Abstract
The biological production of two-carbon compounds (ethylene glycol (EG) and glycolate) has been studied for the sustainable supply of the compounds to the polymer, cosmetic, textile, and medical industries. Here, we demonstrated the bioconversion of xylose to either ethylene glycol (EG) or glycolate using engineered Corynebacterium glutamicum, a well-known industrial amino acid producer. A synthetic ribulose 1-phosphate (Ru1P) pathway involving heterologous d-tagatose 3-epimerase and l-fuculose kinase/aldolase reactions was introduced in C. glutamicum. Subsequently, heterologous expression of Escherichia coli YqhD reductase with the synthetic Ru1P pathway led to ethylene glycol production from xylose. Additional pathway engineering in C. glutamicum by mutating ald, which encodes an aldehyde dehydrogenase, abolished the by-product formation of glycolate during xylose conversion to EG at a yield of 0.75 mol per mol. In addition, the bioconversion of xylose to glycolate was achieved, and the almost maximum molar yield was 0.99 mol per mol xylose in C. glutamicum via the Ru1P pathway. Thus, the synthetic Ru1P pathway in C. glutamicum led bioconversion of xylose to either ethylene glycol or glycolate with high molar yields.
Introduction
Ethylene glycol, a two-carbon diol compound, can be commercially produced from ethylene or CO in the petrochemical industry and is used as a polymer precursor (i.e., polyethylene terephthalate) and antifreezing agent. Glycolate, a two-carbon α-hydroxyl acid with alcohol and carboxyl moieties, is chemically produced by energy-intensive carbonylation of CO/H2-derived formaldehyde, enzymatic oxidation of ethylene glycol, or hydrolyzation of glycolonitrile. Glycolate is widely used as a skin care product in the cosmetic industry, a rinsing agent in the textile industry, and as a precursor for biopolymers, e.g., poly(lactate-co-glycolate)1 or glycolate-based polyester.2 Although current chemical processes for the two-carbon chemicals are cheap and fast, low selectivity limits the energy-saving process. Thus, a biological process with high selectivity may be an alternative environmentally friendly process with a low energy demand. It is necessary to develop efficient microbial cell factories that use renewable carbon sources and produce target chemicals with a high yield.3−5 Glucose and xylose are the most abundant sugars from lignocellulosic materials that are composed of cellulose, hemicellulose (mostly xylan), and lignin.
Metabolic engineering of microbial strains has been studied to supply sustainable ethylene glycol or glycolate from glucose and xylose. In Escherichia coli, a synthetic pathway for either ethylene glycol or glycolate production from xylose via synthetic xylose 1-phosphate has been designed by expressing xylulose 1-phosphate aldolase, resulting in 2.1 g/L ethylene glycol at a yield of 0.47 mol/mol or 4.3 g/L glycolate at a yield of 0.9 mol/mol, respectively.6 Ethylene glycol and glycolate were co-produced by the engineered E. coli strains, with a yield of 0.93 mol/mol as two-carbon compounds. By optimizing the E. coli strain with FucO reductase, 20 g/L of ethylene glycol was only produced with 0.37 g/(L h) at a molar yield of 0.94 mol/mol.7
Another synthetic ribulose 1-phosphate (Ru1P) pathway has been used to produce glycolate from xylose, as a sole source of carbon, via heterologous d-tagatose 3-epimerase and l-fuculose kinase/aldolase reactions, resulting in either 40 g/L ethylene glycol or 44 g/L glycolate from 100 g/L xylose.8 Furthermore, pathway engineering in the glyoxylate shunt resulted in 40 g/L glycolate from 65 g/L xylose, with a yield of 0.63 g/g (or 1.21 mol/mol). Recently, the Dahms pathway, which is an oxidative xylose degradation pathway via xylonolactone and xylonite intermediates, has been adopted to convert xylose to ethylene glycol, with a yield of 0.29 g/g (or 0.7 mol/mol),9 and glycolate, with a yield of 0.46 g/g (or 0.91).10,11 In silico simulation and synthetic small rRNA-based metabolic engineering of E. coli have increased the titer, productivity, and yield of a product on xylose (108.2 g/L; 0.36 g/g; 2.25 g/L/h).12 In addition, high-level production of 65.5 g/L glycolate and 10.6 g/L acetate in a fed-batch culture was achieved from 0.77 g/g glucose by balancing the TCA cycle and glyoxylate shunt.13 Besides sole bioconversion from xylose in E. coli, bioconversion of xylose to glycolate via the Dahms pathway has been combined to produce poly(lactate-co-glycolate) and its copolymers,1 and glucose has been redirected to produce poly(lactate-co-glycolate-co-3-hydroxybutyrate) via the glyoxylate shunt and glyoxylate reductase.14
Corynebacterium glutamicum is a well-known industrial producer of amino acids (e.g., 2 200 000 tons l-lysine per year). Recently, metabolically engineered C. glutamicum was grown on alternative carbon sources like xylose15−17 to produce industrially relevant chemicals.4,18,19 Previously, ethylene glycol (3.5 g/L) has been produced from glucose at a yield of 0.25 mol/mol (or 0.10 g/g) via the serine biosynthetic pathway using engineered C. glutamicum.20 In addition, 5.3 g/L glycolate (a yield of 0.18 g/g [0.43 mol/mol]), from a mixture of glucose and acetate, has been produced in engineered C. glutamicum.21 Besides ethylene glycol and glycolate production, C. glutamicum harboring a high-copy number plasmid with xdh encoding xylose dehydrogenase showed a high conversion rate of xylonate from xylose at 0.75 g/(L h).22 Xylonate (6.23 g/L) from 20 g/L xylan has also been reported to be produced in C. glutamicum.23 The use of C. glutamicum ΔiolR (gene for the transcriptional repressor IolR) resulted in a high level of xylonate production at a molar yield of 1.0 and volumetric productivity of 3.98 g/(L h).24 However, there are no reports on the conversion of xylose to ethylene glycol and glycolate using C. glutamicum via a synthetic Ru1P pathway or other xylose-utilizing pathways.
In this study, we engineered a C. glutamicum strain capable of converting xylose to either ethylene glycol at a yield of 0.75 mol/mol (0.31 g/g) or glycolate at a yield of 0.99 mol/mol (0.51 g/g) via the synthetic Ru1P pathway. To increase productivity, a xylose uptake IolT transporter was expressed in the engineered C. glutamicum. In addition, to eliminate the by-product (glycolate) for the sole production of ethylene glycol, the native ald gene encoding aldehyde dehydrogenase was manipulated using CRISPR-Cas12a to have a nonsense mutation. Furthermore, fed-batch fermentation was conducted to produce either ethylene glycol or glycolate from xylose to improve the titer and demonstrate the bioconversion of xylose using the engineered C. glutamicum strains.
Results and Discussion
Synthetic Pathway Design for Ethylene Glycol and Glycolate from Xylose in C. glutamicum
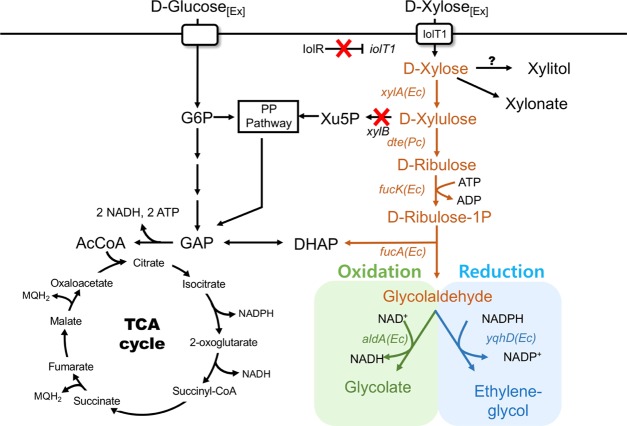
The wild-type (WT) C. glutamicum expressing a heterologous xylose isomerase is capable of using xylose as the sole carbon source.15 For this reason, the xylB encoding xylulose kinase was deleted and C. glutamicum ΔxylB (SL-1) was used as the parental host strain to convert xylose to either ethylene glycol or glycolate via synthetic glycolate pathways (Figure 1). No xylose consumption was exhibited by SL-1 (data not shown). The synthetic ethylene glycol and glycolate pathways via Ru1P consist of 4 common enzymatic steps involving xylose isomerase (I; xylA from E. coli), d-tagatose 3-epimerase (E; dte from P. cichorii), l-fuculose kinase (K; fucK from E. coli), l-fuculose aldolase (A; fucA from E. coli), and a target-specific enzymatic reaction as either reduction by an aldehyde reductase (R; yqhD from E. coli, O; fucO from E. coli) for ethylene glycol or oxidation by an aldehyde dehydrogenase (D; aldA from E. coli) for glycolate. Both pathways were designed to convert xylose to the target product with a theoretical molar yield of 1 mol/mol (C-mole product/C-mole substrate).
Figure 1.
Bioconversion of xylose to ethylene glycol and glycolate using engineered C. glutamicum. (A) Xylose was converted to either ethylene glycol or glycolate via synthetic metabolic pathways in C. glutamicum. The SL-1 strain (C. glutamicum ΔxylB [encoding xylulose kinase]) was used as the parental strain for the bioconversion. To convert xylose to either ethylene glycol or glycolate, five enzymatic steps (shown in colored arrows) were used by expressing genes encoding xylose isomerase (xylA) from E. coli, d-tagatose 3-epimerase (dte) from P. cichorii, l-fuculokinase (fucK) from E. coli, l-fuculose phosphate aldolase (fucA) from E. coli, aldehyde dehydrogenase (aldA) from E. coli, and aldehyde reductase (yqhD) from E. coli in SL-1. Red “X” represents gene deletion in C. glutamicum. Abbreviations: G6P, glucose 6-phosphate; DHAP, dihydroxyacetone phosphate; PP pathway, pentose phosphate pathway; Xu5P, xylulose 5-phosphate; GAP, glyceraldehyde 3-phosphate; and AcCoA, acetyl-CoA.
Bioconversion of Xylose as the Sole Carbon to Either Ethylene Glycol or Glycolate
To produce ethylene glycol from xylose, the plasmid pIEAKR, pIEKAR, pIEAOK, or pIEKAO was transformed into SL-1. SL-1 strains harboring the plasmid were cultivated with 2% (w/v) xylose as the sole carbon source. As a result, no ethylene glycol was detected in the medium. Because of no xylose consumption, the strains were not able to grow from xylose. This could be due to inadequate ATP supply and imbalance of NAD(P)H/NAD(P)+ from the catabolic metabolism of dihydroxyacetone phosphate (DHAP), which is a product of l-fuculose phosphate aldolase (FucA; Figure 2A). In addition, the plasmid with pIEKAD (or pIEAKD) for the synthetic glycolate pathway was transformed into SL-1. SL-1 pIEKAD or pIEAKD were cultivated with 2% (w/v) xylose as the sole carbon source. As a result, 2.4 g/L glycolate was obtained from 5.4 g/L xylose using both strains; the final OD600 values were 3.8 ± 0.1 or 3.6 ± 0.1 (Figure 2B). The conversion yields were less than 0.47 g/g, and only 30% initial xylose was used for growth and glycolate conversion. Unlike recombinant E. coli harboring the synthetic Ru1P pathway,8 SL-1 pIEKAD or SL-1 pIEAKD did not completely convert xylose to glycolate when xylose was provided as the sole carbon source for 120 h. In these C. glutamicum strains, only small fractions of xylose were metabolized and converted to either ethylene glycol or glycolate. In addition, the lower cell growth rates and xylose conversions were observed in ethylene glycol-producing strains than in glycolate-producing strains. This could be due to the fact that strain SL-1 did not provide enough chemical reducing power for xylose conversion in the presence of only xylose as the sole carbon.
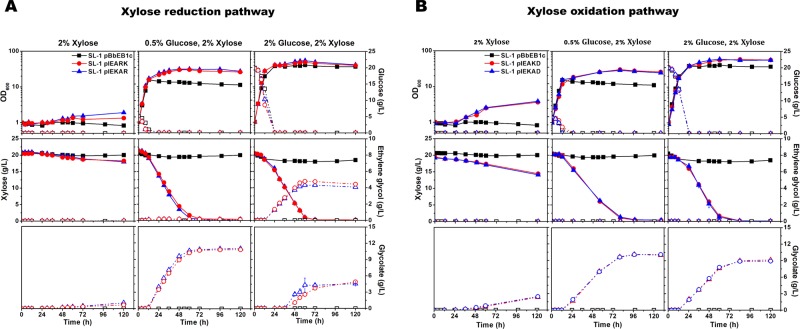
Figure 2.
Time course of growth, xylose consumption, and ethylene glycol and glycolate production in engineered C. glutamicum. Growth and carbon source consumption in recombinant C. glutamicum for the production of ethylene glycol (A) and glycolate (B). Optical densities at 600 nm (OD600; solid symbol; solid line) and glucose (open symbol; dashed line) concentrations [upper row], xylose (solid symbol; solid line) and ethylene glycol (open symbol; dashed line) concentrations [middle row], and glycolate (open symbol; dashed line) concentrations [lower row] in the medium were measured for C. glutamicum SL-1 pBbEB1c (black square in A and B), SL-1 pIEARK (red circle in A), SL-1 pIEKAR (blue triangle in A), SL-1 pIEAKD (red circle in B), and SL-1 pIEKAD (blue triangle in B). Either xylose (2% wt/vol) as the sole carbon source [left panel], a mixture of xylose (2% wt/vol) and glucose (0.5% wt/vol) [middle panel], or a mixture of xylose (2% wt/vol) and glucose (2% wt/vol) [right panel] in CgXII medium was used. The data represent mean values of triplicate cultivations, and the error bars represent standard deviations. See Table 1 for the strains used.
Bioconversion of Xylose to Either Ethylene Glycol or Glycolate with an Additional Carbon Source
To increase the bioconversion capability, an additional 0.5% (w/v) glucose or 2% (w/v) glucose was supplemented for the cell growth with the supply of ATP and recycling of NA(P)D+. Although the recombinants (SL-1 pIEARK and SL-1 pIEKAR) with 0.5% (w/v) glucose showed a final OD600 of 30.0 ± 0.8 and 31.3 ± 0.8, respectively, less than 0.2 g/L ethylene glycol production was observed. Interestingly, 10.78 g/L glycolate (SL-1 pIEARK) and 10.97 g/L glycolate (SL-1 pIEKAR) were produced as by-products. Because the final step for ethylene glycol conversion requires NADPH reduction, 2% glucose was supplied along with 2% xylose for better cell growth and reducing potential. Then, the recombinants (SL-1 pIEARK and SL-1 pIEKAR) with 2% (w/v) glucose attained a final OD600 of 49.5 ± 0.1 and 55.4 ± 0.3, respectively. After glucose was completely depleted, SL-1 harboring either pIEARK or SL-1 pIEKAR produced similar levels of ethylene glycol (4.7 ± 0.1 g/L ethylene glycol from 19.7 ± 0.1 g/L xylose in 60 h with a yield of 0.25 g/g and 0.59 mol/mol) (Figure 2A). Since ethylene glycol has not been produced yet from glucose in C. glutamicum,(20) we believed that ethylene glycol was converted from xylose in a mixture of glucose and xylose as carbon sources.
Under these conditions, undesired glycolate production, caused by native aldehyde dehydrogenase activity, was dramatically reduced by producing ethylene glycol converted from glycolaldehyde as the same precursor. Particularly, overexpression of yqhD encoding a major glycolaldehyde reductase in E. coli resulted in a favorable reaction against native aldehyde dehydrogenase in C. glutamicum. We also tested another aldehyde reductase (fucO encoding lactaldehyde reductase from E. coli), but we did not observe enhanced ethylene glycol production (Figure S2). However, the overexpression of fucO contributed positively to ethylene glycol production in E. coli via the synthetic Ru1P or xylulose 1-phosphate pathway.7,8 Although xylonate was not produced, xylitol (0.1 g/L at 60 h; 0.4 g/L at 120 h) was produced because of unspecific xylose reductase. Nonetheless, to convert xylose to ethylene glycol, C. glutamicum could not provide a sufficient amount of reduction equivalents in the presence of xylose as the sole carbon.
For glycolate production, the recombinants (SL-1 pIEKAD and SL-1 pIEAKD) with 0.5% (w/v) glucose attained a final OD600 of 24.8 ± 0.5 and 26.3 ± 0.8, respectively. After the glucose was completely depleted, SL-1 pIEKAD produced 10.12 ± 0.1 g/L glycolate from 19.5 ± 0.1 g/L xylose in 96 h, with a yield of 0.99 mol/mol (0.51 g/g; Figure 2B). Similarly, SL-1 pIEAKD produced 10.14 ± 0.1 g/L glycolate from 20 g/L xylose, with a yield of 0.99 mol/mol (0.51 g/g).
Since glycolate has not been produced from glucose in C. glutamicum,21 we also believed that glycolate was converted from xylose in a mixture of glucose and xylose as carbon sources. Thus, the molar yields of the recombinant C. glutamicum strains reached the theoretical maximum value with the synthetic glycolate pathway, although the 13C-labeled xylose could be fed for an accurate calculation of the actual yield. To increase the glycolate production rate, cell biomass was increased by cultivating the cells with 2% (w/v) glucose (the final OD600 was 56.6 ± 1.4 and 57.3 ± 2.0 for SL-1 pIEKAD and SL-1 pIEAKD, respectively). However, the production rate was not enhanced (0.12 g/(L h) at 76 h, 0.5% (w/v) glucose and 2% (w/v) xylose; 0.13 g/(L h) at 76 h, 2% (w/v) glucose and 2% (w/v) xylose). The molar yield of glycolate was decreased to 0.92 mol/mol (0.47 g/g). No xylonate was detected. This decrease was due to the by-product formation of xylitol; 0.9 g/L xylitol was measured when the cells were grown with 2% (w/v) glucose and 2% (w/v) xylose when compared with 0.2 g/L xylitol from the cells grown with 0.5% (w/v) glucose and 2% (w/v) xylose. Interestingly, SL-1 harboring the empty vector with 2% (w/v) glucose and 2% (w/v) xylose also consumed 2.2 g/L xylose (14.9 mM) and produced 1.5 g/L (10.13 mM) xylitol. Because the engineered C. glutamicum, exhibiting the Weimberg pathway, has been reported to accumulate small amounts of xylitol when grown on a mixture of glucose and xylose,25 we assumed that C. glutamicum WT, under conditions in which xylose metabolism is not efficient, could enable the oxidation of xylose to xylitol.
Pathway Engineering with an IolT Transporter for Ethylene Glycol and Glycolate
To increase the conversion rate of either ethylene glycol or glycolate, a metabolic engineering strategy to increase the rate of xylose uptake should be considered. Deletion of the iolR gene in C. glutamicum with the xylose isomerase had improved xylose uptake and the cell growth rate by 1.6-fold when compared with WT.26 In addition, the myo-inositol/proton symporter IolT1 has been shown to contribute to faster xylose uptake and cell growth. Therefore, we constructed the SL-1R strain (iolR::P9Ter) to derepress a iol gene cluster, including iolT1. To accelerate strain development, mutant strains were constructed using CoryneCR12-STOP (Figure S2). Then, SL-1 and SL-1R strains were transformed with either pIEARKT or pIEAKDT for ethylene glycol or glycolate, respectively (Table 1).
Table 1. Bacterial Strains and Plasmids Used in this Study.
| strain or plasmid | relevant characteristics | reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F- (80d lacZ M15) (lacZYA-argF) U169 hsdR17(r– m+) recA1 endA1 relA1 deoR96 | (40) |
| C. glutamicum WT | ATCC 13032, wild-type strain | ATCC |
| C. glutamicum SL-1 | ATCC 13032, ΔxylB (in-frame deletion) | this study |
| C. glutamicum SL-1R | SL-1, iolR::P9Ter (nonsense mutation) | this study |
| C. glutamicum SL-1A | SL-1, ald::C49Ter (nonsense mutation) | this study |
| Plasmids | ||
| pK19mobSacB-xylB | KmR; pK19mobsacB derivative containing a crossover PCR product covering the upstream and downstream regions of xylB | this study |
| pBbEB1c-RFP | ColE1 (Ec), pBL1 (Cg), Cmr, Ptrc, BglBrick sites, rfp, CoryneBrick vector | (36) |
| pBbEB1c | ColE1 (Ec), pBL1 (Cg), Cmr, Ptrc, BglBrick sites, CoryneBrick empty vector | (36) |
| pBbEB1c-XylA | pBbEB1c-derived vector carrying xylA of E. coli codon-optimized for C. glutamicum | (41) |
| pUC57-dte(co), -fucK(co),-fucA(co), -aldA(co), -yqhD(co), -fucO(co) | pUC57 vectors containing a single gene: dte originated from P. cichorii and fucK, fucA, aldA, yqhD, and fucO originated from E. coli. All genes were codon-optimized (co) for C. glutamicum | this study |
| pIEKAD | pBbEB1c vector carrying codon-optimized xylA-dte-fucK-fucA-aldA for C. glutamicum, Cmr Symbols: I, isomerase (xylA); E, epimerase (dte); K, kinase (fucK); A, aldolase (fucA); D, dehydrogenase (aldA) | this study |
| pIEAKD | pBbEB1c vector carrying codon-optimized xylA-dte-fucA-fucK-aldA for C. glutamicum, Cmr | this study |
| pIEAKDT | pIEAKD-derived vector carrying iolT1 of C. glutamicum for C. glutamicum | this study |
| pIEAKR | pBbEB1c vector carrying codon-optimized xylA-dte-fuckA-fucK- yqhD for C. glutamicum, Cmr Symbols: I, isomerase (xylA); E, epimerase (dte); K, kinase (fucK); A, aldolase (fucA); R, reductase (yqhD) | this study |
| pIEKAR | pBbEB1c vector carrying codon-optimized xylA-dte-fucK-fucA-yqhD for C. glutamicum, Cmr | this study |
| pIEARK | pBbEB1c vector carrying codon-optimized xylA-dte-fucA-yqhD-fucK for C. glutamicum, Cmr | this study |
| pIEARKT | pIEARK-derived vector carrying iolT1 of C. glutamicum for C. glutamicum | this study |
| pIEKAO | pBbEB1c vector carrying codon-optimized xylA-dte-fucK-fucA-fucO for C. glutamicum, Cmr Symbols: I, isomerase (xylA); E, epimerase (dte); K, kinase (fucK); A, aldolase (fucA); O, oxidoreductase (fucO) | this study |
| pIEAOK | pBbEB1c vector carrying codon-optimized xylA-dte-fucA-fucO-fucK for C. glutamicum, Cmr | this study |
| pJYS1Ptac | pBL1tsoriVC.g. Kmr pSC101 oriVE.c. PlacM-FnCpf1 Ptac-RecT lacIq | (39) |
| pJYS2_crtYf | rep oriVC.g. Spr pMB1 oriVE.c. Pj23119-crRNA-crtYf targeting the crtYf gene | (39) |
| pcrRNA-iolR | pJYS2 derivative, Pj23119-crRNA-iolR targeting iolR | this study |
| pcrRNA-ald | pJYS2 derivative, Pj23119-crRNA-ald targeting ald | this study |
As a result, the SL-1 pIEARKT strain only showed improved ethylene glycol production (5.7 g/L) at a molar yield of 0.69 mol/mol (0.29 g/g) when compared with the control (SL-1 pIEARK; 4.7 ± 0.1 g/L; 0.59 mol/mol [0.25 g/g]; Figure 3A). None of the SL-1R strains harboring pIEARK, pIEARKT, pIEAKD, or pIEAKDT showed better performance than the control. Interestingly, SL-1R yielded a significant amount of xylonate (5.6 g/L for SL-1R pIEARKT and 10.17 g/L for SL-1R pIEAKDT), which lowered the production titer and yield of ethylene glycol and glycolate (Figure 3A,B). This could result from the de-depression of the iolG gene encoding for myo-inositol dehydrogenase. The finding was consistent with that of a previous study in which C. glutamicum ΔiolR (the gene for the transcriptional repressor IolR) accumulated high amounts of xylonate via the oxidation pathway of xylose with native myo-inositol dehydrogenase (IolG).24 Thus, faster xylose uptake rates were achieved by a nonsense mutation in iolR and overexpression of iolT1 in the SL-1R-derived strains (Figure 3). However, faster xylose uptakes were exhibited, and higher xylonate formation was achieved. Since IolR has been predicted to regulate 14 genes including the iolG and iolT1 genes,27 the de-regulation of the IolR regulons in the SL-1R mutants must be followed, which remains unknown. Therefore, the synthetic Ru1P pathway must be optimized and faster xylose consumption and no xylonate formation should be considered to improve the production titer and yield of either ethylene glycol or glycolate by deleting the iolG gene.
Figure 3.
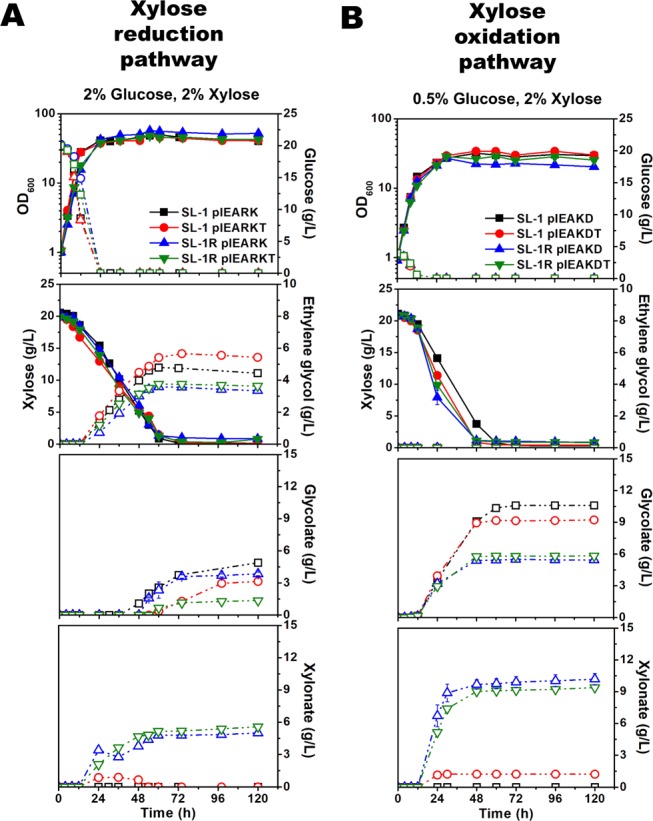
Effects of IolT transport overexpression in either ethylene glycol- or glycolate-producing C. glutamicum strains. Growth and carbon source consumption in recombinant C. glutamicum for the production of ethylene glycol (A) and glycolate (B). Optical densities at 600 nm (OD600; solid symbol; solid line) and glucose (open symbol; dashed line) concentrations, xylose (solid symbol; solid line) and ethylene glycol (open symbol; dashed line) concentrations, and glycolate (open symbol; dashed line) concentrations in the medium were measured. For the production of ethylene glycol (A), C. glutamicum SL-1 pIEARK (black square in A), SL-1 pIEARKT (red circle in A), SL-1R pIEARK (blue triangle in A), and SL-1R pIEARKT (green inversed triangle in A) were cultivated in the CgXII medium with a mixture of xylose (2% wt/vol) and glucose (2% wt/vol). For the production of glycolate (B), C. glutamicum SL-1 pIEAKD (black square in A), SL-1 pIEAKDT (red circle in B), SL-1R pIEAKD (blue triangle in A), and SL-1R pIEAKDT (green inversed triangle in B) were cultivated in the CgXII medium with a mixture of xylose (2% wt/vol) and glucose (2% wt/vol). The data represent mean values of triplicate cultivations, and the error bars represent standard deviations. See Table 1 for the strains used.
Nonsense Mutation of Aldehyde Dehydrogenase Reduced Glycolate Production during Ethylene Glycol Production
To minimize by-product formation and enhance ethylene glycol conversion, the SL-1 pIEARKT strain was further metabolically developed by inactivating the gene encoding aldehyde dehydrogenase (Ald). We searched for a major enzyme for Ald. The protein sequence identity between AldA in E. coli, which has been used for glycolate production in this study, and the Ald encoded by ald (cg3096) was 33%. In addition, Ald encoded by ald is capable of oxidizing formaldehyde to formate.28 Thus, the SL-1A mutant was constructed by creating a nonsense mutation in ald (ald::C49Ter; Table 1 and Figure S2). As a result, glycolate formation was completely eliminated in the SL-1A pIEARKT strain during cell culture and xylose conversion with 2% (w/v) glucose and 2% (w/v) xylose (Figure 4). Similarly, no glycolate production was reported in E. coli ΔxylB ΔaldA.7 However, no significant increase in ethylene glycol conversion was achieved in SL-1A pIEARKT (5.8 g/L ± 0.02 at a yield of 0.75 mol/mol (0.31 g/g)). Instead, ethylene glycol degradation was abolished, and the final OD600 (44.0 ± 1.9) was slightly increased in SL-1A pIEARKT when compared with that in SL-1 pIEARKT (41.3 ± 0.4). Besides the glycolate and ethylene glycol conversion, carbon metabolism for glycolaldehyde in C. glutamicum is still unclear. A possible route could be related to the detoxification of formaldehyde to formate.6
Figure 4.
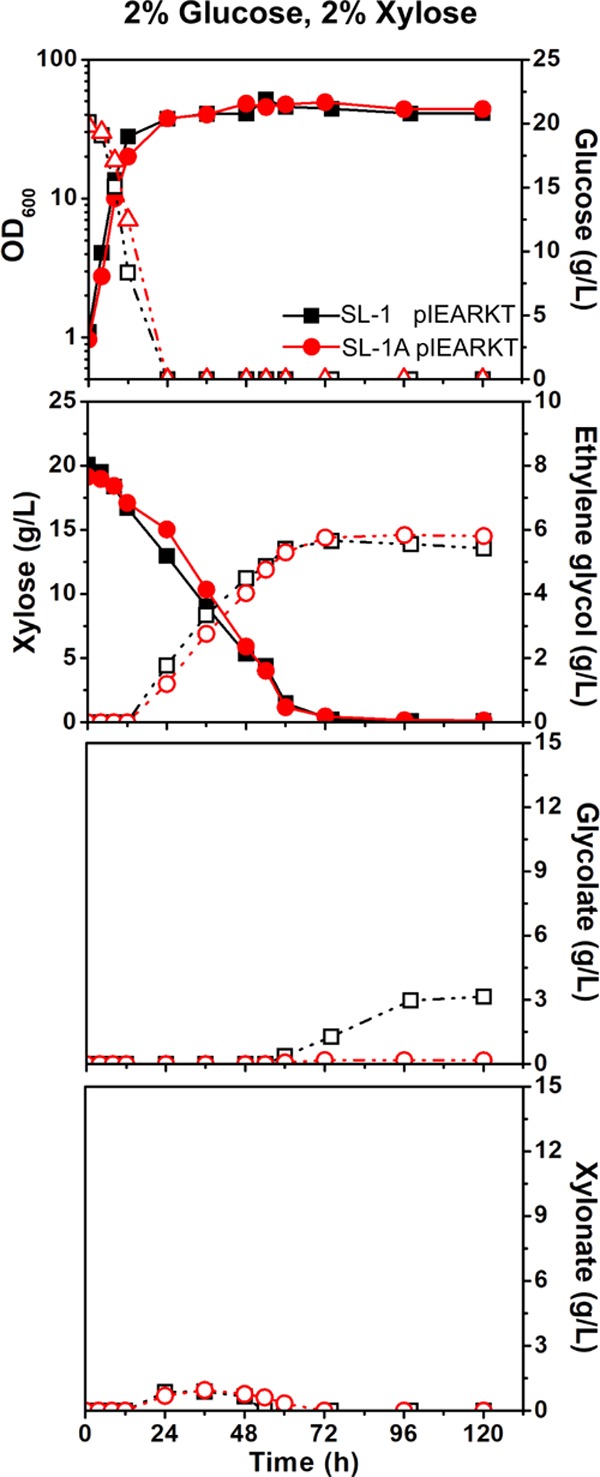
Effects of nonsense mutation of ald encoding aldehyde dehydrogenase in ethylene glycol-producing C. glutamicum strains. Growth and carbon source consumption in recombinant C. glutamicum for the production of ethylene glycol. Optical densities at 600 nm (OD600; solid symbol; solid line) and glucose (open symbol; dashed line) concentrations, xylose (solid symbol; solid line) and ethylene glycol (open symbol; dashed line) concentrations, and glycolate and xylonate (open symbol; dashed line) concentrations in the medium were measured. C. glutamicum SL-1 pIEARKT (black square) and SL-1A pIEARKT (red circle) were cultivated in the CgXII medium with a mixture of xylose (2% wt/vol) and glucose (2% wt/vol). The data represent mean values of triplicate cultivations, and the error bars represent standard deviations. See Table 1 for the strains used.
Fed-Batch Fermentation for Ethylene Glycol and Glycolate from Xylose
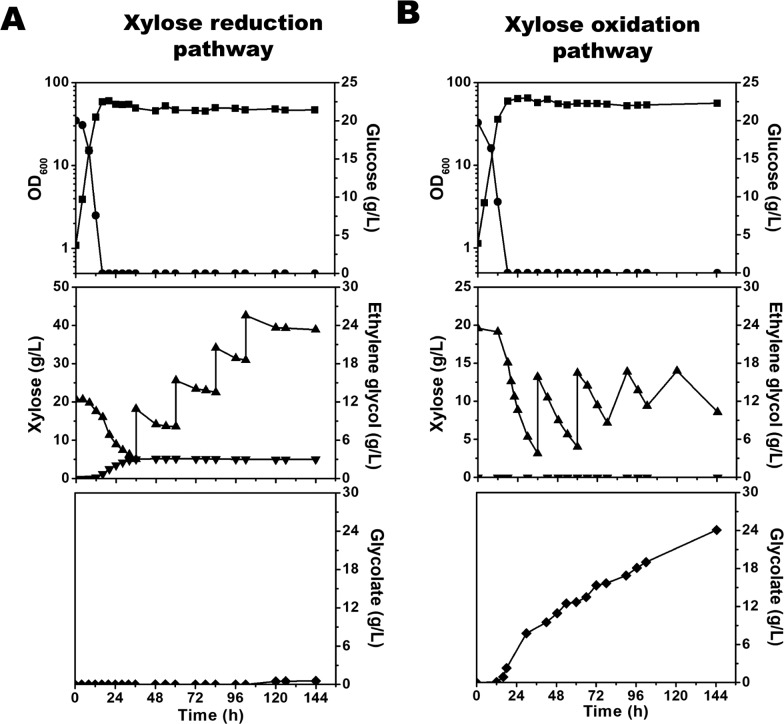
The engineered C. glutamicum strain SL-1A pIEARKT for ethylene glycol resulted in 5.8 ± 0.02 g/L ethylene glycol from 18.6 ± 0.16 g/L xylose at a yield of 0.75 mol/mol (0.31 g/g) in the flask culture. Also, SL-1 pIEAKD produced 10.14 ± 0.1 g/L glycolate from 19.9 ± 0.06 g/L xylose at a yield of 0.99 mol/mol (0.51 g/g) in the flask culture. To obtain a higher production titer using the strains, fed-batch fermentation was performed using 2% (w/v) glucose and 2% (w/v) xylose minimal medium and the engineered C. glutamicum strains. For the production of ethylene glycol, SL-1A pIEARKT was used, and an OD600 of 58.3 at 16 h was attained after the glucose was used completely (Figure 5A). No glycolate was detected in the flask culture. However, no bioconversion of xylose to ethylene glycol was achieved in the absence of glucose. Xylose was accumulated during the fed-batch culture, and the titer of ethylene glycol was not increased after the glucose was depleted. This could be due to the imbalance of NADPH regeneration via DHAP as a three-carbon intermediate of xylose and no NADPH synthesis in the pentose phosphate pathway from xylose (the xylB mutant). In the previous fed-batch fermentation, 108.18 g/L ethylene glycol was produced from xylose via the modulated Dahms pathway using the engineered E. coli (no xylA mutant), in which the NADPH pool could be balanced with the production of ethylene glycol.12 Thus, it may be important to further engineer C. glutamicum for recycling NADPH pools.
Figure 5.
Fed-batch culture for the bioconversion of ethylene glycol and glycolate from xylose using recombinant C. glutamicum. (A) Growth profiles (square), glucose consumption (circle), xylose consumption (triangle), and ethylene glycol production (diamond) are shown for SL-1A pIEARKT in 2% (w/v) glucose and 2% (w/v) xylose in CgXII minimal medium using fed-batch fermentation. (B) Growth profiles (square), glucose consumption (circle), xylose consumption (triangle), and glycolate production (diamond) are shown for SL-1 pIEAKD in 2% (w/v) glucose and 2% (w/v) xylose in CgXII minimal medium using fed-batch fermentation.
For glycolate production, the SL-1 pIEAKD strain reached an OD600 of 56.5 at 24 h, when the glucose was completely depleted (Figure 5B). Then, xylose was rapidly consumed and glycolate was produced (8.3 g/L at 36 h). The final OD600 was 56.2. Subsequently, four additional xylose feedings were added to the fermenter, producing 24.1 g/L glycolate with a yield of 0.48 g/g (0.94 mol/mol) at 144 h. The levels of none of the organic acids were measured, and less than 0.7 g/L xylitol was detected in the fed-batch culture. The xylose conversion rate in the fed-batch fermentation (0.31 mmol/gDW/h) was increased by 1.4-fold when compared with the flask culture (0.23 mmol/gDW/h).
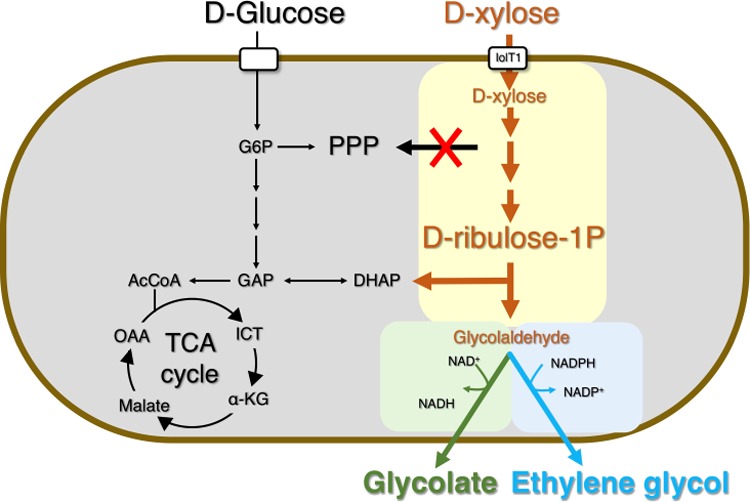
In summary, we have reported the bioconversion of xylose to ethylene glycol and glycolate using recombinant C. glutamicum via the synthetic Ru1P pathway with a molar yield of 0.31 (ethylene glycol) and 0.99 (glycolate; without the reductive pathway from glyoxylate). On the basis of the calculation of the theoretical maximum yield using different xylose utilization pathways,6 synthetic xylulose 1-phosphate, Ru1P pathways, and Dahms pathway, a molar yield of 1.0 for ethylene glycol and 1.0 for glycolate (without the reductive pathway from glyoxylate) and 2.0 for glycolate (with an additional reductive pathway from glyoxylate) can be achieved.8,29 Although the bioconversion of xylose to glycolate was successfully achieved at an almost maximum molar yield of 0.99, dynamic control and fine-tuned control of the valve during the pentose phosphate pathway and synthetic two-carbon pathways in C. glutamicum may be essential to obtain high titers of ethylene glycol and glycolate.
In addition, exploring a reconstructed metabolic network and flux balance analysis30 and evolving microbial cells,31 might help improve the final titer, rate, and yield of ethylene glycol and glycolate from glucose and xylose in the lignocellulosic hydrolysates.32 The rapid strain development of C. glutamicum could be achieved using the CRISPR-Cas systems including Coryne-CRISPR inference33 and CoryneCR12-STOP for systematic gene repression and gene deletion in C. glutamicum. Subsequently, automated strain development using a robotic platform could be combined for system metabolic engineering.
Methods
Bacterial Strains and Culture Conditions
In this study, E. coli DH5α34 and C. glutamicum ATCC 13032 (wild-type) were used (Table 1). The E. coli strain was grown in lysogeny broth (LB) containing 10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl at 37 °C on a rotary shaker at 200 rpm. When appropriate, the medium was supplemented with 25 μg/mL chloramphenicol, 50 μg/mL kanamycin, or 100 μg/mL spectinomycin. C. glutamicum ATCC 13032 and its derivatives were cultivated in brain heart infusion salt (BHIS) medium at 30 °C on a rotary shaker at 120 rpm.35 When appropriate, the medium was supplemented with 7.5 μg/mL chloramphenicol, 25 μg/mL kanamycin, and/or 50 μg/mL spectinomycin.
For flask cultivation, the C. glutamicum strains were precultivated in BHIS medium overnight and then incubated aerobically in CgXII defined medium (50 mL medium in a 250 mL baffled Erlenmeyer flask) containing either xylose as the sole carbon source or a mixture of glucose and xylose at 30 °C on a rotary shaker at 120 rpm.36,37 For both ethylene glycol and glycolate production, cell cultivation was performed in the presence of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG).
Construction of Plasmids and Recombinant Strains
For the bioconversion of xylose to both ethylene glycol and glycolate, dte encoding d-tagatose 3-epimerase from Pseudomonas cichorii and xylA (xylose isomerase), fucK (l-fuculokinase), and fucA (l-fuculose phosphate aldolase) from E. coli were synthesized (Genscript) with codon optimization for C. glutamicum, and the synthetic genes were cloned into a CoryneBrick vector15 (Table 1). In addition, yqhD encoding aldehyde reductase and aldA encoding aldehyde dehydrogenase from E. coli were codon-optimized and synthesized (Genscript) for ethylene glycol and glycolate production, respectively.
The resulting plasmids were introduced into C. glutamicum by electroporation, and strain validation was performed using colony PCR and DNA sequencing.35,38 For Corynebacterial CRISPR-based Recombineering using Cas12a to generate the STOP codon (CoryneCR12-STOP), crRNA plasmids carrying crRNAs were constructed from pJYS2_crtYf.39 Target crRNAs for the recombineering were cloned into the standard vector using a primer containing a target-specific protospacer region. A 59 bp single-stranded oligodeoxynucleotide (lagging strand ssODN) was designed to insert the STOP codons (TAG and TAA) for the target gene as a nonsense mutation. Target crRNAs and ssODN were cotransferred into the competent corynebacterial cells harboring pJYS1Ptac. The oligo primers used for the cloning and ssODNs for the recombineering are listed in Tables S1 and S2.
Quantification and Analysis of Metabolites using High-Performance Liquid Chromatography
Glucose, xylose, and organic acids, including ethylene glycol and glycolate, in the supernatant were quantified using high-performance liquid chromatography (HPLC), as described previously33,38 Briefly, the culture supernatant was passed through a syringe filter (0.45 μm pore size). The concentrations of glucose and organic acids were detected with HPLC (Agilent 1260 equipped with a diode array detector and a refractive index detector and a Hi-Plex H, 7.7 × 300 mm, column; Agilent Technologies, Waldbronn, Germany) under the following conditions: sample volume, 10 μL; mobile phase, 5 mM H2SO4; flow rate, 0.6 mL/min; and column temperature, 65 °C.
Fed-Batch Culture Conditions
Fed-batch fermentation was conducted in a 5 L jar fermenter (MARADO-05S-XS; CNS Inc., Daejeon, South Korea) containing 2 L culture medium. The seed culture was prepared in a 250 mL Erlenmeyer flask containing 100 mL CgXII medium with 2% (w/v) glucose and 2% (w/v) xylose at 30 °C on a rotary shaker at 120 rpm. The seed culture was transferred to a fermenter at an OD600 of 1. The fermenter contained 2 L CgXII medium with 2% (w/v) glucose and 2% (w/v) xylose. When required, 7.5 μg/mL chloramphenicol was used in the main cultivation. For induction of the target gene in the CoryneBrick vectors, 1 mM IPTG was added. The culture pH was adjusted to 7 by adding either 3 N NaOH or 3 N HCl. Filtered air was constantly supplied at 2 L/min, and the dissolved oxygen (DO) level was maintained at more than 30% by automatically controlling the agitation speed between 400 and 800 rpm. The feeding solution contained 700 g/L xylose. Pulse feeding solutions were added at 10 g/L using the DO-stat feeding strategy, where the feeding solution was added when the DO level was 80%.
Acknowledgments
The authors thank Mieun Lee, Jae Hyun Park, and Yoojin Lee at Sungkyunkwan University for technical support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b02805.
Oligonucleotides used for gene cloning in this study (Table S1); single-stranded oligonucleotides used for the generation of CoryneCR12-STOP (Table S2); time course of growth, xylose consumption, and ethylene glycol production by the engineered C. glutamicum with fucO expression (Figure S1); sequence chromatograms of the editing sites at iolR and ald of C. glutamicum and sequence verification for the mutants (Figure S2) (PDF)
Author Contributions
S.S.L. and H.M.W. designed the experiments, analyzed the data, and wrote the manuscript. S.S.L. performed the experiments. J.-i.C. and H.M.W. guided the scope of the project and provided critical input for the manuscript. All authors read and approved the final manuscript.
This study was supported by the Basic Science Research Program (2017R1A2B2002566) through the National Research Foundation of Korea. In addition, this study was partially supported by the Golden Seed Project (213008-05-3-WT911) grant, funded by the Ministry of Agriculture and Ministry of Oceans and Fisheries. H.M.W was supported by the Technology Innovation Program funded by the Ministry of Trade, Industry and Energy (No. 20000158).
The authors declare no competing financial interest.
Supplementary Material
References
- Choi S. Y.; Park S. J.; Kim W. J.; Yang J. E.; Lee H.; Shin J.; Lee S. Y. One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. Nat Biotechnol 2016, 34, 435–440. 10.1038/nbt.3485. [DOI] [PubMed] [Google Scholar]
- Matsumoto K.; Ishiyama A.; Sakai K.; Shiba T.; Taguchi S. Biosynthesis of glycolate-based polyesters containing medium-chain-length 3-hydroxyalkanoates in recaombinnt Escherichia coli expressing engineered polyhydroxyalkanoate synthase. J. Biotechnol. 2011, 156, 214–217. 10.1016/j.jbiotec.2011.07.040. [DOI] [PubMed] [Google Scholar]
- Song C. W.; Lee J.; Lee S. Y. Genome engineering and gene expression control for bacterial strain development. Biotechnol. J. 2015, 10, 56–68. 10.1002/biot.201400057. [DOI] [PubMed] [Google Scholar]
- Becker J.; Rohles C. M.; Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab. Eng. 2018, 50, 122–141. 10.1016/j.ymben.2018.07.008. [DOI] [PubMed] [Google Scholar]
- Gong Z.; Nielsen J.; Zhou Y. J. Engineering Robustness of Microbial Cell Factories. Biotechnol. J. 2017, 12, 1700014 10.1002/biot.201700014. [DOI] [PubMed] [Google Scholar]
- Cam Y.; Alkim C.; Trichez D.; Trebosc V.; Vax A.; Bartolo F.; Besse P.; François J. M.; Walther T. Engineering of a Synthetic Metabolic Pathway for the Assimilation of (d)-Xylose into Value-Added Chemicals. ACS Synth. Biol. 2015, 5, 607–618. 10.1021/acssynbio.5b00103. [DOI] [PubMed] [Google Scholar]
- Alkim C.; Cam Y.; Trichez D.; Auriol C.; Spina L.; Vax A.; Bartolo F.; Besse P.; François J. M.; Walther T. Optimization of ethylene glycol production from (d)-xylose via a synthetic pathway implemented in Escherichia coli. Microb. Cell Fact. 2015, 14, 127 10.1186/s12934-015-0312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B.; Li Z. J.; De Mey M.; Lim C. G.; Zhang H.; Hoeltgen C.; Stephanopoulos G. Efficient utilization of pentoses for bioproduction of the renewable two-carbon compounds ethylene glycol and glycolate. Metab. Eng. 2016, 34, 80–87. 10.1016/j.ymben.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Liu H.; Ramos K. R. M.; Valdehuesa K. N. G.; Nisola G. M.; Lee W.-K.; Chung W.-J. Biosynthesis of ethylene glycol in Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 97, 3409–3417. 10.1007/s00253-012-4618-7. [DOI] [PubMed] [Google Scholar]
- Cabulong R. B.; Lee W.-K.; Bañares A. B.; Ramos K. R. M.; Nisola G. M.; Valdehuesa K. N. G.; Chung W.-J. Engineering Escherichia coli for glycolic acid production from D-xylose through the Dahms pathway and glyoxylate bypass. Appl. Microbiol. Biotechnol. 2018, 102, 2179–2189. 10.1007/s00253-018-8744-8. [DOI] [PubMed] [Google Scholar]
- Liu M.; Ding Y.; Xian M.; Zhao G. Metabolic engineering of a xylose pathway for biotechnological production of glycolate in Escherichia coli. Microb. Cell Fact. 2018, 17, 51 10.1186/s12934-018-0900-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae T. U.; Choi S. Y.; Ryu J. Y.; Lee S. Y. Production of ethylene glycol from xylose by metabolically engineered Escherichia coli. AIChE J. 2018, 64, 4193–4200. 10.1002/aic.16339. [DOI] [Google Scholar]
- Deng Y.; Ma N.; Zhu K.; Mao Y.; Wei X.; Zhao Y. Balancing the carbon flux distributions between the TCA cycle and glyoxylate shunt to produce glycolate at high yield and titer in Escherichia coli. Metab. Eng. 2018, 46, 28–34. 10.1016/j.ymben.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Li Z.-J.; Qiao K.; Shi W.; Pereira B.; Zhang H.; Olsen B. D.; Stephanopoulos G. Biosynthesis of poly(glycolate- co -lactate- co -3-hydroxybutyrate) from glucose by metabolically engineered Escherichia coli. Metab. Eng. 2016, 35, 1–8. 10.1016/j.ymben.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Kang M. K.; Lee J.; Um Y.; Lee T. S.; Bott M.; Park S. J.; Woo H. M. Synthetic biology platform of CoryneBrick vectors for gene expression in Corynebacterium glutamicum and its application to xylose utilization. Appl. Microbiol. Biotechnol. 2014, 98, 5991–6002. 10.1007/s00253-014-5714-7. [DOI] [PubMed] [Google Scholar]
- Radek A.; Krumbach K.; Gatgens J.; Wendisch V. F.; Wiechert W.; Bott M.; Noack S.; Marienhagen J. Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of D-xylose containing substrates. J. Biotechnol. 2014, 192, 156–160. 10.1016/j.jbiotec.2014.09.026. [DOI] [PubMed] [Google Scholar]
- Kim D.; Woo H. M. Deciphering bacterial xylose metabolism and metabolic engineering of industrial microorganisms for use as efficient microbial cell factories. Appl. Microbiol. Biotechnol. 2018, 102, 9471–9480. 10.1007/s00253-018-9353-2. [DOI] [PubMed] [Google Scholar]
- Baritugo K.-A.; Kim H. T.; David Y.; Choi J.-i.; Hong S. H.; Jeong K. J.; Choi J. H.; Joo J. C.; Park S. J. Metabolic engineering of Corynebacterium glutamicum for fermentative production of chemicals in biorefinery. Appl. Microbiol. Biotechnol. 2018, 102, 3915–3937. 10.1007/s00253-018-8896-6. [DOI] [PubMed] [Google Scholar]
- Joo Y.-C.; You S. K.; Shin S. K.; Ko Y. J.; Jung K. H.; Sim S. A.; Han S. O. Bio-Based Production of Dimethyl Itaconate From Rice Wine Waste-Derived Itaconic Acid. Biotechnol. J. 2017, 12, 1700114 10.1002/biot.201700114. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Huang J.; Wu Y.; Liu D. Metabolic engineering of Corynebacterium glutamicum for the de novo production of ethylene glycol from glucose. Metab. Eng. 2016, 33, 12–18. 10.1016/j.ymben.2015.10.013. [DOI] [PubMed] [Google Scholar]
- Zahoor A.; Otten A.; Wendisch V. F. Metabolic engineering of Corynebacterium glutamicum for glycolate production. J. Biotechnol. 2014, 192, 366–375. 10.1016/j.jbiotec.2013.12.020. [DOI] [PubMed] [Google Scholar]
- Choi J. W.; Yim S. S.; Jeong K. J. Development of a high-copy-number plasmid via adaptive laboratory evolution of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2017, 102, 873–883. 10.1007/s00253-017-8653-2. [DOI] [PubMed] [Google Scholar]
- Yim S. S.; Choi J. W.; Lee S. H.; Jeon E. J.; Chung W.-J.; Jeong K. J. Engineering of Corynebacterium glutamicum for Consolidated Conversion of Hemicellulosic Biomass into Xylonic Acid. Biotechnol. J. 2017, 12, 1700040 10.1002/biot.201700040. [DOI] [PubMed] [Google Scholar]
- Tenhaef N.; Brüsseler C.; Radek A.; Hilmes R.; Unrean P.; Marienhagen J.; Noack S. Production of d-xylonic acid using a non-recombinant Corynebacterium glutamicum strain. Bioresour. Technol. 2018, 268, 332–339. 10.1016/j.biortech.2018.07.127. [DOI] [PubMed] [Google Scholar]
- Radek A.; Müller M.-F.; Gätgens J.; Eggeling L.; Krumbach K.; Marienhagen J.; Noack S. Formation of xylitol and xylitol-5-phosphate and its impact on growth of d -xylose-utilizing Corynebacterium glutamicum strains. J. Biotechnol. 2016, 231, 160–166. 10.1016/j.jbiotec.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Brüsseler C.; Radek A.; Tenhaef N.; Krumbach K.; Noack S.; Marienhagen J. The myo-inositol/proton symporter IolT1 contributes to d-xylose uptake in Corynebacterium glutamicum. Bioresour. Technol. 2017, 249, 953–961. 10.1016/j.biortech.2017.10.098. [DOI] [PubMed] [Google Scholar]
- Pauling J.; Rottger R.; Tauch A.; Azevedo V.; Baumbach J. CoryneRegNet 6.0--Updated database content, new analysis methods and novel features focusing on community demands. Nucleic Acids Res. 2011, 40, D610–D614. 10.1093/nar/gkr883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witthoff S.; Mühlroth A.; Marienhagen J.; Bott M. C1Metabolism in Corynebacterium glutamicum: an Endogenous Pathway for Oxidation of Methanol to Carbon Dioxide. Appl. Environ. Microbiol. 2013, 79, 6974–6983. 10.1128/AEM.02705-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S.; Park J.; Heo Y. B.; Woo H. M. Case study of xylose conversion to glycolate in Corynebacterium glutamicum: Current limitation and future perspective of the CRISPR-Cas systems. Enzyme Microb. Technol. 2020, 132, 109395 10.1016/j.enzmictec.2019.109395. [DOI] [PubMed] [Google Scholar]
- Lee S. Y.; Woo H. M.; Lee D.-Y.; Choi H. S.; Kim T. Y.; Yun H. Systems-level analysis of genome-scalein silico metabolic models using MetaFluxNet. Biotechnol. Bioprocess Eng. 2005, 10, 425–431. 10.1007/BF02989825. [DOI] [Google Scholar]
- Lee J.; Saddler J. N.; Um Y.; Woo H. M. Adaptive evolution and metabolic engineering of a cellobiose- and xylose- negative Corynebacterium glutamicum that co-utilizes cellobiose and xylose. Microb. Cell Fact. 2016, 15, 20 10.1186/s12934-016-0420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S.; Mishra G.; Roy A. Metabolic Engineering of Bacteria for Renewable Bioethanol Production from Cellulosic Biomass. Biotechnol. Bioprocess Eng. 2019, 1–22. 10.1007/s12257-019-0134-2. [DOI] [Google Scholar]
- Park J.; Shin H.; Lee S. M.; Um Y.; Woo H. M. RNA-guided single/double gene repressions in Corynebacterium glutamicum using an efficient CRISPR interference and its application to industrial strain. Microb. Cell Fact. 2018, 17, 4 10.1186/s12934-017-0843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Eggeling L.; Reyes O.. Experiments. In Handbook of Corynebacterium glutamicum, 1st ed.; Eggeling L., Bott M., Eds.; CRC Press: Boca Raton, 2005; Chapter 23, pp 535–566. [Google Scholar]
- Kang M.-K.; Eom J.-H.; Kim Y.; Um Y.; Woo H. M. Biosynthesis of pinene from glucose using metabolically-engineered Corynebacterium glutamicum. Biotechnol. Lett. 2014, 36, 2069–2077. 10.1007/s10529-014-1578-2. [DOI] [PubMed] [Google Scholar]
- Jo S.; Yoon J.; Lee S.-M.; Um Y.; Han S. O.; Woo H. M. Modular pathway engineering of Corynebacterium glutamicum to improve xylose utilization and succinate production. J. Biotechnol. 2017, 258, 69–78. 10.1016/j.jbiotec.2017.01.015. [DOI] [PubMed] [Google Scholar]
- Yoon J.; Woo H. M. CRISPR interference-mediated metabolic engineering of Corynebacterium glutamicum for homo-butyrate production. Biotechnol. Bioeng. 2018, 115, 2067–2074. 10.1002/bit.26720. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Qian F.; Yang J.; Liu Y.; Dong F.; Xu C.; Sun B.; Chen B.; Xu X.; Li Y.; Wang R.; Yang S. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 2017, 8, 15179 10.1038/ncomms15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Dusch N.; Puhler A.; Kalinowski J. Expression of the Corynebacterium glutamicum panD gene encoding L-aspartate-alpha-decarboxylase leads to pantothenate overproduction in Escherichia coli. Appl. Environ. Microbiol. 1999, 65, 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.