Abstract
Background
The early-life microbiota exerts a profound and lifelong impact on host health. Longitudinal studies in humans have been informative but are mostly based on the analysis of fecal samples and cannot shed direct light on the early development of mucosa-associated intestinal microbiota and its impact on GI function. Using piglets as a model for human infants, we assess here the succession of mucosa-associated microbiota across the intestinal tract in the first 35 days after birth.
Results
Although sharing a similar composition and predicted functional profile at birth, the mucosa-associated microbiome in the small intestine (jejunum and ileum) remained relatively stable, while that of the large intestine (cecum and colon) quickly expanded and diversified by day 35. Among detected microbial sources (milk, vagina, areolar skin, and feces of sows, farrowing crate, and incubator), maternal milk microbes were primarily responsible for the colonization of the small intestine, contributing approximately 90% bacteria throughout the first 35 days of the neonatal life. Although maternal milk microbes contributed greater than 90% bacteria to the large intestinal microbiota of neonates upon birth, their presence gradually diminished, and they were replaced by maternal fecal microbes by day 35. We found strong correlations between the relative abundance of specific mucosa-associated microbes, particularly those vertically transmitted from the mother, and the expression levels of multiple intestinal immune and barrier function genes in different segments of the intestinal tract.
Conclusion
We revealed spatially specific trajectories of microbial colonization of the intestinal mucosa in the small and large intestines, which can be primarily attributed to the colonization by vertically transmitted maternal milk and intestinal microbes. Additionally, these maternal microbes may be involved in the establishment of intestinal immune and barrier functions in neonates. Our findings strengthen the notion that studying fecal samples alone is insufficient to fully understand the co-development of the intestinal microbiota and immune system and suggest the possibility of improving neonatal health through the manipulation of maternal microbiota.
Keywords: Mucosal microbiota, Spatiotemporal colonization, Early life, Maternal microbial transmission, Immunologic maturation, Microbiota-host interaction
Background
In the gastrointestinal (GI) tract, the mucosal surface uniquely serves as a conduit between the host immune system and the external environment, orchestrating a variety of physiological processes such as nutrient absorption and immune development [1, 2]. In this context, an extremely dense and diverse bacterial community resides on the mucosal surface and plays a critical role in host physiology and health [3]. The proper establishment of the intestinal microbiota in early life is well known to facilitate immune maturation [4, 5]. Colonization of neonatal, but not adult, germ-free mice with conventional intestinal microbiota has a positive long-term influence on the subsequent development of host innate and adaptive immunity [6, 7]. Conversely, aberrant microbial colonization during infancy is associated with a number of childhood diseases [8–10] and increases disease risk in later life [11–13].
Succession and maturation of the infant GI microbiome have been extensively studied [7, 14–16]. To date, most of these studies are, however, based on the use of fecal samples. Given that the maturation of the intestinal immune system significantly depends on mucosa-associated microbes [17–19] and that substantial differences exist between mucosa-associated and fecal microbiota [20–24], it is critically important to study the development of intestinal mucosa-associated microbiota in early life.
Pigs, sharing many common features in the gastrointestinal physiology, microbiology, genetics, and diet with humans, are an excellent animal model and have been widely used in biomedical research [25, 26]. In particular, in contrast to rodents, the sow-piglet dyad has been suggested as a more promising model for the human mother-infant dyad to study the development of the GI tract functions [26–29]. Recent studies have also shown the metagenomic profile and inter-individual variability to be more similar between pigs and humans than between mice and humans, making pigs a superior model for human GI microbiota research [30, 31]. Furthermore, confounding variables that are impractical to control in human studies could be controlled or avoided in swine experimentation. Thus, neonatal piglets are well suitable for in-depth studies of the initial colonization and development of infant intestinal mucosal microbiome.
The influence of the birth mode [32, 33], antibiotics [32, 34, 35], and nutrition [8, 36] on the development of infant GI microbiota has been well studied. However, little is known about the impact and contribution of different microbial sources from the mother and the environment, although existing evidence suggests that neonatal microbes are likely to come from the mother and immediate rearing environment [15, 37]. Recent studies suggested that the vagina, milk, and areolar skin of a mother contain diverse bacterial communities and are important sources of infant GI microbiota [38]. The neonatal environment also exerts a sustained influence on the development of infant intestinal microbiota [37, 39, 40]. However, no longitudinal studies have been conducted to analyze the relative contributions of these microbial sources to the colonization of neonatal GI.
In this study, we comprehensively surveyed the spatial and temporal development of mucosa-associated bacterial community and its correlation with the host gene expression along the GI tract in healthy neonatal piglets. In addition, relative contributions of different microbial sources from the mother (vagina, areolar skin, feces, and milk of the sow) and the neonatal environment (farrowing crate and incubator) were also assessed.
Results
Structure of the mucosa-associated microbiota is relatively stable in the small, but not the large, intestine in early life
After quality filtering and assembly, 13,768,547 16S rRNA gene sequences were obtained from 367 mucosal bacterial DNA samples of Landrace and Rongchang piglets from birth to 35 days (average of 37,516 sequences/sample, Additional file 1: Figure S1). To avoid biases generated by the differences in sequencing depth, we rarefied each sample to a depth of 27,848 sequences/sample prior to performing the following analyses. Rarefaction curves of Chao1 and Shannon diversity indices calculated at the OTU level (Additional file 2: Figure S2) reached a plateau, suggesting that the majority of microbial diversity had been sufficiently captured.
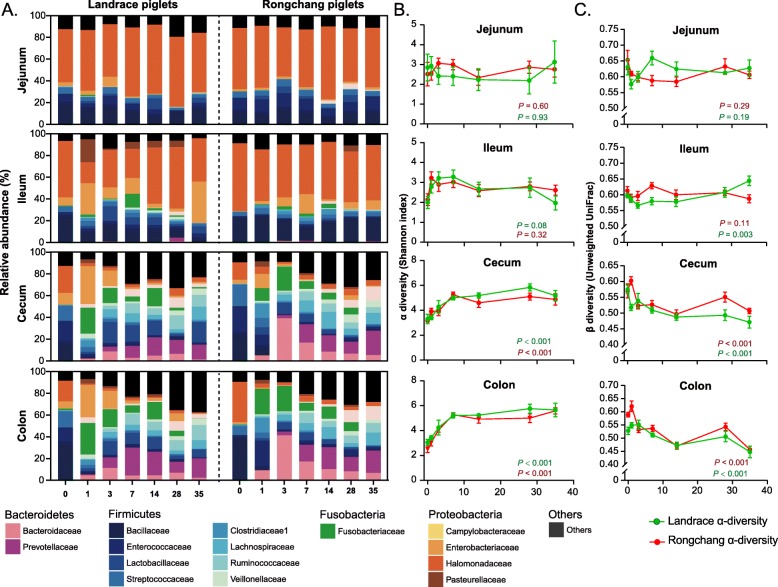
Fecal microbiota is known to gradually increase in the richness and diversity with age [41, 42]. In pigs, mucosa-associated microbiota in the small intestine (jejunum and ileum) of piglets was dominated by Halomonadaceae, whereas that in the large intestine (cecum and colon) was much more diverse with no obvious dominant bacterial taxa in the first 35 days of life (Fig. 1a). Overall, the small intestine harbored mostly Proteobacteria (Halomonadaceae and Enterobacteriaceae) and a small fraction of Firmicutes (Bacillaceae, Enterococcaceae, and Streptococcaceae), while the large intestine consisted of multiple families of Firmicutes, Bacteroidetes, Fusobacteria, and much reduced abundance of Proteobacteria. Moreover, we observed distinct trajectories of bacterial succession and maturation in different intestinal segments for both breeds. As expected, the microbiota experienced a dramatic shift in both the small and large intestines on the first day after birth. To our surprise, the microbiota in the small intestine quickly became stabilized from day 3 to day 35, while those in the large intestine (cecum and colon) failed to achieve the equilibrium until after 7–14 days (Fig. 1a). An abrupt disappearance of Bacillaceae and Enterococcaceae was observed in the cecum and colon shortly after birth, followed by a gradual increase in Lactobacillaceae, Lachnospiraceae, Ruminococcaceae, and Veillonellaceae as piglets aged.
Fig. 1.
Development of the mucosa-associated microbiota in the small and large intestines of Rongchang and Landrace piglets during the first 35 days after birth. a Stacked area plot displaying the changes in the relative abundance (%) of the 15 most abundant bacterial families with age. b The shift of α-diversity (Shannon index) with age across 4 intestinal segments. c The shift of β-diversity (unweighted UniFrac distance) with age across 4 intestinal segments. The β-diversity at each time point is the average distance of one sample to all other samples at that time point
PERMANOVA analyses revealed that mucosa-associated intestinal microbiome was primarily shaped by biogeographic location (R2 = 0.357, P < 0.001, weighted UniFrac, Table 1). Although the breed had a significant effect (P < 0.001) on the GI microbiota structure as measured by unweighted UniFrac and Bray-Curtis metrics, but not weighted UniFrac (P = 0.064, Table 1), extremely small R2 values (ranging between 0.011 and 0.012) indicated that the breed effect is negligible. Indeed, similar results were observed between two breeds at birth as well (Additional file 3: Figure S3A), suggesting that breed had little influence on the phylogenetic composition of the mucosa-associated microbiome. Therefore, samples between the two breeds within each time point and intestinal segment were grouped together in subsequent analyses.
Table 1.
Factors contributing to the variation in intestinal mucosa-associated microbiota
| Items | Weighted UniFrac | Unweighted UniFrac | Bray-Curtis | |||
|---|---|---|---|---|---|---|
| R2 | P value* | R2 | P value* | R2 | P value* | |
| Intestinal segment | 0.357 | < 0.001 | 0.108 | < 0.001 | 0.229 | < 0.001 |
| Age | 0.136 | < 0.001 | 0.110 | < 0.001 | 0.123 | < 0.001 |
| Weaning | 0.019 | < 0.001 | 0.032 | < 0.001 | 0.021 | < 0.001 |
| Breed | 0.006 | 0.064 | 0.011 | < 0.001 | 0.012 | < 0.001 |
| Sex | 0.002 | 0.525 | 0.004 | 0.061 | 0.003 | 0.315 |
*PERMANOVA was performed, and P values in italics represent statistical significance (P < 0.05)
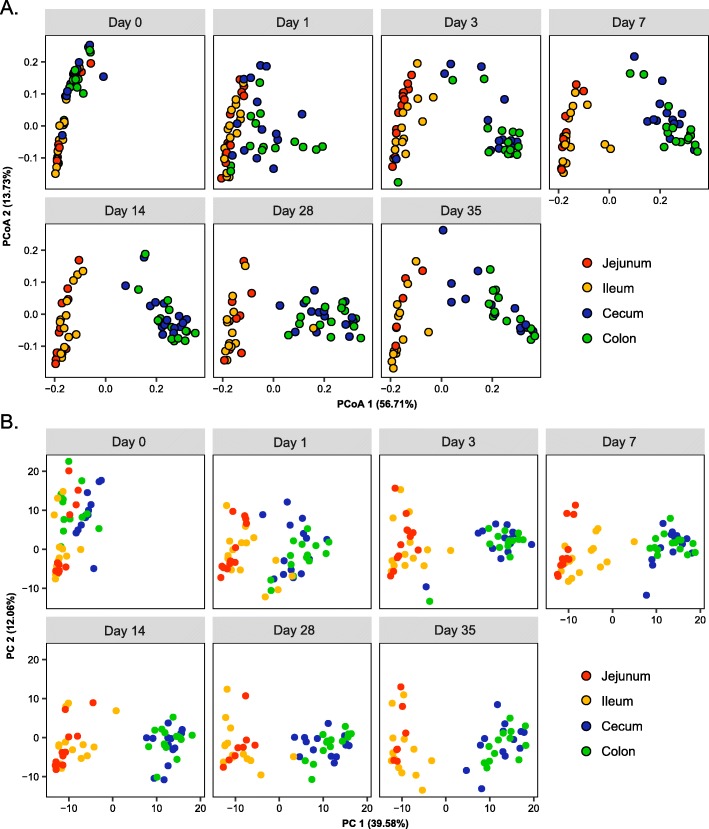
Consistently, no obvious changes in the α-diversity of mucosa-associated microbiota occurred in the small intestine across different ages, while the α-diversity of the large intestinal microbiota was significantly increased in the first 7–14 days before becoming stabilized (P < 0.001, Fig. 1b), except for a transient decline observed on day 1 (Additional file 3: Figure S3B). The β-diversity analysis (unweighted UniFrac) showed a similar trend. The mucosa-associated microbiota remained relatively stable in the small intestine, while diverged from the day 0 microbiota rapidly in the large intestine in the first week before becoming more or less stabilized (Fig. 1c). To further reveal the mature pattern of mucosa-associated microbiota in the small and large intestines, PCoA was performed based on the phylogenetic metrics (weighted and unweighted Unifrac, Fig. 2a, Additional file 4: Figure S4A) or taxonomic metric (Bray-Curtis, Additional file 4: Figure S4B). Nearly identical patterns were yielded from three metrics. While the small intestinal microbiota of different ages were clustered largely together, the large intestine was similar in the microbiota composition to the small intestine at birth, but became progressively divergent with age, resulting in two rather distinct microbial communities between the small and large intestines by day 35 (Fig. 2a; Additional file 4: Figure S4, Table 2). Together, these observations suggested a rather different succession pattern of mucosa-associated microbiota between the small and large intestines.
Fig. 2.
β-diversity of the mucosa-associated microbiota across different intestine locations. a PCoA of phylogenetic community composition based on weighted UniFrac distance. Population-level PERMANOVA statistics are detailed in Table 1. b Principal components analysis (PCA) of predicted functional genes of mucosa-associated microbiota at KEGG level 3
Table 2.
Intestinal location-dependent influence of age on the mucosa-associated microbial community
| Items | Weighted UniFrac | Unweighted UniFrac | Bray-Curtis | |||
|---|---|---|---|---|---|---|
| R2 | P value | R2 | P value | R2 | P value | |
| Jejunum | 0.086 | 0.404 | 0.089 | 0.167 | 0.084 | 0.392 |
| Ileum | 0.076 | 0.208 | 0.094 | < 0.001 | 0.109 | 0.005 |
| Cecum | 0.426 | < 0.001 | 0.286 | < 0.001 | 0.32 | < 0.001 |
| Colon | 0.463 | < 0.001 | 0.312 | < 0.001 | 0.339 | < 0.001 |
PERMANOVA was performed to test the effect of days after birth on mucosa-associated microbiota in different intestinal segments. P values in italics represent statistical significance (P < 0.05)
Mucosa-associated microbiota in the large intestine is predicted to be progressively divergent in functions from that in the small intestine
The difference in the composition of mucosa-associated microbiota between the small and large intestines suggested a distinct functional maturation process of the microbiome. To investigate how the functional profiles of the mucosa-associated microbiome change during early life, PICRUSt analysis [43] was performed. Similar to our earlier observations on the bacterial community structure, the microbiota function was predicted to be similar between the small and large intestines at birth (Fig. 2b). While the bacterial function was relatively stable in the small intestine across the ages, the large intestinal microbiota became progressively divergent as the animals aged (Fig. 2b). In the large intestine, 259 functional pathways were identified to be significantly altered along with piglet development. Specifically, the most highly enriched pathways were predicted to be involved in genetic information processing such as regulation of transcriptional factors, nucleotide excision repair, lysine biosynthesis, homologous recombination, and mismatch repair (Additional file 9: Table S2). In contrast, only 4 pathways were significantly altered (P < 0.05) during small intestinal development, although approximately an equal number of functional pathways were identified in both (small intestine, N = 263; large intestine, N = 264). Moreover, out of 261 most prevalent KEGG pathways that were present in at least 50% samples, 241 were significantly different between the small and large intestines (P < 0.05, Additional file 10: Table S3). In particular, the small intestinal microbiome was associated with enrichment of the pathways in biodegradation and metabolism of xenobiotics, whereas the pathways involved in glycan biosynthesis and metabolism, replication and repair, energy metabolism, translation, and carbohydrate metabolism were more abundant in the large intestine. Together, these data suggested that, in addition to the structural differences, functional profiles of the bacterial community are also vastly different between the small and large intestines. While it was relatively stable in the small intestine, the microbiota function shifted towards an adult-like configuration in the large intestine.
Maternal milk and fecal microbiota are the major contributors of neonatal intestinal mucosa
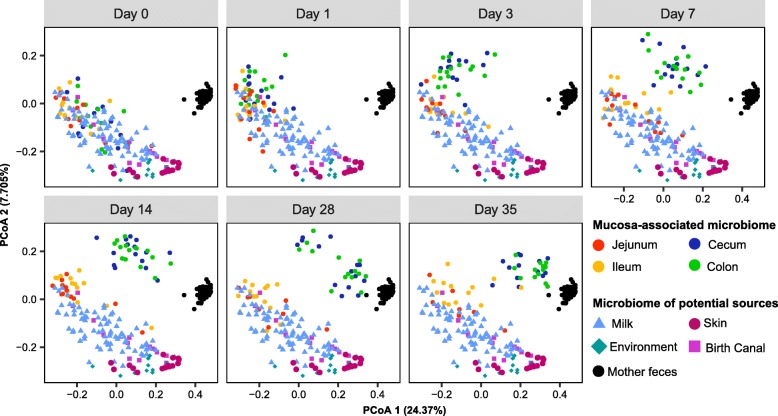
To analyze the potential sources of the microbial assembly in infant piglet intestinal mucosa, fecal and milk samples as well as the areolar skin and vaginal swabs were collected from sows. Farrowing crates and incubators were also swabbed as environmental samples. PCoA using unweighted UniFrac distance showed close clustering of the milk and small intestine samples, while the large intestinal microbiota was clustered with the small intestine at birth, but gradually diverged with age, resembling more and more the fecal microbiota of sows (Fig. 3). PCoA using weighted Unifrac and the Bray-Curtis metrics showed similar clustering patterns (Additional file 5: Figure S5), implying that the maternal milk and fecal microbiome might serve as microbial reservoirs for vertical transmission.
Fig. 3.
β-diversity of the mucosa-associated microbiota, milk, skin, vagina and feces of sows, and environmental microbiota. PCoA of phylogenetic community composition based on unweighted UniFrac distance
SourceTracker [44] was performed to further analyze the relative contributions of different microbial sources from the mother and birth environment on the initial colonization of neonatal GI. Obviously, maternal milk was the primary contributor of the microbiota in the small intestine, accounting for approximately 90% of the microbiota throughout the first 35 days, even after the introduction of solid creep feed on day 7 (Fig. 4). In contrast, although more than 90% of the large intestinal microbiota of piglets also originated from maternal milk, its contribution gradually declined with age (Fig. 4). Milk microbes contributed approximately 20% of the microbiota in the large intestine on day 3, which was further diminished to less than 5% in the colon and 10% in the cecum. On the other hand, the contribution of maternal fecal microbiota became progressively more prominent with age. Albeit with virtually no contributions at birth, fecal microbes of sows contributed 7–20% of the microbiota in piglet’s large intestine between days 7 and 35 (Fig. 4). Similarly, pairwise β-diversity comparisons showed that milk microbiota closely resembled that of the small intestine initially, while fecal microbiota became progressively similar to the large intestinal microbiota (Additional file 6: Figure S6). Maternal vaginal microbiota contributed 6–16% of mucosa-associated microbiota in the ileum, cecum, and colon on day 1; however, such an effect occurred only transiently and quickly diminished by day 35 (Fig. 4). Similarly, the neonatal birth environment contributed 2–10% of mucosal microbiota in the large intestine within the first 2 weeks, and its contribution further diminished with age (Fig. 4).
Fig. 4.
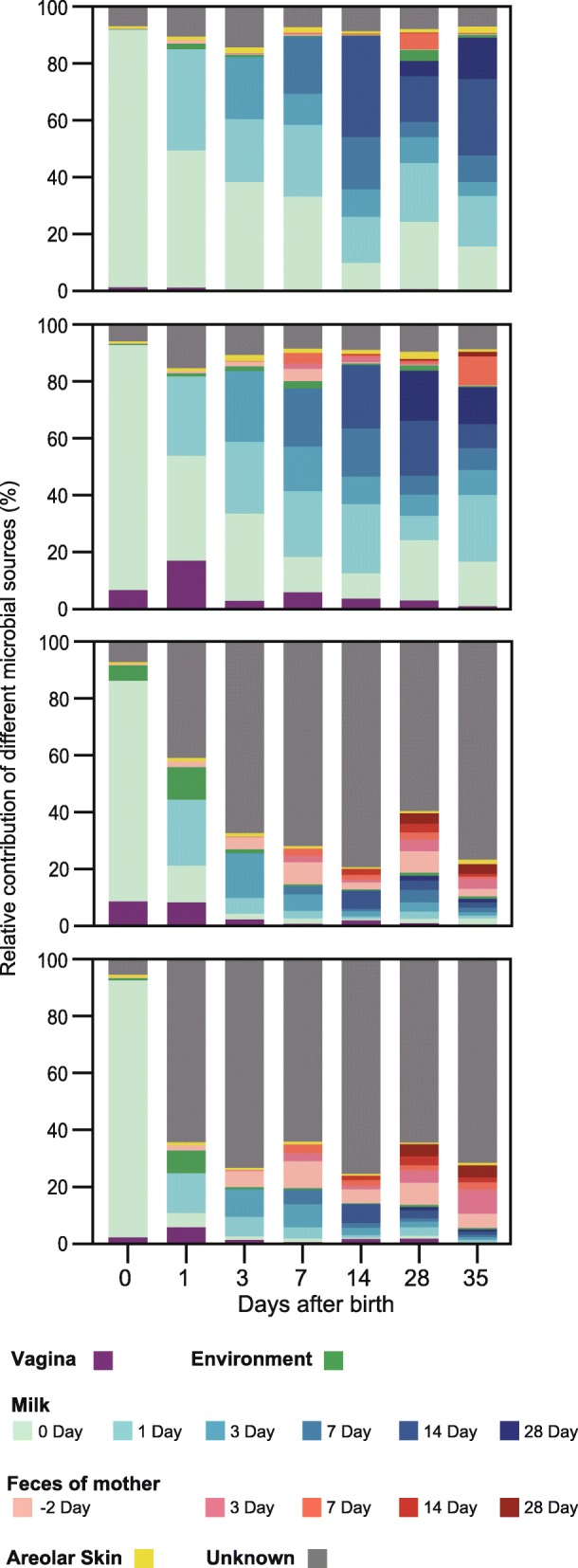
Dynamic contributions of different microbial sources to the neonatal gut mucosal microbiota during the first 35 days. The proportion of microbiota from the jejunum, ileum, cecum, and colon samples of piglets are estimated to originate from different maternal and environmental sources (colored regions), using bacterial source-tracking
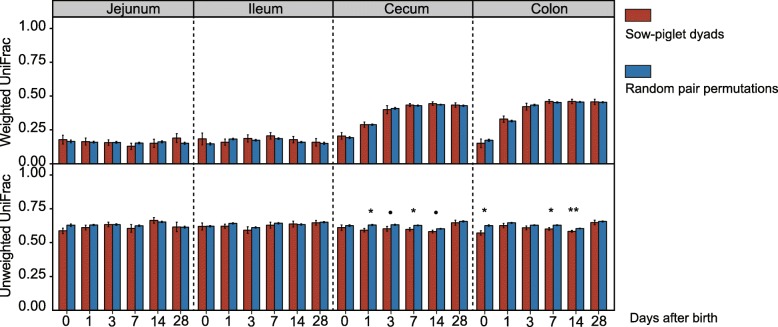
To further confirm bacterial transmission from sows or environment to infant piglets, we hypothesized that a piglet’s intestinal mucosa-associated microbiota more resembles its mother’s microbiota than a random sow’s. A closer resemblance in unweighted Unifrac distance between sow milk microbiota and piglet’s microbiota in the large intestine was observed among sow-piglet dyads than random pairs (P < 0.05; Fig. 5). Similar results were also found between the vaginal and environmental microbiota and piglet’s large intestinal microbiota (P < 0.05; Additional file 7: Figure S7), further corroborating the occurrence of microbial transmission.
Fig. 5.
Distance comparison of microbial communities between true sow-piglet dyads and random pairs for milk microbiota (values are means ± SE; significance between the intestinal segments was determined by pairwise Kruskal-Wallis test; *P < 0.05; **P < 0.01; ***P < 0.001)
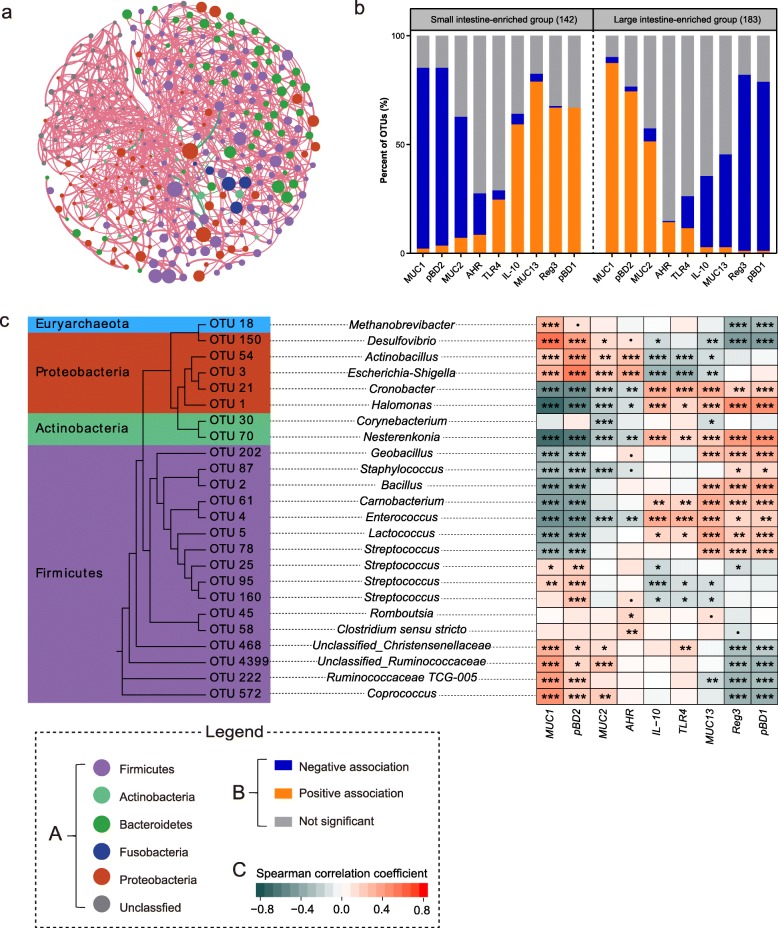
Next, we sought to identify specific OTUs transmitted from sow and birth environment to piglets. A total of 24 OTUs were significantly shared among sow-piglet dyads as compared to random pairs and thus identified as transmitted bacterial taxa (Additional file 11: Table S4). Of the 24 OTUs, 16 belonged to Firmicutes and 5 belonged to Proteobacteria. Genus-level annotation of these OTUs revealed that Corynebacterium, Bacillus, Lactococcus, Staphylococcus, Romboutsia, Ruminococcaceae TCG-005, Escherichia-Shigella, Streptococcus, and Halomonas were likely transferred from the maternal and birth environment to the piglet’s intestine (Additional file 11: Table S4). A species (OTU468) of Christensenellaceae, the most heritable taxon previously reported [45], was likely to be transferred from the feces of sows to the large intestine of piglets. Interestingly, half of these bacterial transmissions (19/38) occurred through milk and the vagina within the first 3 days, while the other half occurred through maternal feces and environment in the following several weeks. More importantly, most of these bacterial transmissions (36/38) occurred in the ileum, cecum, and colon (Additional file 11: Table S4). Although the underlying mechanism remains unclear, this intestinal segment-specific transmission is likely due to the difference in the ability of microbes to colonize different ecological niches in the GI [46, 47].
Maternally transmitted microbes are involved in the regional expression of intestinal immune and functional genes
To examine whether mucosa-associated bacteria are associated with functional development of the neonatal GI, nine genes known to be involved in immune and barrier functions were selected and quantified in the jejunum, ileum, and colon samples at different ages. Among them, porcine β-defensin 1 (PBD1), PBD2, and regenerative III protein (RegIII) are the major host defense peptides against infections [48, 49], while mucin (MUC) 1, MUC2, and MUC13 are the important members of the mucin family forming the protective mucus layer along the intestine [50]. Aryl hydrocarbon receptor (AHR) and Toll-like receptor 4 (TLR4) are important receptors mediating host inflammatory and immune response [51, 52], and interleukin (IL)-10 is a well-known anti-inflammatory cytokine [53].
As expected, most intestinal genes were differentially expressed in the jejunum, ileum, and colon of piglets (P < 0.05; Additional file 12: Table S5), consistent with the idea of regional specialization and maturation of the intestinal immune and barrier functions as animals age [54]. For instance, the expression level of MUC2 was significantly elevated in the colon with age but remained relatively stable in the jejunum and ileum (Additional file 13: Table S6). In contrast, a steady increase in the expression of TLR4 and IL-10 was observed across three intestinal segments (P < 0.05; Additional file 12: Table S5). Furthermore, the expression of intestinal genes was significantly correlated with the relative abundance of a number of OTUs (P < 0.05; Additional file 14: Table S7). Surprisingly, most of these OTUs showed a positive correlation with each other (Fig. 6a), implying a possible existence of the microbial consortia that may promote mutual growth and guide the expression of intestinal immune and barrier function genes. Based on the differential abundance between small and large intestine, these OTUs were stratified into two groups, the small intestine-enriched group and the large intestine-enriched group (Additional file 14: Table S7). The small intestine-enriched OTUs were mostly associated with highly abundant Proteobacteria, but not Bacteroidetes (Additional file 14: Table S7). Intriguingly, an opposite correlation pattern was observed between the two groups and intestinal gene expressions. For instance, the small intestinal group showed a negative correlation with the expression of MUC1, which became positive for the large intestinal group (Fig. 6b; Additional file 14: Table S7). Nearly identical correlation patterns were observed between the two groups and the expressions of remaining genes (P < 0.05, Fig. 6b; Additional file 14: Table S7). Importantly, all vertically transmitted bacteria were significantly correlated with the expression levels of intestinal immune and barrier function genes (Fig. 6c), implying that maternally transmitted bacteria are critically involved in the maturation of immune and barrier functions in the neonatal GI.
Fig. 6.
Involvement of mucosa-associated microbiota in the spatial expression of intestinal functional genes. a Network plots of OTUs (represented by nodes) that are significantly associated with the expression of intestinal genes. Significant correlative associations between OTUs were determined based on the SPIEC-EASI pipeline. Edge color represents positive (pink) and negative (green) correlations, and the edge thickness is equivalent to the magnitude of the correlation coefficient. SPIEC-EASI correlations with a magnitude of < 0.05 were not shown. The diameter of each node is proportional to the average abundance of each OTU across all samples, while each filled color corresponds to a bacterial phylum. b Spatial correlation pattern between the small and large intestine-enriched OTUs with the expression level of the intestinal function genes. The full list of significant correlative associations is presented in Additional file 14: Table S7. c A list of bacterial OTUs that are transmitted from sow and birth environment to the piglets showing a significant correlation with the expression of intestinal functional genes. The left panel is the phylogenetic tree of transmitted OTUs, whereas the right panel is the heat map of Spearman’s rank correlation coefficient between 24 transmitted OTUs and expression levels of intestinal genes (•P < 0.1, *P < 0.05, **P < 0.01, ***P < 0.001). The background color corresponds to the phyla to which the OTU belongs. Spearman’s rank correlation coefficient is indicated using a color gradient: red indicates positive correlation; cyan, negative correlation
Discussion
The GI microbiota in early life has long-term implications in host physiology and health [6, 55]. Because the GI microbiota varies greatly along the intestinal tract and undergoes substantial changes with age, it is important to understand initial colonization and succession of microbiota in different segments of the neonatal GI. However, most studies on the development of infant microbiome were conducted only with stool samples due to technical and ethical limitations [14, 15, 56]. Here, using mucosa-associated microbiota from the small and large intestines of piglets as well as maternal and environmental microbiota, we systematically studied the development and origin of piglet’s GI microbiota and its impact on intestinal innate immune and barrier functions.
Fecal microbiota undergoes progressive succession before maturation [57]. Unexpectedly, we observed for the first time distinct succession patterns of the mucosa-associated microbiota between the small and large intestines. Despite a similar initial composition and predicted functional profile at birth, mucosal microbiota in the small intestine remains relatively stable, while that in the large intestine undergoes progressive expansion and diversification as soon as 24 h after birth and continuously shifts in the first 35 days of age. These temporal and spatial dynamics resemble the development of the lumen-associated microbiota, which showed similar initial structure across different intestinal segments (duodenum, jejunum, ileum, cecum, colon, and rectum) on postnatal day 1, but evolved and quickly differentiated at later intervals [58]. Similarly, the changes of the lumen-associated microbial community in the small intestine are negligible during the nursing (postnatal day 7 to day 35) and weaning periods (postnatal day 120 to day 180); in contrast, that in the large intestine undergoes considerable changes. However, we did not observe either similar composition or changes in the microbiota of the small intestine at the taxonomic level as previously reported [58]. Nevertheless, it should be noted that different intestinal sites, luminal digesta and mucosa, were sampled in two studies. Indeed, GI microbiota differs radically in their functional potential, density, and composition from the mucosa to the lumen [23, 59]. Second, it is likely that microbial compositions vary according to the genetic background of pig and many other factors [30]. Lastly, the small cohort (N = 5) and substantial inter-individual variation in the lumen-associated microbiota in the research conducted by Liu et al. [58] might also lead to the contrary observations. Collectively, these observations suggested a spatiotemporal developmental and maturation pattern of the mucosa-associated bacterial community.
In this study, we detected diverse microbiota in different segments of the intestine and different body sites at birth, consistent with earlier detection of bacteria within the first-pass meconium [60, 61], placenta [62], and amniotic fluid [63] from healthy term infants. The first meconium microbiota is thought to reflect the in utero environment since the fetus could swallow amniotic fluid in the womb. Indeed, the meconium has been shown to harbor a microbial community resembling those in the amniotic fluid and placenta [60, 63, 64]. Furthermore, maternal transmission of bacteria to the fetal GI during gestation has been observed in murine models [65], reinforcing the idea that microbial colonization of the fetus may occur before birth.
We have also revealed that mucosa-associated intestinal microbiota is primarily derived from vertical transmission of the microbes in maternal milk and the GI. Milk contributes greater than 90% of mucosa-associated microbiota in the small intestine of the neonates in the first 35 days after birth. The contribution of the milk microbes to the neonatal mucosal microbiota is gradually declined in the large intestine, but with a concomitant increase in the contribution of maternal fecal microbes. In agreement with our study, up to 30% of the infant fecal microbes were previously reported to come from milk and declined as the infant ages [38]. Perhaps not coincidently, the significance of the milk microbiota is underscored by the observation that the GI microbiota is drastically different between formula-fed and breastfed infants [32, 66]. A more recent study involving 25 mother-infant pairs concluded that maternal GI microbiome is also a major source of neonatal GI microbes [67].
We found that vaginal microbes of sows colonize neonatal GI mucosa at day 1, consistent with a number of earlier studies showing distinct GI microbiota between infants delivered vaginally and by cesarean section [15, 33]. However, this “vaginal seeding” is likely transient after birth. We observed that the contribution of vaginal microbiota became diminished quickly in a few days, which is in line with the results showing that the influence of birth mode failed to be detected in 6 weeks [15, 56, 67]. Indeed, we found that the transfer of maternal vaginal and milk microbes mostly occurred within 24 h shortly after birth, whereas that of maternal GI and environment microbes mostly occurred after 7 days. On the other hand, the initial acquisition from the mother could prepare the newborns for host-microbial symbiosis. Indeed, paralleling the frequent occurrence of the maternal bacterial seeding within 3 days after birth, a decrease at days 1 and 3 in microbial diversity has been observed. Similar observations have been made in mice and human infants [15, 68], suggesting a selective effect of the pioneer microbes from maternal milk and vagina on the infant GI microbiota colonization [69], while the higher bacterial diversity at birth could be explained by the in utero colonization of the neonates.
The infant’s intestinal microbiota is believed to guide the development and maturation of the intestinal immunity and barrier function [5, 70]. Aberrant mucosal defense and barrier integrity of germ-free mice can be rescued by transplantation with normal microbiota [71, 72]. In this study, we demonstrated that the relative abundances of a number of mucosa-associated microbes are significantly correlated with the expression of multiple intestinal genes known to be involved in innate immunity and barrier function. Importantly, these bacteria can be clustered into two groups enriched in either the small or large intestine, showing a strong correlation with the spatial expression pattern of the intestinal functional genes. In line with our observation, induction of TH17 cells requires the colonization of segmented filamentous bacteria (SFB) in the terminal ileum, which subsequently leads to increased expression of the genes associated with inflammation and antimicrobial defense [17]. Furthermore, microbes are known to differentially colonize along the intestinal mucosa surface [47, 73] and has a profound impact on local expressions of a multitude of host genes [74, 75]. Our findings have further supported the notion on the involvement of mucosa-associated microbiota in the site-specific development and maturation of intestinal mucosal immunity and barrier function.
Importantly, we have revealed that most maternally transmitted bacteria show a strong correlation with the expression of intestinal functional genes, suggesting a significant involvement of maternally derived microbes in the maturation of intestinal function. In agreement with this, human milk microbes have been shown to contribute to the immune development and maturation [76, 77], and cesarean increases the risk of allergic and autoimmune diseases in the offspring [78–80], while breastfeeding has been linked to the enhanced immunity and decreased risks for illnesses such as obesity [81, 82]. Our results have further supported the rationale for maternal bacterial seeding such as vaginal delivery and breastfeeding. Further studies on the roles of maternally derived microbes in infant health are warranted.
Conclusion
We revealed for the first time that mucosa-associated microbiota in the small intestine (jejunum and ileum) remains relatively stable in early life, while those in the large intestine (cecum and colon) become quickly diversified in both the structure and function. Moreover, the neonatal GI microbiota is primarily shaped by the microbes from maternal milk and feces. We further demonstrated a strong temporal and spatial correlation between maternally derived microbiota and the expression pattern of host immune and functional genes along the intestinal mucosal surface, suggesting a potentially critical involvement of maternal microbiota in the site-specific development and maturation of intestinal mucosal immunity and barrier function. A better understanding of the succession of mucosa-associated intestinal microbiota throughout the neonatal GI may lead to new approaches to precise therapeutic manipulations.
Materials and methods
Experimental design and sample collection
Healthy, third-parity purebred Rongchang sows (n = 8) and Landrace sows (n = 8) with a similar expected delivery date were selected for this study. Each sow was individually housed in a different environmentally controlled room under standard management with access to a common non-medicated diet. To avoid cross-contamination, the building was decontaminated prior to the beginning of the trial and each breed was housed on a separate side of the building. After delivery, newborn piglets were co-housed with sows by litter and ear-notched for individual identification. Suckling piglets were offered a common creep feed ad libitum at day 7 and weaned at day 28. All piglets remained in nursing pens for another week till day 35, while sows were removed from the piglets at day 28.
One piglet from each litter was randomly selected and euthanized with Zoletil 50® (Virbac, Carros, France) at birth and at days 1, 3, 7, 14, 28, and 35 (Additional file 1: Figure S1), while ensuring half males and half females for each breed at each time point. Mucosa-associated microbiota was collected from a middle section of the jejunum, ileum, cecum, and colon of each piglet for bacterial DNA isolation as previously described [83] and adjacent intestinal segments were also collected for RNA extraction. A composite sample of milk was collected from each sow at the same time points after farrowing and continued until weaning. In addition, fresh fecal samples were taken from sows 2 days before farrowing and at days 3, 7, 14, and 28 after farrowing (Additional file 1: Figure S1). Additionally, vaginal and areolar skin swabs were taken from sows immediately after giving birth. A vaginal swab was taken by swirling a sterile cotton swab (Kangjie Medical Devices Co., Jiangsu, China) near the mid-vaginal canal six times, while areolar skin around the teat (approximate 10 cm in diameter) was swabbed with saline-lubricated sterile swabs. Farrowing crates and incubators were also sampled with sterile saline-soaked swabs. Each of these three specimens was sampled in triplicate for microbial analysis. All samples were immediately snap-frozen in liquid nitrogen and stored at − 80 °C for further analysis.
Bacterial DNA extraction and 16S r RNA gene sequencing
Microbial DNA was extracted from the intestinal mucosa-associated microbiota, feces, and swabs using QIAamp DNA Stool Mini Kit (Qiagen, Duesseldorf, Germany). Microbial DNA was extracted from the milk using a DNeasy PowerFood Microbial Kit (Qiagen). Both procedures were carried out according to the manufacturer’s instructions, with an addition of a bead-beating step using 0.25 g of 0.15 mm garnet beads and 0.25 g of 0.1 mm zirconia beads. DNA was quantified with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, DE, USA), and the integrity was checked by 1% agarose gel electrophoresis. Amplification of the V3-V4 region of bacterial 16S rRNA genes was carried out as previously described [84]. Briefly, bar-coded universal primers 341F and 806R were designed for PCR amplification with initial denaturation at 95 °C for 5 min and 27 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 72 °C for 45 s, followed by a final extension at 72 °C for 10 min. The PCR products were gel purified, quantified via NanoDrop™ 2000 spectrophotometer (Thermo Scientific), pooled at equal molar ratios, and sequenced on Illumina HiSeq 2500.
Sequence analysis
Raw sequence data from a total of 556 samples were processed using QIIME (version 1.8.0). Sequences were de-noised using denoise_wrapper.py. High-quality sequences were clustered into distinct operational taxonomic units (OTUs) using UCLUST with a 97% threshold of the pairwise identity. Chimeric sequences were removed using identify_chimeric_seqs.py. The OTU table was filtered using filter_otus_from_otu_table.py. The most abundant sequence was picked for each OTU and taxonomically assigned using the SILVA reference database (Version 111) [85]. Representative OTUs were aligned using PyNAST [86] to build a phylogenetic tree with FastTree [87], which was used subsequently to estimate the α- and β-diversity. Microbial community distances were calculated using Bray-Curtis, weighted, and unweighted UniFrac distance metrics [88].
Gene expression and qPCR
RNA isolation and quantification of intestinal segments were performed as previously described [89] using the primers listed in Additional file 8: Table S1. All reactions were run in triplicate. Relative gene expression was calculated according to the ΔΔCt method [90] using porcine β-actin as the reference gene.
Data analysis and statistics
Statistical analysis was performed using SPSS 22.0 (SPSS, Chicago, IL, USA) and R programming. For normally distributed continuous variables, the mean values were examined using an unpaired Student’s t test or one-way ANOVA with Tukey’s post hoc test. The α-diversity was calculated using Chao1 and Shannon diversity indices and compared using Kruskal-Wallis tests with Benjamini-Hochberg correction. Principal coordinates analysis (PCoA) was performed on the Bray-Curtis, unweighted, and weighted UniFrac distance metrics to visualize the relationships between the samples. Permutational multivariate analysis of variance (PERMANOVA) using the adonis function in vegan with 9999 permutations was performed to analyze the distance metrics for factors that shape mucosa-associated microbiota.
PICRUSt analysis was applied to infer putative metagenomes from the 16S rRNA gene profiles [43]. Differences in the abundance of KEGG pathways between the groups were analyzed using STAMP software [91] and Welch’s t test with Benjamini-Hochberg correction. To estimate the sources of microbial communities observed at different intestinal segments and days after birth, we used SourceTracker (v1.0), a Bayesian approach for bacterial source tracking [44]. Samples collected from different maternal body sites and the birth environment were designated as sources, and samples from the intestinal mucosa of the piglets were tagged as sinks. SourceTracker R package (Version 1.0) was used to perform the analysis with default parameters. OTU sharing was defined as the percentage of mother-infant dyads in which a given OTU was found in both members. Permutation testing with randomly shuffled mother-infant pairings was used to assess the significance of OTU sharing as previously described [38]. Fisher’s exact test was employed to test the association between frequencies within sow-piglet dyads as described [92]. Because both tests ignore the OTUs that are present in all samples, Spearman’s rank correlation test between the relative abundance of OTUs in the milk or vagina and those in piglets were performed as well. Prevalent OTUs (> 20% in either the source or sink samples) were identified as potential transmitted OTUs if both OTU sharing permutation test, and Fisher’s exact test were significant or if Spearman’s rank correlation test was significant (P < 0.05) for an OTU present in all individuals. The phylogenetic tree of transmitted OTUs was constructed with RAxML [93]. Correlations between the intestinal gene expression levels and relative abundance of OTUs were tested with Spearman correlation [94, 95]. SPIEC-EASI [96] was applied for the inference of microbial ecological networks among the OTUs that were significantly associated with gene expression. The networks were visualized using Gephi [97].
Supplementary information
Additional file 1: Figure S1. Experimental design and workflow of sample collection. Each tick denotes a time-point of sample collection.
Additional file 2: Figure S2. Rarefaction curves based on the Chao1 and Shannon index at increasing sequencing depth of intestinal mucosal, maternal and environmental samples.
Additional file 3: Figure S3. (A) Negligible influence of breed on the mucosa-associated microbiota at birth. Average weighted UniFrac, unweighted UniFrac and Bray-Curtis distance between individuals at birth within and between Rongchang and Landrace piglets. (B) Shift of α diversity of mucosa-associated microbiota with age across four intestinal segments based on Chao1 index (Values are Means ± SE; significance between groups was determined by Kruskal-Wallis test).
Additional file 4: Figure S4. PCoA of phylogenetic community composition based on unweighted UniFrac distance (A), and taxonomic community composition based on Bray–Curtis (B).
Additional file 5: Figure S5. β-diversity of the mucosa-associated microbiota, milk, skin, vagina and feces of sows, and environmental microbiota. PCoA of phylogenetic community composition based on weighted UniFrac (A), and taxonomic community composition based on Bray–Curtis (B).
Additional file 6: Figure S6. Distance comparison of microbial communities between mucosa-associated intestinal microbiota with that of maternal milk and feces. Values are Means ± SE; significance between intestinal segments was determined by pairwise Kruskal-Wallis test; * P < 0.05; ** P < 0.01; *** P < 0.001.
Additional file 7: Figure S7. Distance comparison of microbial communities between true compared with random sow-piglet pairs for fecal, vaginal and environmental microbiota (Values are Means ± SE; significance between intestinal segments was determined by pairwise Kruskal-Wallis test; * P < 0.05; ** P < 0.01; *** P < 0.001).
Additional file 8: Table S1. Primers used in the study.
Additional file 9: Table S2. Shift of PICRUSt-predicted KEGG pathways of mucosa-associated microbiota in small intestine and large intestine (P values by Welch’s t-test with a Benjamini–Hochberg FDR correction were computed by STAMP).
Additional file 10: Table S3. Different PICRUSt-predicted KEGG pathways of mucosa-associated microbiota between two clusters, small intestinal cluster (including the large intestinal samples at birth) and large intestinal cluster (except the large intestinal samples at birth). P values by Welch’s t-test with a Benjamini–Hochberg FDR correction were computed by STAMP.
Additional file 11: Table S4. Candidate transmitted OTUs identified from mother and environment.
Additional file 12: Table S5. Relative expression of intestinal function-related genes using the whole statistical model.
Additional file 13: Table S6. Contrasts for MUC1, MUC2 and pBD1 expression analyzed within each intestinal segment.
Additional file 14: Table S7. Correlation of the relative abundance of OTUs and expression of genes involved in the development of intestinal barrier function.
Acknowledgements
We would like to thank Chongqing Academy of Animal Science for providing all resources in animal trials. We also thank Dr. Chenhao Li at Genome Institute of Singapore; Dr. Xuan He at the University of California, Davis; and Dr. Grace M. Aldrovandi at the University of California, Los Angeles for their expert help with data analysis.
Abbreviations
- GI
Gastrointestinal
- SFB
Segmented filamentous bacteria
- PERMANOVA
Permutational multivariate analysis of variance
- PCoA
Principal coordinates analysis
- PBD
Porcine β-defensin
- RegIII
Regenerative III
- MUC
Mucin
- AHR
Aryl hydrocarbon receptor
- TLR4
Toll-like receptor 4
- IL
Interleukin
Authors’ contributions
HL, XZ, and CH participated in the overall study design. HL conducted the animal experiment with help from XZ, HY, LS, ZL, and FY. HL also carried all analyses and drafted the manuscript. GZ, NL, and PT provided comments for the manuscript. SQ directed the overall research project. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31420103908) and the H2020 European Research Council (Feed-a-Gene, No. 633531).
Availability of data and materials
The data generated or analyzed during this study are included in this published article, its supplementary information files, and publicly available repositories. Raw 16S rRNA gene sequences and study metadata were deposited in the National Center for Biotechnology Information—NCBI repository (BioProject accession number: PRJNA524979 [98] and PRJNA480348 [99]).
Ethics approval and consent to participate
All animal procedures reported in this study were approved by the China Agricultural University Institutional Animal Care and Use Committee under Protocol number CAU20161110-2.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12915-019-0729-2.
References
- 1.Bellmann S, Carlander D, Fasano A, Momcilovic D, Scimeca JA, Waldman WJ, et al. Mammalian gastrointestinal tract parameters modulating the integrity, surface properties, and absorption of food-relevant nanomaterials. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2015;7:609–622. doi: 10.1002/wnan.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohman L, Tornblom H, Simren M. Crosstalk at the mucosal border: importance of the gut microenvironment in IBS. Nat Rev Gastroenterol Hepatol. 2015;12:36–49. doi: 10.1038/nrgastro.2014.200. [DOI] [PubMed] [Google Scholar]
- 3.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. New Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 4.El Aidy Sahar, van Baarlen Peter, Derrien Muriel, Lindenbergh-Kortleve Dicky J, Hooiveld Guido, Levenez Florence, Doré Joël, Dekker Jan, Samsom Janneke N, Nieuwenhuis Edward E S, Kleerebezem Michiel. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunology. 2012;5(5):567–579. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- 5.Gensollen Thomas, Iyer Shankar S., Kasper Dennis L., Blumberg Richard S. How colonization by microbiota in early life shapes the immune system. Science. 2016;352(6285):539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. 2016;22:713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 7.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramanian Sathish, Huq Sayeeda, Yatsunenko Tanya, Haque Rashidul, Mahfuz Mustafa, Alam Mohammed A., Benezra Amber, DeStefano Joseph, Meier Martin F., Muegge Brian D., Barratt Michael J., VanArendonk Laura G., Zhang Qunyuan, Province Michael A., Petri Jr William A., Ahmed Tahmeed, Gordon Jeffrey I. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen A, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:152r–307r. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 11.Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond). 2011;35:522–529. doi: 10.1038/ijo.2011.27. [DOI] [PubMed] [Google Scholar]
- 12.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 14.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016;22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renz Harald, Brandtzaeg Per, Hornef Mathias. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nature Reviews Immunology. 2011;12(1):9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- 17.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2010;2:19. doi: 10.1186/1757-4749-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20:50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guilloteau P, Zabielski R, Hammon HM, Metges CC. Nutritional programming of gastrointestinal tract development. Is the pig a good model for man? Nutr Res Rev. 2010;23:4–22. doi: 10.1017/S0954422410000077. [DOI] [PubMed] [Google Scholar]
- 27.Heinritz SN, Mosenthin R, Weiss E. Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutr Res Rev. 2013;26:191–209. doi: 10.1017/S0954422413000152. [DOI] [PubMed] [Google Scholar]
- 28.Buddington RK, Sangild PT, Hance B, Huang EY, Black DD. Prenatal gastrointestinal development in the pig and responses after preterm birth. J Anim Sci. 2012;90(Suppl 4):290–298. doi: 10.2527/jas.54604. [DOI] [PubMed] [Google Scholar]
- 29.Calder PC, Krauss-Etschmann S, de Jong EC, Dupont C, Frick JS, Frokiaer H, et al. Early nutrition and immunity - progress and perspectives. Br J Nutr. 2006;96:774–790. doi: 10.1079/BJN20061881. [DOI] [PubMed] [Google Scholar]
- 30.Xiao L, Estellé J, Kiilerich P, Ramayo-Caldas Y, Xia Z, Feng Q, et al. A reference gene catalogue of the pig gut microbiome. Nat Microbiol. 2016;1:16161. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 31.Merrifield CA, Lewis M, Claus SP, Beckonert OP, Dumas ME, Duncker S, et al. A metabolic system-wide characterisation of the pig: a model for human physiology. Mol Biosyst. 2011;7:2577–2588. doi: 10.1039/c1mb05023k. [DOI] [PubMed] [Google Scholar]
- 32.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343r–382r. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulfer A, Blaser MJ. Risks of antibiotic exposures early in life on the developing microbiome. Plos Pathog. 2015;11:e1004903. doi: 10.1371/journal.ppat.1004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz S, Chapkin RS, Friedberg I, Ivanov IV, Davidson LA, Goldsby JS, et al. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol. 2012;13:r32. doi: 10.1186/gb-2012-13-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merrifield CA, Lewis MC, Berger B, Cloarec O, Heinzmann SS, Charton F, et al. Neonatal environment exerts a sustained influence on the development of the intestinal microbiota and metabolic phenotype. ISME J. 2016;10:145–157. doi: 10.1038/ismej.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. Jama Pediatr. 2017;171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin H, Pei Z, Martinez KA, Rivera-Vinas JI, Mendez K, Cavallin H, et al. The first microbial environment of infants born by C-section: the operating room microbes. Microbiome. 2015. 10.1186/s40168-015-0126-1. [DOI] [PMC free article] [PubMed]
- 40.Brooks B, Olm MR, Firek BA, Baker R, Thomas BC, Morowitz MJ, et al. Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat Commun. 2017. 10.1038/s41467-017-02018-w. [DOI] [PMC free article] [PubMed]
- 41.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Wei, Chow Kin-hoe, Fleming Elizabeth, Oh Julia. Selective colonization ability of human fecal microbes in different mouse gut environments. The ISME Journal. 2018;13(3):805–823. doi: 10.1038/s41396-018-0312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI, Borisy GG. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci. 2017;114:E9105–E9114. doi: 10.1073/pnas.1711596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sang Y, Blecha F. Porcine host defense peptides: expanding repertoire and functions. Dev Comparative Immunol. 2009;33:334–343. doi: 10.1016/j.dci.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016;16:639–649. doi: 10.1038/nri.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gribar SC, Sodhi CP, Richardson WM, Anand RJ, Gittes GK, Branca MF, et al. Reciprocal expression and signaling of TLR4 and TLR9 in the pathogenesis and treatment of necrotizing enterocolitis. J Immunol. 2009;182:636–646. doi: 10.4049/jimmunol.182.1.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Are A, Aronsson L, Wang S, Greicius G, Lee YK, Gustafsson JA, et al. Enterococcus faecalis from newborn babies regulate endogenous PPARgamma activity and IL-10 levels in colonic epithelial cells. Proc Natl Acad Sci U S A. 2008;105:1943–1948. doi: 10.1073/pnas.0711734105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 55.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017. 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed]
- 56.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8:343r–381r. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Zheng Z, Yu L, Wu S, Sun L, Wu S, et al. Examination of the temporal and spatial dynamics of the gut microbiome in newborn piglets reveals distinct microbial communities in six intestinal segments. Sci Rep-UK. 2019. 10.1038/s41598-019-40235-z. [DOI] [PMC free article] [PubMed]
- 59.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen R, Scott KP, Khan S, Martin JC, Berry SH, Stevenson M, et al. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. Plos One. 2015;10:e133320. doi: 10.1371/journal.pone.0133320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagpal R, Tsuji H, Takahashi T, Kawashima K, Nagata S, Nomoto K, et al. Sensitive quantitative analysis of the meconium bacterial microbiota in healthy term infants born vaginally or by cesarean section. Front Microbiol. 2016. 10.3389/fmicb.2016.01997. [DOI] [PMC free article] [PubMed]
- 62.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237r–265r. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep-UK. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. Plos One. 2014;9:e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 66.Ardeshir A, Narayan NR, Mendez-Lagares G, Lu D, Rauch M, Huang Y, et al. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med. 2014;6:120r–252r. doi: 10.1126/scitranslmed.3008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–145. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, et al. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 2013;7:1112–1115. doi: 10.1038/ismej.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pacheco AR, Barile D, Underwood MA, Mills DA. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci. 2015;3:419–445. doi: 10.1146/annurev-animal-022114-111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chung Hachung, Pamp Sünje J., Hill Jonathan A., Surana Neeraj K., Edelman Sanna M., Troy Erin B., Reading Nicola C., Villablanca Eduardo J., Wang Sen, Mora Jorge R., Umesaki Yoshinori, Mathis Diane, Benoist Christophe, Relman David A., Kasper Dennis L. Gut Immune Maturation Depends on Colonization with a Host-Specific Microbiota. Cell. 2012;149(7):1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastro Hepat. 2018;15:197–205. doi: 10.1038/nrgastro.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sommer F, Nookaew I, Sommer N, Fogelstrand P, Bäckhed F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015. 10.1186/s13059-015-0614-4. [DOI] [PMC free article] [PubMed]
- 75.Lichtman JS, Alsentzer E, Jaffe M, Sprockett D, Masutani E, Ikwa E, et al. The effect of microbial colonization on the host proteome varies by gastrointestinal location. Isme J. 2016;10:1170–1181. doi: 10.1038/ismej.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Diaz-Ropero MP, Martin R, Sierra S, Lara-Villoslada F, Rodriguez JM, Xaus J, et al. Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J Appl Microbiol. 2007;102:337–343. doi: 10.1111/j.1365-2672.2006.03102.x. [DOI] [PubMed] [Google Scholar]
- 77.Chiu Y, Tsai J, Lin S, Chotirosvakin C, Lin M. Characterisation of bifidobacteria with immunomodulatory properties isolated from human breast milk. J Funct Foods. 2014;7:700–708. doi: 10.1016/j.jff.2013.12.015. [DOI] [Google Scholar]
- 78.Blustein J, Attina T, Liu M, Ryan AM, Cox LM, Blaser MJ, et al. Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. Int J Obes (Lond). 2013;37:900–906. doi: 10.1038/ijo.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Almqvist C, Cnattingius S, Lichtenstein P, Lundholm C. The impact of birth mode of delivery on childhood asthma and allergic diseases--a sibling study. Clin Exp Allergy. 2012;42:1369–1376. doi: 10.1111/j.1365-2222.2012.04021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Black M, Bhattacharya S, Philip S, Norman JE, McLernon DJ. Planned repeat cesarean section at term and adverse childhood health outcomes: a record-linkage study. Plos Med. 2016;13:e1001973. doi: 10.1371/journal.pmed.1001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin RM, Kramer MS, Patel R, Rifas-Shiman SL, Thompson J, Yang S, et al. Effects of promoting long-term, exclusive breastfeeding on adolescent adiposity, blood pressure, and growth trajectories: a secondary analysis of a randomized clinical trial. Jama Pediatr. 2017;171:e170698. doi: 10.1001/jamapediatrics.2017.0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Victora CG, Bahl R, Barros AJ, Franca GV, Horton S, Krasevec J, et al. Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet. 2016;387:475–490. doi: 10.1016/S0140-6736(15)01024-7. [DOI] [PubMed] [Google Scholar]
- 83.Looft T, Allen HK, Cantarel BL, Levine UY, Bayles DO, Alt DP, et al. Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. Isme J. 2014;8:1566–1576. doi: 10.1038/ismej.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Behrendt L, Larkum AW, Trampe E, Norman A, Sorensen SJ, Kuhl M. Microbial diversity of biofilm communities in microniches associated with the didemnid ascidian Lissoclinum patella. Isme J. 2012;6:1222–1237. doi: 10.1038/ismej.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. Plos One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. Isme J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu H, Hou C, Wang G, Jia H, Yu H, Zeng X, et al. Lactobacillus reuteri I5007 modulates intestinal host defense peptide expression in the model of IPEC-J2 cells and neonatal piglets. Nutrients. 2017;9:559. doi: 10.3390/nu9060559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 91.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grönlund MM, Gueimonde M, Laitinen K, Kociubinski G, Grönroos T, Salminen S, et al. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exper Allergy. 2007;37:1764–1772. doi: 10.1111/j.1365-2222.2007.02849.x. [DOI] [PubMed] [Google Scholar]
- 93.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 94.Flemer B, Lynch DB, Brown JMR, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2016:2015–309595. [DOI] [PMC free article] [PubMed]
- 95.Huang Y, Ma SF, Espindola MS, Vij R, Oldham JM, Huffnagle GB, et al. Microbes are associated with host innate immune response in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;196:208–219. doi: 10.1164/rccm.201607-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Sparse and compositionally robust inference of microbial ecological networks. Plos Comput Biol. 2015;11:e1004226. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Web and Social Nedia. 2014; https://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154.
- 98.Liu H, Zeng X, Zhang G, Hou C, Li N, Yu H, Shang L, Zhang X, Paolo T, Yang F, Liu Z, Qiao S, Biehl LM, Vogel W, Dörfel D, Hamprecht A, Seifert H, Autenrieth IB, Peter S. Maternal milk and fecal microbes guide the spatiotemporal development of mucosa-associated microbiota and barrier function in the neonatal gut. NCBI repository with study accession number PRJNA524979.
- 99.Liu H, Zeng X, Zhang G, Hou C, Li N, Yu H, Shang L, Zhang X, Paolo T, Yang F, Liu Z, Qiao S. Biehl LM, Vogel W, Dörfel D, Hamprecht A, Seifert H, Autenrieth IB, Peter S. Maternal milk and fecal microbes guide the spatiotemporal development of mucosa-associated microbiota and barrier function in the neonatal gut. NCBI repository with study accession number PRJNA480348.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Experimental design and workflow of sample collection. Each tick denotes a time-point of sample collection.
Additional file 2: Figure S2. Rarefaction curves based on the Chao1 and Shannon index at increasing sequencing depth of intestinal mucosal, maternal and environmental samples.
Additional file 3: Figure S3. (A) Negligible influence of breed on the mucosa-associated microbiota at birth. Average weighted UniFrac, unweighted UniFrac and Bray-Curtis distance between individuals at birth within and between Rongchang and Landrace piglets. (B) Shift of α diversity of mucosa-associated microbiota with age across four intestinal segments based on Chao1 index (Values are Means ± SE; significance between groups was determined by Kruskal-Wallis test).
Additional file 4: Figure S4. PCoA of phylogenetic community composition based on unweighted UniFrac distance (A), and taxonomic community composition based on Bray–Curtis (B).
Additional file 5: Figure S5. β-diversity of the mucosa-associated microbiota, milk, skin, vagina and feces of sows, and environmental microbiota. PCoA of phylogenetic community composition based on weighted UniFrac (A), and taxonomic community composition based on Bray–Curtis (B).
Additional file 6: Figure S6. Distance comparison of microbial communities between mucosa-associated intestinal microbiota with that of maternal milk and feces. Values are Means ± SE; significance between intestinal segments was determined by pairwise Kruskal-Wallis test; * P < 0.05; ** P < 0.01; *** P < 0.001.
Additional file 7: Figure S7. Distance comparison of microbial communities between true compared with random sow-piglet pairs for fecal, vaginal and environmental microbiota (Values are Means ± SE; significance between intestinal segments was determined by pairwise Kruskal-Wallis test; * P < 0.05; ** P < 0.01; *** P < 0.001).
Additional file 8: Table S1. Primers used in the study.
Additional file 9: Table S2. Shift of PICRUSt-predicted KEGG pathways of mucosa-associated microbiota in small intestine and large intestine (P values by Welch’s t-test with a Benjamini–Hochberg FDR correction were computed by STAMP).
Additional file 10: Table S3. Different PICRUSt-predicted KEGG pathways of mucosa-associated microbiota between two clusters, small intestinal cluster (including the large intestinal samples at birth) and large intestinal cluster (except the large intestinal samples at birth). P values by Welch’s t-test with a Benjamini–Hochberg FDR correction were computed by STAMP.
Additional file 11: Table S4. Candidate transmitted OTUs identified from mother and environment.
Additional file 12: Table S5. Relative expression of intestinal function-related genes using the whole statistical model.
Additional file 13: Table S6. Contrasts for MUC1, MUC2 and pBD1 expression analyzed within each intestinal segment.
Additional file 14: Table S7. Correlation of the relative abundance of OTUs and expression of genes involved in the development of intestinal barrier function.
Data Availability Statement
The data generated or analyzed during this study are included in this published article, its supplementary information files, and publicly available repositories. Raw 16S rRNA gene sequences and study metadata were deposited in the National Center for Biotechnology Information—NCBI repository (BioProject accession number: PRJNA524979 [98] and PRJNA480348 [99]).