Abstract
Background
Grain weight is an important yield component. Selection of advanced lines with heavy grains show high grain sink potentials and strong sink activity, which is an increasingly important objective in wheat breeding programs. Rice OsGS3 has been identified as a major quantitative trait locus for both grain weight and grain size. However, allelic variation of GS3 has not been characterized previously in hexaploid wheat.
Results
We cloned 2445, 2393, and 2409 bp sequences of the homologs TaGS3-4A, TaGS3-7A, and TaGS3-7D in wheat ‘Changzhi 6406’, a cultivar that shows high grain weight. The TaGS3 genes each contained five exons and four introns, and encoded a deduced protein of 170, 169, and 169 amino acids, respectively. Phylogenetic analysis of plant GS3 protein sequences revealed GS3 to be a monocotyledon-specific gene and the GS3 proteins were resolved into three classes. The length of the atypical Gγ domain and the cysteine-rich region was conserved within each class and not conserved between classes. A single-nucleotide polymorphism in the fifth exon (at position 1907) of TaGS3-7A leads to an amino acid change (ALA/THR) and showed different frequencies in two pools of Chinese wheat accessions representing extremes in grain weight. Association analysis indicated that the TaGS3-7A-A allele was associated with higher grain weight in the natural population. The TaGS3-7A-A allele was favoured in global modern wheat cultivars but the allelic frequency varied among different wheat-production regions of China, which indicated that this allele is of potential utility to improve wheat grain weight in certain wheat-production areas of China.
Conclusions
The novel molecular information on wheat GS3 homologs and the KASP functional marker designed in this study may be useful in marker-assisted breeding for genetic improvement of wheat.
Keywords: TaGS3, Kernel traits, Phylogenetic tree, Functional marker
Background
Wheat (Triticum aestivum, AABBDD) provides more than 20% of the calories and 25% of the protein in the human diet (FAOSTAT; http://Faostat.fao.org/site/339/default.aspx and http://www.fao.org/worldfoodsituation/wfs-home/csdben). With the ongoing increase in population size, climate change, and reduction in the available land area, annual gains in crop yields of ~ 2% are required to meet food security [1, 2]. The grain yield of wheat and other cereals is a polygenic trait that is influenced by environmental and genetic interactions at all stages of plant growth [3].
In rice, genes controlling grain weight have been extensively studied [4, 5]. Grain Size 3 (GS3), which encodes a transmembrane protein containing an atypical Gγ domain and a cysteine-rich region, was the first-characterized quantitative trait locus (QTL) that negatively regulates grain length [6, 7] A C–A mutation in the second exon of GS3 (A allele) is associated with enhanced grain length in Oryza sativa but is absent in other Oryza species [7]. Pyramiding GL3.3 (Grain Length 3.3) and GW7 (Grain Width 7) enhances yield in an indica hybrid rice background and simultaneously improves yield and grain quality [8]. Grain Size 5 (GS5) encodes a putative serine carboxypeptidase and functions as a positive regulator of grain size and yield [9] Analysis of natural variation indicates that mutation of GS5 is associated with enhanced grain length [10]. Grain width 5 (GW5) encodes a novel nuclear protein of 144 amino acids that is localized to the nucleus and affects grain width [11]. A 1212-bp deletion in rice ‘Nipponbare’ was selected during japonica domestication, which functions most likely through influencing the expression levels of OsGW5. The GW5 protein is localized to the plasma membrane and can physically interact with and repress the kinase activity of rice GSK2, resulting in accumulation of unphosphorylated OsBZR1 and DLT [12]. A recently reported gene, GLW7 (GRAIN LENGTH AND WEIGHT ON CHROMOSOME 7), also showed a differential expression level in regulating grain weight by allelic variation in the 5′ untranslated region [13]. In addition, Grain Width 6 (GW6) encodes a GNAT-like protein that harbors intrinsic histone acetyltransferase activity, and elevated expression of this gene enhances grain length and weight by enlarging spikelet hulls and accelerating grain filling [14]. Grain Width 2 (GW2) encodes a previously unknown RING-type protein with E3 ubiquitin ligase, which negatively regulates cell division by degrading its substrate(s) by means of the ubiquitin–proteasome pathway [15]. DA2 (orthologous to GW2) is an E3 ubiquitin ligase that physically interacts with DA1 in vitro and in vivo in Arabidopsis [16]. Grain Length 3 (GL3) encodes a protein phosphatase with Kelch-like domains (PPKL) family Ser/Thr phosphatase, which shows epistatic interaction with GS3 and OsGSK3 to modulate brassinosteroid signaling to produce extra-long grains in rice [17–19]. The Kelch domains are essential for OsPPKL1 (GL3) biological function [20]. DENSE AND ERECT PANICLE 1 (DEP1) encodes a highly cysteine-rich G protein gamma subunit protein. Gain-of-function mutation of DEP1 enhances meristematic activity, resulting in reduction in the length of the inflorescence internodes, an increased number of grains per panicle, and a consequent increase in grain yield [21].
In wheat, the large genome size and complex genome composition renders map-based cloning of functional genes difficult, especially for complex traits. However, the close relationship between wheat and rice provides the opportunity for researchers to clone homologous functional genes with a common ancestor, in particular for development-related traits [22–24]. On the basis of this theory, a number of grain weight-related genes have been cloned and characterized in natural populations and/or bi-parental populations. Orthologs of TaGW2 have been widely characterized in wheat. Two single-nucleotide polymorphisms (SNPs) forming a Hap-6A-A haplotype for TaGW2-6A are associated with broader grains and higher 1000-grain weight [25]. An additional haplotype, Hap-6B-1 for TaGW2-6B, shows stronger effects on grain weight than TaGW2-6A and this was further validated by gene-editing mutant analysis [26, 27]. A number of reverse genetic studies have shown that TaGW2 has a negative influence on grain size in hexaploid and tetraploid wheat [28, 29]. Homologs of GW5 also exert many effects on grain weight and grain size; the haplotype TaGS5-3A-T, based on the SNP T/G in the sixth exon, is significantly correlated with larger grain size, higher 1000-kernel weight, and lower plant height, spike length, and internode length below the spike, which is under strong positive selection in Chinese modern wheat breeding [2, 30]. TaGW6, an ortholog of rice GW6, encodes a novel indole-3-acetic acid-glucose hydrolase. Variation among wheat homologs of TGW6 on chromosome arms 3AL and 4AL individually explain ~ 17% of TGW variation, and the low-expression alleles are associated with low auxin content and high TGW [31, 32]. TaGW7 is a homolog of a gene encoding a TONNEAU1-recruiting motif protein, which is mainly expressed in young spike tissues, was shown to negatively regulate grain weight and grain width using CRISPR-Cas9 gene editing analysis. This gene regulates seed size probably effects genes through cell division pathways [33, 34]. Notably, a number of grain trait-related genes (e.g., TaGW2, TaGL3, and TaGW8) were mapped in the QTL region that is likely to harbpr the causative genes [35–37].
TaGS3-1D has been cloned in hexaploid wheat and a 40-bp indel predicted to generate a new isoform that is associated with grain weight [38]. However, the hexaploid wheat genome consists of three subgenomes, of which the D genome generally shows the lowest genetic diversity [39, 40]. Therefore, we were interested in isolating the two homologous TaGS3 genes in the A and B subgenomes and determining the functional variation for potential use in wheat improvement. For this purpose, we cloned the TaGS3 genes, investigated the evolution of GS3 genes in plants, designed genetic markers for TaGS3, validated its effects in a natural population, and evaluated its distribution in major wheat-production areas of China.
Results
Isolation of TaGS3 in wheat
In wheat, three copies of TaGS3 on chromosome arms 4AL (TraesCS4A02G474000), 7AS (TraesCS7A02G017700), and 7DS (TraesCS7D02G015000) were searched in the Chinese Spring RefSeq v1.0 genome database. Primers were designed based on specific regions for each sequence (Table 1). The primers amplified PCR products 2445, 2393, and 2409 bp in length for TaGS3-4A (GenBank accession: KY888174), TaGS3-7A (GenBank accession: KY888186), and TaGS3-7D (GenBank accession: KY888197) in wheat accession ‘Changzhi 6406’ (a cultivar that exhibits high grain weight). The lengths of the predicted coding sequences of the three genes were 513, 510, and 510 bp, encoding putative proteins of 170, 169, and 169 amino acids, respectively. TaGS3-4A, TaGS3-7A, and TaGS3-7D showed similarity in exon–intron structure to that of OsGS3, which consisted of five exons and four introns (Fig. 1). The degree of similarity between the coding region of OsGS3 and that of TaGS3-4A, TaGS3-7A, and TaGS3-7D was 45.92, 43.94, and 44.87%, respectively. In parallel, the similarity of the deduced amino acid sequences was 45.26, 44.83, and 42.24%, respectively.
Table 1.
Primers used in this study
| Primer | Sequence (5′-3′) | explanation | predicted PCR product Size |
|---|---|---|---|
| TaGS3-4A-F | CGATCCTTCTCTGCGGCAAG | primers for amplifying TaGS3-4A | 2435 bp |
| TaGS3-4A-R | CCTACAGACCGACGACTTCCTG | ||
| TaGS3-7A-F | CGACTTCCTGTCTCCTTCCGG | primers for amplifying TaGS3-7A | 2389 bp |
| TaGS3-7A-R | TCATGCCCGTCAAAAACACCAG | ||
| TaGS3-7D-F | GACGACTTCCTGTCTCCTACTTCC | primers for amplifying TaGS3-7D | 2409 bp |
| TaGS3-7D-R | TCATGCCCGTCAAAAACACCAG | ||
| TaGS3-7A-1907-F1 | FAM-GCGCCGGTTGCTCCTCATCTTGCG | KASP marker for distinguishing A/G allele | |
| TaGS3-7A-1907-F2 | HEX-GCGCCGGTTGCTCCTCATCTTGCA | ||
| TaGS3-7A-1907-CR | AGGGACGCCRCCGCAGCACACGGT |
Fig. 1.
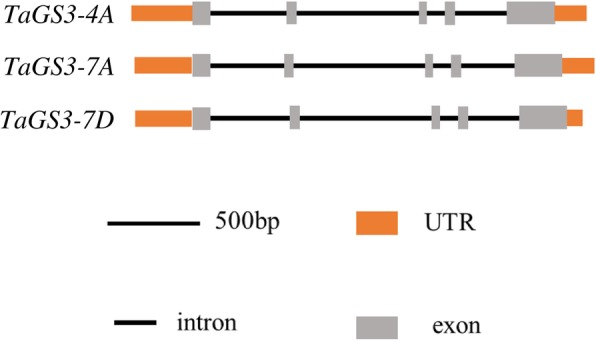
Predicted exon–intron structure of the three TaGS3 homologues in Changzhi6406
Phylogenetic analysis of GS3 in plants
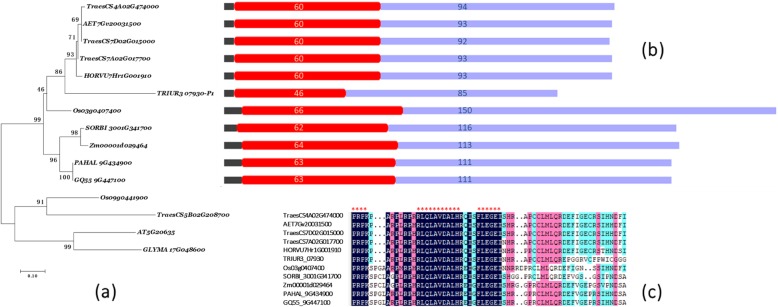
A phylogenetic tree was reconstructed for 14 deduced amino sequences for GS3 homologs from Aegilops tauschii, Hordeum vulgare, Panicum hallii, Setaria italica, Triticum aestivum, Zea mays, Triticum urartu, Sorghum bicolor, Glycine max, and Arabidopsis thaliana. A reported atypical Gγ domain gene, DEP1, was used as the outgroup. In the phylogenetic tree the sequences were resolved into two main groups, which comprised the GS3 orthologs and DEP1 orthologs (Fig. 2a). The GS3 orthologous group consisted of ten sequences that were all derived from monocotyledons, whereas the DEP1-orthologous group comprised sequences derived from monocotyledons and dicotyledons.
Fig. 2.
A Phylogenetic analysis of GS3 orthologous in plants using deduced amino acid sequences. B Domain structure of GS3 orthologous in corresponding to left phylogenetic tree. Red solid box represented the conserved atypical Gγ domain and light purple represented cysteine-rich region. The number represented the length of each domain/region. c) Multiple alignment of GS3 homologous sequences. Conserved amino acids are indicated by red asterisk. TraesCS4A02G474000 (Triticum aestivum), TraesCS7A02G017700 (Triticum aestivum), TraesCS7D02G015000 (Triticum aestivum), AET7Gv20031500 (Aegilops tauschii), HORVU7Hr1G001910 (Hordeum vulgare), TRIUR3_07930 (Triticum urartu), Os03g0407400 (GS3, Oryza sativa Japonica Group), SORBI_3001G341700 (Sorghum bicolor), Zm00001d029464 (Zea mays), PAHAL_9G434900 (Panicum hallii FIL2), GQ55_9G447100 (Panicum hallii HAL2), Os09g0441900 (DEP1, Oryza sativa Japonica Group), TraesCS5B02G208700 (Triticum aestivum), GLYMA_17G048600 (Glycine max), AT5G20635 (Arabidopsis thaliana)
In the GS3 orthologs group, the relationships among the genes were congruent with the species phylogeny, except that the relationship between TaGS3-7A in wheat subgenome A and TuGS-7A in Triticum urartu was more distant than that with GS3 of Hordeum vulgare. The similarity of the conserved domain among the sequences was 85.14% overall and the sequences were of similar length (60–66 amino acids) except for that of T. urartu (46), whereas the length of the cysteine-rich region was variable and was divided into three classes. Class I consisted of Triticeae species with a conserved length of 92–94 amino acids (85 in Triticum urartu was an exception); Class II included Oryza species of which the longest sequence was 150 amino acids; and Class III consisted of Panicoideae species with a conserved length of 111–116 amino acids. Comparison of the atypical Gγ subunit conserved domain among the GS3 ortholog sequences revealed that all sequences contained an identical amino acids fragment “PRP--RLQLAVDALHR--FLEGEI” (“--” represents a non-conserved region and the number of “-” does not reflect the number of amino acids).
Polymorphism of TaGS3-4A and TaGS3-7A among wheat accessions
Sequences for TaGS3-4A and TaGS3-7A from ten wheat accessions were obtained. To avoid possible false variation generated by sequencing error, an advanced analysis for detection of real variants resulted in a minor allele frequency > 0.1 (real SNPs must be detected at least twice in ten sequences). In total 17 and 18 variations for TaGS3-4A and TaGS3-7A was detected by multiple alignment, respectively. There were 3, 1, 6, 0, 3, 0, 0, 0, 0, 2, and 2 variations located in the 5′ upstream region, first exon, first intron, second exon, second intron, third exon, third intron, fourth exon, fourth intron, fifth exon, and the 3′ downstream region for TaGS3-4A, respectively. In parallel, 4, 0, 2, 0, 4, 0, 0, 1, 3, 3, and 1 variable sites were detected in the corresponding regions for TaGS3-7A (Tables 2 and 3). Among them, both three SNPs causing changes causing amino acid change were determined for each of TaGS3-4A and TaGS3-7A, which were located in the first exon (1) and fifth exon (2), and in the fourth exon (1) and fifth exon (2), respectively. The variant (GC/AT) at position 70 for TaGS3-4A was located in an atypical Gγ subunit, of which the GC allele was harbored by 80 and 60% of accessions grouped into heavy and light grain weight pools, respectively, which indicated this locus was not associated with grain weight. In addition, the frequency of allele A at position 1907 (amino change: ALA/THR) was 0.6 in the heavy grain weight accessions compared with zero in the light grain weight accessions. Therefore, this allele was considered more likely to be associated with grain weight in wheat.
Table 2.
Allelic variation of TaGS3-4A. The position was accounted from ATG (1)
| position | location | Changzhi 6406 | Sankecun | Shannong 7859 | Kangdingxiaomai | Xingyi 4 | Youmangbaifu | Dongnong 101 | Gansu 96 | Fuzhuang 30 | Bima 4 | amino acid change |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| − 306 | 5’UTR | T | C | C | T | T | T | C | T | T | T | – |
| −129 | 5’UTR | TG | CG | TG | TG | TG | TG | CA | CG | TG | TG | – |
| −53 | 5’UTR | G | A | G | G | G | G | A | A | G | G | – |
| 70 | 1st exon | GC | AT | GC | GC | GC | GC | AT | AT | GC | GC | A/M |
| 200 | 1st intron | G | G | G | C | G | G | G | G | C | C | – |
| 250 | 1st intron | T | T | T | – | T | T | T | T | – | – | – |
| 331 | 1st intron | T | A | T | T | T | T | A | A | T | T | – |
| 354 | 1st intron | ACT | G | ACTG | ACTG | G | ACTACTG | G | G | G | G | – |
| 378 | 1st intron | A | G | A | A | A | A | G | G | A | A | – |
| 397 | 1st intron | T | T | T | T | C | T | C | T | T | T | – |
| 633 | 2nd intron | C | T | T | T | C | C | T | T | T | T | – |
| 712 | 2nd intron | A | T | T | A | A | A | T | T | A | A | – |
| 1140 | 2nd intron | T | T | T | T | C | T | T | C | T | T | – |
| 1747 | 5th exon | T | C | T | C | T | T | T | T | T | T | L/P |
| 1897 | 5th exon | C | C | T | C | T | T | C | C | C | C | A/V |
| 1976 | 3’UTR | C | C | G | C | G | G | C | C | C | C | – |
| 2004 | 3’UTR | C | G | C | C | C | C | G | G | C | C | – |
Table 3.
Allelic variation of TaGS3-7A. The position was accounted from ATG (1)
| position | location | Changzhi 6406 | Sankecun | Shannong 7859 | Kangdingxiaomai | Xingyi 4 | Youmangbaifu | Dongnong 101 | Gansu 96 | Fuzhuang 30 | Bima 4 | amino acid change |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| − 285 | 5’UTR | T | T | T | C | C | C | T | C | C | C | – |
| −244 | 5’UTR | (CCG)6 | (CCG)6 | (CCG)6 | (CCG)3 | (CCG)5 | (CCG)6 | (CCG)6 | (CCG)3 | (CCG)3 | (CCG)3 | – |
| − 210 | 5’UTR | – | – | – | C | – | G | C | C | C | C | – |
| − 159 | 5’UTR | C | C | C | T | T | T | T | T | T | T | – |
| 417 | 1st intron | GCTAGC.....TAGTAGCT | GCTAGC.....TAGTAGCT | GCTAGC.....TAGTAGCT | GCTAGCTACTGTAGTAGCT | GCTAGC.....TAGTAGCT | GCTAGC.....TAGTAGCT | GCTAGC.....TAGTAGCT | GCTAGCTACTGTAGTAGCT | GCTAGCTACTGTAGTAGCT | GCTAGCTACTGTAGTAGCT | – |
| 479 | 1st intron | T | T | T | – | – | – | – | – | – | – | – |
| 582 | 2nd intron | C | C | C | T | T | T | T | T | T | T | – |
| 785 | 2nd intron | G | G | G | C | G | G | G | C | C | C | – |
| 1022 | 2nd intron | C | C | C | T | T | T | T | T | T | T | – |
| 1117 | 2nd intron | T | T | T | C | T | T | T | T | C | C | – |
| 1435 | 4th exon | T | T | C | T | T | T | T | C | T | T | I/T |
| 1515 | 4th intron | G | G | G | A | G | G | G | A | A | A | – |
| 1523 | 4th intron | A | A | A | G | A | A | A | A | G | G | – |
| 1543 | 4th intron | A | A | A | G | A | A | A | A | G | G | – |
| 1814 | 5th exon | G | G | G | C | G | G | G | C | C | C | G/R |
| 1907 | 5th exon | A | A | A | G | G | G | G | G | G | G | T/A |
| 1948 | 5th exon | C | C | C | T | C | C | C | T | T | T | – |
| 2000 | 3’UTR | TT | CT | TT | AT | CT | TT | TT | A | AT | AT | – |
Molecular marker design and validation
Based on the results of SNP mining, a Kompetitive allele-specific PCR (KASP) marker was designed for the SNP at position 1907 (A/G). The calls for the TaGS3-7A-A allele of the potential light grain weight group were clustered along with the x-axis, whereas the calls of the TaGS3-7A-G allele of the potential heavy grain weight group were clustered along with the y-axis (Fig. 3). Thus, the clustering results clearly distinguished the two alleles, which was used to validate its effects on grain weight in the natural population.
Fig. 3.
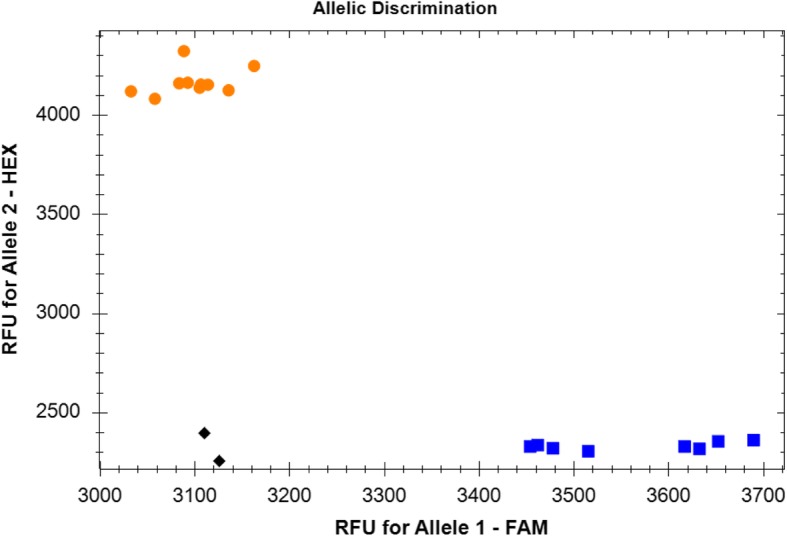
Scatter plots for KASP assays for TaGS3-7A showing clustering of varieties on the X- (FAM) and Y- (HEX) axes. Varieties colored blue have the FAM-type allele, for which the clustered samples carried TaGS3-7A-G allele showed small grain weight value and grain size; varieties colored orange have the HEX-type allele, the clustered samples carried TaGS3-7A-A allele showed larger grain weight value and grain size; black dots represent the NTC (no template control)
The association between genotypes and grain traits was analyzed based on a mini-core collection (MCC) of 224 wheat accessions for this marker (Additional file 1: Table S1). On the basis of the SNP calls using the KASP marker, significant differences for kernel width, kernel weight, and kernel thickness was observed for the TaGS3-7A-G and TaGS3-7A-A alleles, whereas no significant difference for kernel length was observed (Table 4). In addition, the allele TaGS3-7A-A with superior traits with a rare frequency of 13.107%, which was significantly less than that for TaGS3-7A-G. However, the frequency of TaGS3-7A-A in cultivars (18.085%) was significantly higher than that in landraces (0.980%).
Table 4.
Association analysis of kernel traits between TaGS3-7A-G and TaGS3-7A-A genotypes. note: * p<0.05, ** p<0.01
| Trait | Allele | N | Mean | Sig |
|---|---|---|---|---|
| TKW_E1 | TaGS3-7A-G | 116 | 36.40 ± 0.69 | 0.010** |
| TaGS3-7A-A | 20 | 40.96 ± 1.28 | ||
| TKW_E2 | TaGS3-7A-G | 140 | 32.76 ± 0.61 | 0.075 |
| TaGS3-7A-A | 21 | 35.77 ± 1.44 | ||
| TKW_E3 | TaGS3-7A-G | 145 | 34.58 ± 0.61 | 0.004** |
| TaGS3-7A-A | 24 | 39.11 ± 1.22 | ||
| GL_E1 | TaGS3-7A-G | 147 | 0.639 ± 0.005 | 0.27 |
| TaGS3-7A-A | 24 | 0.654 ± 0.013 | ||
| GL_E2 | TaGS3-7A-G | 143 | 0.643 ± 0.005 | 0.089 |
| TaGS3-7A-A | 22 | 0.665 ± 0.011 | ||
| GL_E3 | TaGS3-7A-G | 145 | 0.668 ± 0.005 | 0.134 |
| TaGS3-7A-A | 24 | 0.686 ± 0.010 | ||
| GW_E1 | TaGS3-7A-G | 147 | 0.303 ± 0.003 | 0.013* |
| TaGS3-7A-A | 24 | 0.321 ± 0.006 | ||
| GW_E2 | TaGS3-7A-G | 143 | 0.301 ± 0.002 | 0.007** |
| TaGS3-7A-A | 22 | 0.316 ± 0.005 | ||
| GW_E3 | TaGS3-7A-G | 145 | 0.306 ± 0.002 | 0.020* |
| TaGS3-7A-A | 24 | 0.318 ± 0.006 | ||
| GT_E1 | TaGS3-7A-G | 147 | 0.267 ± 0.002 | 0.010** |
| TaGS3-7A-A | 24 | 0.281 ± 0.004 | ||
| GT_E2 | TaGS3-7A-G | 143 | 0.278 ± 0.002 | 0.005** |
| TaGS3-7A-A | 22 | 0.292 ± 0.004 | ||
| GT_E3 | TaGS3-7A-G | 145 | 0.289 ± 0.002 | 0.083 |
| TaGS3-7A-A | 24 | 0.298 ± 0.004 | ||
| GL/GW_E1 | TaGS3-7A-G | 147 | 2.119 ± 0.019 | 0.221 |
| TaGS3-7A-A | 24 | 2.053 ± 0.059 | ||
| GL/GW_E2 | TaGS3-7A-G | 143 | 2.146 ± 0.016 | 0.456 |
| TaGS3-7A-A | 22 | 2.113 ± 0.044 | ||
| GL/GW_E3 | TaGS3-7A-G | 145 | 2.190 ± 0.016 | 0.784 |
| TaGS3-7A-A | 24 | 2.178 ± 0.053 |
Distribution of the TaGS3-7A-A allele in the major wheat-production areas of China
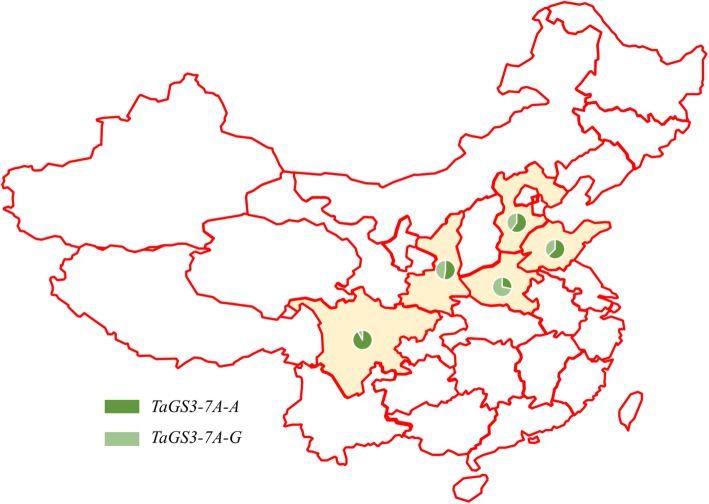
In total, 238 wheat cultivars grown in the main wheat-production areas of China were analyzed to determine the frequency of the TaGS3-7A-A allele (Additional file 1: Table S2). The TaGS3-7A-A allele was detected in 58.4% of all tested cultivars. Significant differences in the frequency of TaGS3-7A-A were observed among the five province areas. The TaGS3-7A-A allelic frequency exceeded 50% in all provinces except Henan (i.e., 92.0% in Sichuan, 62.5% in Shandong, 60% in Hebei, and 52.6% in Shaanxi). The TaGS3-7A-A allele was detected in 50% of the cultivars in Shaanxi province. However, a significantly lower frequency of TaGS3-7A-A was detected among cultivars from Henan (28.23%) compared with the other provinces.
Discussion
Structure and evolution of GS3
A number of genes that encode G proteins in rice have been cloned and show a variety of functions in the regulation of organ development [10, 21]. GS3, which encodes heterotrimeric G proteins that contain an atypical Gγ domain, is a major QTL for grain size [6, 10]. However, the evolutionary history of GS3 in plants has not been examined previously. In the present study, we performed a phylogenetic analysis of protein sequences using the rice GS3 sequence as a query. There was no GS3 homologs in dicotyledon plant, indicated that GS3 homologs was a monocotyledon-specific gene family. We also determined that the wheat genome contains three GS3 orthologous genes on chromosome arms 7AS, 7DS, and 4AL based on the released ‘Chinese Spring’ reference genome sequence [22]. The molecular evolution of GS3 was consistent with their species evolution relationship. However, the phylogenetic relationship between TaGS3-7A and TuGS3-7A was more distant than that between TaGS3-7A and other Triticeae species, which probably because TuGS3-7A was located on 4AL/7BS translocation that is occurred at polyploidizing progress during diploid wheat to polyploid wheat [41–44].
All GS3 orthologous genes showed a conserved atypical Gγ domain with high similarity, whereas the length of the cysteine-rich region was variable. In particular, the length of the cysteine-rich region in Triticeae species was ~ 60% and ~ 80% the length of that in rice and Panicoideae species, respectively. Although the conserved domain usually determines the function of genes universally, the length of the C-terminal tail may affect degradation of the GS3 protein in rice [45]. Therefore, evolution of GS3 has led to differences in the length of GS3 in monocotyledons and likely fine-tune of the protein function.
Polymorphism of GS3 and its association with grain weight
In the present study, the isolated wheat TaGS3-4A, TaGS3-7A, and TaGS3-7D genes were predicted to contain five exons and four introns, which are identical to GS3 genes in rice. A large number of variations for TaGS3-7A and TaGS3-4A were detected. Only one variant located in the GS3 Gγ-subunit was detected in TaGS3-4A and this variant seemed to be not associated with grain weight in the different phenotype pools. Nevertheless, the non-conserved domain of GS3 is involved in positive regulation of grain length in rice [46]. Additional polymorphic loci motifs of (AT) n in the fourth intron and (TCC) n in the fifth exon of GS3 were mainly associated with medium to short grains among Chinese rice accessions because 13 accessions harboring the TGC mutation produced long grains, which suggested that the C-A mutation did not completely explain the accessions with long or short grains in Chinese rice germplasm. In maize, two polymorphisms in the fifth exon (not located within the conserved region) and the promoter region are significantly associated with kernel length and 1000-kernel weight In similarity, we also found that variations in the conserved domain region was not associated with grain weight in consideration of allelic frequency between two pools with different phenotype. However, an additional SNP located in the cysteine-rich region that showed a difference in frequencyand thus applied for marker development. This variation probably effect Gβγ dimers from autoinhibition through alternation of the structure of cysteine-rich region [45, 47]. The cysteine-rich region plays a variety of roles in exercise its function. GAST1-like proteins are involved in redox reactions via their cysteine-rich domain and GASA4 mutated by replacement of conserved cysteines with alanines lost its redox activity and the capability to promote gibberellin responses [48]. In addition, the C-terminal cysteine-rich region is sufficient for rice OsDep1 to confer cadmium tolerance to yeast cells [49]. Collectively, the afore-mentioned findings suggest that variation in the non-conserved domains of GS3 may also affect the phenotype.
In previous studies, one QTL for grain weight was indicated to be associated with TaGS3-7D in wheat and all variation between parental lines was contained in the second intron. However, subgenomes A and B show broader genetic diversity compared with that of D subgenome in hexaploid wheat, leading to assessment of the sequence diversity of TaGS3 and development of a functional marker for TaGS3 for use in breeding [39, 50]. However, we observed that none of the reported major QTLs for grain weight/size were located in the terminal region of chromosome arm 7AS in the vicinity of TaGS3-7A, which was probably because cultivars often carry a weak allelic variation of functional genes, which could provide balance for maintaining the enhancement of grain weight and plant fitness [45, 51, 52]. However, variations with weak effects on phenotype was usually behind high threshold in QTL mapping. Therefore, application of natural accessions with extreme phenotype as a sampling was an alternatively method for detection of functional allelic variation in wheat [31, 53, 54].
In particular, modern cultivars grown in the main wheat-production areas of China showed a different allele frequency of TaGS3-7A-A and exceeded more than 50% of cultivars in most provinces (Fig. 4). The highest frequency of TaGS3-7A-A and TaGS5-3A-T alleles were detected in southwestern China, which indicated that the alleles were probably strongly selected in this region [2]. In addition, the lowest frequency of the TaGS3-7A-A allele was observed in Henan province, which suggested that there remains potential for improvement of grain weight in some wheat-production areas by utilization of the TaGS3-7A-A allele.
Fig. 4.
Distribution of the TaGS3-7A-A and TaGS3-7A-G allele in the main wheat production areas of China
Cloning of grain weight-related genes and characterization of allelic variation in wheat are essential for molecular breeding. In addition, the functional marker for TaGS3-7A developed in the present study is potentially useful for wheat yield improvement.
Conclusions
In the present study, we cloned homologs of TaGS3 in hexaploid wheat, systematically evaluated the evolutionary progress of GS3 in plant species, and designed a functional marker for TaGS3-7A. Wheat carries three TaGS3 homologs, for which the amino acid sequences contain a similar atypical Gγ domain and a cysteine-rich region that differed in length from that of rice. On the basis of sequence comparison between two extreme phenotypic pools, a KASP marker was designed and shown to have significant genetic effects on grain weight, kernel width, and kernel thickness. The present results provide novel molecular information on GS3 and the designed functional marker may be useful in genetic improvement programs for wheat.
Methods
Plant materials
Wheat (Triticum aestivum) ‘Changzhi 6406’, which is characterized by a high grain weight in previous, was used for isolation of sequences for the three GS3 homologs. Ten wheat accessions, divided into two pools, were used for TaGS3-4A and TaGS3-7A gene cloning; one pool comprised accessions with high grain weights (‘Changzhi 6406’, ‘Sankecun’, ‘Shannong 7859’, ‘Kangdingxiaomai’, and ‘Xingyi 4’) and the second pool consisted of accessions with low grain weight (‘Youmangbaifu’, ‘Dongnong 101’, ‘Gansu 96’, ‘Fuzhuang 30’, and ‘Bima 4’) [2].
The MCC germplasm collection, comprising 224 wheat accessions representing high genetic diversity in China, were used for validation of the association of the genetic marker and kernel-related traits (kernel length, kernel width, kernel thickness, and 1000-kernel weight) in three independent environments (E1: Luoyang at 2002; E2 Luoyang at 2005; E3 Shunyi at 2010) [2].
A panel of 238 modern wheat cultivars grown in the main wheat-production areas of China and released in the twenty-first century were used to investigate the allelic frequency and distribution of TaGS3-7A-A. These accessions including the 75 accessions in southwest of China and another previously characterized 163 accessions in the central plateau and north of the Yellow River [35].
Gene isolation and analysis
TaGS3 homologous gene sequences were isolated using a comparative genomics method. GS3 gene sequences for rice and wheat (GenBank accession numbers DQ355996 and KF687956, respectively) were used to search for orthologs in the wheat genome using the Ensembl Plants online resource (http://plants.ensembl.org/Triticum_aestivum/Tools/Blast?db=core). Primer sets to target a specific region for each sequence were designed with Oligo 7 software and synthesized by Tsingke Biological Technology Co., Ltd. (Beijing, China).
Since TaGS3 gene containing high GC content and repetitive sequence, Taq DNA polymerase with GC Buffer (probably lose some fidelity. Takara, RR02AG) were selected for PCR reaction. Amplification of the gene sequences by PCR was performed in a volume of 30 μl, which contained 15 μl of 2× GC buffer I, 10 μM dNTPs, 20 μmol of each primer, 1 U LA Taq™ and ~ 100 ng template DNA. All reagents were obtained from Takara Biotechnology Co., Ltd. (http://www.takara.com.cn). The reaction protocol was 94 °C for 5 min, followed by 35 cycles of 94 °C for 45 s, 58 °C for 45 s, and 72 °C for 2 min 30 s, and final extension at 72 °C for 10 min.
The PCR products were separated by electrophoresis on 2% agarose gels. All bands were cleaned and cloned into the pEASY®-T5 Zero vector (Beijing TransGen Biotech Co., Ltd.; http://www.transgen.com.cn) and transformed into DH5α competent Escherichia coli cells using the heat shock method. Single clones were sequenced by Tsingke Biotech Co., Ltd. Sequence alignment and assembly was performed using DNAMAN software (http://www.lynnon.com).
All sequenced clones were aligned against the wheat reference genome (IWGSC v1.0) to confirm their chromosomal locations.
Phylogenetic analysis
The rice GS3 (Os03g0407400) ortholog sequence was used to search genomic databases for common higher plants (e.g., rice, maize, Arabidopsis, soybean, sorghum, and medicago) in Ensembl Plants website (http://plants.ensembl.org/Triticum_aestivum/Tools/Blast?db=core). The gene OsDep1, which encodes a G protein, was used as the paralogous outgroup. Alignment of the deduced amino acids sequences was performed using MUSCLE in MEGA 7.0 software [55]. A phylogenetic tree was constructed using the neighbor-joining method, Poisson model, uniform rate, and partial deletion (site coverage off 50%) as implemented in MEGA [56]. A bootstrap analysis was performed to test the robustness of the phylogenetic construction with 1000 replications.
SNP mining and development of a gene-specific KASP marker
Single-nucleotide polymorphisms that caused a putative amino acid change and differed in frequency between the grain weight accessions were used for marker development.
For the KASP genotyping procedure, the 10 μl PCR mixture contained 100 ng template DNA, 5 μl KASP Master mix, 4 mM MgCl2, and 1.4 μl primer mixture, which comprised 46 μl ddH2O, 30 μl common primer (10 μM), and 12 μl of each tailed primer (10 μM). The PCR cycling protocol consisted of a hot start at 95 °C for 15 min, followed by ten touchdown cycles (95 °C for 20 s; touchdown at 65 °C initially and decreasing by − 1 °C per cycle for 60 s), followed by 30 additional cycles of annealing (95 °C for 10 s; 57 °C for 60 s). The extension step (each three cycles of annealing: 95 °C for 10 s; 57 °C for 60 s) was repeated three times to obtain the best genotype clustering results [35, 57]. The last 37 °C for 1 min was set up for collection of HEX and FAM fluorescence signal value. Final signal detection was applied under Bio-Rad CFX Connect™ PCR system and allele discrimination plots (clustering) generated with Bio-Rad CFX Manager™ software. The primers were synthesized by Tsingke Biological Technology Co., Ltd., and the developed KASP marker was used to screen all 224 MCC germplasm and 238 modern wheat cultivars in China.
Statistical analysis
One-way analysis of variance was performed using IBM SPSS Statistics for Windows version 22.0 (IBM Corporation, Armonk, NY, USA) to determine the significance of differences in grain traits between the two alleles. Phenotypic data recorded in three independent environments (E1: Luoyang at 2002; E2: Luoyang at 2005; E3: Beijing at 2010) were derived from a previous study [26]. Accessions either with indistinct genotyping signal or lacking phenotype data were excluded from analyses.
Supplementary information
Additional file 1: Table S1. Allelic variation of TaGS3-7A in Chinese mini-core collection based on Kompetitive allele-specific PCR (KASP) marker designed for the SNP at position 1907 (A/G). Table S2. Allelic variation of TaGS3-7A in main wheat production areas (Hebei, Shandong, Shaanxi, Henan and Sichuan) in China based on Kompetitive allele-specific PCR (KASP) marker designed for the SNP at position 1907 (A/G).
Acknowledgments
We gratefully acknowledge Prof. Xueyong Zhang for kindly providing the mini-core germplasm collection.
Abbreviations
- ALA
Alanine
- bp
Base pair
- GS3
Grain shape 3
- KASP
Kompetitive Allele-Specific PCR
- MEGA
Molecular Evolutionary Genetic Analysis
- PCR
Polymerase Chain Reaction
- QTL
Quantitative trait locus
- SNP
Single Nucleotide Polymorphism
- THR
Threonine
- UTR
Untranslated region
Authors’ contributions
ZL, TC, JY and QW conceived the study. JY designed the experiments. JY, YZ, WH and YC performed gene cloning, SNP mining and KASP marker design. YZ, HZ and XW conducted the phylogenetic analysis. GG performed the allele frequency analysis. JY and ZL wrote the manuscript. All authors read and approved the manuscript.
Funding
This research was supported by the Special Fund for Henan Agricultural Research System (S2010–01-G03) and this funding played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and additional files). The kernel traits of mini-core collection (MCC) germplasms are derived from the following public publication: Ma L, Li T, Hao C, Wang Y, Chen X, Zhang X: TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol J 2016, 14(5):1269–1280. 10.1111/pbi.12492.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jian Yang, Email: Yangjian_1988@outlook.com.
Yanjie Zhou, Email: Zhouyj199203@163.com.
Yu’e Zhang, Email: 1225538150@qq.com.
Weiguo Hu, Email: hnhuweiguo@163.com.
Qiuhong Wu, Email: qhwu@genetics.ac.cn.
Yongxing Chen, Email: yxchen@genetics.ac.cn.
Xicheng Wang, Email: Wxicheng@163.com.
Guanghao Guo, Email: guanghaoguo@163.com.
Zhiyong Liu, Email: zyliu@genetics.ac.cn.
Tingjie Cao, Email: caotingjie893@163.com.
Hong Zhao, Email: xmszhhong@126.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12863-019-0800-6.
References
- 1.Lopes M. S., Reynolds M. P., Manes Y., Singh R. P., Crossa J., Braun H. J. Genetic Yield Gains and Changes in Associated Traits of CIMMYT Spring Bread Wheat in a “Historic” Set Representing 30 Years of Breeding. Crop Science. 2012;52(3):1123. doi: 10.2135/cropsci2011.09.0467. [DOI] [Google Scholar]
- 2.Ma Lin, Li Tian, Hao Chenyang, Wang Yuquan, Chen Xinhong, Zhang Xueyong. TaGS5-3A,a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnology Journal. 2015;14(5):1269–1280. doi: 10.1111/pbi.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.SLAFER G A. Genetic basis of yield as viewed from a crop physiologist's perspective. Annals of Applied Biology. 2003;142(2):117–128. doi: 10.1111/j.1744-7348.2003.tb00237.x. [DOI] [Google Scholar]
- 4.Zuo J, Li J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet. 2014;48:99–118. doi: 10.1146/annurev-genet-120213-092138. [DOI] [PubMed] [Google Scholar]
- 5.Li Na, Xu Ran, Li Yunhai. Molecular Networks of Seed Size Control in Plants. Annual Review of Plant Biology. 2019;70(1):435–463. doi: 10.1146/annurev-arplant-050718-095851. [DOI] [PubMed] [Google Scholar]
- 6.Fan Chuchuan, Xing Yongzhong, Mao Hailiang, Lu Tingting, Han Bin, Xu Caiguo, Li Xianghua, Zhang Qifa. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theoretical and Applied Genetics. 2006;112(6):1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 7.Fan Chuchuan, Yu Sibin, Wang Chongrong, Xing Yongzhong. A causal C–A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theoretical and Applied Genetics. 2008;118(3):465–472. doi: 10.1007/s00122-008-0913-1. [DOI] [PubMed] [Google Scholar]
- 8.Xia Duo, Zhou Hao, Liu Rongjia, Dan Wenhan, Li Pingbo, Wu Bian, Chen Junxiao, Wang Lingqiang, Gao Guanjun, Zhang Qinglu, He Yuqing. GL3.3, a Novel QTL Encoding a GSK3/SHAGGY-like Kinase, Epistatically Interacts with GS3 to Produce Extra-long Grains in Rice. Molecular Plant. 2018;11(5):754–756. doi: 10.1016/j.molp.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Li Yibo, Fan Chuchuan, Xing Yongzhong, Jiang Yunhe, Luo Lijun, Sun Liang, Shao Di, Xu Chunjue, Li Xianghua, Xiao Jinghua, He Yuqing, Zhang Qifa. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nature Genetics. 2011;43(12):1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 10.Takano-Kai Noriko, Jiang Hui, Kubo Takahiko, Sweeney Megan, Matsumoto Takashi, Kanamori Hiroyuki, Padhukasahasram Badri, Bustamante Carlos, Yoshimura Atsushi, Doi Kazuyuki, McCouch Susan. Evolutionary History ofGS3, a Gene Conferring Grain Length in Rice. Genetics. 2009;182(4):1323–1334. doi: 10.1534/genetics.109.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng Jianfeng, Gu Suhai, Wan Xiangyuan, Gao He, Guo Tao, Su Ning, Lei Cailin, Zhang Xin, Cheng Zhijun, Guo Xiuping, Wang Jiulin, Jiang Ling, Zhai Huqu, Wan Jianmin. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Research. 2008;18(12):1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Chen J, Zheng X, Wu F, Lin Q, Heng Y, Tian P, Cheng Z, Yu X, Zhou K, et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants. 2017;3:17043. doi: 10.1038/nplants.2017.43. [DOI] [PubMed] [Google Scholar]
- 13.Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y, et al. OsSPL13 controls grain size in cultivated rice. Nat Genet. 2016;48:447. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- 14.Song XJ, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc Natl Acad Sci. 2015;112(1):76–81. doi: 10.1073/pnas.1421127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Xian-Jun, Huang Wei, Shi Min, Zhu Mei-Zhen, Lin Hong-Xuan. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genetics. 2007;39(5):623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 16.Xia Tian, Li Na, Dumenil Jack, Li Jie, Kamenski Andrei, Bevan Michael W., Gao Fan, Li Yunhai. The Ubiquitin Receptor DA1 Interacts with the E3 Ubiquitin Ligase DA2 to Regulate Seed and Organ Size in Arabidopsis. The Plant Cell. 2013;25(9):3347–3359. doi: 10.1105/tpc.113.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia D, Zhou H, Liu R, Dan W, Li P, Wu B, Chen J, Wang L, Gao G, Zhang Q, et al. GL3.3, a Novel QTL Encoding a GSK3/SHAGGY-like Kinase, Epistatically Interacts with GS3 to Form Extra-long Grains in Rice. Mol Plant. 2018;11(5):754–56. [DOI] [PubMed]
- 18.Qi Peng, Lin You-Shun, Song Xian-Jun, Shen Jin-Bo, Huang Wei, Shan Jun-Xiang, Zhu Mei-Zhen, Jiang Liwen, Gao Ji-Ping, Lin Hong-Xuan. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Research. 2012;22(12):1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Xiuying, Zhang Jia-Qi, Zhang Xiaojun, Zhou Jun, Jiang Zhisheng, Huang Peng, Tang Zhengbin, Bao Yongmei, Cheng Jinping, Tang Haijuan, Zhang Wenhua, Zhang Hongsheng, Huang Ji. Rice qGL3/OsPPKL1 Functions with the GSK3/SHAGGY-Like Kinase OsGSK3 to Modulate Brassinosteroid Signaling. The Plant Cell. 2019;31(5):1077–1093. doi: 10.1105/tpc.18.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Wang J., Huang J., Lan H., Wang C., Yin C., Wu Y., Tang H., Qian Q., Li J., Zhang H. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proceedings of the National Academy of Sciences. 2012;109(52):21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Xianzhong, Qian Qian, Liu Zhengbin, Sun Hongying, He Shuyuan, Luo Da, Xia Guangmin, Chu Chengcai, Li Jiayang, Fu Xiangdong. Natural variation at the DEP1 locus enhances grain yield in rice. Nature Genetics. 2009;41(4):494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- 22.Appels REK, Feuillet C. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Sci. 2018;361(6403):eaar7191. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 23.Brenchley R, Spannagl M, Pfeifer M, Barker GLA, D’Amore R, Allen AM, McKenzie N, Kramer M, Kerhornou A, Bolser D, et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nat. 2012;491:705. doi: 10.1038/nature11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurata Nori, Moore Graham, Nagamura Yoshiaki, Foote Tracie, Yano Masahiro, Minobe Yuzo, Gale Mike. Conservation of Genome Structure Between Rice and Wheat. Bio/Technology. 1994;12(3):276–278. doi: 10.1038/nbt0394-276. [DOI] [Google Scholar]
- 25.Su Zhenqi, Hao Chenyang, Wang Lanfen, Dong Yuchen, Zhang Xueyong. Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2010;122(1):211–223. doi: 10.1007/s00122-010-1437-z. [DOI] [PubMed] [Google Scholar]
- 26.Qin L, Hao C, Hou J, Wang Y, Li T, Wang L, Ma Z, Zhang X. Homologous haplotypes, expression, genetic effects and geographic distribution of the wheat yield gene TaGW2. BMC Plant Biol. 2014;14(1):107. doi: 10.1186/1471-2229-14-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Yi, Li Da, Zhang Dingbo, Zhao Xiaoge, Cao Xuemin, Dong Lingli, Liu Jinxing, Chen Kunling, Zhang Huawei, Gao Caixia, Wang Daowen. Analysis of the functions ofTaGW2homoeologs in wheat grain weight and protein content traits. The Plant Journal. 2018;94(5):857–866. doi: 10.1111/tpj.13903. [DOI] [PubMed] [Google Scholar]
- 28.Sestili Francesco, Pagliarello Riccardo, Zega Alessandra, Saletti Rosaria, Pucci Anna, Botticella Ermelinda, Masci Stefania, Tundo Silvio, Moscetti Ilaria, Foti Salvatore, Lafiandra Domenico. Enhancing grain size in durum wheat using RNAi to knockdown GW2 genes. Theoretical and Applied Genetics. 2018;132(2):419–429. doi: 10.1007/s00122-018-3229-9. [DOI] [PubMed] [Google Scholar]
- 29.Hong Yantao, Chen Longfei, Du Li-pu, Su Zhenqi, Wang Jinfen, Ye Xingguo, Qi Lin, Zhang Zengyan. Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Functional & Integrative Genomics. 2014;14(2):341–349. doi: 10.1007/s10142-014-0380-5. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Zhang X, Chen F, Cui D. A single-nucleotide polymorphism of TaGS5 gene revealed its association with kernel weight in Chinese bread wheat. Front Plant Sci. 2015;6:1166. doi: 10.3389/fpls.2015.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanif M, Gao F, Liu J, Wen W, Zhang Y, Rasheed A, Xia X, He Z, Cao S. TaTGW6-A1, an ortholog of rice TGW6, is associated with grain weight and yield in bread wheat. Mol Breed. 2015;36(1):1. doi: 10.1007/s11032-015-0425-z. [DOI] [Google Scholar]
- 32.Hu M-J, Zhang H-P, Cao J-J, Zhu X-F, Wang S-X, Jiang H, Wu ZY, Lu J, Chang C, Sun G-L, et al. Characterization of an IAA-glucose hydrolase gene TaTGW6 associated with grain weight in common wheat (Triticum aestivum L.) Mol Breeding. 2016;36(3):25. doi: 10.1007/s11032-016-0449-z. [DOI] [Google Scholar]
- 33.Wang W, Pan Q, Tian B, He F, Chen Y, Bai G, Akhunova A, Trick HN, Akhunov E. Gene editing of the wheat homologs of TONNEAU1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J. 2019. [DOI] [PMC free article] [PubMed]
- 34.Ma Jian, Ding Puyang, Qin Peng, Liu Ya-Xi, Xie Quan, Chen Guangdeng, Li Wei, Jiang Qiantao, Chen Guoyue, Lan Xiu-Jin, Wei Yu-Ming, Liu Chunji, Zheng You-Liang. Structure and expression of the TaGW7 in bread wheat (Triticum aestivum L.) Plant Growth Regulation. 2017;82(2):281–291. doi: 10.1007/s10725-017-0258-3. [DOI] [Google Scholar]
- 35.Yang Jian, Zhou Yanjie, Wu Qiuhong, Chen Yongxing, Zhang Panpan, Zhang Yu’e, Hu Weiguo, Wang Xicheng, Zhao Hong, Dong Lingli, Han Jun, Liu Zhiyong, Cao Tingjie. Molecular characterization of a novel TaGL3-5A allele and its association with grain length in wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2019;132(6):1799–1814. doi: 10.1007/s00122-019-03316-1. [DOI] [PubMed] [Google Scholar]
- 36.Zhai H, Feng Z, Du X, Song Y, Liu X, Qi Z, Song L, Li J, Li L, Peng H, et al. A novel allele of TaGW2-A1 is located in a finely mapped QTL that increases grain weight but decreases grain number in wheat (Triticum aestivum L.) Theor Appl Genet. 2018;131(3):539–553. doi: 10.1007/s00122-017-3017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan X, Zhao L, Ren Y, Dong Z, Cui D, Chen F. Genome-wide association study revealed that the TaGW8 gene was associated with kernel size in Chinese bread wheat. Sci Rep. 2019;9(1):2702. doi: 10.1038/s41598-019-38570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Yingjun, Liu Jindong, Xia Xianchun, He Zhonghu. TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Molecular Breeding. 2014;34(3):1097–1107. doi: 10.1007/s11032-014-0102-7. [DOI] [Google Scholar]
- 39.Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, Crossman CC, Deal KR, et al. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics. 2010;11(1):702. doi: 10.1186/1471-2164-11-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Shichen, Wong Debbie, Forrest Kerrie, Allen Alexandra, Chao Shiaoman, Huang Bevan E., Maccaferri Marco, Salvi Silvio, Milner Sara G., Cattivelli Luigi, Mastrangelo Anna M., Whan Alex, Stephen Stuart, Barker Gary, Wieseke Ralf, Plieske Joerg, Lillemo Morten, Mather Diane, Appels Rudi, Dolferus Rudy, Brown-Guedira Gina, Korol Abraham, Akhunova Alina R., Feuillet Catherine, Salse Jerome, Morgante Michele, Pozniak Curtis, Luo Ming-Cheng, Dvorak Jan, Morell Matthew, Dubcovsky Jorge, Ganal Martin, Tuberosa Roberto, Lawley Cindy, Mikoulitch Ivan, Cavanagh Colin, Edwards Keith J., Hayden Matthew, Akhunov Eduard. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnology Journal. 2014;12(6):787–796. doi: 10.1111/pbi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devos K. M., Dubcovsky J., Dvořák J., Chinoy C. N., Gale M. D. Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theoretical and Applied Genetics. 1995;91(2):282–288. doi: 10.1007/BF00220890. [DOI] [PubMed] [Google Scholar]
- 42.Berkman Paul J., Skarshewski Adam, Manoli Sahana, Lorenc Michał T., Stiller Jiri, Smits Lars, Lai Kaitao, Campbell Emma, Kubaláková Marie, Šimková Hana, Batley Jacqueline, Doležel Jaroslav, Hernandez Pilar, Edwards David. Sequencing wheat chromosome arm 7BS delimits the 7BS/4AL translocation and reveals homoeologous gene conservation. Theoretical and Applied Genetics. 2011;124(3):423–432. doi: 10.1007/s00122-011-1717-2. [DOI] [PubMed] [Google Scholar]
- 43.Hernandez Pilar, Martis Mihaela, Dorado Gabriel, Pfeifer Matthias, Gálvez Sergio, Schaaf Sebastian, Jouve Nicolás, Šimková Hana, Valárik Miroslav, Doležel Jaroslav, Mayer Klaus F.X. Next-generation sequencing and syntenic integration of flow-sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content. The Plant Journal. 2011;69(3):377–386. doi: 10.1111/j.1365-313X.2011.04808.x. [DOI] [PubMed] [Google Scholar]
- 44.Akpinar Bala Ani, Biyiklioglu Sezgi, Alptekin Burcu, Havránková Miroslava, Vrána Jan, Doležel Jaroslav, Distelfeld Assaf, Hernandez Pilar, Budak Hikmet. Chromosome-based survey sequencing reveals the genome organization of wild wheat progenitorTriticum dicoccoides. Plant Biotechnology Journal. 2018;16(12):2077–2087. doi: 10.1111/pbi.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun S, Wang L, Mao H, Shao L, Li X, Xiao J, Ouyang Y, Zhang Q. A G-protein pathway determines grain size in rice. Nat Commun. 2018;9(1):851. doi: 10.1038/s41467-018-03141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao H., Sun S., Yao J., Wang C., Yu S., Xu C., Li X., Zhang Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proceedings of the National Academy of Sciences. 2010;107(45):19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urano Daisuke, Miura Kotaro, Wu Qingyu, Iwasaki Yukimoto, Jackson David, Jones Alan M. Plant Morphology of Heterotrimeric G Protein Mutants. Plant and Cell Physiology. 2016;57(3):437–445. doi: 10.1093/pcp/pcw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubinovich Lior, Weiss David. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. The Plant Journal. 2010;64(6):1018–1027. doi: 10.1111/j.1365-313X.2010.04390.x. [DOI] [PubMed] [Google Scholar]
- 49.Kunihiro S, Saito T, Matsuda T, Inoue M, Kuramata M, Taguchi-Shiobara F, Youssefian S, Berberich T, Kusano T. Rice DEP1, encoding a highly cysteine-rich G protein gamma subunit, confers cadmium tolerance on yeast cells and plants. J Exp Bot. 2013;64(14):4517–27. [DOI] [PMC free article] [PubMed]
- 50.Gutierrez-Gonzalez JJ, Mascher M, Poland J, Muehlbauer GJ. Dense genotyping-by-sequencing linkage maps of two synthetic W7984×Opata reference populations provide insights into wheat structural diversity. Sci Rep. 2019;9(1):1793. doi: 10.1038/s41598-018-38111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Yipu, Xu Mingliang. CCT family genes in cereal crops: A current overview. The Crop Journal. 2017;5(6):449–458. doi: 10.1016/j.cj.2017.07.001. [DOI] [Google Scholar]
- 52.Sakuma Shun, Golan Guy, Guo Zifeng, Ogawa Taiichi, Tagiri Akemi, Sugimoto Kazuhiko, Bernhardt Nadine, Brassac Jonathan, Mascher Martin, Hensel Goetz, Ohnishi Shizen, Jinno Hironobu, Yamashita Yoko, Ayalon Idan, Peleg Zvi, Schnurbusch Thorsten, Komatsuda Takao. Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proceedings of the National Academy of Sciences. 2019;116(11):5182–5187. doi: 10.1073/pnas.1815465116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sajjad M, Ma X, Habibullah Khan S, Shoaib M, Song Y, Yang W, Zhang A, Liu D. TaFlo2-A1, an ortholog of rice Flo2, is associated with thousand grain weight in bread wheat (Triticum aestivum L.) BMC Plant Biol. 2017;17(1):164. doi: 10.1186/s12870-017-1114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Yingjun, Xia Xianchun, He Zhonghu. The seed dormancy allele TaSdr-A1a associated with pre-harvest sprouting tolerance is mainly present in Chinese wheat landraces. Theoretical and Applied Genetics. 2016;130(1):81–89. doi: 10.1007/s00122-016-2793-0. [DOI] [PubMed] [Google Scholar]
- 55.Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar Sudhir, Stecher Glen, Tamura Koichiro. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rasheed A, Wen W, Gao F, Zhai S, Jin H, Liu J, Guo Q, Zhang Y, Dreisigacker S, Xia X, et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor Appl Genet. 2016;129(10):1843–1860. doi: 10.1007/s00122-016-2743-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Allelic variation of TaGS3-7A in Chinese mini-core collection based on Kompetitive allele-specific PCR (KASP) marker designed for the SNP at position 1907 (A/G). Table S2. Allelic variation of TaGS3-7A in main wheat production areas (Hebei, Shandong, Shaanxi, Henan and Sichuan) in China based on Kompetitive allele-specific PCR (KASP) marker designed for the SNP at position 1907 (A/G).
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and additional files). The kernel traits of mini-core collection (MCC) germplasms are derived from the following public publication: Ma L, Li T, Hao C, Wang Y, Chen X, Zhang X: TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol J 2016, 14(5):1269–1280. 10.1111/pbi.12492.