Infection of the host with Mycobacterium avium subsp. paratuberculosis results in chronic and progressive enteritis that traverses both subclinical and clinical stages. The mechanism(s) for the shift from an asymptomatic subclinical disease state to advanced clinical disease is not fully understood.
KEYWORDS: Mycobacterium avium subsp. paratuberculosis, cattle, immunity
ABSTRACT
Infection of the host with Mycobacterium avium subsp. paratuberculosis results in chronic and progressive enteritis that traverses both subclinical and clinical stages. The mechanism(s) for the shift from an asymptomatic subclinical disease state to advanced clinical disease is not fully understood. In the present study, naturally infected dairy cattle were divided into subclinical and clinical infection groups, along with noninfected control cows of similar parity, to study host immune responses in different stages of infection. Both infection groups had higher levels of secretion of gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin-2 (IL-2) than control cows, whereas only clinical cows had increased secretion of IL-10, IL-12, and IL-18 upon stimulation of peripheral blood mononuclear cells (PBMCs) with antigen. Conversely, secretion of IL-17Α was decreased for clinical cows compared to subclinical and control cows. Proinflammatory cytokine genes were upregulated only for subclinical cows, whereas increased IL-10 and IL-17 gene expression levels were observed for both infection groups. Increased CD4+, CD8+, and γδ T cell receptor-positive (TCR+) T cells were observed for subclinical cows compared to clinical cows. Although clinical cows expressed antigen-specific immune responses, the profile for subclinical cows was one of a dominant proinflammatory response to infection. We reason that a complex coordination of immune responses occurs during M. avium subsp. paratuberculosis infection, with these responses shifting as the host transitions through the different stages of infection and disease (subclinical to clinical). A further understanding of the series of events characterized by Th1/Th2/Th17 responses will provide mechanisms for disease progression and may direct insightful intervention strategies.
INTRODUCTION
Johne’s disease (paratuberculosis), caused by infection with Mycobacterium avium subsp. paratuberculosis, has become widespread in U.S. dairy herds in recent years (1) and has worldwide presence in cattle, sheep, goats, as well as other ruminant and nonruminant species. The actual cost of this disease to the average producer is still a relative unknown (2, 3), but much of this uncertainty stems from the hierarchy of disease states within a herd, with few cases of clinical disease and the majority of infected animals maintaining a subclinical infection status (4). Although economic losses have been associated with both disease states, greater losses are associated with clinical disease (5–7).
Paratuberculosis in cattle is characterized by a protracted period of asymptomatic subclinical infection during which the host seemingly controls the disease by thwarting bacterial replication within macrophages (8, 9). A majority of infected animals remain in an asymptomatic disease state throughout their productive life span, dominated by Th1-mediated host immunity. As infection advances and the disease state becomes clinical, host immunity shifts to a more Th2-dominated immunity. However, this paradigm is an oversimplification, as a significant overlap of Th1/Th2 immunity may exist in infected animals, even those that are eventually culled because of advanced clinical disease (10, 11). Despite stalwart research efforts, there remains a basic lack of understanding as to the stimulus that results in clinical disease. Stressors as well as host genetics appear to play some role in the transition, but dysregulation of host immunity is at the forefront of causative factors (12–15). While antigen-presenting cells (APCs) such as dendritic cells and macrophages are the first defense against this intracellular pathogen, adaptive immune responses defined by T cell-mediated immunity are critical for the control of bacterial replication in the host (16). The paradigm of Th1/Th2 adaptive immunity in M. avium subsp. paratuberculosis infection is still being unraveled, and Th17-mediated immunity is still relatively undefined.
The present study was conducted to further characterize host immune responses to M. avium subsp. paratuberculosis infection in subclinical and clinical stages of disease in naturally infected dairy cattle. Results from this study will allow a more comprehensive view of participant immune markers that may differentiate between subclinical and clinical infection and provide information on immune dysregulation allowing animals to succumb to a more advanced state.
RESULTS
Subclinical and clinical disease increases proinflammatory cytokine secretion.
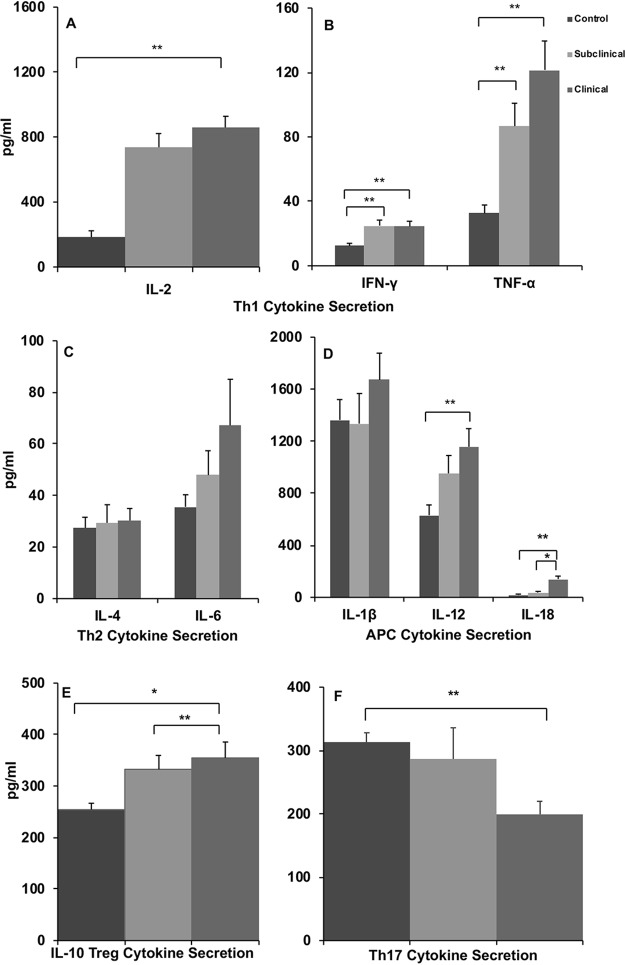
In the present study, host immune responses were compared for noninfected control cows and cows naturally infected with M. avium subsp. paratuberculosis, in both subclinical and clinical stages of disease. Cytokine secretion was used to characterize the influence of the disease state on activated peripheral blood mononuclear cells (PBMCs). Measurement of cytokine secretion in cell-free supernatants after stimulation of cells with a whole-cell sonicate of M. avium subsp. paratuberculosis (MPS) demonstrated significant (P < 0.05) increases in interleukin-2 (IL-2), gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α) for infected animals, regardless of status, compared to noninfected controls (Fig. 1A and B). There were no significant effects of infection status on the secretion of IL-4, IL-6, or IL-1β, although increased secretion of IL-1β and IL-6, both proinflammatory cytokines, trended for cows in the clinical disease group (clinical cows) (Fig. 1C and D). Clinical cows further demonstrated higher levels of secretion (P < 0.05) of IL-12 and IL-18 after MPS stimulation of cells in culture than did control cows (Fig. 1D). Additionally, the level of secretion of IL-10, a key regulatory cytokine, was higher (P < 0.01) in clinical cows and intermediate (P < 0.04) for subclinical cows (Fig. 1E). Interestingly, IL-10 secretion yielded a similar pattern for nonstimulated (NS) cultures, with higher levels of constitutive secretion of IL-10 for clinical cows than for controls (data not shown). Finally, clinical disease resulted in reduced (P < 0.05) IL-17A secretion by PBMCs, an effect that was notable in MPS cultures (Fig. 1F) as well as NS and concanavalin A (ConA)-stimulated cell cultures compared to secretion by cells from noninfected control cows (data not shown).
FIG 1.
Secretion of Th1-mediated interleukin-2 (IL-2) (A), interferon gamma (IFN-γ) (B), and tumor necrosis factor alpha (TNF-α) (B); Th2-mediated interleukin-4 (IL-4) and interleukin-6 (IL-6) (C); APC-mediated interleukin-1β (IL-1β), interleukin-12 (IL-12), and interleukin-18 (IL-18) (D); Treg-mediated interleukin-10 (IL-10) (E); and Th17-mediated interleukin-17A (IL-17A) (F) cytokines (picograms per milliliter). PBMCs were isolated from control noninfected cows and cows in subclinical and clinical stages of disease and stimulated for 24 h in vitro with a whole-cell sonicate of Mycobacterium avium subsp. paratuberculosis (MPS). Cell-free supernatants were harvested and then analyzed by using an ELISA or Aushon Biosystems bovine multiplex custom arrays. Data are expressed as means ± SEM (*, P < 0.01; **, P < 0.05).
Upregulation of cytokine gene expression in cows with subclinical disease.
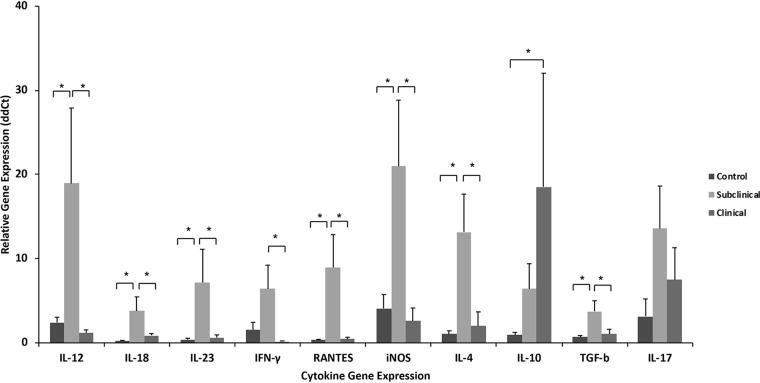
Gene expression can provide key information on the ability of cells to respond to infection. Despite this, gene expression does not always align itself with protein secretion due to posttranslational modifications. In the present study, the expression of cytokine genes proffered a somewhat different pattern from that of secreted cytokines, with greater upregulation noted for cows in the subclinical treatment group (Fig. 2). Significant upregulation (P < 0.05) of IFN-γ, IL-12, IL-18, RANTES, IL-4, IL-23, transforming growth factor β (TGF-β), and inducible nitric oxide synthase (iNOS) genes was noted in PBMCs isolated from subclinical cows and cultured for 24 h with MPS compared to the control and clinical groups. In contrast, upregulation of IL-10 and IL-17 genes was noted in stimulated cells for both infected groups compared to control noninfected cows, with only IL-10 gene expression being significantly (P < 0.05) upregulated for clinical cows. Similar patterns of cytokine gene expression were noted after stimulation of cells with ConA across infection treatment groups (data not shown).
FIG 2.
Relative gene expression analysis for cytokines was performed using custom TaqMan gene expression assays for bovine IL-4, IL-10, IL-12, IL-17A, IL-18, IL-23, IFN-γ, TGF-β, iNOS, and RANTES. PBMCs were isolated from control noninfected cows and cows in subclinical and clinical stages of disease and stimulated for 24 h in vitro with a whole-cell sonicate of Mycobacterium avium subsp. paratuberculosis (MPS), followed by RNA extraction. A eukaryotic 18S rRNA endogenous control (FAM-MGB probe, non-prime limited) was used as an internal control to normalize RNA content between samples. Nonstimulated cell controls for each cow were used as the calibrator. All reactions were performed in triplicate, and data were analyzed with the method. Data are expressed as means ± SEM (*, P < 0.05).
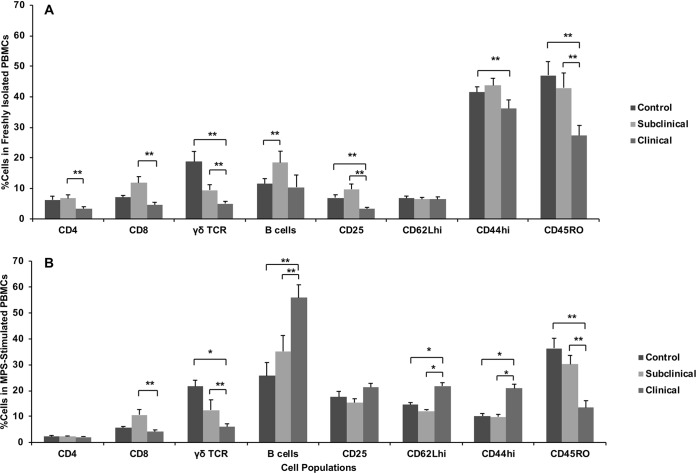
Subclinical disease results in increased T cell subpopulations in freshly isolated PBMCs.
Dissimilar and, at times, divergent results were observed in cell subpopulations for cows in different stages of infection. The first comparison is the stratification of cell subpopulations in freshly isolated PBMCs compared to PBMCs that had been stimulated with MPS antigen for 144 h (Fig. 3). Evaluating the cell subpopulations in freshly isolated PBMCs allowed us to more directly evaluate effects of in vivo infection status. Interestingly, PBMCs isolated from clinical cows were comprised of lower (P < 0.05) percentages of CD4+ and CD8+ cell subpopulations than those from subclinically infected cows (Fig. 3A). Similarly, percentages of γδ T cell receptor-positive (TCR+) cells were lower (P < 0.05) for both infected groups, but the lowest percentage was noted for cows with clinical infection status. An increase (P < 0.05) in the percentage of B cells was observed for subclinical cows compared to control noninfected cows. The percentages of PMBCs that expressed activation markers, CD44hi and CD45RO, were also lower (P < 0.05) for clinical cows than for the control and subclinical cows.
FIG 3.
CD4+, CD8+, γδ TCR+, B cell-positive, CD25+, CD62Lhi, CD44hi, and CD45RO+ lymphocyte populations within total PBMCs isolated from control noninfected cows and cows in subclinical and clinical stages of disease. Cells were either freshly isolated (A) or stimulated in vitro for 6 days with a whole-cell sonicate of Mycobacterium avium subsp. paratuberculosis (MPS) (B). Mononuclear cells, based on forward- and side-scatter characteristics, were analyzed for cell surface marker expression. Cell subpopulations are expressed as a percentage of the total mononuclear cell population to determine effects of infection status on population shifts. Data are expressed as means ± SEM (*, P < 0.01; **, P < 0.05).
Stimulation of PBMCs with M. avium subsp. paratuberculosis antigen decreases CD4+ T cells but increases B cells.
Ex vivo culture of PBMCs with MPS for 6 days allowed for antigen recall responses to occur, and changes in percentages of some cell populations were observed, as noted in Fig. 3B. Although effects of infection status remained relatively similar for CD8+ and γδ TCR+ T cell subpopulations, exposure to the antigen preparation resulted in an overall decline in CD4+ T cells for all treatment groups and dispelled any effects due to infection status. In contrast, the B cell population increased (P < 0.05) dramatically for clinical cows compared to the other treatment groups. Additionally, clinical disease resulted in greater numbers of PBMCs expressing CD62Lhi and CD44hi upon stimulation of cells with antigen. There were no observed differences due to infection status of cows in the percentage of monocytes (CD14) or natural killer (NK) (CD335) cells present in freshly isolated PBMCs or after antigen stimulation of cells (data not shown).
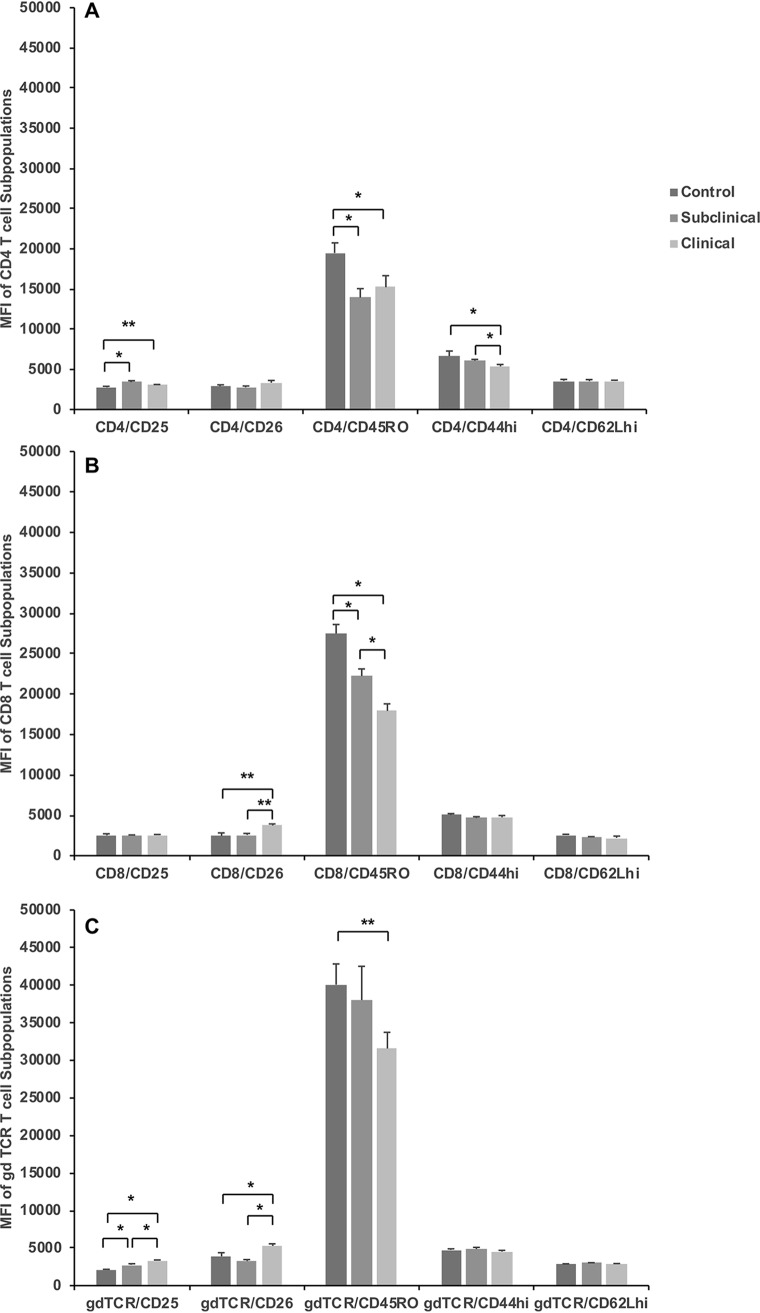
Clinical disease markedly decreases expression of CD45RO on T cells.
Key differences due to infection status of cows were observed when activation marker expression was assessed within T cell subpopulations stimulated for 144 h with M. avium subsp. paratuberculosis antigen (Fig. 4). Within the CD4+ T cell population, the CD25 expression level was higher (P < 0.01) for subclinical cows, with an intermediate response (P = 0.07) noted for clinical cows (Fig. 4A). In contrast, a decline in CD45RO (P < 0.01) expression was observed for infected cows regardless of status, and decreased CD44hi (P < 0.01) expression was noted for cows that were clinically affected. A decrease in CD45RO expression was also noted on CD8+ T cells (P < 0.0001) after stimulation of PBMCs with MPS for naturally infected cows (Fig. 4B). Although subclinically infected cows had the highest (P < 0.05) proportion of CD8+ T cells, cows with clinical infection had higher (P < 0.05) expression levels of CD26 on CD8+ T cells than did subclinical and control animals (Fig. 4B). The most interesting impact of infection status was observed for γδ TCR+ T cells and the expression of activation markers within that subpopulation (Fig. 4C). Infection with M. avium subsp. paratuberculosis resulted in a stair-step-type reduction in the number of γδ TCR+ T cells within the total PBMCs, with the subclinical cows having an intermediate percentage of cells (12.52%) and the clinical cows having the lowest percentage of cells (6.06%), compared to control noninfected cows (21.63%). Conversely, the expression level of CD25 on the γδ TCR+ subpopulation was higher for clinical cows (P < 0.0001) and intermediate for subclinical cows (P < 0.0002), compared to control cows. Additionally, CD26 expression was increased (P < 0.01) on γδ TCR+ T cells isolated from clinical cows (Fig. 4C). Once again, a reduction (P < 0.05) in CD45RO+ expression on γδ TCR+ T cells was observed for clinical cows.
FIG 4.
Mean fluorescence intensities (MFI) of expression of activation markers on lymphocyte populations of CD4+ (A), CD8+ (B), and γδ TCR+ (C) T cells within total PBMCs isolated from control noninfected cows and cows in subclinical and clinical stages of disease. Cells were stimulated in vitro for 6 days with a whole-cell sonicate of Mycobacterium avium subsp. paratuberculosis (MPS). Mononuclear cells, based on forward- and side-scatter characteristics, were analyzed for cell surface marker expression. Activation markers expressed on CD4, CD8, and γδ TCR+ T cell subsets are expressed as MFI. Data are expressed as means ± SEM (*, P < 0.01; **, P < 0.05).
Expression of CD40, CD80, and CD86 was also assessed on CD14+ cells (data not shown). Although no differences were noted for freshly isolated PBMCs, trending increases (P < 0.08) in CD80 and CD86 expression were demonstrated for CD14+ cells from subclinical cows after stimulation with antigen compared to cells from either control or clinical cows (data not shown).
DISCUSSION
In this study, subclinical and clinical cow statuses were stratified using standard diagnostic tools for paratuberculosis, including antigen-specific IFN-γ responses, M. avium subsp. paratuberculosis-specific antibody, and fecal shedding of M. avium subsp. paratuberculosis. Although cattle classified as having clinical disease demonstrated higher serum antibody levels (sample/positive [S/P] ratio of 2.73) and greater fecal shedding (6,305 CFU/g) than subclinical cows (S/P ratio of 0.21 and 3 CFU/g; respectively), antigen-specific IFN-γ responses (0.23 [MPS-nonstimulated Abs450nm]) were similar for cows regardless of infection status, confirming that a definitive association of Th1 and Th2 immune responses with infection status is more complicated than previously thought. Nevertheless, antigen-specific IFN-γ responses generally precede the development of antibody responses or fecal shedding of M. avium subsp. paratuberculosis.
Although these basic diagnostic tools are helpful, aligning results of cytokine secretion and expression with host immunity would allow us to better interpret how the infection status of cattle may impact the ability to control infection. Similar to the above-mentioned whole-blood IFN-γ responses, both subclinical and clinical cows had equivalent responses for antigen-specific IL-2, IFN-γ, and TNF-α secretion in culture supernatants, suggesting that Th1 responsiveness was not impacted by the stage of infection. Despite this, elevated levels of IL-6 were also noted in cows with a clinical infection status. This is interesting, as IL-6 plays a role in driving CD4+ T cells to a Th2 phenotype (17), and high levels of serum antibody to M. avium subsp. paratuberculosis in clinical cows provide evidence of a Th2 immune response. These results suggest that as disease progresses, cows will gain the capability of invoking Th2-mediated immune responses but maintain the ability to mount Th1-mediated immunity as well.
Further indication that a strong Th1-mediated immune response is still apparent in clinical cows is the elevation in the levels of the proinflammatory cytokines IL-12 and IL-18 observed here. Both IL-12 and IL-18 are engaged in critical roles in the induction of IFN-γ in mycobacterial infections (18, 19), and increases in the levels of these cytokines in the present study align with increased secretion of IFN-γ. These cytokines are secreted predominantly by APCs, and elevated levels seem contradictory to dogma that macrophage function is diminished in clinical disease, thus allowing for intracellular replication of M. avium subsp. paratuberculosis (20–22). Secretion of these cytokines in culture suggests that cows with a clinical infection status retain the ability to respond to antigenic stimulation, and the greater bacterial burden of the host may even elicit heightened responses, as we note here, compared to subclinical cows.
The upregulation of these proinflammatory cytokines during advanced disease is problematic for the host, eventually resulting in damage to the tissue(s) at the site of infection (23). A role of IL-10 in tempering proinflammatory responses becomes more critical in animals with chronic inflammatory responses to pathogens, such as cows in the clinical stage of paratuberculosis (24). The IL-10 level was more elevated in clinical cows in the present study, indicating that a regulatory immune response was in play. It is also important to note that despite its regulatory role, IL-10 may inhibit macrophage function, allowing intracellular replication of M. avium subsp. paratuberculosis (25). Additionally, sources of IL-10 include not only regulatory T cells (Tregs) but also Th2 T cells and macrophages, and IL-10 expression is tightly controlled by both itself and IFN-γ (26). It is clear from recent studies that although macrophages are present in higher numbers in the intestinal tissue of clinical cows, the macrophage phenotype may be confounding the ability of the host to clear the pathogen (27–30). The balance between unbridled proinflammatory responses and activation of macrophages may be the determining factor in the host advancing to full-blown clinical disease.
Except for increased gene expression for IL-10 observed in clinical cows, all other cytokine genes, primarily proinflammatory in nature, responded to antigen stimulation with greater expression in cells from subclinical cows. Subclinical cows often secreted low or equivalent levels of these cytokines compared to cows with clinical disease. Gene transcription and protein secretion often do not align for a myriad of reasons, but for cytokines in particular, this is largely due to posttranscriptional control (31). In addition, there are different pathways for cytokine secretion by innate and adaptive immune cells, depending upon the cell type, the cytokine, and the signal for release, and this may have impacted the measurement of secreted cytokines in the present study (32). Additionally, increased cytokine release could be due to priming of cells in vivo by a higher bacterial burden in the clinical cows, whereas at a subclinical stage of infection, cytokines may be sequestered by the cell until needed (32). If this is the case, in the present study, the upregulation of proinflammatory genes observed in cells from subclinical cows would suggest an immune system “at the ready,” with the ability to elicit robust Th1-mediated responses.
The effect of M. avium subsp. paratuberculosis infection status on lymphocyte subsets has been previously reported for a variety of ruminant species but remains a salient point in this study, as flow cytometric analyses were performed concurrent with cytokine secretion/expression analyses. It is interesting to note that cells freshly isolated from clinical cows demonstrated reduced numbers of CD4, CD8, and γδ TCR T cells compared to subclinical cows, and this pattern remained consistent for CD8 and γδ TCR T cells after antigen stimulation in vitro. These results are similar to those observed in naturally infected sheep stratified postmortem into different disease states, with no differences noted in numbers of CD4+ T cells within total PBMCs but an increased number of CD8+ T cells observed in paucibacillary sheep (29). More importantly, γδ TCR T cells were reduced for both paucibacillary and multibacillary sheep compared to asymptomatic and noninfected controls, consistent with results that we observed for subclinical and clinical cows here. The consistent decrease in memory T cells (CD45RO+) from naturally infected cows, but particularly for clinical cows, is an interesting observation. A decrease in memory T cells has been observed in patients with active tuberculosis (33) but has not been well defined for mycobacterial infection in cattle. The lower frequency of memory T cells noted in clinical cows than in subclinical cows in the present study suggests a shift in antigen recognition and response that may correlate with advanced clinical disease.
Another interesting observation was the expansion of the B cell population for clinical cows upon stimulation with antigen. This was in contrast to the higher number of B cells in freshly isolated PBMCs observed for cows with a subclinical status. We previously demonstrated that cows in the clinical stage of disease had higher total numbers of B cells and that the CD5 marker could discriminate different B cell subpopulations between subclinical and clinical cows (34). B cells may take on both effector and regulatory roles in the host and, as such, produce a plethora of cytokines, such as IFN-γ, IL-12, IL-2, IL-13, IL-4, IL-10, and TGF-β (35). Therefore, a greater presence of B cells and a heightened response to antigen may explain the disparate levels of secreted cytokines observed between cows with subclinical and clinical infection statuses.
Finally, reduced secretion of IL-17A for cows in the clinical stage of disease is thought provoking. Similar findings were reported by Park et al. (36), who compared the expression levels of Th17-derived cytokines within peripheral blood mononuclear cells across cows in various clinical stages of paratuberculosis. Expression of IL-17A was downregulated in naturally infected cows with low, medium, and high M. avium subsp. paratuberculosis-specific antibody responses compared to noninfected and enzyme-linked immunosorbent assay (ELISA)-negative controls. In contrast, responses to Mycobacterium bovis vaccination and/or infection have demonstrated an upregulation of IL-17A secretion, but responses were variable depending upon the bacterial burden, pathological lesion score, and timing of sampling after infection and vaccination (37). Lockhart et al. (38) demonstrated that the primary source of IL-17 is γδ T cells, rather than CD4 T cells, in response to Mycobacterium tuberculosis infection. This is a pivotal observation, as IL-17 has been thought to primarily be produced by the Th17 subset of CD4 T cells. Studies in mice challenged with M. tuberculosis and Pseudomonas aeruginosa demonstrated that IL-17A-producing γδ TCR+ T cells provide critical immunity to the lung, perhaps through the recruitment of neutrophils or the production of antimicrobial peptides early in the infection period (39, 40). In the present study, we found that clinical cows had reduced numbers of γδ TCR+ T cells, corresponding to reduced IL-17A secretion by cultured cells. Furthermore, a model has been proposed that suggests a plasticity of helper T cells in states of chronic inflammation that allows Th17 cells to revert to hybrid Th17/Th1 cells or to Th1 cells that do not produce IL-17 (41). It is interesting to note that TGF-β is involved in the differentiation of T cells to both Th17 and Treg phenotypes, suggesting that a lack of IL-23 may shift away from the induction of proinflammatory IL-17 activity and toward a more regulatory IL-10 response. Additionally, IL-6 also plays a critical role in the induction of IL-17 via differentiation of Th17 cells (42); however, the elevated IL-6 secretion observed in the present study for clinical cows is inconsistent with reduced IL-17A secretion. Porcherie et al. (43) mimicked bovine mastitis using a mouse model and found that mammary glands challenged with Escherichia coli had increased IL-6 and decreased IL-17A levels. These effects correlated with higher bacterial numbers in the gland and increased IL-10 levels, similar to results observed in the present study. Although IL-17 is notable for roles in protection against extracellular pathogens and in autoimmune disorders, it has not been shown to be critical for host defense against intracellular pathogens such as mycobacteria (44). Indeed, it seems more likely that Th17 and Th1 cells work in a synergistic manner to achieve the highest level of protective immunity, and populations of these cell types may flux with the stage of disease. With this in mind, it seems reasonable to surmise that IL-17 may well be a critical mediator of the loss of effective immunity for clinical cows.
In summary, it is clear that the singular measure of cytokines would not serve to present a complete picture of the dynamic host immunity to M. avium subsp. paratuberculosis infection since there is a significant overlap of responses. However, the combined data for cytokine secretion and shifts in immune cell subpopulations provide a more comprehensive analysis. The profiles obtained suggest that even in clinical disease, cows maintain an active immune response, but proinflammatory responses are tempered by regulatory control. In contrast, subclinical cows demonstrated a robust proinflammatory response that would provide critical help to clear the pathogen but seems tightly controlled based upon the level of infection. Additional studies into the mechanisms of innate and adaptive immunity during the course of infection will be helpful to further define a basis of understanding for this complex disease.
MATERIALS AND METHODS
Animals.
Holstein dairy cows ranged in age from 4 to 9 years in this experiment and were placed into three groups, consisting of 6 noninfected healthy cows, 6 cows naturally infected with M. avium subsp. paratuberculosis but asymptomatic (i.e., subclinical), and 6 cows with the clinical form of Johne’s disease. Infection was monitored bacteriologically by fecal shedding of M. avium subsp. paratuberculosis using standard culture methods on Herrold’s egg yolk agar medium containing mycobactin J, amphotericin, nalidixic acid, and vancomycin (Becton, Dickinson, Sparks, MD), as previously described (45). Serological tests were used to further characterize the status of infected animals. Serum was harvested from whole blood and assayed for the presence of M. avium subsp. paratuberculosis antibodies by a commercial ELISA (Herdchek; Idexx, Westbrook, ME), and bovine IFN-γ was measured in plasma using the Bovigam test kit (Prionics, La Vista, NE) according to the manufacturer’s instructions. Animals categorized as having clinical disease had ELISA antibody titers with an S/P ratio averaging 2.73, and fecal shedding averaged 6,305 CFU of M. avium subsp. paratuberculosis/g of feces. Cows in the subclinical treatment group were ELISA negative and averaged <3 CFU of M. avium subsp. paratuberculosis/g of feces. Infected animals in both the subclinical and clinical stages of infection had positive antigen-specific IFN-γ results (Abs450nmMPS-Abs450nmNS = 0.23 ± 0.05 and 0.22 ± 0.05, respectively). All animals were housed in American Association for Accreditation of Laboratory Animal Care-accredited facilities, and all animal-related procedures were approved by the IACUC (National Animal Disease Center, Ames, IA). Infected cows were housed separately on-site from healthy control cows to prevent cross-contamination between groups.
Cell culture.
Peripheral blood mononuclear cells (PBMCs) were isolated from the buffy coat fractions of blood. PBMCs were resuspended in complete medium (RPMI 1640 [Gibco, Grand Island, NY] with 10% fetal calf serum [Atlanta Biologics, Atlanta, GA], 100 U of penicillin G sodium [Gibco] per ml, 100 μg of streptomycin sulfate [Gibco] per ml, 0.25 μg of amphotericin B [Gibco] per ml, and 2 mM l-glutamine [Gibco]). Cells were cultured at 2.0 × 106 cells/ml in replicate 48-well flat-bottomed plates (Corning Incorporated, Corning, NY) for 1, 3, or 6 days at 39°C in 5% CO2 in a humidified atmosphere. Duplicate wells were set up for each animal for the following in vitro treatments: medium only (nonstimulated [NS]), concanavalin A (ConA) (10 μg/ml; Sigma), pokeweed mitogen (PWM) (10 μg/ml; Sigma), and a whole-cell sonicate preparation of M. avium subsp. paratuberculosis (MPS) (10 μg/ml). For cytokine analyses, one set of plates was removed at 1 and 3 days and centrifuged at 400 × g for 5 min. Supernatants were removed without disturbing the cells in culture and stored at −20°C prior to cytokine measurement. A replicate set of plates was incubated for either 3 days (NS, ConA, and PWM) or 6 days (NS and MPS), and cells were harvested for flow cytometric analyses.
Cytokine analyses of cell culture supernatants.
Bovine IL-10 was quantified as previously described (46), using anti-bovine IL-10 antibodies (MCA2110 and MCA2111B; Serotec, Raleigh, NC) and a bovine IL-10 standard (0.3125 to 20 ng/ml; Kingfisher Biotech, St. Paul, MN). Similarly, bovine IL-12 in cell culture supernatants was measured by an ELISA using anti-bovine IL-12 antibodies (MCA1782EL and MCA2173B; Serotec) and a bovine IL-12 standard (55 to 1,500 U/ml; Kingfisher Biotech). Further cytokine analyses of the culture supernatant for IFN-γ, IL-1β, IL-2, IL-4, IL-6, and tumor necrosis factor alpha (TNF-α) concentrations were performed using Searchlight custom bovine arrays (Cira custom bovine 6-plex array kit; Aushon Biosystems, Billerica, MA) according to the manufacturer’s instructions. Concentrations (picograms per milliliter) of each cytokine were quantified in samples using Searchlight array software (Aushon Biosystems) by reference to a standard curve for each cytokine. Measurement of bovine IL-17A (Kingfisher Biotech, Inc., St. Paul, MN) and IL-18 (Biotang, Inc., Lexington, MA) was also performed according to the manufacturers’ instructions.
Flow cytometric analysis.
For flow cytometric analyses, culture plates were removed at 3 days (NS, ConA, PWM) and 6 days (NS, MPS) of incubation, cells within each well were gently resuspended, and 50 μl of the cell suspension was added to wells of 96-well round-bottom plates (Corning Incorporated, Corning, NY) containing 50 μl of primary monoclonal antibodies to CD4, CD8, and γδ TCR+ T cells; B cells; and CD5, CD14, CD25, CD26, CD44, CD62L, CD80, CD86, CD335, and CD45RO (Table 1). Cells were then incubated at 4°C for 30 min. After incubation, plates were centrifuged at 1,250 rpm for 2 min at 4°C, and the supernatant was decanted. One hundred microliters of a secondary antibody cocktail consisting of fluorescein-conjugated anti-mouse IgM (Southern Biotech, Birmingham, AL), R-phycoerythrin-conjugated goat F(ab)2 anti-mouse IgG2a (Southern Biotech, Birmingham, AL), and peridinin chlorophyll protein complex-conjugated rat anti-mouse IgG1 (Becton, Dickinson, San Jose, CA) diluted 1:312, 1:625, and 1:42, respectively, in phosphate-buffered saline (PBS) with 1% fetal calf serum and 0.04% sodium azide was then added to the designated wells, and the plate was centrifuged again at 1,250 rpm for 2 min at 4°C. The cells were then suspended in 200 μl of BD FacsLyse (BD Biosciences, San Jose, CA) for immediate flow cytometric analysis. Samples were evaluated using 30,000 events per sample using a FACScan flow cytometer (Cell Quest software; Becton, Dickinson). Mononuclear cells, based on forward- and side-scatter characteristics, were analyzed for cell surface marker expression (FlowJo; TreeStar, Inc., San Carlos, CA). Cell subpopulations were expressed as a percentage of the total mononuclear cell population to determine effects of infection status on population shifts, whereas activation markers expressed on CD4, CD8, and γδ TCR+ T cell subsets were articulated as mean fluorescence intensities (MFI).
TABLE 1.
Primary antibodiesa
| Antigen | MAb clone | Isotype | Working MAb concn (μg/ml)b | Specificity |
|---|---|---|---|---|
| CD4 | GC50A1 | IgM | 14 | T helper cell |
| CD8 | BAQ111A | IgM | 14 | Cytotoxic/suppressor T cell |
| N12 | CACT61A | IgM | 14 | γδ cell receptor |
| B cell | BAQ155 | IgG1 | 7 | Total B cell |
| CD25 | CACT116A | IgG1 | 15 | IL-2 receptor |
| CACT108A | IgG2a | 15 | ||
| LCTB2A | IgG3 | 15 | ||
| CD26 | CACT114A | IgG2b | 15 | Activation marker |
| CD45RO | GC42A1 | IgG1 | 10 | Memory/activation marker |
| CD44 | BAG40 | IgG3 | 10 | Activation marker |
| CD62L | DREG-5 | IgG1 | 10 | Activation marker |
| CD80 | ILA159A | IgG1 | 10 | Costimulatory signal marker |
| CD86 | IT2.2 | IgG2b | 5 | Costimulatory signal marker |
From VMRD, Inc. (Pullman, WA). MAb, monoclonal antibody.
Diluted in PBS with 1% fetal calf serum and 0.04% sodium azide.
RNA extraction and reverse transcription.
RNA was extracted from PBMCs after 24 h of culture with in vitro treatments (NS and MPS), as described above. Briefly, cells from duplicate wells were harvested after centrifugation of plates at 1,500 rpm for 5 min and lysed with 350 μl of RLT buffer (RNeasy lysis buffer; Qiagen, Valencia, CA). RNA was isolated using an RNeasy minikit (Qiagen) according to the manufacturer’s directions and eluted from the column with 40 μl of RNase-free water (Ambion, Austin, TX). Total RNA (500 ng) was reverse transcribed using SuperScript III (Invitrogen, Carlsbad, CA) with 150 ng of random hexamers, 10 mM deoxynucleoside triphosphates (dNTPs), and 40 U of RNaseOut (Invitrogen), according to the manufacturer’s directions. Samples were heated to 65°C for 5 min and then reverse transcribed at 50°C for 60 min. The resulting cDNAs were stored at −80°C until used for real-time PCR.
Cytokine gene expression.
Real-time PCR was performed using custom TaqMan gene expression assays for bovine IL-4, IL-10, IL-12, IL-17A, IL-18, IL-23, IFN-γ, TGF-β, iNOS, and RANTES (Life Technologies, Grand Island, NY) according to the manufacturer’s directions for relative quantitation. Target sequences for the above-mentioned cytokines are presented in Table 2. Briefly, 4 μl of the cDNA template was added to a 20-μl reaction mixture containing TaqMan universal PCR master mix and a gene expression assay working stock consisting of forward and reverse primers and a 6-carboxyfluorescein (FAM)-MGB probe. A eukaryotic 18S rRNA endogenous control (FAM-MGB probe, non-prime limited; Invitrogen) was used as an internal control to normalize RNA content between samples. The NS sample was used as the calibrator. All reactions were performed in triplicate, and data were analyzed with the method.
TABLE 2.
Real-time PCR bovine cytokine gene target sequencesa
| Cytokine | Target sequence | Assay ID |
|---|---|---|
| IL-4 | CTTGGCAAGCAAGACCTGTTCTGTG | Bt03211898_m1 |
| IL-10 | CTGGATGACTTTAAGGGTTACCTGG | Bt03212725_m1 |
| IL-12A | GCTACAGAAGGCCAGACAAACTCTA | Bt03213918_g1 |
| IL-17A | ACTTCATCTATGTCACTGCTACTGC | Bt03210251_m1 |
| IL-18 | ATTGTTTCCTTTAAGGAAATGAATC | Bt03212733_m1 |
| IL-23 | AACAGTCAGTCCTGCTTGCAAAGAA | Bt04284624_m1 |
| IFN-γ | ATTGGAAAGATGAAAGTGACAAAAA | Bt03212722_g1 |
| TGF-β | ACCCGCAGAGAGGAAATAGAGGGCT | Bt04259486_m1 |
| iNOS | CAGCCCCCGTCCAGTCCAGTGACAC | Bt03249590_m1 |
| RANTES | CTCCATGGCAGCAGTTGTCTTTATC | Bt03216832_m1 |
Gene expression assays from Life Technologies (Grand Island, NY).
Statistics.
Data were analyzed using the PROC Mixed procedure of the Statistical Analysis System (SAS Institute, Cary, NC). Values are reported as least-square means ± standard errors of the means (SEM). When significant effects (P < 0.05) due to infection or in vitro treatment were observed, a means comparison was conducted using the Tukey-Kramer post hoc test.
ACKNOWLEDGMENTS
We thank Margaret Walker and Bruce Pesch for their technical experience and expertise and animal caretakers for their support of the animals within this research project.
This research was supported by federal funding through the USDA-ARS.
REFERENCES
- 1.Lombard JE, Gardner IA, Jafarzadeh SR, Fossler CP, Harris B, Capsel RT, Wagner BA, Johnson WO. 2013. Herd-level prevalence of Mycobacterium avium subsp. paratuberculosis infection in United States dairy herds in 2007. Prev Vet Med 108:234–238. doi: 10.1016/j.prevetmed.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Tiwari A, Vanleeuwen JA, Dohoo IR, Keefe GP, Weersink A. 2008. Estimate of the direct production losses in Canadian dairy herds with subclinical Mycobacterium avium subspecies paratuberculosis infection. Can Vet J 49:569–576. [PMC free article] [PubMed] [Google Scholar]
- 3.Raizman EA, Fetrow JP, Wells SJ. 2009. Loss of income from cows shedding Mycobacterium avium subspecies paratuberculosis prior to calving compared with cows not shedding the organism on two Minnesota dairy farms. J Dairy Sci 92:4929–4936. doi: 10.3168/jds.2009-2133. [DOI] [PubMed] [Google Scholar]
- 4.Whitlock RH, Buergelt C. 1996. Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet Clin North Am Food Anim Pract 12:345–356. doi: 10.1016/S0749-0720(15)30410-2. [DOI] [PubMed] [Google Scholar]
- 5.Hendrick SH, Kelton DF, Leslie KE, Lissemore KD, Archambault M, Duffield TF. 2005. Effect of paratuberculosis on culling, milk production and milk quality in dairy herds. J Am Vet Med Assoc 227:1302–1308. doi: 10.2460/javma.2005.227.1302. [DOI] [PubMed] [Google Scholar]
- 6.Johnson-Ifearulundu YJ, Kaneene JB, Sprecher DJ, Gardiner JC, Lloyd JW. 2000. The effect of subclinical Mycobacterium paratuberculosis infection on days open in Michigan, USA, dairy cows. Prev Vet Med 46:171–181. doi: 10.1016/S0167-5877(00)00145-8. [DOI] [PubMed] [Google Scholar]
- 7.Biet F, Boschiroli ML. 2014. Non-tuberculous mycobacterial infections of veterinary relevance. Res Vet Sci 97(Suppl):S69–S77. doi: 10.1016/j.rvsc.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Stabel JR. 2006. Host responses to Mycobacterium avium subsp. paratuberculosis: a complex arsenal. Anim Health Res Rev 7:61–70. doi: 10.1017/S1466252307001168. [DOI] [PubMed] [Google Scholar]
- 9.Arsenault RJ, Maattanen P, Daigle J, Potter A, Griebel P, Napper S. 2014. From mouth to macrophage: mechanisms of innate immune subversion by Mycobacterium avium subsp. paratuberculosis. Vet Res 45:54. doi: 10.1186/1297-9716-45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begg DJ, de Silva K, Carter N, Plain KM, Purdie A, Whittington RJ. 2011. Does a Th1 over Th2 dominancy really exist in the early stages of Mycobacterium avium subspecies paratuberculosis infections? Immunobiology 216:840–846. doi: 10.1016/j.imbio.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Vazquez P, Garrido JM, Juste RA. 2013. Specific antibody and interferon-gamma responses associated with immunopathological forms of bovine paratuberculosis in slaughtered Friesian cattle. PLoS One 8:e64568. doi: 10.1371/journal.pone.0064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lugton IW. 2004. Review of the possible links between the clinical expression of paratuberculosis and deficiency of macro and micronutrients. Aust Vet J 82:490–496. doi: 10.1111/j.1751-0813.2004.tb11167.x. [DOI] [PubMed] [Google Scholar]
- 13.Karcher EL, Beitz DC, Stabel JR. 2008. Parturition invokes changes in peripheral blood mononuclear cell populations in Holstein dairy cows naturally infected with Mycobacterium avium subsp. paratuberculosis. Vet Immunol Immunopathol 124:50–62. doi: 10.1016/j.vetimm.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Kirkpatrick BW, Shi X, Shook GE, Collins MT. 2011. Whole-genome association analysis of susceptibility to paratuberculosis in Holstein cattle. Anim Genet 42:149–160. doi: 10.1111/j.1365-2052.2010.02097.x. [DOI] [PubMed] [Google Scholar]
- 15.Pribylova-Dziedzinska R, Slana I, Lamka J, Pavlik I. 2014. Influence of stress connected with moving to a new farm on potentially MAP-infected mouflons. ISRN Microbiol 2014:450130. doi: 10.1155/2014/450130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leite FL, Eslabao LB, Pesch B, Bannantine JP, Reinhardt TA, Stabel JR. 2015. ZAP-70, CTLA-4, and proximal T cell receptor signaling in cows infected with Mycobacterium avium subsp. paratuberculosis. Vet Immunol Immunopathol 167:15–21. doi: 10.1016/j.vetimm.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 17.Diehl S, Rincon M. 2002. The two faces of IL-6 on Th1/Th2 differentiation. Mol Immunol 39:531–536. doi: 10.1016/s0161-5890(02)00210-9. [DOI] [PubMed] [Google Scholar]
- 18.Sugawara I, Yamada H, Kaneko H, Mizuno S, Takeda K, Akira S. 1999. Role of interleukin-18 (IL-18) in mycobacterial infection in IL-18-gene-disrupted mice. Infect Immun 67:2585–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendez-Sampiero P. 2010. Role of interleukin-12 family cytokines in the cellular response to mycobacterial disease. Int J Infect Dis 14:e366–e371. doi: 10.1016/j.ijid.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 20.Hostetter J, Steadham E, Haynes J, Bailey T, Cheville N. 2003. Phagosomal maturation and intracellular survival of Mycobacterium avium subspecies paratuberculosis in J774 cells. Comp Immunol Microbiol Infect Dis 26:269–283. doi: 10.1016/S0147-9571(02)00070-X. [DOI] [PubMed] [Google Scholar]
- 21.Weiss DJ, Evanson OA, de Souza C, Abrahamsen MS. 2005. A critical role of interleukin-10 in the response of bovine macrophages to infection by Mycobacterium avium subsp. paratuberculosis. Am J Vet Res 66:721–726. doi: 10.2460/ajvr.2005.66.721. [DOI] [PubMed] [Google Scholar]
- 22.Souza CD, Evanson OA, Weiss DJ. 2006. Mitogen activated protein kinase p38 pathway is an important component of the anti-inflammatory response in Mycobacterium avium subsp. paratuberculosis-infected bovine monocytes. Microb Pathog 41:59–66. doi: 10.1016/j.micpath.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Aho AD, McNulty AM, Coussen PM. 2003. Enhanced expression of interleukin-1alpha and tumor necrosis factor receptor-associated protein 1 in ileal tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis. Infect Immun 71:6479–6486. doi: 10.1128/iai.71.11.6479-6486.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer SS, Cheng G. 2012. Role of interleukin-10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol 32:23–63. doi: 10.1615/CritRevImmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain T, Shah SZ, Zhao D, Sreevatsan S, Zhou X. 2016. The role of IL-10 in Mycobacterium avium subsp. paratuberculosis infection. Cell Commun Signal 14:29. doi: 10.1186/s12964-016-0152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabat R, Grütz G, Warszawska K, Kirsch S, Witte E, Wolk K, Geginat J. 2010. Biology of interleukin-10. Cytokine Growth Factor Rev 21:331–344. doi: 10.1016/j.cytogfr.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Thirunavukkarasu S, de Silva K, Begg DJ, Whittington RJ, Plain KM. 2015. Macrophage polarization in cattle experimentally exposed to Mycobacterium avium subsp. paratuberculosis. Pathog Dis 73:ftv085. doi: 10.1093/femspd/ftv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernández M, Benavides J, Castaño P, Elguezabal N, Fuertes M, Muñoz M, Royo M, Ferreras MC, Pérez V. 2017. Macrophage subsets within granulomatous intestinal lesions in bovine paratuberculosis. Vet Pathol 54:82–93. doi: 10.1177/0300985816653794. [DOI] [PubMed] [Google Scholar]
- 29.Gillan S, O’Brien R, Hughes AD, Griffin JFT. 2010. Identification of immune parameters to differentiate disease states among sheep infected with Mycobacterium avium subsp. paratuberculosis. Clin Vaccine Immunol 17:108–117. doi: 10.1128/CVI.00359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenvey CJ, Shircliff AL, Bannantine JP, Stabel JR. 2019. Phenotypes of macrophages present in the intestine are impacted by stage of disease in cattle naturally infected with Mycobacterium avium subsp. paratuberculosis. PLoS One 14:e0217649. doi: 10.1371/journal.pone.0217649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson P. 2008. Post-transcriptional control of cytokine production. Nat Immunol 9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 32.Stanley AC, Lacy P. 2010. Pathways for cytokine secretion. Physiology (Bethesda) 25:218–229. doi: 10.1152/physiol.00017.2010. [DOI] [PubMed] [Google Scholar]
- 33.Marin ND, Paris SC, Rojas M, Garcia LF. 2012. Reduced frequency of memory T cells and increased Th17 responses in patients with active tuberculosis. Clin Vaccine Immunol 19:1667–1676. doi: 10.1128/CVI.00390-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stabel JR, Khalifeh MS. 2008. Differential expression of CD5 on B lymphocytes in cattle infected with Mycobacterium avium subsp. paratuberculosis. Vet Immunol Immunopathol 126:211–219. doi: 10.1016/j.vetimm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Lund FE. 2008. Cytokine-producing B lymphocytes—key regulators of immunity. Curr Opin Immunol 20:332–338. doi: 10.1016/j.coi.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park HE, Park HT, Jung YH, Yoo HS. 2018. Gene expression profiles of immune-regulatory genes in whole blood of cattle with a subclinical infection of Mycobacterium avium subsp. paratuberculosis. PLoS One 13:e0196502. doi: 10.1371/journal.pone.0196502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters WR, Maggioli MF, Palmer MV, Thacker TC, McGill JL, Vordermeier HM, Berney-Meyer L, Jacobs WR Jr, Larsen MH. 2016. Interleukin-17A as a biomarker for bovine tuberculosis. Clin Vaccine Immunol 23:168–180. doi: 10.1128/CVI.00637-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lockhart E, Green AM, Flynn JL. 2006. IL-17 production is dominated by γδ T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol 177:4662–4669. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 39.Umemura M, Okamoto-Yoshida Y, Yahagi A, Touyama S, Nakae S, Iwakura Y, Matsuzaki G. 2016. Involvement of IL-17A-producing TCR γδ T cells in late protective immunity against pulmonary Mycobacterium tuberculosis infection. Immun Inflamm Dis 4:401–412. doi: 10.1002/iid3.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bayes HK, Ritchie ND, Evans TJ. 2016. Interleukin-17 is required for control of chronic lung infection caused by Pseudomonas aeruginosa. Infect Immun 84:3507–3516. doi: 10.1128/IAI.00717-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuwabara T, Ishikawa F, Kondo M, Kakiuchi T. 2017. The role of IL-17 and related cytokines in inflammatory autoimmune diseases. Mediators Inflamm 2017:3908061. doi: 10.1155/2017/3908061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. 2007. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol 8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 43.Porcherie A, Gilbert FB, Germon P, Cunha P, Trotereau A, Rossignol C, Winter N, Berthon P, Rainard P. 2016. IL-17A is an important effector of the immune response of the mammary gland to Escherichia coli infection. J Immunol 196:803–812. doi: 10.4049/jimmunol.1500705. [DOI] [PubMed] [Google Scholar]
- 44.Curtis MM, Way SS. 2009. Interleukin-17 in host defense against bacterial, mycobacterial and fungal pathogens. Immunology 126:177–185. doi: 10.1111/j.1365-2567.2008.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stabel JR. 1997. An improved method for cultivation of Mycobacterium paratuberculosis from bovine fecal samples and comparison to three other methods. J Vet Diagn Invest 9:375–380. doi: 10.1177/104063879700900406. [DOI] [PubMed] [Google Scholar]
- 46.Stabel JR, Waters WR, Bannantine JP, Palmer MV. 2013. Disparate host immunity to Mycobacterium avium subsp. paratuberculosis antigens in calves inoculated with M. avium subsp. paratuberculosis, M. avium subsp. avium, M. kansasii, and M. bovis. Clin Vaccine Immunol 20:848–857. doi: 10.1128/CVI.00051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]