acceleration. We also evaluated the evidence for sex-specific associations between SDB and epigenetic age acceleration. Epidemiological studies have suggested sex-based differences in the associations between SDB and health outcomes [5,19], potentially related to differences in age of onset and duration of exposure to SDB, modulation by hormonal factors, or differences in the physiological mechanisms underlying SDB in men and women. Researchers have advocated the need for studies to further elucidate sex differences in the natural history of SDB. Finally, to further study potential prospective evidence for the relationship between SDB and epigenetic age acceleration, we performed secondary analyses using data from the Framingham Heart Study (FHS), in which sleep study was performed 2–10 years prior to DNAm measurement.
2. Materials and methods
2.1. Study design and sample
MESA is a multisite prospective study designed to investigate the prevalence, correlates, and progression of subclinical CVD in a racially/ethnically diverse sample [20]. At baseline (Exam 1, 2000–2002), 6814 adults aged 45–84 years and free of clinical CVD were enrolled, with participants representing four ethnic groups (non-Hispanic White, African American, Hispanic American, and Asian (primarily Chinese) American). These analyses used data from two MESA ancillary studies – “MESA Sleep” and “MESA Epigenomics”, conducted during MESA Exam 5 (2010–2012). Institutional Review Board approval was obtained at each of six MESA study sites (Columbia University, Johns Hopkins University, Northwestern University, University of California-Los Angeles, University of Minnesota, Wake Forest University) and written informed consent was obtained from all participants. The study sample analyzed here consisted of 622 individuals who participated in both the MESA Sleep and the MESA Epigenomics ancillary studies of non-Hispanic White, African American, and Hispanic American ancestry.
2.2. Predictors: SDB traits
At MESA Exam 5, 2261 MESA Sleep study participants underwent in-home polysomnography (PSG) using a 15-channel monitor (Compumedics Somte´ System; Compumedics Ltd., Abbotsford, Victoria, Australia) as described before [21]. Sleep data were centrally scored by trained PSG technicians blinded to other data. We used three correlated SDB traits from PSG: Apnea-hypopnea index (AHI), the average number of all apneas plus hypopneas accompanied by at least a 3% drop in oxygen saturation per hour of sleep; Hypoxemia, the percentage of sleep time that oxygen saturation is lower than 90% (Per90); and Arousal index, the average number of cortical arousals per hour of sleep.
2.3. Outcomes: epigenetic age acceleration measures
DNAm data were obtained from the MESA Epigenomics study that randomly selected 1264 MESA Exam 5 participants. Methods for collection and assays for DNAm were described [22]. DNAm in monocytes was measured using the Illumina HumanMethylation450 BeadChip. Based on DNAm data, we used two well-defined estimators to estimate the epigenetic age of each blood sample. Epigenetic age estimators are typically built by regressing a transformed version of chronological age on a set of CpGs using a supervised machine learning method (e.g., a penalized regression model). The penalized regression model automatically selects the most informative CpGs for the age prediction/estimation model. It yields both a set of CpGs and a corresponding mathematical model incorporating the DNAm levels into an age estimate. First, we used the approach based on 513 CpGs to define DNAm-PhenoAge, which is also known as the “Levine's clock” [12]. Second, we used the approach based on 353 CpGs to define DNAm-Age, which is also known as the “Horvath's clock” [10]. The two measures are highly correlated, but the DNAm-PhenoAge was developed while taking into account 10 clinical characteristics (e.g., glucose, C-reactive protein levels, white blood cell counts) and has been found to be a better predictor of morbidity and mortality, compared to DNAm-Age, which was developed based on chronological age alone [12]. The DNAm-PhenoAge estimator is said to measure “extrinsic” aging whereas the DNAm-Age estimator is said to measure “intrinsic” aging. Further details of these methods were published elsewhere [11].
DNAm-PhenoAge acceleration and DNAm-Age acceleration were calculated as the residuals from regressing the respective epigenetic age on chronological age. A relatively high correlation between epigenetic age and chronological age is expected. Those whose epigenetic age is different from expected are informative. Positive age acceleration indicates higher biological age compared to chronological age (i.e., the underlying tissue ages faster than expected on the basis of chronological age), and the opposite holds for negative age acceleration.
2.4. Covariates
We included sex, race/ethnicity, household income, study site, body mass index (BMI), as well as smoking and drinking status as potential confounders, following prior studies’ approach [15,17]. While the two epigenetic age acceleration measures were independent of chronological age, chronological age was still included as a covariate to obtain unbiased estimates of standard errors [23]. A few covariates were explored in sensitivity analyses, including leukocyte subset counts (basophils, neutrophils, monocytes, lymphocytes, and eosinophils), hypertension, diabetes, and depression.
2.5. Primary statistical analyses in MESA
We presented the overall sample characteristics and compared the characteristics by sex. Linear regression models were used to estimate the associations between SDB traits and the two epigenetic age acceleration measures. Model 1 included only the specific SDB trait. Model 2 adjusted for chronological age, sex, race/ethnicity, household income, site, BMI, as well as smoking and drinking status. We tested for improved model fit (compared with a linear function of the specific SDB trait) when using quadratic or cubic functions of the trait. Because no significant divergence from linearity was observed, the associations of the SDB traits with each outcome were tested with the predictors modeled as continuous variables. To evaluate potential sex-specific relationships between SDB traits and epigenetic age acceleration, we performed sex-stratified analyses.
In prespecified sensitivity analyses, we examined the relationships between SDB traits and epigenetic age acceleration with alternative measures of AHI and hypoxemia. For AHI, we used (1) the average number of all apneas plus hypopneas accompanied by at least a 4% drop in oxygen saturation per hour of sleep, (2) the average number of all obstructive apneas and hypopneas with at least a 3% (or 4%) drop in oxygen saturation or arousals per hour of sleep, and (3) a binary variable indicating moderate or severe SDB (AHI >= 15). For hypoxemia, we used (1) binary variables indicating that more than 5% (or 10%) of total sleep time when oxygen saturation was lower than 90%, (2) a binary variable indicating that more than 1% of total sleep time when oxygen saturation was below 80%, (3) minimum oxygen saturation during sleep, and (4) log-transformed percentage of sleep time that oxygen saturation was lower than 90%. We also examined models that included morbidities that may be on the causal pathway (hypertension, diabetes, and depression) and white blood cell counts. In post-hoc analyses, we examined the relationships between SDB traits and epigenetic age acceleration by comparing the SDB traits between those whose epigenetic age (after rounding) was older than expected to those whose epigenetic age was as expected or younger than expected, using logistic regression adjusting for covariates. We also bootstrapped the regression models using 1000 bootstrap samples to get 95% bias corrected accelerated confidence intervals (CIs).
All analyses were performed using R version 3.5.1 (https://www.r-project.org/). All tests were two-sided, with a significance level of 5%.
2.6. Secondary analyses in FHS
There were 699 individuals from the Offspring cohort of FHS who participated in the Sleep Heart Health Study (SHHS), the first visit of which took place between 1995 and 1998, and the second visit of which took place between 2001 and 2003. Both visits of SHHS included in-home PSG as previously described and used a similar montage as MESA other than the absence of nasal pressure signals and was scored by the same central Sleep Reading Center [24]. In this manuscript, we included data from 530 individuals who participated in SHHS and had DNAm obtained in whole blood during 2005–2008 [25,26]. To minimize the gap between the SDB and the DNAm measures, the SDB measures were taken from the second SHHS visit when available, and otherwise, from the first SHHS visit. Across FHS individuals, the sleep studies were performed between 2 and 10 years prior to the DNAm measurements. Statistical analyses in FHS were conducted similar to that in MESA, estimating the associations of AHI, Per90, and arousal index with epigenetic age acceleration in FHS, adjusting for chronological age, sex, BMI, smoking, drinking, and white blood cell counts. Age, lifestyle factors, and comorbidities, were computed at the time of blood draw for the methylation study. Immune cells proportions were computed by applying the Houseman algorithm on the methylation data [27]. Mixed models were used to account for family structure.
2.7. Data statement
The data that support the findings of this study are available from MESA and FHS but restrictions apply to the availability of these data, which were used under agreement for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the studies.
3. Results
3.1. MESA sample description
Descriptive statistics of the MESA sample are reported in Table 1. The sample had a mean chronological age of 68.7 years (standard deviation (SD): 9.2 years); 46.0% were White, 21.4% African American, and 32.6% Hispanic; 53.2% were women. The means of AHI, Per90, and arousal index were 19.6, 4.2, and 22.0, respectively. Compared to women, men had significantly higher income, higher proportions of former/current smokers and drinkers, higher proportions with impaired fasting glucose, lower depression score, higher AHI and arousal index. Men had greater DNAm-Age acceleration than women, consistent with prior literature documenting that men have higher epigenetic aging rates than women in blood, saliva, and brain tissue [28]. This might be reflecting the fact that men have higher mortality at all ages than women [29], and are exhibiting faster age acceleration. The SDB traits were all correlated (Table S1). Both DNAm-PhenoAge and DNAm-Age were correlated with chronological age, with coefficients of 0.77 and 0.80 (Fig. S1). Both DNAm-PhenoAge acceleration and DNAm-Age acceleration were approximately normally distributed and the residuals from regressing each on chronological age were uniformly scattered above and below x-axis, indicating the adequacy of linear regression for chronological age adjustment.
Table 1.
Sample description, by sex, MESA.
| Overall | Women | Men | ||
|---|---|---|---|---|
| (n = 622) | (n = 331) | (n = 291) | ||
| Sociodemographics | ||||
| Chronological age, years (mean (sd)) | 68.7 (9.2) | 68.5 (9.4) | 69.0 (9.1) | |
| Race/ethnicity (%) | ||||
| White | 46.0 | 44.7 | 47.4 | |
| African American | 21.4 | 23.9 | 18.6 | |
| Hispanic | 32.6 | 31.4 | 34.0 | |
| Family income (mean (sd))+ | 9.1 (3.4) | 8.4 (3.4) | 9.9 (3.3) | |
| Lifestyle factors | ||||
| BMI (%) | ||||
| Normal (BMI < 25) | 19.3 | 17.2 | 21.7 | |
| Overweight (25 <= BMI < 30) | 37.2 | 33.5 | 41.4 | |
| Obese (BMI >= 30) | 43.5 | 49.2 | 36.9 | |
| Smoking status (%) | ||||
| Never | 39.1 | 47.1 | 30.0 | |
| Former | 52.7 | 46.2 | 60.0 | |
| Current | 8.2 | 6.6 | 10.0 | |
| Drinking alcohol: Yes (%) | 46.2 | 39.9 | 53.4 | |
| Morbidities | ||||
| Hypertension: Yes (%) | 58.8 | 58.9 | 58.8 | |
| Diabetes (%) | ||||
| Normal | 57.1 | 62.2 | 51.2 | |
| Impaired fasting glucose | 22.5 | 17.2 | 28.5 | |
| Diabetes | 20.4 | 20.5 | 20.3 | |
| Depression (mean (sd)) | 8.3 (7.3) | 9.3 (8.0) | 7.2 (6.3) | |
| Immune cells, *10E3/uL | ||||
| Basophils (mean (sd)) | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | |
| Neutrophils (mean (sd)) | 3.6 (1.3) | 3.6 (1.2) | 3.7 (1.3) | |
| Monocytes (mean (sd)) | 0.5 (0.2) | 0.4 (0.1) | 0.5 (0.2) | |
| Lymphocytes (mean (sd)) | 1.9 (2.1) | 1.9 (0.7) | 1.9 (2.9) | |
| Eosinophils (mean (sd)) | 0.2 (0.1) | 0.2 (0.1) | 0.2 (0.1) | |
| SDB traits | ||||
| AHI (median [IQR]) | 14.1 [6.6, 25.7] | 11.0 [4.9, 20.4] | 17.3 [8.1, 31.3] | |
| Per90 (median [IQR]) | 0.7 [0.1, 3.5] | 0.5 [0.1, 2.6] | 0.9 [0.1, 4.5] | |
| Arousal index (median [IQR]) | 18.9 [13.7, 26.8] | 17.3 [12.4, 24.4] | 21.1 [15.6, 29.4] | |
| Epigenetic age acceleration, years | ||||
| DNAm-PhenoAge acceleration (mean (sd)) | −0.0 (5.9) | −0.1 (5.9) | 0.1 (6.0) | |
| DNAm-Age acceleration (mean (sd)) | 0.0 (5.1) | −0.6 (4.9) | 0.7 (5.3) | |
Definition of abbreviations: MESA = Multi-Ethnic Study of Atherosclerosis; AHI = apnea-hypopnea index; Per90 = percentage of sleep time that oxygen saturation is lower than 90%; BMI = body mass index; SDB = sleep disordered breathing; DNAm = DNA methylation.
+Income was rated on a 15-item scale (1 = less than $5k, 15 = $15k or more).
3.2. Results of primary analyses in MESA
3.2.1. Total sample analyses
Regression results are presented in Table 2. Analyses from the adjusted model (Model 2) revealed that higher AHI was significantly associated with greater DNAm-PhenoAge acceleration (β = 0.03; 95% CI [0.001, 0.06]). Therefore, a one-unit increase in AHI was associated with 0.03 years (or 11 days) of DNAm-PhenoAge acceleration. Alternatively, 1-SD AHI was associated with 215 days of DNAm-PhenoAge acceleration. Arousal index was significantly associated with DNAm-Age acceleration (β = 0.04; 95% CI [0.01, 0.07]). A one-unit increase in arousal index was associated with 0.04 years (or 15 days) of DNAm-Age acceleration, representing 321 days of DNAm-Age acceleration for 1-SD increase in arousal index. On the other hand, the direction of the effect estimates for Per90 on both epigenetic age acceleration measures was as expected (positive) but not statistically significant.
Table 2.
Effect estimates from linear regression relating sleep disordered breathing traits to epigenetic age acceleration, MESA.
| DNAm-PhenoAge acceleration |
DNAm-Age acceleration |
|||||||
|---|---|---|---|---|---|---|---|---|
| Model 1 | Model 2 | Model 1 | Model 2 | |||||
| Beta | [95% CI] | Beta | [95% CI] | Beta | [95% CI] | Beta | [95% CI] | |
| AHI | 0.03** | [0.01, 0.06] | 0.03* | [0.001, 0.06] | 0.03* | [0.004, 0.05] | 0.02 | [−0.01, 0.04] |
| Per90 | 0.03 | [−0.01, 0.08] | 0.03 | [−0.02, 0.08] | 0.04 | [−0.001, 0.08] | 0.02 | [−0.02, 0.06] |
| Arousal index | 0.03 | [−0.01, 0.07] | 0.03 | [−0.01, 0.08] | 0.05* | [0.02, 0.09] | 0.04* | [0.01, 0.07] |
Definition of abbreviations: MESA = Multi-Ethnic Study of Atherosclerosis; AHI = apnea-hypopnea index; Per90 = percentage of sleep time that oxygen saturation is lower than 90%; SDB = sleep disordered breathing; BMI = body mass index; DNAm = DNA methylation; CI = confidence interval.
***p < 0.001; **p < 0.01; *p < 0.05.
Model 1 included only the specific SDB trait of interest. Model 2 controlled for chronological age, sex, race/ethnicity, income, study site, BMI, smoking, and drinking.
3.2.2. Sex-stratified analyses
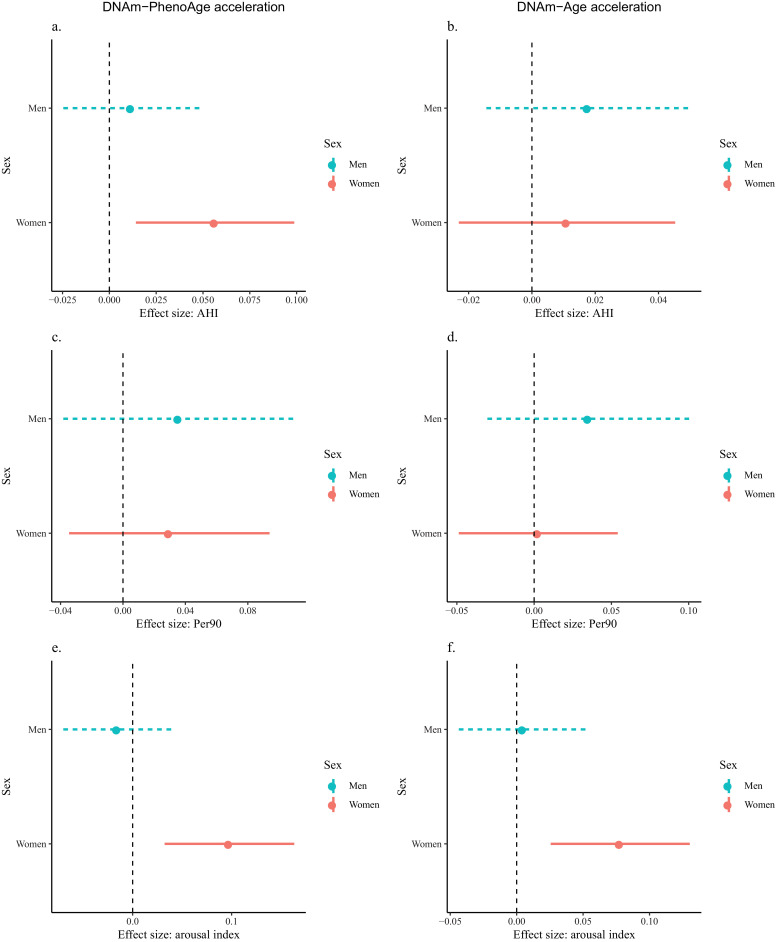
In sex interaction analyses, the interaction between sex and arousal index was statistically significant (p-value = 0.03) in the model for DNAm-PhenoAge acceleration while other interaction tests had p-values greater than 0.05. Stratified analyses show that among women, higher AHI was associated with greater DNAm-PhenoAge acceleration (β = 0.06; 95% CI [0.01, 0.10]). In contrast, no association was observed among men (β = 0.01; 95% CI [−0.02, 0.05]) (Fig. 1). Similarly, among women, higher arousal index was significantly associated with both greater DNAm-PhenoAge acceleration (β = 0.10; 95% CI [0.03, 0.16]) and greater DNAm-Age acceleration (β = 0.08; 95% CI [0.03, 0.13]), but not in men (DNAm-PhenoAge acceleration: β = −0.02; 95% CI [−0.07, 0.04]; DNAm-Age acceleration: β = 0.00; 95% CI [−0.04, 0.05]). The associations between Per90 and epigenetic age acceleration measures did not reach statistical significance for women or men.
Fig. 1.
Sex-stratified analyses on the associations between sleep disordered breathing (SDB) traits and epigenetic age acceleration in the Multi-Ethnic Study of Atherosclerosis (MESA). Regression analyses adjusted for chronological age, race/ethnicity, income, study site, BMI, smoking, and drinking. AHI was associated with DNAm-PhenoAge acceleration only in women (a). Arousal index was associated with DNAm-PhenoAge acceleration and DNAm-Age acceleration only in women (e and f).
3.2.3. Sensitivity analyses
Results did not differ substantively when we explored the association between AHI and epigenetic age acceleration using alternative continuous measures of AHI (e.g., average number of all apneas plus hypopneas with at least a 4% drop in oxygen saturation per hour of sleep). There was no evidence that the binary AHI indicator was associated with epigenetic age acceleration. For hypoxemia, none of the alternative measures (e.g., minimum oxygen saturation) was associated with either epigenetic age acceleration measure. Including hypertension, diabetes, depression, and white blood cell counts did not change the results appreciably (Table S2). When evaluating the epigenetic age acceleration outcomes as binary variables, we observed that greater SDB severity was associated with higher odds of epigenetic age acceleration, but the associations did not reach statistical significance (Table S3). For instance, the association between AHI and DNAm-PhenoAge acceleration had an adjusted odds ratio of 1.01 and the associated 95% CI was [0.998, 1.02]. The 95% bias corrected accelerated CIs were very similar to those from the main analyses (Table S4).
3.3. Results of secondary analyses in FHS
The FHS sample had a mean chronological age of 67.6 years (SD: 8.6 years), which was younger than the MESA sample (Table S5). All FHS participants were White and half were women. Adjusted analyses on the FHS data showed positive associations between AHI and arousal index with DNAm-PhenoAge acceleration (AHI: β = 0.02; 95% CI [−0.01, 0.06]; arousal index: β = 0.03; 95% CI [−0.02, 0.08]). The effect sizes (magnitudes of the adjusted linear regression coefficients) were similar to those from MESA, but the coefficients were not statistically significant. Per90 was significantly associated with DNAm-PhenoAge acceleration (β = 0.06; 95% CI [0.02, 0.11]). Associations between the SDB traits and DNAm-Age acceleration were smaller in magnitude compared to those between the SDB traits and DNAm-PhenoAge acceleration (Table S6). The results from FHS seemed to be driven by men (Fig. S2). To increase power, we meta-analyzed estimated associations across MESA and FHS, using a fixed-effect inverse-variance weighted meta-analysis. Results show that all SDB traits were significantly positively associated with both epigenetic age acceleration measures, except for the association between AHI and DNAm-Age acceleration (β = 0.03; 95% CI [−0.003, 0.03]; p-value = 0.11) (Table S7).
4. Discussion
To our knowledge, the findings in this study are the first reported empirical evidence documenting the relationships between SDB traits and epigenetic age acceleration in a diverse sample of US adults. This study shows that, overall, more severe SDB was associated with greater epigenetic age acceleration. Specifically, in the MESA cohort, higher AHI was associated with greater DNAm-PhenoAge acceleration, and higher arousal index was associated with greater DNAm-Age acceleration. The associations between Per90 and epigenetic age acceleration measures were positive yet not statistically significant. The secondary analyses from FHS show that overnight hypoxemia (Per90) was associated with increased epigenetic age acceleration few years after the sleep study. Meta-analysis across MESA and FHS confirmed that greater SDB severity was associated with greater epigenetic age acceleration. Sex-stratified analyses revealed that there might be sex-specific relationships between SDB traits and epigenetic age acceleration, but uncertainty remained: in MESA the associations of AHI and arousal index with epigenetic age acceleration were stronger in women than in men; in FHS the associations of AHI and Per90 with epigenetic age acceleration were stronger in men than in women.
Individuals with SDB might be more vulnerable to advanced epigenetic age for a few reasons. The hallmarks of SDB – successive episodes of partial or complete airflow obstruction during sleep leading to intermittent hypoxemia and repetitive arousals and sleep fragmentation – stimulate oxidative stress, inflammation, sympathetic nervous system activation, and endothelial dysfunction, which might contribute to accelerated aging [3]. Reactive oxygen species (ROS) can damage surrounding tissues and alter cellular functions, and consequently promote inflammation [30]. Indeed, intense local and systematic inflammation have been found in individuals with SDB, indicated by elevated levels of inflammation markers such as leukocytes, C-reactive protein, and IL-6 [31]. Taken together, increased generation and propagation of ROS and initiation and aggravation of inflammatory processes in the pathogenesis of SDB might mimic an aging context and decrease individuals’ ability to respond to stressors [3].
The extrinsic DNAm-PhenoAge acceleration was significantly associated with AHI and arousal index whereas the intrinsic DNAm-Age acceleration was significantly associated with arousal index only in MESA. To note, all associations had estimated effects in the hypothesized directions. It is possible that other associations existed but were not detected at the appropriate statistical level due to the limited power from the relatively small sample. This is partially supported by the results from the secondary FHS analyses and the meta-analyses. On the other hand, prior research has found that, compared to DNAm-Age acceleration, DNAm-PhenoAge acceleration is more highly correlated with clinical measures (e.g., C-reactive protein), morbidity (e.g., age-related dementia), and mortality [11]. DNAm-PhenoAge acceleration is argued to capture the wear-and-tear in tissues and to especially reflect the aging mechanism of the immune system. SDB might correlate with DNAm-PhenoAge acceleration through multiple mechanisms that cause physiological dysfunction in tissues. DNAm-PhenoAge acceleration is also associated with lifestyle and demographic variables, such as physical activity, BMI, smoking status, diet, educational level, and income [11], which might differ between women and men. By contrast, DNAm-Age acceleration exhibits weaker associations with lifestyle factors and markers of inflammation.
Prior studies have reported inconclusive findings regarding the role of sex on the associations between sleep and health outcomes. Early studies of younger individuals from the Sleep Heart Health Study reported that SDB was associated with increased risks of incident heart failure and mortality among men, but not women [4,5]. However, a recent study from the Atherosclerosis Risk in the Communities suggests that older women are more susceptible to the negative health effects of SDB. Specifically, it showed that SDB was associated with higher levels of high-sensitivity troponin T and incident heart failure among women but not men, and that women with SDB have poorer survival than men [19]. We did not find independent evidence for the sex-specific results, as they were different between MESA and FHS. But here we discuss a few possible reasons for the different patterns that we observed in MESA and FHS. The stronger associations of AHI and arousal index with epigenetic aging we observed among women in comparison to men in the MESA analyses may arise from the sex differences in pathogenesis of SDB. Women are more likely to have rapid eye movement (REM) dominant SDB and REM AHI is generally higher among women than men [32]. SDB during REM sleep is associated with longer and more frequent apneas and deeper desaturation. Thus, the more severe apneas during REM sleep experienced by women might explain the stronger associations between SDB and epigenetic age acceleration in women than men. Sex differences in the timing of SDB development might also contribute to the sex-specific relationships in that men usually develop SDB earlier in life while women usually develop SDB after menopause [19]. Women might have less time to adapt to SDB compared to men, which might make them more susceptible to SDB induced damages. On the other hand, the stronger associations of AHI and Per90 with epigenetic aging we observed among men in comparison to women in the FHS analyses might arise from the fact that the FHS sample was younger at the time of sleep measurement than the MESA sample, consistent with prior evidence documenting that among younger individuals, SDB was more detrimental to men's cardiovascular health and mortality risk, whereas among older individuals, SDB was more detrimental to women's health. Additionally, the FHS participants were mostly White whereas the MESA participants were racially/ethnically diverse. The follow-up time also varied substantially for the FHS participants and SDB may influence epigenetic age acceleration differently according to age and duration of follow-up, all of which might have contributed to the different patterns we observed between the two cohorts.
Only one study has evaluated the relationships between sleep characteristics (sleep quality and duration, but not SDB) and epigenetic age acceleration, reporting that insomnia was associated with extrinsic epigenetic aging [17]. An association between arousal index and accelerated aging (measured by shortening of leukocyte telomere length over a 10-year period) was also reported in MESA [33]. Our study contributes to the literature by providing innovative information on the associations between SDB traits and epigenetic age acceleration. This study has a number of strengths: gold-standard and standardized PSG allowed us to objectively quantify SDB traits; two well-established measures of epigenetic age acceleration were used to capture fast biological aging; comprehensive data allowed us to control for multiple potential confounders; and our study cohort was racially/ethnically diverse and included both women and men.
Several limitations must also be acknowledged. Although several SDB traits were evaluated and prior research has shown that sleep apnea classification remains largely stable over several months to years [19], the SDB traits were derived from a single night of PSG, which did not capture variation over time. Our primary analyses were based on the MESA cohort, which provided the ability to examine a multi-ethnic cohort with state-of-the-art polysomnography. However, the cross-sectional analyses precluded assessment of causality. While our secondary analyses included prospectively collected epigenetic information, the population differences between FHS and MESA and the different study designs may limit comparisons. Additionally, our analyses did not adjust for multiple comparisons as we were making only a few planned biologically-informed comparisons and “chance” was not expected to serve as the first-order explanation [34]. We however performed exploratory Bonferroni correction, after which only the association between Per90 and DNAm-PhenoAge acceleration in FHS remained statistically significant. This could be due to limited power from the relatively small samples. Future studies involving bigger samples are needed. Moreover, even with inclusion of two cohorts, our results might not generalize to populations of other (younger) age groups or other racial/ethnic groups not included in the samples. Future efforts are warranted to further investigate the sex-specific relationships between SDB traits and epigenetic age acceleration and to elucidate the underlying mechanisms. It would be of interest for future research to examine whether the relationships between SDB traits and epigenetic age acceleration are modified by other factors such as socioeconomic status, and whether epigenetic age acceleration mediates the relationships between SDB traits and morbidity and mortality, which might help understand mechanisms of health inequalities between different groups.
Taken together, our cross-sectional and prospective analyses suggest an association of SDB severity with epigenetic age acceleration. One of the most promising features of DNAm biomarkers is that epigenetic changes are reversible, indicating that epigenetic age estimators might be useful to identifying and validating anti-aging interventions. SDB, one of the most common yet under recognized sleep disorders, might carry unique prospects in terms of delaying aging progression and preventing age-related disorders. If causal, our data suggest that improving AHI and sleep fragmentation may positively affect age-related chronic diseases, as well as extend the human health span and lifespan.
Declaration of Competing Interest
All other authors declare no competing interests.
Acknowledgments and funding sources
XL was funded by National Heart, Lung, and Blood Institute (NHLBI) grants R01 HL137192, R01AG042463, and R35HL135818. SR and TS were partially funded by R35 HL135818. MESA: This research was supported by the Multi-Ethnic Study of Atherosclerosis (MESA). MESA and the MESA SHARe project are conducted and supported by the NHLBI in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. MESA Sleep was funded by R01HL098433. Funding support for the Sleep Polysomnography dataset was provided by (NHLBI) grant HL56984. The MESA Epigenomics Studies were funded by NHLBI R01HL101250, NIDDK R01 DK103531-01, R01DK103531, National Institute on Aging (NIA) R01 AG054474, and NHBLI R01 HL13500901 to Wake Forest University Health Sciences. FHS: The Framingham Heart Study is funded by National Institutes of Health contract N01-HC-25195. The laboratory work for this investigation was funded by the Division of Intramural Research, NHLBI. The analytical component of this project was funded by the Division of Intramural Research, NHLBI, and the Center for Information Technology, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders of the study had no role in the design of the study, data collection, analysis, interpretation, writing of the report, or in the decision to submit the report for publication. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
SR receives grants from the National Institute of Health, American Sleep Medicine Foundation, and Jazz Pharmaceuticals. She received consulting fees from Jazz Pharmaceuticals.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.020.
Appendix. Supplementary materials
References
- 1.Young T., Palta M., Dempsey J., Skatrud J., Weber S., Badr S. The occurrence of sleep-disordered breathing among middle-aged adults. N Engl J Med. 1993;328(17):1230–1235. doi: 10.1056/NEJM199304293281704. [DOI] [PubMed] [Google Scholar]
- 2.Rosenzweig I., Glasser M., Polsek D., Leschziner G.D., Williams S.C.R., Morrell M.J. Sleep apnoea and the brain: a complex relationship. Lancet Respir Med. 2015;3(5):404–414. doi: 10.1016/S2213-2600(15)00090-9. [DOI] [PubMed] [Google Scholar]
- 3.Gaspar L.S., Álvaro A.R., Moita J., Cavadas C. Obstructive sleep apnea and hallmarks of aging. Trends Mol Med. 2017;23(8):675–692. doi: 10.1016/j.molmed.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Punjabi N.M., Caffo B.S., Goodwin J.L. Sleep-disordered breathing and mortality: a prospective cohort study. PLoS Med. 2009;6(8) doi: 10.1371/journal.pmed.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottlieb Daniel J., Gayane Y., Newman Anne B. et al. prospective study of obstructive sleep apnea and incident coronary heart disease and heart failure. Circulation. 2010;122(4):352–360. doi: 10.1161/CIRCULATIONAHA.109.901801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redline S., Yenokyan G., Gottlieb D.J. Obstructive sleep apnea–hypopnea and incident stroke. Am J Respir Crit Care Med. 2010;182(2):269–277. doi: 10.1164/rccm.200911-1746OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jylhävä J., Pedersen N.L., Hägg S. Biological age predictors. EBioMedicine. 2017;21:29–36. doi: 10.1016/j.ebiom.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson T.E. Recent results: biomarkers of aging. Exp Gerontol. 2006;41(12):1243–1246. doi: 10.1016/j.exger.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Hannum G., Guinney J., Zhao L. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:3156. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S., Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 12.Levine M.E., Lu A.T., Quach A. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dugué P.-.A., Bassett J.K., Joo J.E. DNA methylation-based biological aging and cancer risk and survival: pooled analysis of seven prospective studies. Int J Cancer. 2018;142(8):1611–1619. doi: 10.1002/ijc.31189. [DOI] [PubMed] [Google Scholar]
- 14.Horvath S., Ritz B.R. Increased epigenetic age and granulocyte counts in the blood of Parkinson's disease patients. Aging. 2015;7(12):1130–1142. doi: 10.18632/aging.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marioni R.E., Shah S., McRae A.F. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quach A., Levine M.E., Tanaka T. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging. 2017;9(2):419–437. doi: 10.18632/aging.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll J.E., Irwin M.R., Levine M. Epigenetic aging and immune senescence in women with insomnia symptoms: findings from the Women's Health Initiative Study. Biol Psychiatry. 2017;81(2):136–144. doi: 10.1016/j.biopsych.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tempaku P.F., Mazzotti D.R., Hirotsu C. The effect of the severity of obstructive sleep apnea syndrome on telomere length. Oncotarget. 2016;7(43):69216–69224. doi: 10.18632/oncotarget.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Querejeta R.G., Susan R., Brian C. Sex-specific association of sleep apnea severity with subclinical myocardial injury, ventricular hypertrophy, and heart failure risk in a community-dwelling cohort. Circulation. 2015;132(14):1329–1337. doi: 10.1161/CIRCULATIONAHA.115.016985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bild D.E., Bluemke D.A., Burke G.L. Multi-Ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 21.Chen X., Wang R., Zee P. Racial/ethnic differences in sleep disturbances: the multi-ethnic study of atherosclerosis (MESA) Sleep. 2015 doi: 10.5665/sleep.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Ding J., Reynolds L.M. Methylomics of gene expression in human monocytes. Hum Mol Genet. 2013;22(24):5065–5074. doi: 10.1093/hmg/ddt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sofer T., Zheng X., Gogarten S.M. A fully adjusted two-stage procedure for rank-normalization in genetic association studies. Genet Epidemiol. 2019;43(3):263–275. doi: 10.1002/gepi.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redline S., Sanders M.H., Lind B.K. Methods for obtaining and analyzing unattended polysomnography data for a multicenter study. Sleep Heart Health Research grOup. Sleep. 1998;21(7):759–767. [PubMed] [Google Scholar]
- 25.Quan S.F., Howard B.V., Iber C. The sleep heart health study: design, rationale, and methods. Sleep. 1997;20(12):1077–1085. [PubMed] [Google Scholar]
- 26.Kannel W.B., Feinleib M., McNamara P.M., Garrison R.J., Castelli W.P. An investigation of coronary heart disease in families: the Framingham Offspring Study. Am J Epidemiol. 1979;110(3):281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 27.Koestler D.C., Christensen B.C., Karagas M.R. Blood-based profiles of DNA methylation predict the underlying distribution of cell types. Epigenetics. 2013;8(8):816–826. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath S., Gurven M., Levine M.E. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17(1):171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Case A., Paxson C. Sex differences in morbidity and mortality. Demography. 2005;42(2):189–214. doi: 10.1353/dem.2005.0011. [DOI] [PubMed] [Google Scholar]
- 30.Lavie L., Lavie P. Molecular mechanisms of cardiovascular disease in OSAHS: the oxidative stress link. Eur Respir J. 2009;33(6):1467–1484. doi: 10.1183/09031936.00086608. [DOI] [PubMed] [Google Scholar]
- 31.Hatipoğlu U., Rubinstein I. Inflammation and obstructive sleep apnea syndrome pathogenesis: a working hypothesis. Respiration. 2003;70(6):665–671. doi: 10.1159/000075218. [DOI] [PubMed] [Google Scholar]
- 32.Basoglu O.K., Tasbakan M.S. Gender differences in clinical and polysomnographic features of obstructive sleep apnea: a clinical study of 2827 patients. Sleep Breath Schlaf Atm. 2018;22(1):241–249. doi: 10.1007/s11325-017-1482-9. [DOI] [PubMed] [Google Scholar]
- 33.Carroll J.E., Irwin M.R., Seeman T.E. Obstructive sleep apnea, nighttime arousals, and leukocyte telomere length: the Multi-Ethnic Study of Atherosclerosis. Sleep. 2019 doi: 10.1093/sleep/zsz089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothman K.J. No adjustments are needed for multiple comparisons. Epidemiol Camb Mass. 1990;1(1):43–46. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.