Abstract
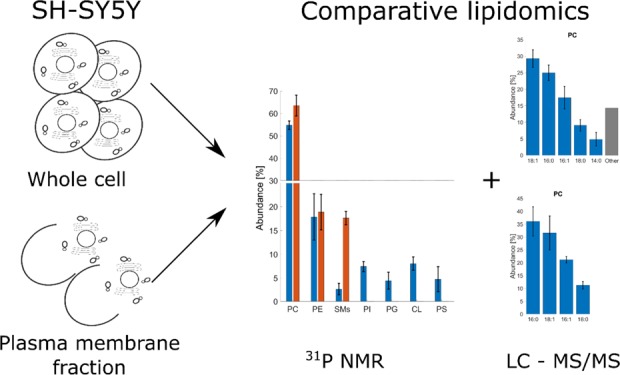
Global lipid analysis still lags behind proteomics with respect to the availability of databases, experimental protocols, and specialized software. Determining the lipidome of cellular model systems in common use is of particular importance, especially when research questions involve lipids directly. In Parkinson’s disease research, there is a growing awareness for the role of the biological membrane, where individual lipids may contribute to provoking α-synuclein oligomerisation and fibrillation. We present an analysis of the whole cell and plasma membrane lipid isolates of a neuroblastoma cell line, SH-SY5Y, a commonly used model system for research on this and other neurodegenerative diseases. We have used two complementary lipidomics methods. The relative quantities of PC, PE, SMs, CL, PI, PG, and PS were determined by 31P NMR. Fatty acid chain composition and their relative abundances within each phospholipid group were evaluated by liquid chromatography–tandem mass spectrometry. For this part of the analysis, we have developed and made available a set of Matlab scripts, LipMat. Our approach allowed us to observe several deviations of lipid abundances when compared to published reports regarding phospholipid analysis of cell cultures or brain matter. The most striking was the high abundance of PC (54.7 ± 1.9%) and low abundance of PE (17.8 ± 4.8%) and SMs (2.7 ± 1.2%). In addition, the observed abundance of PS was smaller than expected (4.7 ± 2.7%), similar to the observed abundance of PG (4.5 ± 1.8%). The observed fatty acid chain distribution was similar to the whole brain content with some notable differences: a higher abundance of 16:1 PC FA (17.4 ± 3.4% in PC whole cell content), lower abundance of 22:6 PE FA (15.9 ± 2.2% in plasma membrane fraction), and a complete lack of 22:6 PS FA.
Introduction
Lipids play a critical role in many cell processes including regulation of transcription,1 protein and metabolites distribution,2 energy metabolism,3 cell apoptosis induction,4 and protein folding and misfolding.5,6 However, in many cases, we still lack basic knowledge of the cell lipid composition, and how it varies as a function of the subcellular compartment or cell state. This deficiency is in part caused by the great diversity displayed by lipid species in cells. The combination of headgroups and fatty acid chains (FA), each with different lengths and levels of saturation, give rise to thousands of possible lipid species in the phospholipid class alone. The analytical problem is exacerbated when accurate quantitative data is also sought.7,8
The lipidomic gap in knowledge is evident for the neuroblastoma cell line SH-SY5Y. This cell line is of human origin, catecholaminergic, easy to maintain, and reported to be differentiable into a neuron, like a phenotype.9 These properties have led SH-SY5Y to be the cellular model of choice when studying neurodegenerative diseases like Alzheimer’s disease, amyotrophic lateral sclerosis, and, in particular, Parkinson’s disease (PD).9,10 Lipids are implicated in all these diseases,11,12 and for PD, this connection is particularly strong.13 α-Synuclein, a key protein of PD pathology, is thought to be a regulator of vesicle recycling in presynaptic terminals through reversibly contacting the lipid membrane in a carefully timed fashion.14 Moreover, its misfolding is influenced by particular lipids and the physical state of the membrane, which in turn affects its rate of oligomerization.15 Given the diverse and robust evidence for the role of lipids in α-Synuclein pathology, it is then disconcerting to realize that lipid the composition of one of the vital cell models used in PD research, SH-SY5Y, is still not yet known in detail.
We, therefore, focus on the determination of the phospholipid content of the whole cell and plasma membrane (PM)-enriched SH-SY5Y cell isolates. First, we have determined the abundance of individual phospholipid headgroups by 31P NMR using approaches similar to those developed by Bosco et al.16 Then, we have determined the FA composition of individual lipid headgroups by LC–MS/MS with an iterative exclusion technique.7 By this method, we have generated large amounts of data, which is required to be processed on a batch-to-batch basis. To achieve this in an expedient and informative way, we have automated the approach for liquid chromatography–tandem mass spectrometry (LC–MS/MS) by developing a script in Matlab. This script, designated as LipMat, is able to (i) build libraries of predicted lipid fragmentations based on information collected from the literature and based on user input, (ii) find possible lipid species by comparing intact m/z values and by scoring MS2 fragments, and (iii) do semiquantitative FA abundance analysis from MS1 spectra chromatograms.
Results and Discussion
It has been shown that the phospholipid composition in the eukaryotic cell is highly variable. It differs between organisms and cell types or between healthy and diseased states of a cell.17−21 The current state of knowledge regarding the lipidome of eukaryotic cells starts to be insufficient when specific cell types are considered and is often completely absent in many cases. Recent reports concerning the importance of phospholipids in the development and progress of PD22−25 has led to our current goal: to perform a mutually supportive NMR and LC–MS/MS phospholipid analysis of a cell model system commonly used in PD research, the SH-SY5Y neuroblastoma cell line.
The phospholipid content of the whole cell was isolated using a method we have previously established.26 For PM isolation, we have employed a modified method of sucrose gradient separation27,28 (Figure S1). Upon assessing the purity of the PM fraction using Western blot, we observed contamination of calnexin (a marker for the endoplasmic reticulum29,30). However, there was a much lower presence of β-actin (cytoskeletal marker31) and almost no nucleophosmin (nuclear marker;32Figure S2). We therefore concluded that our approach provided us with PM-enriched samples.
We then used 31P NMR to analyze the phospholipid content of whole cell SH-SY5Y lipid isolates (Figure 1). To ensure consistency of measurement, we have used the CUBO solvent (named after the primary authors of the original work, Culeddu and Bosco) for all solution NMR experiments. The CUBO solvent is a ternary mixture of dimethylformamide, trimethylamine, and guanidine hydrochloride, and it has a well-documented good chemical shift reproducibility and chemical stability for work with phospholipids.33−36
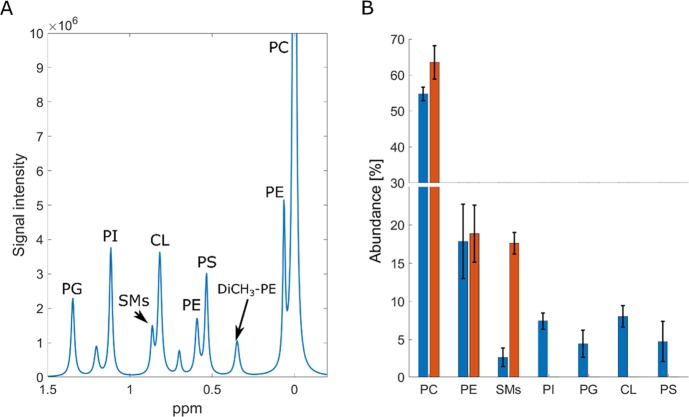
Figure 1.
31P NMR phospholipid analysis of SH-SY5Y whole cell and plasma membrane lipid isolates. (A) Example of lipid assignment after deconvolution of spectra. The highest abundant lipid, PC, is used for axis calibration, and it is set to 0 ppm. (B) Abundance of identified phospholipids in the whole cell isolates (blue) and PM isolate (red) of SH-SY5Y. For plasma PM samples, we have only observed the three most abundant lipid species (PC, PE, and SMs). This was caused by the low sample mass used for analysis.
In accordance with other studies, the most abundant phospholipid was PC. However, the abundance was higher than that reported for human erythrocytes (∼30%)20 or yeast (∼15%).37 With 54.7 ± 1.9% of total abundance, PC is by far the most common phospholipid in SH-SY5Y. This is also in agreement with lipidomics analysis of human plasma where PC showed up as the most abundant phospholipid. However, this study did not further explore the FA profiles of lipid species quantified.8 We have observed some difference in the fraction of PE and PS when we compare the SH-SY5Y cell to brain matter. For whole brain extracts, the reported content of PE and PS is ∼30% of brain phospholipids,38−42 yet for SH-SY5Y, the amount of PE and PS is 17.8 ± 4.8 and 4.7 ± 2.6%, respectively. Another interesting result is the relatively low abundance of SMs (2.7 ± 1.2%) when compared to CL (8.0 ± 1.4%). The only source of CL in the eukaryotic cell is the mitochondria, while almost all of the SMs is present only in the PM. This suggests that SH-SY5Y is especially energetically active. Moreover, based on the amount of SMs, SH-SY5Y appears to have a lower PM surface than differentiated neurons in vivo, as the only source of SMs in cells is PM. Both of these observations reflect the carcinogenic properties of the nondifferentiated SH-SY5Y cell line.
We also strived to secure PM-specific quantitative data using NMR. However, after PM isolation from the SH-SY5Y cell, we did not obtain sufficient quantities of lipids for a complete 31P NMR analysis. We have observed just three of the most abundant phospholipids in the PM-enriched fraction: PC, PE, and SMs. Although not complete with respect to minor lipid species, they suggest that almost all SMs in SH-SY5Y is localized in the PM. It is also apparent and not unexpected that 31P NMR, although potentially straightforward and informative, suffers from lack of sensitivity when little material is available. In our case, working with samples containing ∼30 μg of lipids on a modern 600 MHz instrument fitted with a cryogenically cooled probe did not detect lipid species of low relative abundance. Increasing the number of scans did not seem to affect this, suggesting that the weakest resonances were not detectable at any receiver gain setting.
The detection limit of mass spectrometry is significantly lower than that of NMR,43 and we therefore proceeded with LC–MS/MS analysis of the FA chains of all phospholipids identified by 31P NMR. Prior to the main experiments, the liquid chromatography conditions were optimized to obtain the elution of phospholipid as narrow peaks. We used an iterative exclusion protocol7 to filter out abundant ions as well as background signals. Our custom-built LipMat script written in Matlab was then used for further processing (Figure 2).
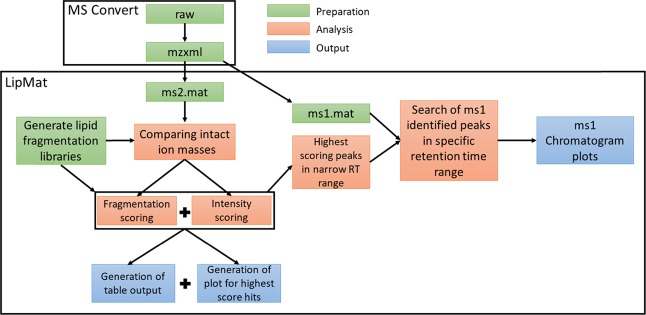
Figure 2.
Schematic of LipMat processing. For file preparation, MS convert can be used to convert the raw file into mzxml.57 LipMat can further process mzxml files by extracting MS2 and MS1 spectra. The lipid fragmentation library is generated using the user input specifying the length of FA chains and number of double bonds. The analysis starts with comparing intact ion masses with the library. In the case of a hit, the MS2 spectrum is loaded and scored based on the presence of corresponding lipid fragments and their intensity. Afterward, retention time is analyzed, and the MS1 spectrum is searched for the elution peak of assigned lipid species. Generated outputs include lipid fragmentation figures, chromatogram plots, and table output with identified lipid species and their respective scoring.
The LipMat identifies the presence of individual lipids by comparing MS2 fragmentation spectra with the literature and in silico prediction (see the Experimental Section for more information and Figure S3). The lipid species deemed robustly detected by using a scoring function (sum of eqs 1 and 2; Experimental Section) were then selected for MS1 search of the intact lipid m/z (Table S2). Because of the high overlap of m/z in different lipid species, only the area of retention time, where the lipid species was identified, was searched in this way. An additional retention offset of 1 min was added, so it would be possible to observe complete elution of a given peak. This leads to a reconstruction of chromatograms (Figure S4) for the elution of individual lipid species and to relative quantification of FA within each headgroup (Figure S5).
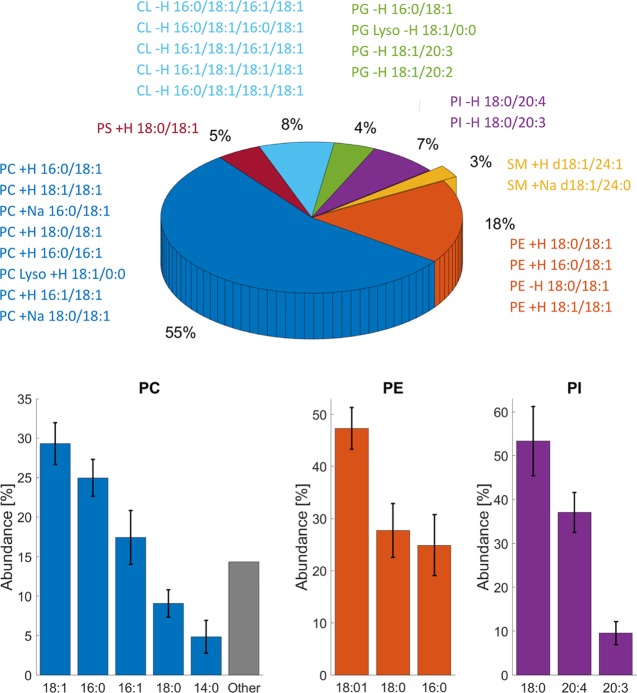
Using a combination of 31P NMR and the LC–MS/MS method described herein, we have successfully identified phospholipid abundances as well as the FA chain composition for whole cell isolates (Figure 3) and plasma membrane-enriched samples (Figure 4). The most abundant lipid, PC, was well ionized, and we had a low detection limit for positive mode MS runs. The diversity of PC FAs was low, as almost 75% of all PC lipids in the SH-SY5Y cell are composed of two of the four FA chains (18:1, 16:0, 16:1, 18:0, or 14:0). Similar observations were also made for the PM-enriched isolate where 16:0 FAs were particularly abundant. We also noted that the 16:1 PC FA chain (17.4 ± 3.4% in the whole cell sample) deviates from other reports of brain composition, where 16:1 FA accounts for fewer than 3% of the PC content.38,44,45
Figure 3.
Analysis of headgroup abundance and fatty acid distribution in the whole cell lipid isolate of SH-SY5Y. The pie plot represents phospholipid abundance of lipid headgroups as measured by 31P NMR. The text around represents lipid species identified by LC–MS/MS. Bar plots for PC, PE, and PI represent individual FA abundance within each headgroup as evaluated by the LipMat script. Only lipid species with Stotal > 30 were taken into account. For the rest of the lipid headgroups (PS, PG, SM, and CL), we were unable to identify relative FA chain abundance.
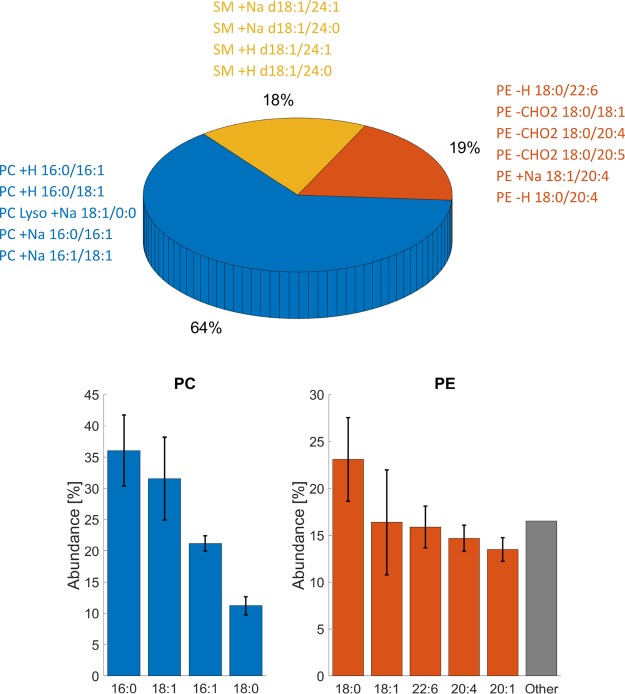
Figure 4.
Analysis of headgroup abundance and fatty acid distribution of the PM lipid fraction of SH-SY5Y. The pie plot represents phospholipid abundance of lipid headgroups as measured by 31P NMR. The text around represents lipid species identified by LC–MS/MS. Bar plots for PC and PE represent individual FA abundance within each headgroup as evaluated by the LipMat script. Only lipid species with Stotal > 30 were taken into account. For SMs, we were unable to identify relative FA chain abundance.
SMs was the second most abundant lipid detected in the positive mode. Even at lower concentrations, we could observe a diverse array of SMs hits. However, we were unable to identify the exact SMs FA acid composition because most observed SM lipid species lacked FA-specific fragments. The two lipid species that could be identified were SM d18:1/24:1 and SM d18:1/24:0, and these were both observed in whole cell and PM fraction samples.
In the whole cell samples, all the PE FA chains were 18:1, 18:0, or 16:0. However, in the PM-enriched samples, we observed a higher variability of different PE species (Figure 4) and disproportional FA lengths. One PE FA had a length of 18:0, while others were composed of long unsaturated chains (22:6, 20:4, or 20:1).
The PG lipid abundance was too low to perform FA distribution analysis. However, four FA chains were identified repeatedly within the PG lipid headgroup (16:0, 18:1, 20:3, and 20:4). Both PI and PS were much worse ionizers, and we therefore detected only a few lipid species from each headgroup repeatedly, namely, PI 18:0/20:4, PI 18:0/20:3, and PS 18:0/18:1. However, we did not identify any docosahexaenoic acid FA (22:6) in the whole cell sample regardless of the fact that this FA is reported in high abundance (>25%) of brain matter for both PS and PE.38,44,45 Yet, PE 22:6 was present in the PM sample with an abundance of 15.9 ± 2.2%. This suggests that all 22:6 PE FAs are located in the plasma membrane and that the presence of this FA was masked by other more abundant PE FAs in the whole cell samples. This underscores the importance of sample fractionation in order to provide a complete and reliable picture of the cell lipidome.
Conclusions
We have analyzed the phospholipid composition of SH-SY5Y cells by NMR and LC–MS/MS using the customizable and highly flexible LipMat script for Matlab software. Compared to the phospholipid composition of the brain, there exist notable differences, which should be taken into account when SH-SY5Y is used as a model for studying neurodegeneration or brain lipid metabolism. The most striking differences are a relatively low abundance of PE and PS, higher occurrence of 16:1 PC FA, and missing 22:6 FA for PS.
Experimental Section
Reagents and Chemicals
Lipids were purchased from Avanti Polar Lipids Inc. (Alabaster, Alamaba, US). These were all used without further purification. The following primary antibodies were used: β-actin (AC-15) and Na+/K+ ATPase α (H-3) from Santa Cruz Biotechnology (Texas, USA), nucleophosmin (FC-61991) from Thermo Fisher Scientific (Massachusetts, USA), and calnexin from Abcam (Cambridge, UK). Lipase inhibitors (FIDI, U73122, and D609) were acquired from Tocris Bioscience Ltd. (Bristol Somerset, UK). Growth medium, solvents, and fine chemicals were purchased from Sigma Aldrich (Germany) and used as described in the separate experimental sections, vide infra.
SH-SY5Y Cultivation
SH-SY5Y cells (a generous gift from Prof. Kari Fladmark) were cultured in Dulbecco’s modified Eagle’s medium with high glucose supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin in a humidified atmosphere at 310 K and 5% CO2.
Whole Cell Lipid Extraction
Whole cell lipid extraction was performed based on a modified version of a previously published method.26 An overview of whole cell lipid sample preparation and plasma membrane isolation is provided in Supplementary Figure S1. Briefly, SH-SY5Y cells were harvested into phosphate-buffered saline (PBS) and pelleted by centrifugation (900g, 5 min, and 277 K). The supernatant was removed, and the pellet resuspended using 1 mL of PBS. To this, 330 μL of guanidine chloride and thiourea mix (3:0.75 molar ratio) was added, and the final mixture was vortexed and stored at 193 K until further use. The frozen material was freeze-dried and stored at 253 K until lipid extraction. For the extraction, the powdered material was resuspended in an organic extraction mixture (3:1 dichloromethane/methanol with 0.5 mg/mL triethylammonium chloride (TEAC)). Milli-Q water was added (1:1 by volume), and the mixture was transferred into a separating funnel where it was diluted with methanol until a monophasic solution was formed. The solution was agitated briefly and made biphasic again by the addition of more dichloromethane. The denser organic phase was collected using a separating funnel. The water fraction was washed three more times with dichloromethane. Collected organic fractions were pooled and concentrated in vacuo and stored in the form of a dry lipid layer under nitrogen in the dark at 253 K until analysis.
Plasma Membrane-Enriched Lipid Sample Preparation
The protocol used in this study was modified from two existing protocols.27,28 Cells from 5 × 150 mm plates were harvested by trypsinization, pelleted and washed twice with ice-cold PBS, and centrifuged (900g, 4 min, and 277 K). Hypotonic buffer (2 mL, 10 mM Tris, and pH 7.8) with FUD (0.1 mL of a concentrated stock of F191 (200 μM), U73122 (1 mM), and D609 (1.5 mM) in a solution of DMSO/water (1:9)) was layered over the cell pellet and removed immediately. The cell pellet was then resuspended using 6 mL of hypotonic buffer and rested on ice for 4 min. After the incubation, 15 mL of H2O was mixed into the suspension. The resulting cell suspension was subsequently incubated for 3 min on ice before the cells were passed through a hypodermic syringe 12 times and afterward centrifuged (1000g, 1 min, and 277 K). The pellet was washed with washing buffer (10 mM Tris, pH 7.5, 2.5 mM MgCl2, 2.5 mM CaCl2, 10 mM NaCl, and FUD) and centrifuged (1000g, 1 min, and 277 K). The supernatants from the two centrifugation steps were pooled and diluted with hypotonic buffer. The plasma membrane was isolated by loading the homogenate onto a sucrose gradient consisting of 30% (w/v) and 45% (w/v) sucrose solutions and centrifuged (3270g, 30 min, and 277 K). The PM-enriched fraction was collected and diluted with hypotonic buffer and centrifuged (3270g, 20 min, and 277 K). The pellet was resuspended in 500 μL of GTCU (6 M guanidinium chloride/1.5 M thiourea). The resuspension was freeze-dried within the same day, and the resulting material was then stored at 253 K until lipid extraction could be performed using the same approach as for the whole cell samples.
Western Blot Analysis
Samples collected from PM isolations were resuspended in 1X RIPA buffer (150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, and pH 8.0), sonicated, and centrifuged (17,000g, 5 min, and 277 K). The total protein concentration was measured using a standard BCA assay,46 and the protein composition of the samples was analyzed using 10% SDS-polyacrylamide gels and subsequently transferred to a nitrocellulose membrane. The membrane was blocked for 1 h with 7% dry milk in PBS with Tween-20 (0.05%, v/v) and incubated overnight at 277 K with primary antibodies to Na+/K+ ATPase (1:5000), nucleophosmin (1:10000), calnexin (1:1000), and β-actin (1:1000). The primary antibodies were detected using HRP-conjugated secondary antibodies (1:10000, 1 h, and 293 K). Protein bands were visualized using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher, USA), and images were collected with a Molecular Imager ChemiDoc XRS + imaging system and the ImageLab software version 3.0 (Bio-Rad, USA).
Solution-Phase 31P NMR
Dried lipid films were dissolved in the Culeddu-Bosco “CUBO” solvent system.16 Data acquisition was similar to our previously published work26 using a Bruker BioSpin NEO600 spectrometer equipped with a cryogenic probe operating at 300 K for all data. 31P spectra were acquired at 242.93 MHz using inverse-gated proton decoupling with 3072 scans per sample and a spectral width of 54.16 ppm. An overall recovery delay of 8 s was used, a setting that ensured full relaxation of the 31P nuclei between scans. Data were processed using an exponential line broadening window function of 1.0 Hz prior to Fourier transform followed by manual phase correction and automatic baseline correction. The spectra were calibrated by setting the most abundant phospholipid signal in the sample, phosphatidylcholine (PC), to 0 ppm. All peaks were then deconvoluted. The peak assignment to individual phospholipids was done according to a previously published work.26,33,47 The analyses were done using TopSpin 4.0.1. A total of four independent whole cell sample and three plasma membrane sample 31P spectra were collected.
LC–MS/MS Experimental Setup
Accurate mass LC–MS and MS/MS was performed on a Thermo Q-Exactive mass spectrometer and a Dionex Ultimate 3000 UPLC (Thermo Fisher, USA). Dry lipid mixtures from the whole cell and PM-enriched extractions were dissolved in a mixture of water, dichloromethane, and acetonitrile (2:2:1) with 10 mM ammonium acetate. The analytes were then separated on a UPLC C18 column (1.7 μm particle size, Waters, USA) at 318 K at a rate of 0.4 mL/min; an injection volume of 20 μL was used. Mobile phase A consisted of 40% acetonitrile and 60% water, mobile phase B consisted of 10% acetonitrile and 90% isopropanol, and both phases were supplemented with 10 mM ammonium acetate. Lipid separation was then achieved using a multistep gradient from 40 to 100% of solvent B in 17 min (full gradient provided in Table S1).
Ions were monitored both in positive and negative Full MS/data-dependent Top 5 mode. The full MS scan range was 300–2000 m/z with a resolution of 140,000 at m/z = 200. Data-dependent peaks were fragmented using a normalized collision energy of 24, and the resulting MS2 spectrum was collected with a resolution of 17,500 at m/z = 200, an isolation window of 0.4 m/z, and the dynamic exclusion parameter set to “auto”. Each run was repeated three times for each mode using dynamic exclusion of previously analyzed ions.7 Three biologically independent samples of SH-SY5Y whole cell samples were analyzed three times for each mode (positive or negative), that is, 18 LC–MS/MS runs in total. The PM has been analyzed with just two iterative exclusions, a total of 12 LC–MS/MS runs.
Lipid Analysis by LipMat Scripts
For MS/MS analysis, we have built highly flexible customizable scripts using Matlab 2017b software. LipMat script was tested on Avanti Lipid MAPS standards: 12:0-13:0 PC, 17:0-14:1 PC, 17:1 LPC, 12:0-13:0 PE, 17:0-14:1 PE, 17:0-20:4 PE, 12:0-13:0 PG, 12:0-13:0 PI, 17:0-14:1 PI, 17:0-20:4 PI, 12:0-13:0 PS, 17:0-14:1 PS, 17:0-20:4 PS, and cardiolipin mix 1. The LipMat script and fragmentation patterns of Avanti standards are freely available on GitHub: https://github.com/MarJakubec/LipMat.
Lipid Fragmentation Libraries
We have searched the literature for experimental48−54 and theoretical55,56 lipid fragmentation patterns of the phospholipid groups identified by 31P NMR. These were phosphatidylcholine (PC), phosphoethanolamine (PE), phosphatidylinositol (PI), phosphoglycerol (PG), phosphatidylserine (PS), sphingomyelin (SMs), and cardiolipin (CL). For positive ion mode and all mentioned lipids (M), except CL, we have included adducts of alkali metals [M + Met]+, where Met = Na+, K+, or Li+, or nonmetal adducts [M + Non]+ , where Non = H+, NH4+, or TEAC. For negative mode, we have included [M + Neg]– adducts, where Neg is loss of H+, and gain of Cl–, CHCOO–, and CH3COO–. CLs have a double negative charge, which leads to the formation of different adducts: [CL + H+]+, [CL + Met]+, [CL – H+ + 2Met]+, and [CL – 2H+ + 3Met]+ for the positive mode and [CL – H+]–, [CL – 2H+ + Met]–, and [CL – 2H+]2– for the negative mode.
Peak and Intensity Scoring
The precursor m/z values are compared with the intact masses of adducts from the libraries. For each instance where a hit is found, the script proceeds with a peak (Speak) and intensity (SIntesity) scoring for each hit. The total score (Stotal) is the sum of the negative log of Speak and the negative log of Sintensity. The peak score (Speak) provides the largest sum of the total hit score, and it composes of a hypergeometric distribution (eq 1) used previously by other authors to describe mass spectrometric peak distributions.55 This function reflects the probability that any overlap of theoretical and experimental MS2 peaks is random.
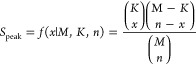 |
1 |
where x is the number of hits, K is the number of theoretical MS2 fragments, n is the number of observed MS2 fragments, and M is the size of the scanning range divided into bins, where the number of bins is the size of the scanning range divided by the ppm error. The ppm error is set by the user, typically to 5 ppm.
The second component of the final score is the intensity score (eq 2). This is a minor part of the final score and reflects the intensity component of identified peaks.55 Experimental MS2 data and the number of identified peaks are used as the input for this value. The same number of peaks is randomly selected from the spectra, and if the total intensity of the resulting random spectra is lower than the total intensity of identified peaks, then one point is added to the intensity score ratio. The process is repeated for all possible combinations of randomly generated spectra, and the ratio is then used to calculate Sintensity. However, to save computing power, if there are more than six identified peaks in the spectra, then only 500 random combinations of peaks are selected.
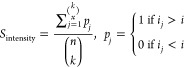 |
2 |
where x is the number of hits, k is the number of theoretical MS2 fragments, n is the number of observed MS2 fragments, and i is the intensity of the identified fragments
Construction of Lipid-Specific Elution Profiles
After the identification of possible lipid compositions, the LipMat scripts will start to scan the MS1 spectrum. First, it will filter out nondistinguishable lipid species. These are lipid species with the same m/z ± a user-determined defined ppm error, which are eluted in the same retention time ± user determined retention time offset. These lipid species are written to the LipMat output file and subject to manual checking by the expert user. From these nondistinguishable species, only the ones with the highest score are processed further.
The area of each defined and distinguishable lipid species in the MS1 spectra is then calculated. The abundance of each unique FA is calculated as a sum of the area for each FA occurrence. The MS1 chromatogram for up to five of the most abundant lipid species associated with each headgroup category and bar plots for up to five of the most abundant FA are plotted and written to the LipMat output files.
Acknowledgments
The authors gratefully acknowledge the Norwegian Research Council grant no. NFR240063 for providing infrastructure support through the Norwegian NMR Platform (NNP; grant no. 226244/F50), Bergen Research Foundation (BFS-NMR-1), and Sparebankstiftinga Sogn og Fjordane (509-42/16). The authors would like to thank the staff at the NNP Bergen node for facilitating the NMR experimental work, Kristoffer Dyrkorn for helpful discussion, and Kari Fladmark for providing the SH-SY5Y cell line.
Glossary
Abbreviations
- PC
phosphatidylcholine
- PE
phosphoethanolamine
- PI
phosphatidylinositol
- PG
phosphoglycerol
- PM
plasma membrane
- PS
phosphatidylserine
- SMs
sphingomyelin
- TEAC
triethylammonium chloride
- CL
cardiolipin
- FA
fatty acid chain
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b03463.
Lipid isolation methodology, purity of plasma membrane fraction, MS/MS fragmentation and identification, MS1 chromatogram reconstruction, an example of FA abundance of PC lipids, LC multistep gradient settings, and an example of LipMat output results (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Nakamura M. T.; Cheon Y.; Li Y.; Nara T. Y. Mechanisms of Regulation of Gene Expression by Fatty Acids. Lipids 2004, 39, 1077–1083. 10.1007/s11745-004-1333-0. [DOI] [PubMed] [Google Scholar]
- Wenk M. R.; De Camilli P. Protein-Lipid Interactions and Phosphoinositide Metabolism in Membrane Traffic: Insights from Vesicle Recycling in Nerve Terminals. Proc. Natl. Acad. Sci. 2004, 101, 8262–8269. 10.1073/pnas.0401874101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A.; Boveris A. The Mitochondrial Energy Transduction System and the Aging Process. Am. J. Physiol.: Cell Physiol. 2007, 292, C670–C686. 10.1152/ajpcell.00213.2006. [DOI] [PubMed] [Google Scholar]
- Green D. R.; Kroemer G. The Pathophysiology of Mitochondrial Cell Death. Science 2004, 305, 626–629. 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- Brown M. F. Curvature Forces in Membrane Lipid-Protein Interactions. Biochemistry 2012, 9782. 10.1021/bi301332v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aisenbrey C.; Borowik T.; Byström R.; Bokvist M.; Lindström F.; Misiak H.; Sani M. A.; Gröbner G.. How Is Protein Aggregation in Amyloidogenic Diseases Modulated by Biological Membranes? In European Biophysics Journal; Springer-Verlag, 2008; Vol. 37, pp 247–255, 10.1007/s00249-007-0237-0. [DOI] [PubMed] [Google Scholar]
- Koelmel J. P.; Kroeger N. M.; Gill E. L.; Ulmer C. Z.; Bowden J. A.; Patterson R. E.; Yost R. A.; Garrett T. J. Expanding Lipidome Coverage Using LC-MS/MS Data-Dependent Acquisition with Automated Exclusion List Generation. J. Am. Soc. Mass Spectrom. 2017, 28, 908–917. 10.1007/s13361-017-1608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampler E.; Criscuolo A.; Zeller M.; El Abiead Y.; Schoeny H.; Hermann G.; Sokol E.; Cook K.; Peake D. A.; Delanghe B.; et al. A Novel Lipidomics Workflow for Improved Human Plasma Identification and Quantification Using RPLC-MSn Methods and Isotope Dilution Strategies. Anal. Chem. 2018, 90, 6494–6501. 10.1021/acs.analchem.7b05382. [DOI] [PubMed] [Google Scholar]
- Xicoy H.; Wieringa B.; Martens G. J. M. The SH-SY5Y Cell Line in Parkinson’s Disease Research: A Systematic Review. Mol. Neurodegener. 2017, 12, 10–11. 10.1186/s13024-017-0149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalevich J.; Langford D. Considerations for the Use of SH-SY5Y Neuroblastoma Cells in Neurobiology. Methods Mol. Biol. 2013, 1078, 9–21. 10.1007/978-1-62703-640-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen W. A.; Fu W.; Keller J. N.; Markesbery W. R.; Appel S.; Smith R. G.; Kasarskis E.; Mattson M. P. Protein Modification by the Lipid Peroxidation Product 4-Hydroxynonenal in the Spinal Cords of Amyotrophic Lateral Sclerosis Patients. Ann. Neurol. 1998, 44, 819–824. 10.1002/ana.410440518. [DOI] [PubMed] [Google Scholar]
- Sayre L. M.; Zelasko D. A.; Harris P. L. R.; Perry G.; Salomon R. G.; Smith M. A. 4-Hydroxynonenal-Derived Advanced Lipid Peroxidation End Products Are Increased in Alzheimer’s Disease. J. Neurochem. 1997, 68, 2092–2097. 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- Parkinson J. An Essay on the Shaking Palsy - 1817. J. Neuropsychiatry Clin. Neurosci. 2002, 223–236. 10.1176/jnp.14.2.223. [DOI] [PubMed] [Google Scholar]
- Lautenschläger J.; Stephens A. D.; Fusco G.; Ströhl F.; Curry N.; Zacharopoulou M.; Michel C. H.; Laine R.; Nespovitaya N.; Fantham M.; et al. C-Terminal Calcium Binding of α-Synuclein Modulates Synaptic Vesicle Interaction. Nat. Commun. 2018, 9, 712. 10.1038/s41467-018-03111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo E.; McLaurin J.; Yip C. M.; George-Hyslop P. S.; Fraser P. E. Alpha-Synuclein Membrane Interactions and Lipid Specificity. J. Biol. Chem. 2000, 275, 34328–34334. 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- Bosco M.; Culeddu N.; Toffanin R.; Pollesello P. Organic Solvent Systems for 31P Nuclear Magnetic Resonance Analysis of Lecithin Phospholipids: Applications to Two-Dimensional Gradient-Enhanced 1H-Detected Heteronuclear Multiple Quantum Coherence Experiments. Anal. Biochem. 1997, 245, 38–47. 10.1006/abio.1996.9907. [DOI] [PubMed] [Google Scholar]
- Cullis P. R.; Hope M. J. Chapter 1 Physical Properties and Functional Roles of Lipids in Membranes. New Compr. Biochem. 1991, 20, 1–41. 10.1016/S0167-7306(08)60329-4. [DOI] [Google Scholar]
- Devaux P. F.Static and Dynamic Asymmetry in Cell Membranes. In Biochemistry; American chemical Societ, 1991; Vol. 30, pp 1163–1173. [DOI] [PubMed] [Google Scholar]
- Fadeel B.; Xue D. The Ins and Outs of Phospholipid Asymmetry in the Plasma Membrane: Roles in Health and Disease. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 264–277. 10.1080/10409230903193307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachowski A. Phospholipids in Animal Eukaryotic Membranes: Transverse Asymmetry and Movement. Biochem. J. 1993, 294, 1–14. 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama T.; Riezman H. Understanding the Diversity of Membrane Lipid Composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- Aliakbari F.; Mohammad-Beigi H.; Rezaei-Ghaleh N.; Becker S.; Dehghani Esmatabad F.; Eslampanah Seyedi H. A.; Bardania H.; Tayaranian Marvian A.; Collingwood J. F.; Christiansen G.; et al. The Potential of Zwitterionic Nanoliposomes against Neurotoxic Alpha-Synuclein Aggregates in Parkinson’s Disease. Nanoscale 2018, 9174. 10.1039/C8NR00632F. [DOI] [PubMed] [Google Scholar]
- Terakawa M. S.; Lin Y.; Kinoshita M.; Kanemura S.; Itoh D.; Sugiki T.; Okumura M.; Ramamoorthy A.; Lee Y.-H. Impact of Membrane Curvature on Amyloid Aggregation. Biochim. Biophys. Acta, Biomembr. 2018, 1741–1764. 10.1016/j.bbamem.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakawa M. S.; Lee Y.-H.; Kinoshita M.; Lin Y.; Sugiki T.; Fukui N.; Ikenoue T.; Kawata Y.; Goto Y. Membrane-Induced Initial Structure of α-Synuclein Control Its Amyloidogenesis on Model Membranes. Biochim. Biophys. Acta, Biomembr. 2018, 1860, 757–766. 10.1016/J.BBAMEM.2017.12.011. [DOI] [PubMed] [Google Scholar]
- O’Leary E. I.; Jiang Z.; Strub M.-P.; Lee J. C. Effects of Phosphatidylcholine Membrane Fluidity on the Conformation and Aggregation of N-Terminally Acetylated α-Synuclein. J. Biol. Chem. 2018, 11195. 10.1074/jbc.RA118.002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furse S.; Jakubec M.; Rise F.; Williams H. E.; Rees C. E. D.; Halskau Ø. Evidence That Listeria Innocua Modulates Its Membrane’s Stored Curvature Elastic Stress, but Not Fluidity, through the Cell Cycle. Sci. Rep. 2017, 7, 8012. 10.1038/s41598-017-06855-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A. E.; Sommer L.; Arntzen M. Ø.; Strahm Y.; Morrice N. A.; Divecha N.; D’Santos C. S. Identification of Nuclear Phosphatidylinositol 4,5-Bisphosphate-Interacting Proteins by Neomycin Extraction. Mol. Cell. Proteomics 2011, 10, M110.003376. 10.1074/mcp.M110.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson P. H. Chapter 8 HeLa Cell Plasma Membranes. Methods Cell Biol. 1974, 7, 157–188. 10.1016/S0091-679X(08)61776-8. [DOI] [PubMed] [Google Scholar]
- Wada I.; Rindress D.; Cameron P. H.; Ou W. J.; Doherty J. J. II; Louvard D.; Bell A. W.; Dignard D.; Thomas D. Y.; Bergeron J. J. SSR α and Associated Calnexin Are Major Calcium Binding Proteins of the Endoplasmic Reticulum Membrane. J. Biol. Chem. 1991, 266, 19599–19610. [PubMed] [Google Scholar]
- Ou W. J.; Cameron P. H.; Thomas D. Y.; Bergeron J. J. M. Association of Folding Intermediates of Glycoproteins with Calnexin during Protein Maturation. Nature 1993, 364, 771–776. 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- Miyamoto S.; Teramoto H.; Coso O. A.; Gutkind J. S.; Burbelo P. D.; Akiyama S. K.; Yamada K. M. Integrin Function: Molecular Hierarchies of Cytoskeletal and Signaling Molecules. J. Cell Biol. 1995, 131, 791–805. 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer R. A.; Lehner C. F.; Eppenberger H. M.; Nigg E. A. Major Nucleolar Proteins Shuttle between Nucleus and Cytoplasm. Cell 1989, 56, 379–390. 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- Murgia S.; Mele S.; Monduzzi M. Quantitative Characterization of Phospholipids in Milk Fat via 31P NMR Using a Monophasic Solvent Mixture. Lipids 2003, 38, 585–591. 10.1007/s11745-003-1500-3. [DOI] [PubMed] [Google Scholar]
- Cremonini M. A.; Laghi L.; Placucci G. Investigation of Commercial Lecithin by 31P NMR in a Ternary CUBO Solvent. J. Sci. Food Agric. 2004, 84, 786–790. 10.1002/jsfa.1683. [DOI] [Google Scholar]
- Massou S.; Augé S.; Tropis M.; Lindley N. D.; Milon A. NMR Analyses of Deuterated Phospholipids Isolated from Pichia Angusta. J. Chim. Phys. Phys.-Chim. Biol. 1998, 95, 406–411. 10.1051/jcp:1998152. [DOI] [Google Scholar]
- Culeddu N.; Bosco M.; Toffanin R.; Pollesello P. 31P NMR Analysis of Phospholipids in Crude Extracts from Different Sources: Improved Efficiency of the Solvent System. Magn. Reson. Chem. 1998, 36, 907–912. . [DOI] [Google Scholar]
- Ejsing C. S.; Sampaio J. L.; Surendranath V.; Duchoslav E.; Ekroos K.; Klemm R. W.; Simons K.; Shevchenko A. Global Analysis of the Yeast Lipidome by Quantitative Shotgun Mass Spectrometry. Proc. Natl. Acad. Sci. 2009, 106, 2136–2141. 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naudí A.; Cabré R.; Jové M.; Ayala V.; Gonzalo H.; Portero-Otín M.; Ferrer I.; Pamplona R. Chapter Five - Lipidomics of Human Brain Aging and Alzheimer’s Disease Pathology. International Review of Neurobiology 2015, 122, 133–189. 10.1016/bs.irn.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Rouser G.; Galli C.; Kritchevsky G. Lipid Class Composition of Normal Human Brain and Variations in Metachromatic Leucodystrophy, Tay-Sachs, Niemann-Pick, Chronic Gaucher’s and Alzheimer’s Diseases. J. Am. Oil Chem. Soc. 1965, 42, 404–410. 10.1007/BF02635576. [DOI] [PubMed] [Google Scholar]
- Rouser G.; Feldman G.; Galli C. Fatty Acid Compositions of Human Brain Lecithin and Sphingomyelin in Normal Individuals, Senile Cerebral Cortical Atrophy, Alzheimer’s Disease, Metachromatic Leucodystrophy, Tay-Sachs and Niemann-Pick Diseases. J. Am. Oil Chem. Soc. 1965, 42, 411–412. 10.1007/BF02635577. [DOI] [PubMed] [Google Scholar]
- Rouser G.; Yamamoto A. Curvilinear Regression Course of Human Brain Lipid Composition Changes with Age. Lipids 1968, 3, 284–287. 10.1007/BF02531202. [DOI] [PubMed] [Google Scholar]
- O’Brien J. S.; Sampson E. L. Lipid Composition of the Normal Human Brain: Gray Matter, White Matter, and Myelin. J. Lipid Res. 1965, 6, 537–544. [PubMed] [Google Scholar]
- Liebisch G.; Binder M.; Schifferer R.; Langmann T.; Schulz B.; Schmitz G. High Throughput Quantification of Cholesterol and Cholesteryl Ester by Electrospray Ionization Tandem Mass Spectrometry (ESI-MS/MS). Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2006, 1761, 121–128. 10.1016/j.bbalip.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Panganamala R. V.; Horrocks L. A.; Geer J. C.; Cornwell D. G. Positions of Double Bonds in the Monounsaturated Alk-1-Enyl Groups from the Plasmalogens of Human Heart and Brain. Chem. Phys. Lipids 1971, 6, 97–102. 10.1016/0009-3084(71)90031-4. [DOI] [PubMed] [Google Scholar]
- O’Brien J. S.; Sampson E. L. Fatty Acid and Fatty Aldehyde Composition of the Major Brain Lipids in Normal Human Gray Matter, White Matter, and Myelin. J. Lipid Res. 1965, 6, 545–551. [PubMed] [Google Scholar]
- Smith P. K.; Krohn R. I.; Hermanson G. T.; Mallia A. K.; Gartner F. H.; Provenzano M. D.; Fujimoto E. K.; Goeke N. M.; Olson B. J.; Klenk D. C. Measurement of Protein Using Bicinchoninic Acid. Anal. Biochem. 1985, 150, 76–85. 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Furse S.; Liddell S.; Ortori C. A.; Williams H.; Neylon D. C.; Scott D. J.; Barrett D. A.; Gray D. A. The Lipidome and Proteome of Oil Bodies from Helianthus Annuus (Common Sunflower). J. Chem. Biol. 2013, 63. 10.1007/s12154-012-0090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F. F.; Turk J. Structural Determination of Sphingomyelin by Tandem Mass Spectrometry with Electrospray Ionization. J. Am. Soc. Mass Spectrom. 2000, 11, 437–449. 10.1016/S1044-0305(99)00150-6. [DOI] [PubMed] [Google Scholar]
- Murphy R. C.; Fiedler J.; Hevko J. Analysis of Nonvolatile Lipids by Mass Spectrometry. Chem. Rev. 2001, 101, 479–526. 10.1021/cr9900883. [DOI] [PubMed] [Google Scholar]
- Pulfer M.; Murphy R. C. Electrospray Mass Spectrometry of Phospholipids. Mass Spectrom. Rev. 2003, 22, 332–364. 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- Han X.; Gross R. W. Structural Determination of Picomole Amounts of Phospholipids via Electrospray Ionization Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 1995, 6, 1202–1210. 10.1016/1044-0305(95)00568-4. [DOI] [PubMed] [Google Scholar]
- Byrdwell W. C.; Perry R. H. Liquid Chromatography with Dual Parallel Mass Spectrometry and 31P Nuclear Magnetic Resonance Spectroscopy for Analysis of Sphingomyelin and Dihydrosphingomyelin. I. Bovine Brain and Chicken Egg Yolk. J. Chromatogr. A 2006, 1133, 149–171. 10.1016/j.chroma.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Hsu F.-F.; Turk J. Characterization of Cardiolipin from Escherichia Coli by Electrospray Ionization with Multiple Stage Quadrupole Ion-Trap Mass Spectrometric Analysis of [M - 2H + Na]- Ions. J. Am. Soc. Mass Spectrom. 2006, 17, 420–429. 10.1016/j.jasms.2005.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F. F.; Turk J.; Rhoades E. R.; Russell D. G.; Shi Y.; Groisman E. A. Structural Characterization of Cardiolipin by Tandem Quadrupole and Multiple-Stage Quadrupole Ion-Trap Mass Spectrometry with Electrospray Ionization. J. Am. Soc. Mass Spectrom. 2005, 16, 491–504. 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Kochen M. A.; Chambers M. C.; Holman J. D.; Nesvizhskii A. I.; Weintraub S. T.; Belisle J. T.; Islam M. N.; Griss J.; Tabb D. L. Greazy: Open-Source Software for Automated Phospholipid Tandem Mass Spectrometry Identification. Anal. Chem. 2016, 88, 5733–5741. 10.1021/acs.analchem.6b00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting M.; Ruttkies C.; Neumann S.; Schmitt-Kopplin P. LipidFrag: Improving Reliability of in Silico Fragmentation of Lipids and Application to the Caenorhabditis Elegans Lipidome. PLoS One 2017, 12, e0172311 10.1371/journal.pone.0172311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.