Abstract
Personalized medicine has largely been enabled by the integration of genomic and other data with electronic health records (EHRs) in the U.S. and elsewhere. Increased EHR adoption across various clinical settings, and the establishment of EHR-linked population-based biobanks provide unprecedented opportunities for the types of translational and implementation research that drive personalized medicine. We review advances in the digitization of health information and the proliferation of genomic research in health systems, and provide insights into emerging paths for the widespread implementation of personalized medicine.
Introduction
The medical community has long recognized that inherent features of disease, and response to therapeutics, may often uniquely cluster in individuals, families, and population groups. Yet, for most of the history of practice of medicine, a broad approach to diagnosis and therapy has been adopted. The term personalized medicine was first given prominence in the late 1990s to early 2000s (Ginsburg and McCarthy, 2001; Jain, 2002), coincident with the sequencing of the human genome. Linking genomic and clinical profiles of individual patients held the promise to understand their disease at a deeper level to develop more targeted therapies (National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease, 2011). Today, the availability of vast amounts of digital data captured in Electronic Health Records (EHRs), combined with the emergence of genomic data in health systems, is opening new research avenues and opportunities for improving health management. Thus, the field of personalized medicine, and overlapping terms like genomic medicine, precision medicine and precision health, seek to use genomic approaches to tailor therapeutics, prevent disease, and promote health. This perspective covers the intersection between genomic research and EHRs, with an emphasis on emerging paths toward the widespread implementation of personalized medicine.
The Rise of Genomics in Medicine
In 2003, the completion of the first human genome sequence opened the door to personalizing the practice of medicine. It was expected that, over time, new ways of using genomic data for predicting and preventing disease, and more targeted and effective use of therapeutics, would emerge. Early personalized medicine efforts focused on genetic variants that predict drug response have resulted in the growth of genome-informed clinical practice guidelines (Caudle et al., 2014). The development of ‘next generation’ sequencing technology (Shendure and Ji, 2008), and its precipitous drop in price in the last decade, has enabled the use of genomic information to inform clinical decision-making not just for certain individuals, but across health systems. There are currently over 5,000 single gene disorders and traits with a known molecular etiology (https://www.omim.org/statistics/geneMap). Since 2009, targeted gene panel and exome sequencing (sequencing of some or all of the protein-coding regions of the genome) have been used for the diagnosis of these individually rare, but collectively common genetic conditions. Today, with over 55,000 commercially available clinical genetic tests (https://www.ncbi.nlm.nih.gov/gtr/) and new tests entering the market daily, genome sequencing is increasingly used in routine clinical care for diagnostic purposes. Other emerging uses of genomic information to inform clinical decision-making exist in areas like cancer detection and treatment (Deng and Nakamura, 2017), pre- and perinatal testing, (Hui and Bianchi, 2017; Peters et al., 2015), inpatient management of critically ill infants (Farnaes et al., 2018; Petrikin et al., 2015), and care of healthy and sick newborns (Holm et al., 2018) and adults (Vassy et al., 2014).
On the research side, early investments in population-scale efforts, like the International HapMap Project (International HapMap, 2003), were leveraged to create catalogs of human genetic variation shared across many individuals. These catalogs were used to design the first generation of microarrays that assayed hundreds of thousands of genetic variants in a single test for low cost. In 2007, the Wellcome Trust Case Control Consortium published one of the first landmark papers that paved the way for how these studies, known as Genome-Wide Association Studies (GWAS), should be performed (Wellcome Trust Case Control, 2007). This study, undertaken in a British population, examined two thousand individuals for each of seven major diseases, compared to three thousand healthy volunteers, to look for variants associated with each disease. The study demonstrated not only novel genomic discoveries, but also how methodology, data, and results should be broadly shared with the scientific community, greatly impacting the speed with which genomic discoveries could be made and disseminated worldwide. As of December 2018, the NHGRI-EBI GWAS catalog (Buniello et al., 2019) contains 3720 published studies for thousands of diseases, biomarkers, and drug responses (https://www.ebi.ac.uk/gwas). Furthermore, studies have grown to very large samples sizes in international research consortia with several recent studies topping 1 million participants (Evangelou et al., 2018; Liu et al., 2019; Nielsen et al., 2018), accelerating the rate of genomic discoveries.
With the increasing accessibility of genomic testing, and a greater understanding of both individual and population-scale genomic variation, efforts to more broadly integrate genomics in health systems for individualizing healthcare are gaining steam. Biobanks of human germline DNA samples are being used to generate genomic data linked to clinical information from EHRs in health systems. These biobanks serve as a rich resource for discovery, translation, and implementation of genomics in medicine. With dense, longitudinal clinical data, EHR-linked biobanks can empower the study of the natural history of disease. With accruing genomic data, they can also facilitate the implementation of individualized strategies for early detection, prevention, and management of disease. Nationwide biobanks are emerging in countries like the United Kingdom (Collins, 2012), Denmark (Agerbo et al., 2015), Estonia (Metspalu et al., 2004), China (Chen et al., 2011), Japan (Nagai et al., 2017) and others (Stark et al., 2019). In 2015, the U.S. launched its own national Precision Medicine Initiative, encompassing a large biobanking effort called the All of Us Research Program (AoURP). AoURP is actively recruiting over 1 million Americans invited to share their genomic, EHR, and other digital data, and be active partners in medical research (Collins and Varmus, 2015).
Digitizing Health Information in Electronic Health Records
To appreciate how EHRs have played a role in enabling personalized medicine, it is important to understand what EHRs contain, and how they can be used for genomic discovery and personalized medicine implementation. EHRs are real-time, patient-centered, digital records of health information and clinical care, generated and maintained by healthcare providers. They are designed to systematically collect patient information and share it across healthcare providers and settings, to help deliver more comprehensive and accurate clinical care. Substantial investment and increased financial incentives to implement EHRs over the last decade have resulted in widespread EHR adoption in the U.S. and other high-income countries (Adler-Milstein and Jha, 2017; Blumenthal and Tavenner, 2010), as well as increasing EHR adoption in low- and middle-income countries (Williams and Boren, 2008). EHRs are widely believed to improve healthcare quality, with benefits including secure long-term storage, improved consistency and standardization, and point-of-care accessibility of patient information. In the U.S., EHR adoption accelerated due to the Meaningful Use program, which was introduced as part of the 2009 Health Information Technology for Economic and Clinical Health Act to provide federal funds to healthcare providers successfully demonstrating meaningful use of EHRs (Centers for Medicare & Medicaid Services, 2010, 2012). As of 2017, over 95% of U.S. hospitals had certified EHR technology, with the lowest rates (93%) occurring in small rural and critical access hospitals, and the highest rates (99%) occurring in large hospitals with over 300 beds (Office of the National Coordinator for Health Information Technology, September 2018). With hundreds of EHR vendors available (Office of the National Coordinator for Health Information Technology, July 2017), EHR systems vary widely across hospitals and other healthcare settings. So, although the majority of hospitals in the U.S. have moved past the implementation stage of EHRs, there remains a need for improved accessibility, standardization, and interoperability between EHRs in different health systems.
In general, EHRs contain a wealth of longitudinal, real-world patient information collected in standard clinical care (Pendergrass and Crawford, 2019; Sutherland et al., 2016). This information includes demographics, medical and surgical history, allergies and medications, diagnoses and procedures, details from patient encounters, and results and reports from various clinical studies (Table 1). EHRs also track other aspects of patient care, including practice management functions such as scheduling, billing, and insurance information. The information recorded in EHRs is a combination of structured and unstructured data. Structured data use a uniform format (within each EHR system) and may also use a controlled vocabulary, constraining users to entering or choosing pre-determined values. Unstructured data do not follow a particular format and allow users to enter free text without constraints. This element allows healthcare providers to include details and context around health information and clinical encounters. Therefore, the same clinical information can be recorded in EHRs in myriad ways depending on the user. The structure of EHR data can significantly impact data usability for research purposes. Structured data are consistent and readily extractable, while unstructured data can require additional tools, such as natural language processing (NLP), to standardize, codify, and extract (Pendergrass and Crawford, 2019). Ultimately, both structured and unstructured data are important in providing a complete story around patients’ clinical data, offering multidimensional insight into health and disease, provider and patient behavior, and healthcare outcomes across populations and health systems.
Table 1.
Structure, categories, and examples of patient-level data available in EHRs
| Structure and Availability of EHR Data |
Categories of Data* | Examples of Data and Data Standards |
|---|---|---|
| Structured | Demographics | Age, gender, race/ethnicity, contact information |
| Vital Signs | Height, weight, heart rate, blood pressure, temperature | |
| Allergies | Environmental and drug allergies, adverse drug reactions | |
| Immunization status | Vaccinations and dates obtained | |
| Prescriptions and medications | Past and current medications, dosage, and frequency; RxNorm codes | |
| Laboratory results | Longitudinal laboratory measures; LOINC codes | |
| Provider order entries | Laboratory tests, imaging, and other studies ordered, referrals to specialists | |
| Diagnosis codes | International Classification of Diseases (ICD) 9 and 10 codes | |
| Procedure codes | Current Procedural Terminology (CPT) codes | |
| Semi-structured or mixed (e.g. unstructured text organized into pre-defined sections) | Problem list | Up-to-date list of important health problems, including diagnoses, symptoms, physical findings, clinical test findings |
| Personal history | Past medical, surgical, obstetric, developmental, social histories | |
| Family history | Medical history of family members | |
| Clinical test reports | Interpretations of laboratory, radiology, pathology, and other tests | |
| Unstructured | Clinical notes | Progress note, consultation, hospital admission note, discharge summary, etc. Free-text narratives of clinical encounters, including provider thought and decision-making processes |
| Other | Imaging data | Radiology: X-rays, ultrasounds, computerized tomography (CT) scans, magnetic resonance imaging (MRI) scans |
| Endoscopy | ||
| Echocardiography and electrocardiography (ECG) | ||
| Scanned documents | Genomic test results; pedigrees; medical records and test results from external sources | |
| Inconsistent/missing from EHRs | Family history | Medical history of family members, including age of onset and severity; Pedigree |
| Medication compliance | Prescriptions filled and taken at correct dose and frequency | |
| Exposures | Tobacco, pollution, radiation, hazardous chemicals (e.g. pesticides) | |
| Lifestyle and behavior | Diet, exercise, consumption of alcohol, tobacco products, recreational drugs, non-prescribed medications, supplements, natural/herbal remedies |
This is a non-exhaustive list of common EHR data categories. Different data structures may exist within categories, and vary between EHRs.
Abbreviations: LOINC, Logical Observation Identifiers Names and Codes
Despite the wealth of data contained in EHRs, barriers persist for integrating genomic and clinical data in health systems to empower the implementation of personalized medicine. One challenge is that genomic test results are typically reported in EHRs as a summary and interpretation of relevant findings (rather than raw data), frequently in the form of scanned paper reports (Shirts et al., 2015), limiting how genomic data is represented and accessed. New standards are emerging to digitize clinical genomic test results in EHRs (see Alterovitz et al. (2015) for a recent primer), requiring the development of EHR infrastructure to interact with clinical laboratories, patients, and providers. Several pilot EHR-based programs embed genomic test results related to drug response as structured data, combined with CDS, to provide point-of-care guidance for providers (Hoffman et al., 2014; Obeng et al., 2016; Weitzel et al., 2014). Some efforts are focused on providing patient-facing genomic test reports (Williams et al., 2018b). Another challenge is that some data elements important for interpreting genomic information are often missing from EHRs, for example, information on patient lifestyle and behavior (e.g. diet, exercise, and environmental exposures), and medication compliance (Table 1). Family health history, which plays a critical role in personalized medicine as an indicator of genetic susceptibility to disease (Guttmacher et al., 2004), can be inconsistent, sparse, or inaccurate as the acquisition of accurate and detailed family history in clinical practice is a timeconsuming and cumbersome process. Innovative patient-facing tools have been developed to improve the collection of family history information in EHRs, such as the web-based U.S. Surgeon General’s My Family Health Portrait (https://familyhistorv.hhs.gov) and others (Li et al., 2019; Orlando et al., 2013), but these have yet to be implemented widely across health systems. Increasingly, personal health records, containing health information generated (e.g. via mobile health devices) and maintained by patients, are being used to supplement data in EHRs (Roehrs et al., 2017). Over time, with improved integration of genomic and other data into EHRs, a fuller picture of personal health may emerge.
In addition to storing digitized health data, EHRs have the ability to support other care-related activities, such as clinical decision support (CDS), either directly or indirectly by interfacing with other health information technology systems. CDS is considered a key functionality of health information technology that builds upon EHRs to provide individuals involved in clinical care processes with pertinent knowledge at appropriate times, in order to improve care quality, patient safety, and health outcomes (see https://www.healthit.gov/topic/safety/clinical-decision-support). Examples of CDS interventions include computerized alerts and reminders, condition-specific order sets, and clinical guidelines, which are intended to enhance (but not replace) clinical decision-making and reduce errors. CDS is increasingly used to support the integration of genomic information into clinical care, for example, drug related CDS systems can be extended to include drug-gene interaction information (Dolin et al., 2018). Newer Artificial Intelligence (AI)-based services, such as those introduced by IBM, Google, Amazon, and other companies looking to invest in healthcare, may provide more sophisticated forms of CDS by gathering and analyzing massive amounts of data across health systems to make personalized, data-driven predictions at the point-of-care. The burgeoning integration of AI into clinical care is a promising research area; however, translating technical success in AI-driven analytics into meaningful clinical impact remains challenging (The Lancet, 2018).
As EHRs are adapting to empower personalized medicine implementation, they are also enabling new paradigms of genomic discovery and biomedical research embedded in health systems. However, EHR data are collected for clinical and billing purposes, not for research, which poses additional challenges to this secondary use. Challenges to using EHRs for research include data inaccuracy, missingness, and bias (Hersh et al., 2013; Sutherland et al., 2016). For example, the absence of a laboratory test in the EHR should not be considered the same as a negative result. Patient factors and provider preferences may influence EHR data elements, contributing to this bias. To address some of these challenges, the field of clinical informatics has evolved to generate tools and techniques for the secondary use of EHR data (Embi and Payne, 2009; Greenes and Shortliffe, 1990), and increasingly sophisticated strategies to effectively leverage EHRs for personalized medicine research are emerging (Pendergrass and Crawford, 2019). For example, by applying NLP approaches, researchers have been able to detect hepatic decompensation from findings described in unstructured text contained in radiology reports (Garla et al., 2011). In addition, various elements of EHRs can be mined creatively, for example using patients’ emergency contact information to infer pedigrees and estimate disease heritability (Polubriaginof et al., 2018). Finally, applications of a form of AI called deep learning focusing on types of medical images typically stored in EHRs, have shown improvement in the analysis of skin cancer images (Esteva et al., 2017) and diagnosis of diabetic retinopathy (Gulshan et al., 2016). As long as the limitations of EHRs are understood, opportunities for using real-world patient data to drive translational genomic research and personalized medicine are immense.
The Intersection of Genomic Research and EHRs
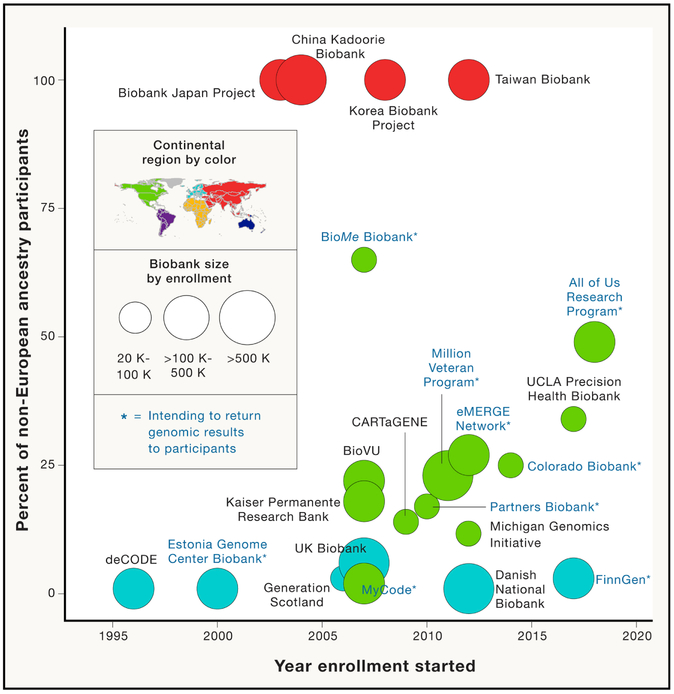
The proliferation of EHR-linked biobanks for personalized medicine research and implementation has been facilitated by the close collaboration between government funders, health systems, and industry partners. Some of the first population-scale biobanks are deCODE genetics in Iceland, which is currently a subsidiary of Amgen, and the Estonian Genome Center at the University of Tartu’s biobank, with many new biobanks emerging worldwide over the last twenty years (Figure 1 and Supplementary Table 1). In the U.S., the National Human Genome Research Institute (NHGRI) set forth a 20-year plan leveraging genomics and EHRs to chart a course for bringing genomics into clinical care (Green et al., 2011). One NHGRI-established program is the Electronic Medical Records and Genomics (eMERGE) Network, launched in 2007, which links biobanks to EHRs at multiple sites to perform genomic research embedded in health systems and establish best practices that replicate across health systems (Gottesman et al., 2013; McCarty et al., 2011). Some biobanks in large academic centers or national health systems partner with industry, for example the collaboration of the Geisinger MyCode Biobank (Dewey et al., 2016b), Mount Sinai BioMe Biobank (https://www.mountsinai.org/about/newsroom/2016/mount-sinai-health-system-launches-collaboration-with-the-regeneron-genetics-center), and UK Biobank (http://www.ukbiobank.ac.uk/2018/01/regeneron-announces-major-collaboration-to-exome-sequence-uk-biobank-genetic-data-more-quickly/) with the Regeneron Genetic Center (RGC; Regeneron Pharmaceuticals Inc) to support exome sequencing of their respective biorepositories, and Vanderbilt University’s BioVU Biobank collaboration with Google to develop infrastructure for data collection and research. In addition, large academic health system or governments often make significant institutional investments, even when the direct downstream economic benefits are not always clear, recognizing the potential for personalized medicine to make real and lasting benefits for health and healthcare.
Figure 1:
Selected population-based biobanks with current or planned genomic data linked to EHRs and ≥ 20,000 enrolled participants. The x-axis indicates the year in which enrollment started (information obtained from personal communication, press releases, recent publications or biobank websites; Supplementary Table 1) and the y-axis indicates the proportion non-European ancestry participants in each biobank. The size of the solid circles indicates sample size of enrolled participants in bins of 20K-100K, >100K-500K, and >500K. Color of the solid circles indicates region of the world in which the biobank is enrolling (per the colored map of the world). Biobanks are labeled by name and those indicated with an asterisk currently have or plan programs to return genomic results to participants.
The models in which health system data may be used to support personalized medicine research are varied, and there are special ethical and scientific (Hersh et al., 2013; Sutherland et al., 2016; Wolford et al., 2018) considerations to take into account when linking genomic data with EHRs. For example, participants in biobanks typically volunteer under broad informed consents, as EHRs are expected to add data types over time, and all possible uses of participants’ data are often unknown at the time of joining. Furthermore, protected health information can be challenging to strip completely from EHRs when synthesizing data for research purposes, particularly from free text and images, requiring special regulation models that may restrict some access (Boyd et al., 2007). Increasingly, biobanks are being built with recontact in mind for future research or for the return of genomic results to participants (Schwartz et al., 2018), and some newer biobanks operate under a model of continuous engagement and partnership with participants (Collins and Varmus, 2015). A cornerstone of genomic research using EHRs is the ability to replicate findings across different health systems, populations, and clinical contexts. To facilitate this, some biobanks have an open sharing model, such as the UK Biobank, who have released genomic and clinical data for nearly 500,000 participants freely to the research community via an application process (https://www.ukbiobank.ac.uk/). Others, require specific collaborations for access, but may make analytical results freely available, such as the Michigan Genomics Initiative PheWeb server (http://pheweb.sph.umich.edu/) and the Stanford Global Biobank Engine (https://biobankengine.stanford.edu/).
Mining EHRs for clinical research has created both rich opportunities and several challenges for the genetics community. In typical cohort-based GWAS, one or a few related diseases or traits (termed phenotypes) are studied at a time. However, access to longitudinal clinical data from large numbers of patients via EHRs means that all variants can be tested against thousands of phenotypes at myriad time points at once. This all-against-all strategy is a powerful means for genomic discovery in both hypothesis-driven and hypothesis-generating ways. It can also help to identify variants that have pleiotropic effects, for example a recent study in the Biobank Japan Project showed over 300 pleiotropic variants linked to 53 biomarkers (Kanai et al., 2018). Furthermore, Mendelian randomization studies using biobanks can be used to better understand the relationship between biomarkers and disease risk (Au Yeung et al., 2018). Alternatively, participants can be stratified by a single variant, a method called Phenome-Wide Association (PheWAS) (Denny et al., 2010), to gain a better understanding of clinical impact and comorbidities associated with that variant. Recent analysis has demonstrated how combining genomic and EHR data can enable partially automation of the diagnosis of genetic disease (Son et al., 2018). As mentioned, EHR data is amenable not only to cross-sectional, but also longitudinal approaches, for example to predict disease progression (Huopaniemi et al., 2014). Novel derived phenotypes can also be extracted from EHRs, such as traits related to brain morphology or function that can be derived from brain images, which have recently been shown to have a heritable component (Elliott et al., 2018). We can expect that EHR-linked biobanks will increasingly be used in genomics-driven drug discovery (Abul-Husn et al., 2018; Dewey et al., 2016a; Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators et al., 2016) and drug repurposing. However, as mentioned above, issues of data missingness, error, harmonization, and ascertainment bias in EHRs have the potential to confound analysis. Strategies combining population-based biobanks with specific disease cohorts will further improve our understanding of diseases impacted by ascertainment bias due to age (e.g. Alzheimer's disease and childhood cancers) or prevalence (e.g. Crohn's disease, ulcerative colitis, and Cystic Fibrosis (CF)). There are also several statistical challenges to overcome, including correctly calibrating models of association, and developing methods that can operate over large samples sizes and data complexity. Method development to handle the computational and statistical challenges in genome-linked EHRs is a very active area of research (Wolford et al., 2018).
There are several pitfalls when considering how findings from this type of research will translate to real-world clinic populations. One of the most obvious is that participants in EHR-linked biobanks are predominantly of European ancestry (Figure 1). Of the approximately 5 million participants enrolled in large population-based biobanks around the world in 2019, 68% are of European ancestry, 22% are East Asian, and 10% are of other non-European ancestry (Supplementary Table 1). This is not surprising, as a similar bias is seen in cohort-based GWAS research (Popejoy and Fullerton, 2016). This is a particularly pernicious problem as (1) differences in disease burden among ancestrally diverse populations are a major cause of health disparities (http://www.cdc.gov/minorityhealth/OMHHE.html) and (2) the vast majority of human genomic variation is expected to be population private (1000 Genomes Project Consortium et al., 2015). The latter is a challenge for interpreting genetic testing, even for very well characterized genetic diseases. For example, in the U.S., non-white newborns are more likely to get a false negative CF genetic diagnosis compared to white babies, mainly because we have less understanding of CF-causing variants in non-white populations (Pique et al., 2017). By focusing the majority of our genomic research efforts on a narrow slice of humanity, we miss the opportunity for discovery of disease-causing variants that have arisen to appreciable frequency in understudied populations. A recent study in a diverse biobank in New York City (the BioMe Biobank) demonstrated that the variant underlying a little-known recessive genetic disease, Steel Syndrome, segregates commonly in populations of Puerto Rican ancestry (Belbin et al., 2017). Studies like this empower a better understanding of the natural progression of diseases, which populations are at risk, and the development of genetic testing and screening. Overall, there is a general consensus that properly powered multi-factorial disease studies will require genetic analysis of individual-level genomic data from hundreds of thousands to millions of individuals across diverse populations. U.S.-based programs like the Million Veteran Program (MVP) (https://www.research.va.gov/mvp/), AoURP (https://allofus.nih.gov/), and Kaiser Permanente Research Bank (https://researchbank.kaiserpermanente.org/), will add to diversity, as too will emerging national personalized medicine programs in global regions like South America, Africa, the Middle East, and South East Asia (Manolio et al., 2015; Stark et al., 2019).
From Research to Clinical Care
Advances in genomic technology and research have led to rising expectations for genomics to widely impact clinical care and public health. However, it can take many years for genomic discoveries to directly benefit patients. The reasons for this include lack of infrastructure to integrate genomics into existing clinical workflows, insufficient evidence of clinical utility, and concerns about cost and reimbursement (Sperber et al., 2017). Additional healthcare provider-related barriers include an unwillingness to adopt new practices, and lack of genomic knowledge and training (Eden et al., 2016; Feero and Green, 2011; Rohrer Vitek et al., 2017). To address these challenges, implementation science, the study and application of appropriate methods to promote the integration of research findings into healthcare policy and practice, is increasingly being applied to genomics. The convergence of implementation science approaches with learning healthcare systems (i.e. healthcare systems that continuously self-study and improve through data capture and analytics embedded in daily practice) seeks to enable faster integration of genomics knowledge and personalized medicine into clinical practice (Chambers et al., 2016). In the U.S., the NHGRI Clinical Sequencing and Evidence-generating Research (CSER) and Implementing Genomics in Practice (IGNITE) Networks both focus on supporting strategies for implementing genomics in healthcare and understanding the impact on health systems (Green et al., 2016; Weitzel et al., 2016). Additionally, both programs have an emphasis on medically underserved populations, promoting the reach of personalized medicine to an increasing proportion of the U.S. population (Amendola et al., 2018). Through these and other programs, various genomic education strategies for healthcare providers have been applied to diverse clinical settings (Rohrer Vitek et al., 2017; Sperber et al., 2017). Efforts to implement genomic medicine in real-world clinical settings are essential to laying the foundation for a personalized medicine transformation of health care.
Pharmacogenomics (PGx), or the use of genomic information to predict drug response, has been one of the earliest success stories for EHR-driven personalized medicine research and implementation. Genetic variants can either alter a drug’s pharmacodynamics, including absorption, distribution, metabolism or elimination, or its pharmacokinetics, for example by modifying a drug target (Relling and Evans, 2015). In the clinic, PGx can help identify drug responders and non-responders, avoid adverse events, and optimize drug dose. For example, interindividual variability in response to clopidogrel, a commonly used antiplatelet drug, is explained in part by the cytochrome P450 enzyme CYP2C19, in which loss-of-function variation increases risk of adverse clinical outcomes (Mega et al., 2010). This has prompted practice guidelines for CYP2C19 genome-directed antiplatelet therapy (Scott et al., 2013). Global initiatives, like the U.S.-based Pharmacogenomic Research Network (https://www.pgrn.org/), have helped spur PGx discoveries that are starting to be used to personalize the selection and dosing of medications in individual patients. Today, although less than 10% of published GWAS have focused on PGx (Giacomini et al., 2017), over 200 drugs have PGx information included in their Food and Drug Administration (FDA)-approved labels, or “black box warnings” that encourage genetic testing where possible, and may include specific actions to be taken based on an individual’s genetic variants (https://www.fda.gov/drugs/scienceresearch/ucm572698.htm). In addition, there are a growing number of peer-reviewed, evidence-based clinical practice guidelines for PGx implementation, such as those published by the Clinical Pharmacogenetics Implementation Consortium (Caudle et al., 2014). Therefore, the promise of PGx for broadly implementing personlized medicine is great, especially as over 90% of individuals are estimated to be impacted by PGx variation (Dunnenberger et al., 2015; Van Driest et al., 2014).
To be used effectively in clinical care, PGx information requires rapid return of results, ideally at the point-of-care. A number of PGx implementation programs have explored models to disseminate PGx-aided clinical decision-making by coupling PGx testing with CDS through EHRs (Herr et al., 2015). This approach requires preemptive clinical PGx testing of prospective or biobank patients, depositing genomic information into EHRs, and alerting prescribers at the point-of-care through CDS when a drug is ordered for a patient with an at-risk genetic variant (Abul-Husn et al., 2014). The use of CDS in this context enables the delivery of evidence-based, genome-guided prescribing recommendations through the EHR with minimal disruption to clinical workflows. Although several pilot programs to implement PGx via EHRs are underway at large medical centers (Dunnenberger et al., 2015), many barriers exist for scaling this model across health systems including effectively integrating PGx data with EHRs, improving PGx knowledge in healthcare providers, and avoiding the overuse of CDS that can lead to burnout (Hicks et al., 2016; Sperber et al., 2017). The lessons from PGx implementation are a harbinger for challenges that have been or will be faced by other programs seeking to integrate genomics broadly in health systems, particularly in arenas where providers lack genomic expertise and support infrastructure (Manolio et al., 2015; Sperber et al., 2017).
Genomic Screening for Preventive Health
Today, there are growing arguments for the use of genomic screening in preventive health, even though there remain some concerns about its widespread implementation in routine clinical care (Evans et al., 2017; Murray, 2018). For the most part, the first indication that an individual harbors a disease-associated variant happens at the time they or a family member exhibit signs or symptoms of the disease. In diseases for which preventive measures are available, typically termed ‘medically actionable’, genomic screening of asymptomatic individuals could have significant public health impact (Berg et al., 2011). For example, an estimated 1 to 2 million people in the U.S. are at increased risk of cancer due to Hereditary Breast and Ovarian Cancer (HBOC) or Lynch syndrome, and most do not know it. Both syndromes have evidence-based guidelines to reduce or prevent cancer risk, and have been prioritized by The Centers for Disease Control and Prevention (CDC) as Tier 1 genomic applications, i.e. having the highest level of evidence to support their implementation (Green et al., 2019; Khoury et al., 2018). In general, a medically actionable genomic result is defined as one having an established association with a disease that has important health implications and for which proven medical interventions or therapies exist to reduce morbidity and mortality (Green et al., 2013; Kalia et al., 2017). Current estimates predict 3-5% of individuals harbor a medically actionable variant (Dewey et al., 2016b), and programs that combine individual screening with cascade testing of family members are poised to capture many more affected individuals (Knowles et al., 2017). Therefore, several research endeavors are using EHR-linked biobanks to explore the clinical utility and demonstrate effective implementation models of genomic screening for preventive health and personalized medicine (Williams et al., 2018a).
In 2007, the Geisinger Health System (GHS), an integrated health system in Pennsylvania and an early adopter of EHRs, launched an EHR-linked biobank, the MyCode Community Health Initiative (Carey et al., 2016), which now has over 200,000 unselected participants enrolled and broadly consented for discovery research. The DiscovEHR collaboration between GHS and the RGC was established in 2014 to combine exome sequence data with de-identified, longitudinal EHR data from MyCode participants. These data are being used to fuel genomic research and genome-guided drug discovery efforts (Abul-Husn et al., 2018; Dewey et al., 2016b), as well as the GHS GenomeFIRST Medicine program, which returns genomic results to MyCode participants (Schwartz et al., 2018). The first 50,000 individuals sequenced through the DiscovEHR collaboration have been screened for genomic variants associated with HBOC (Manickam et al., 2018) and Familial Hypercholesterolemia (FH, another CDC Tier 1 genomic condition) (Abul-Husn et al., 2016), among other conditions. Investigators examined the prevalence of expected pathogenic variants – i.e. known pathogenic variants as per the clinical genetics database ClinVar (Landrum et al., 2014), and predicted loss-of-function variants – in genes known to be associated with FH or HBOC. In each case, the number of individuals harboring expected pathogenic variants was larger than expected, demonstrating that deploying genomic screening across unselected populations is likely to uncover a higher prevalence of genomic conditions than has been previously estimated. Furthermore, these studies showed that FH and HBOC are often underdiagnosed. Compared with previous clinical care, unselected genomic screening identified 5 times more individuals with genomic risk for HBOC (Manickam et al., 2018), half of whom would not have met current screening guidelines (Daly et al., 2017) for referral to genetic counseling and genetic testing. Similarly, less than a quarter of individuals with FH-associated variants would have met pre-sequencing criteria for a clinical diagnosis of FH based on their EHR data (Abul-Husn et al., 2016), which is consistent with current estimates that fewer than 10% of FH cases in the U.S are identified (Knowles et al., 2017). Overall, these data suggest that genomic conditions are likely to be underdiagnosed, and that unselected genomic screening would be beneficial in accurately ascertaining individuals with conditions putting them at higher risk of cardiovascular and cancer-related morbidity and mortality.
To date, there are not clear guidelines or procedures in the U.S. on reporting medically actionable genomic results to research participants, and biobank policies vary on how they address the return of genomic results (Jarvik et al., 2014; Wolf et al., 2012). In the GenomeFIRST Medicine Program, exome sequence data are analyzed for variants in 80 genes for return of results to MyCode participants (Schwartz et al., 2018). These include 59 genes that the American College of Medical Genetics (ACMG) has recommended for return to patients having undergone clinical exome or genome sequencing, even when unrelated to the primary indication for testing (Kalia et al., 2017). Genomic results are clinically confirmed and then deposited into the EHR. By returning genomic results to MyCode participants and following their outcomes, investigators are gaining knowledge about the clinical utility of performing population genomic screening for preventive health (Buchanan et al., 2018). The opportunity for a single test to screen for many diseases at once poses challenges for evaluating its appropriate clinical utility and cost effectiveness. Therefore, a major impact of such research would be to generate sufficient evidence for payers to offer reimbursement of genomic screening, and turn this into a scalable and sustainable model. In the meantime, programs like GenomeFIRST Medicine rely on support from multiple internal and external sources, including industry partnerships, grants, institutional funding, and philanthropy.
Other health systems are quickly following suite to explore population genomic screening initiatives and their impact on clinical care. Color, a consumer-facing genomic testing company launched in 2015, has partnered with a number of U.S. health systems to provide population screening for genes associated with hereditary cancer risk, cardiovascular disease risk, and drug response, as part of the Color Population Health program (https://www.color.com/health-systems). The Alabama Genomic Health Initiative, a collaboration between the University of Alabama at Birmingham and the HudsonAlpha Institute for Biotechnology, is funded by the state of Alabama to offer genomic screening to an unselected population cohort of adults. Using the Illumina Global Screening Array, it is returning results for the ACMG 59 genes to its participants (https://www.uabmedicine.org/aghi). The Yale Center for Genomic Health recently announced their genomic medicine project, partially funded by the state of Connecticut, that intends to recruit 100,000 participants into a research biobank that will offer clinical exome sequencing and the return of actionable genomic results (https://www.genomeweb.com/clinical-sequencing/yale-launch-large-scale-genomic-medicine-project). Other existing EHR-linked biobanks are piloting programs to return results to participants, including the BioMe Biobank and MVP (Supplementary Table 1). Outside the U.S., countries like Estonia, Denmark, Japan, and Qatar are undertaking population-based genomic screening programs offering return of results to participants (Stark et al., 2019). As global investments in the billions of dollars are being made to integrate genomics into healthcare, we should expect that genomic applications of personalized medicine, including PGx and preventive health, will scale broadly in many health systems.
The Future of Personalized Medicine
We are in the midst of an acceleration of personalized medicine, driven by a dramatic drop in price of genome sequencing, and an explosion in the number of companies offering genetic tests. The global genetic testing market is expected to surpass $22 billion by 2024 (https://www.gminsights.com/pressrelease/genetic-testing-market), and direct-to-consumer companies increasingly place genomics into the hands of consumers (Khan and Mittelman, 2018), changing the way patients and providers approach genetic testing. Several more advancements in genomic technology are expected to impact the landscape of genetic testing in the near future. Although the majority of current clinical sequencing uses panels or exomes, there are an increasing number of pilot programs using whole genome sequencing (WGS), which has the potential to capture all classes of genetic variation in one analysis. Genomics England, in partnership with National Health Service (NHS) England, has successfully implemented WGS at scale in direct healthcare, having completed sequencing of 100,000 patients, and their families, with rare diseases or cancer (Turnbull et al., 2018). In the U.S., rapid WGS of acutely ill inpatient infants has been shown to reduce morbidity and cost of hospitalization (Farnaes et al., 2018). However, there is still much to learn about the clinical utility of WGS in clinical settings, and challenges to address, including cost, clinical interpretation, and data storage in health systems (Manolio, 2017). At the time of this writing, a number of long-read sequencing technologies are reducing in cost (i.e. PacBio, 10X Genomics), with the promise of improved understanding of the order and structure of genomic rearrangement, which are hard to capture with next generation technology. If these technologies are successful in entering the clinical domain more broadly, they could increase the diagnostic yield for genetic diseases. Other technical hurdles will be overcome as the scientific community moves toward pan-ethnic reference genomes, large databases of publicly available population-scale variants (Lek et al., 2016), and deepening clinical genetic databases (Landrum et al., 2014; Rehm et al., 2015). However, even as genomic testing improves in accuracy and validity, we expect that we will continue to face challenges in genomic test interpretation, understanding, and communication.
We can also expect genomics might enter new arenas in medicine, for example for predicting lifetime risk for complex diseases. Because complex diseases often involve multiple underlying genetic and environmental factors, genomic prediction for these diseases is more complicated and thought to be less useful clinically (Manolio et al., 2009). However, thanks to breakthroughs from very large GWAS, ‘polygenic risk scores’ (PRS), which aggregate large numbers of genetic variants in a single test for prediction, are gaining accuracy. PRS have been recently shown to predict some complex diseases, such as breast cancer and coronary artery disease, reaching similar accuracy as tests for rare pathogenic variants for those diseases (Khera et al., 2018; Shieh et al., 2016). As GWAS continue to progress, accuracy of these scores are anticipated to increase (Witte et al., 2014). PRS can be performed across multiple diseases using low cost technology, and can provide information in addition to lifestyle and family history, to identify high-risk individuals in the general population. However, many challenges exist for translating these findings into clinical care, particularly as it is not clear how well PRS will work in real-world clinical settings. Several clinical trials are underway to evaluate how PRS affect management for some complex diseases like breast cancer (https://clinicaltrials.gov/ct2/show/NCT03688204). There is however accruing evidence to suggest that PRS may not transfer well across populations (Martin et al., 2017). Early applications of PRS show greater accuracy of prediction in European populations compared to other populations (Martin et al., 2019). This skew has the potential to exacerbate current health disparities. To move personalized medicine forward ethically, predictive analytics for complex diseases must be developed in a way that proffers equitable clinical utility for all populations.
Personalized medicine implementation will increasingly rely on EHRs to store vast amounts of genomic data and appropriately integrate relevant genomic information into clinical care. However, many developing countries lack the robust healthcare and information technology infrastructure to broadly implement EHRs. Even within the U.S., healthcare settings that have not yet adopted EHRs, such as small and rural hospitals, represent patient populations with the least access to genomics and personalized medicine. This disparity poses a challenge to the broad representation of diverse populations in genomic research and personalized medicine implementation. AoURP is making a concerted effort to include diverse populations to advance health for all, through outreach to underserved communities via mobile education and enrollment centers. The Genomic Medicine Alliance is a global academic research network that encourages collaboration and harmonization of genomic research activities between developed and developing countries (Cooper et al., 2014). For personalized medicine to globally impact clinical care, it is becoming more apparent that international, collaborative efforts are needed to supplement the current one-off, local, implementation efforts. The Global Alliance for Genomics and Health (GA4GH), which aims to promote and facilitate data sharing in genomic research (Hayden, 2013), and the Global Genomic Medicine Collaborative (G2MC), which aims to develop and disseminate best practices for global genomic medicine implementation (Manolio et al., 2015), have established the International 100K Cohort Consortium (IHCC) in 2018 to coalesce genomic data from large unselected population cohorts from around the world (https://ihcc.g2mc.org/). These efforts promote universally applicable frameworks to understand how complex genomics information can successfully be implemented in health systems.
Fundamental discoveries elucidating the genomic factors underlying disease over the past decades have yielded powerful engines of knowledge, and an enormous potential to benefit patients. Substantial investments in the coming decade to integrate this complex information into routine clinical care are likely to transform the field of medicine as we know it today. Given that the delivery of healthcare is increasingly dependent on EHRs, we are likely to see an evolution in how patients and providers engage with healthcare data. Emerging paradigms for leveraging genomic information, combined with other biological data, AI, and robotics, in health systems will empower the next era of personalized medicine.
Supplementary Material
Details of 22 global, population-based, EHR-linked biobanks as of January 2019.
Acknowledgments
We would like to thank the following people for helping to assemble information about global biobanks; Tim Assimes, Kathleen Barnes, Natalie Boutin, Archie Campbell, Chien-Hsiung Chen, Nancy Cox, Mark Daly, Eleazar Eskin, Tonu Esko, Dan Geschwind, Chris Gignoux, Sebastien Jacquemont, Anu Jalanko, Krzysztof Kiryluk, Bartha Knoppers, Mitja Kurki, Pui-Yan Kwok, Jee Young Kwon, Clara Lagonchere, Charles Lee, Te-Chang Lee, Yi-Ling Lin, Alicia Martin, Teri Manolio, Alexandra Obadia, Chris O’Donnell, Brad Ozenberg, Aarno Palotie, Cuiping Pan, Broet Philippe, Robb Rowley, Sue Slaugenhaupt, Brian Smith, and Philip Tsao. We would also like to thank Omri Gottesman for helpful comments and Dean Bobo for help constructing Figure 1. Finally, we would like to thank all those who volunteer to participate in biobanks and genomic research. The Center for Genomic Health is supported by funds from the Icahn School of Medicine at Mount Sinai. EEK is supported by grants from NHGRI, NHLBI, and NIDDK; U01HG009080, R01DK110113, U01HG009610, and R01HL104608.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, Liu Y, Kozlitina J, Stender S, Wood GC, et al. (2018). A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med 378, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abul-Husn NS, Manickam K, Jones LK, Wright EA, Hartzel DN, Gonzaga-Jauregui C, O'Dushlaine C, Leader JB, Lester Kirchner H, Lindbuchler DM, et al. (2016). Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science 354. [DOI] [PubMed] [Google Scholar]
- Abul-Husn NS, Owusu Obeng A, Sanderson SC, Gottesman O, and Scott SA (2014). Implementation and utilization of genetic testing in personalized medicine. Pharmgenomics Pers Med 7, 227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler-Milstein J, and Jha AK (2017). HITECH Act Drove Large Gains In Hospital Electronic Health Record Adoption. Health Aff (Millwood) 36, 1416–1422. [DOI] [PubMed] [Google Scholar]
- Agerbo E, Sullivan PF, Vilhjalmsson BJ, Pedersen CB, Mors O, Borglum AD, Hougaard DM, Hollegaard MV, Meier S, Mattheisen M et al. (2015). Polygenic Risk Score, Parental Socioeconomic Status, Family History of Psychiatric Disorders, and the Risk for Schizophrenia: A Danish Population-Based Study and Meta-analysis. JAMA Psychiatry 72, 635–641. [DOI] [PubMed] [Google Scholar]
- Alterovitz G, Warner J, Zhang P, Chen Y, Ullman-Cullere M, Kreda D, and Kohane IS (2015). SMART on FHIR Genomics: facilitating standardized clinico-genomic apps. J Am Med Inform Assoc 22, 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola LM, Berg JS, Horowitz CR, Angelo F, Bensen JT, Biesecker BB, Biesecker LG, Cooper GM, East K, Filipski K et al. (2018). The Clinical Sequencing Evidence-Generating Research Consortium: Integrating Genomic Sequencing in Diverse and Medically Underserved Populations. Am J Hum Genet 103, 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au Yeung SL, Luo S, and Schooling CM (2018). The Impact of Glycated Hemoglobin (HbA1c) on Cardiovascular Disease Risk: A Mendelian Randomization Study Using UK Biobank. Diabetes Care 41, 1991–1997. [DOI] [PubMed] [Google Scholar]
- Belbin GM, Odgis J, Sorokin EP, Yee MC, Kohli S, Glicksberg BS, Gignoux CR, Wojcik GL, Van Vleck T, Jeff JM, et al. (2017). Genetic identification of a common collagen disease in puerto ricans via identity-by-descent mapping in a health system. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Khoury MJ, and Evans JP (2011). Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med 13, 499–504. [DOI] [PubMed] [Google Scholar]
- Blumenthal D, and Tavenner M (2010). The "meaningful use" regulation for electronic health records. N Engl J Med 363, 501–504. [DOI] [PubMed] [Google Scholar]
- Boyd AD, Hosner C, Hunscher DA, Athey BD, Clauw DJ, and Green LA (2007). An 'Honest Broker' mechanism to maintain privacy for patient care and academic medical research. Int J Med Inform 76, 407–411. [DOI] [PubMed] [Google Scholar]
- Buchanan AH, Manickam K, Meyer MN, Wagner JK, Hallquist MLG, Williams JL, Rahm AK, Williams MS, Chen ZE, Shah CK, et al. (2018). Early cancer diagnoses through BRCA1/2 screening of unselected adult biobank participants. Genet Med 20, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E v et al. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47, D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey DJ, Fetterolf SN, Davis FD, Faucett WA, Kirchner HL, Mirshahi U, Murray MF, Smelser DT, Gerhard GS, and Ledbetter DH (2016). The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet Med 18, 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudle KE, Klein TE, Hoffman JM, Muller DJ, Whirl-Carrillo M, Gong L, McDonagh EM, Sangkuhl K, Thorn CF, Schwab M, et al. (2014). Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab 15, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Medicare & Medicaid Services, H.H.S. (2010). Medicare and Medicaid programs; electronic health record incentive program. Final rule. Fed Regist 75, 44313–44588. [PubMed] [Google Scholar]
- Centers for Medicare & Medicaid Services, H.H.S. (2012). Medicare and Medicaid programs; electronic health record incentive program--stage 2. Final rule. Fed Regist 77, 53967–54162. [PubMed] [Google Scholar]
- Chambers DA, Feero WG, and Khoury MJ (2016). Convergence of Implementation Science, Precision Medicine, and the Learning Health Care System: A New Model for Biomedical Research. JAMA 315, 1941–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Chen J, Collins R, Guo Y, Peto R, Wu F, Li L, and China Kadoorie Biobank collaborative, g. (2011). China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol 40, 1652–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, and Varmus H (2015). A new initiative on precision medicine. N Engl J Med 372, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R (2012). What makes UK Biobank special? Lancet 379, 1173–1174. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Brand A, Dolzan V, Fortina P, Innocenti F, Michael Lee MT, Macek M Jr., Al-Mulla F, Prainsack B, Squassina A, et al. (2014). Bridging genomics research between developed and developing countries: the Genomic Medicine Alliance. Per Med 11, 615–623. [DOI] [PubMed] [Google Scholar]
- Daly MB, Pilarski R, Berry M, Buys SS, Farmer M, Friedman S, Garber JE, Kauff ND, Khan S, Klein C, et al. (2017). NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 2.2017. J Natl Compr Canc Netw 15, 9–20. [DOI] [PubMed] [Google Scholar]
- Deng X, and Nakamura Y (2017). Cancer Precision Medicine: From Cancer Screening to Drug Selection and Personalized Immunotherapy. Trends Pharmacol Sci 38, 15–24. [DOI] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, and Crawford DC (2010). PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26, 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey FE, Gusarova V, O'Dushlaine C, Gottesman O, Trejos J, Hunt C, Van Hout CV, Habegger L, Buckler D, Lai KM, et al. (2016a). Inactivating Variants in ANGPTL4 and Risk of Coronary Artery Disease. N Engl J Med 374, 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey FE, Murray MF, Overton JD, Habegger L, Leader JB, Fetterolf SN, O'Dushlaine C, Van Hout CV, Staples J, Gonzaga-Jauregui C, et al. (2016b). Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 354. [DOI] [PubMed] [Google Scholar]
- Dolin RH, Boxwala A, and Shalaby J (2018). A Pharmacogenomics Clinical Decision Support Service Based on FHIR and CDS Hooks. Methods Inf Med 57, e115–e123. [DOI] [PubMed] [Google Scholar]
- Dunnenberger HM, Crews KR, Hoffman JM, Caudle KE, Broeckel U, Howard SC, Hunkler RJ, Klein TE, Evans WE, and Relling MV (2015). Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol 55, 89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden C, Johnson KW, Gottesman O, Bottinger EP, and Abul-Husn NS (2016). Medical student preparedness for an era of personalized medicine: findings from one US medical school. Per Med 13, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, Marchini J, and Smith SM (2018). Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embi PJ, and Payne PR (2009). Clinical research informatics: challenges, opportunities and definition for an emerging domain. J Am Med Inform Assoc 16, 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, and Thrun S (2017). Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, et al. (2018). Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet 50, 1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Powell BC, and Berg JS (2017). Finding the Rare Pathogenic Variants in a Human Genome. JAMA 317, 1904–1905. [DOI] [PubMed] [Google Scholar]
- Farnaes L, Hildreth A, Sweeney NM, Clark MM, Chowdhury S, Nahas S, Cakici JA, Benson W, Kaplan RH, Kronick R, et al. (2018). Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom Med 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feero WG, and Green ED (2011). Genomics education for health care professionals in the 21st century. JAMA 306, 989–990. [DOI] [PubMed] [Google Scholar]
- Garla V, Lo Re V 3rd, Dorey-Stein Z, Kidwai F, Scotch M, Womack J, Justice A, and Brandt C (2011). The Yale cTAKES extensions for document classification: architecture and application. J Am Med Inform Assoc 18, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini KM, Yee SW, Mushiroda T, Weinshilboum RM, Ratain MJ, and Kubo M (2017). Genome-wide association studies of drug response and toxicity: an opportunity for genome medicine. Nat Rev Drug Discov 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg GS, and McCarthy JJ (2001). Personalized medicine: revolutionizing drug discovery and patient care. Trends Biotechnol 19, 491–496. [DOI] [PubMed] [Google Scholar]
- Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, Sanderson SC, Kannry J, Zinberg R, Basford MA, et al. (2013). The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med 15, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ED, Guyer MS, and National Human Genome Research, I. (2011). Charting a course for genomic medicine from base pairs to bedside. Nature 470, 204–213. [DOI] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O'Daniel JM, Ormond KE, et al. (2013). ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 15, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Goddard KA, Jarvik GP, Amendola LM, Appelbaum PS, Berg JS, Bernhardt AA, Biesecker LG, Biswas S, Blout CL, et al. (2016). Clinical Sequencing Exploratory Research Consortium: Accelerating Evidence-Based Practice of Genomic Medicine. Am J Hum Genet 99, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RF, Ari M, Kolor K, Dotson WD, Bowen S, Habarta N, Rodriguez JL, Richardson LC, and Khoury MJ (2019). Evaluating the role of public health in implementation of genomics-related recommendations: a case study of hereditary cancers using the CDC Science Impact Framework. Genet Med 21, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenes RA, and Shortliffe EH (1990). Medical informatics. An emerging academic discipline and institutional priority. JAMA 263, 1114–1120. [DOI] [PubMed] [Google Scholar]
- Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, et al. (2016). Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 316, 2402–2410. [DOI] [PubMed] [Google Scholar]
- Guttmacher AE, Collins FS, and Carmona RH (2004). The family history--more important than ever. N Engl J Med 351, 2333–2336. [DOI] [PubMed] [Google Scholar]
- Hayden EC (2013). Geneticists push for global data-sharing. Nature 498, 16–17. [DOI] [PubMed] [Google Scholar]
- Herr TM, Bielinski SJ, Bottinger E, Brautbar A, Brilliant M, Chute CG, Cobb BL, Denny JC, Hakonarson H, Hartzler AL, et al. (2015). Practical considerations in genomic decision support: The eMERGE experience. J Pathol Inform 6, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh WR, Weiner MG, Embi PJ, Logan JR, Payne PR, Bernstam EV, Lehmann HP, Hripcsak G, Hartzog TH, Cimino JJ, et al. (2013). Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care 51, S30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE, and Hoffman JM (2016). Integrating pharmacogenomics into electronic health records with clinical decision support. Am J Health Syst Pharm 73, 1967–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, Kornegay NM, Yang W, Pui CH, Reiss UM, Gaur AH, et al. (2014). PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet 166C, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm IA, Agrawal PB, Ceyhan-Birsoy O, Christensen KD, Fayer S, Frankel LA, Genetti AA, Krier JB, LaMay RC, Levy HL, et al. (2018). The BabySeq project: implementing genomic sequencing in newborns. BMC Pediatr 18, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L, and Bianchi DW (2017). Noninvasive Prenatal DNA Testing: The Vanguard of Genomic Medicine. Annu Rev Med 68, 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huopaniemi I, Nadkarni G, Nadukuru R, Lotay V, Ellis S, Gottesman O, and Bottinger EP (2014). Disease progression subtype discovery from longitudinal EMR data with a majority of missing values and unknown initial time points. AMIA Annu Symp Proc 2014, 709–718. [PMC free article] [PubMed] [Google Scholar]
- International HapMap, C. (2003). The International HapMap Project. Nature 426, 789–796. [DOI] [PubMed] [Google Scholar]
- Jain KK (2002). Personalized medicine. Curr Opin Mol Ther 4, 548–558. [PubMed] [Google Scholar]
- Jarvik GP, Amendola LM, Berg JS, Brothers K, Clayton EW, Chung W, Evans BJ, Evans JP, Fullerton SM, Gallego CJ, et al. (2014). Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet 94, 818–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, Herman GE, Hufnagel SB, Klein TE, Korf BR, et al. (2017). Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 19, 249–255. [DOI] [PubMed] [Google Scholar]
- Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K et al. (2018). Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet 50, 390–400. [DOI] [PubMed] [Google Scholar]
- Khan R, and Mittelman D (2018). Consumer genomics will change your life, whether you get tested or not. Genome Biol 19, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. (2018). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ, Bowen MS, Clyne M, Dotson WD, Gwinn ML, Green RF, Kolor K, Rodriguez JL, Wulf A, and Yu W (2018). From public health genomics to precision public health: a 20-year journey. Genet Med 20, 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles JW, Rader DJ, and Khoury MJ (2017). Cascade Screening for Familial Hypercholesterolemia and the Use of Genetic Testing. JAMA 318, 381–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, and Maglott DR (2014). ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42, D980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Murray MF, and Giovanni MA (2019). Obtaining a Genetic Family History Using Computer-Based Tools. Curr Protoc Hum Genet 100, e72. [DOI] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, et al. (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 51, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manickam K, Buchanan AH, Schwartz MB, and et al. (2018). Exome sequencing–based screening for brca1/2 expected pathogenic variants among adult biobank participants. JAMA Network Open 1, e182140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA (2017). Incorporating Whole-Genome Sequencing Into Primary Care: Falling Barriers and Next Steps. Ann Intern Med 167, 204–205. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Abramowicz M, Al-Mulla F, Anderson W, Balling R, Berger AC, Bleyl S, Chakravarti A, Chantratita W, Chisholm RL, et al. (2015). Global implementation of genomic medicine: We are not alone. Sci Transl Med 7, 290ps213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, and Kenny EE (2017). Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet 100, 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, and Daly MJ (2019). Hidden 'risk' in polygenic scores: clinical use today could exacerbate health disparities. bioRxiv, 441261. [Google Scholar]
- McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, et al. (2011). The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, et al. (2010). Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA 304, 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metspalu A, Kohler F, Laschinski G, Ganten D, and Roots I (2004). [The Estonian Genome Project in the context of European genome research]. Dtsch Med Wochenschr 129 Suppl 1, S25–28. [DOI] [PubMed] [Google Scholar]
- Murray MF (2018). The Path to Routine Genomic Screening in Health Care. Ann Intern Med 169, 407–408. [DOI] [PubMed] [Google Scholar]
- Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators, Stitziel NO, Stirrups KE, Masca NG, Erdmann J, Ferrario PG, Konig IR, Weeke PE, Webb TR, Auer PL, et al. (2016). Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med 374, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai A, Hirata M, Kamatani Y, Muto K, Matsuda K, Kiyohara Y, Ninomiya T, Tamakoshi A, Yamagata Z, Mushiroda T, et al. (2017). Overview of the BioBank Japan Project: Study design and profile. J Epidemiol 27, S2–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease (2011). In Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease (Washington (DC)). [PubMed] [Google Scholar]
- Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, et al. (2018). Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nature Genetics 50, 1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng AO, Kaszemacher T, Abul-Husn NS, Gottesman O, Vega A, Waite E, Myers K, Cho J, Bottinger EP, Ellis SB, et al. (2016). Implementing Algorithm-Guided Warfarin Dosing in an Ethnically Diverse Patient Population Using Electronic Health Records and Preemptive CYP2C9 and VKORC1 Genetic Testing. Clin Pharmacol Ther 100, 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Office of the National Coordinator for Health Information Technology (July 2017). 'Certified Health IT Developers and Editions Reported by Health Care Professionals Participating in the Medicare EHR Incentive Program,' Health IT Quick-Stat #30. dashboard.healthit.gov/quickstats/pages/FIG-Vendors-of-EHRs-to-Participating-Professionals.php.
- Office of the National Coordinator for Health Information Technology (September 2018). 'Percent of Hospitals, By Type, that Possess Certified Health IT,' Health IT Quick-Stat #52. dashboard.healthit.gov/quickstats/pages/certified-electronic-health-record-technology-in-hospitals.php.
- Orlando LA, Buchanan AH, Hahn SE, Christianson CA, Powell KP, Skinner CS, Chesnut B, Blach C, Due B, Ginsburg GS, et al. (2013). Development and validation of a primary care-based family health history and decision support program (MeTree). N C Med J 74, 287–296. [PMC free article] [PubMed] [Google Scholar]
- Pendergrass SA, and Crawford DC (2019). Using Electronic Health Records To Generate Phenotypes For Research. Curr Protoc Hum Genet 100, e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DG, Yatsenko SA, Surti U, and Rajkovic A (2015). Recent advances of genomic testing in perinatal medicine. Semin Perinatol 39, 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrikin JE, Willig LK, Smith LD, and Kingsmore SF (2015). Rapid whole genome sequencing and precision neonatology. Semin Perinatol 39, 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique L, Graham S, Pearl M, Kharrazi M, and Schrijver I (2017). Cystic fibrosis newborn screening programs: implications of the CFTR variant spectrum in nonwhite patients. Genet Med 19, 36–44. [DOI] [PubMed] [Google Scholar]
- Polubriaginof FCG, Vanguri R, Quinnies K, Belbin GM, Yahi A, Salmasian H, Lorberbaum T, Nwankwo V, Li L, Shervey MM, et al. (2018). Disease Heritability Inferred from Familial Relationships Reported in Medical Records. Cell 173, 1692–1704 e1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, and Fullerton SM (2016). Genomics is failing on diversity. Nature 538, 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL, Nussbaum RL, et al. (2015). ClinGen--the Clinical Genome Resource. N Engl J Med 372, 2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, and Evans WE (2015). Pharmacogenomics in the clinic. Nature 526, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehrs A, da Costa CA, Righi RD, and de Oliveira KS (2017). Personal Health Records: A Systematic Literature Review. J Med Internet Res 19, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer Vitek CR, Abul-Husn NS, Connolly JJ, Hartzler AL, Kitchner T, Peterson JF, Rasmussen LV, Smith ME, Stallings S, Williams MS, et al. (2017). Healthcare provider education to support integration of pharmacogenomics in practice: the eMERGE Network experience. Pharmacogenomics 18, 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MLB, McCormick CZ, Lazzeri AL, Lindbuchler DM, Hallquist MLG, Manickam K, Buchanan AH, Rahm AK, Giovanni MA, Frisbie L, et al. (2018). A Model for Genome-First Care: Returning Secondary Genomic Findings to Participants and Their Healthcare Providers in a Large Research Cohort. Am J Hum Genet 103, 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR, et al. (2013). Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, and Ji H (2008). Next-generation DNA sequencing. Nat Biotechnol 26, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Shieh Y, Hu D, Ma L, Huntsman S, Gard CC, Leung JW, Tice JA, Vachon CM, Cummings SR, Kerlikowske K v et al. (2016). Breast cancer risk prediction using a clinical risk model and polygenic risk score. Breast Cancer Res Treat 159, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirts BH, Salama JS, Aronson SJ, Chung WK, Gray SW, Hindorff LA, Jarvik GP, Plon SE, Stoffel EM, Tarczy-Hornoch PZ, et al. (2015). CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc 22, 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son JH, Xie G, Yuan C, Ena L, Li Z, Goldstein A, Huang L, Wang L, Shen F, Liu H et al. (2018). Deep Phenotyping on Electronic Health Records Facilitates Genetic Diagnosis by Clinical Exomes. Am J Hum Genet 103, 58–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber NR, Carpenter JS, Cavallari LH, L JD, Cooper-DeHoff RM, Denny JC, Ginsburg GS, Guan Y, Horowitz CR, Levy KD, et al. (2017). Challenges and strategies for implementing genomic services in diverse settings: experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Med Genomics 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark Z, Dolman L, Manolio TA, Ozenberger B, Hill SL, Caulfied MJ, Levy Y, Glazer D, Wilson J, Lawler M v et al. (2019). Integrating Genomics into Healthcare: A Global Responsibility. Am J Hum Genet 104, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland SM, Kaelber DC, Downing NL, Goel VV, and Longhurst CA (2016). Electronic Health Record-Enabled Research in Children Using the Electronic Health Record for Clinical Discovery. Pediatr Clin North Am 63, 251–268. [DOI] [PubMed] [Google Scholar]
- The Lancet (2018). Artificial intelligence in health care: within touching distance. Lancet 390, 2739. [DOI] [PubMed] [Google Scholar]
- Turnbull C, Scott RH, Thomas E, Jones L, Murugaesu N, Pretty FB, Halai D, Baple E, Craig C, Hamblin A, et al. (2018). The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ 361, k1687. [DOI] [PubMed] [Google Scholar]
- Van Driest SL, Shi Y, Bowton EA, Schildcrout JS, Peterson JF, Pulley J, Denny JC, and Roden DM (2014). Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther 95, 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassy JL, Lautenbach DM, McLaughlin HM, Kong SW, Christensen KD, Krier J, Kohane IS, Feuerman LZ, Blumenthal-Barby J, Roberts JS, et al. (2014). The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials 15, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel KW, Alexander M, Bernhardt BA, Calman N, Carey DJ, Cavallari LH, Field JR, Hauser D, Junkins HA, Levin PA, et al. (2016). The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel KW, Elsey AR, Langaee TY, Burkley B, Nessl DR, Obeng AO, Staley BJ, Dong HJ, Allan RW, Liu JF, et al. (2014). Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet 166C, 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control, C. (2007). Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams F, and Boren SA (2008). The role of the electronic medical record (EMR) in care delivery development in developing countries: a systematic review. Inform Prim Care 16, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MS, Buchanan AH, Davis FD, Faucett WA, Hallquist MLG, Leader JB, Martin CL, McCormick CZ, Meyer MN, Murray MF, et al. (2018a). Patient-Centered Precision Health In A Learning Health Care System: Geisinger's Genomic Medicine Experience. Health Aff (Millwood) 37, 757–764. [DOI] [PubMed] [Google Scholar]
- Williams MS, Kern MS, Lerch VR, Billet J, Williams JL, and Moore GJ (2018b). Implementation of a patient-facing genomic test report in the electronic health record using a web-application interface. BMC Med Inform Decis Mak 18, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte JS, Visscher PM, and Wray NR (2014). The contribution of genetic variants to disease depends on the ruler. Nat Rev Genet 15, 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SM, Crock BN, Van Ness B, Lawrenz F, Kahn JP, Beskow LM, Cho MK, Christman MF, Green RC, Hall R et al. (2012). Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genet Med 14, 361–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolford BN, Willer CJ, and Surakka I (2018). Electronic health records: the next wave of complex disease genetics. Hum Mol Genet 27, R14–R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of 22 global, population-based, EHR-linked biobanks as of January 2019.