Abstract
Proteus mirabilis is commonly considered to be an opportunistic pathogen causing urinary tract infections (UTIs) in humans. However, some strains of P. mirabilis were found to be associated with food poisoning outbreaks, with the pathogenic mechanism still unclear. In our study, we described a novel strain of P. mirabilis C02011 isolated from patients’ specimens in a food poisoning in China. In order to determine its gastrointestinal pathogenicity, experiments were performed to compare P. mirabilis B02005 strain (isolated from healthy people) and P. mirabilis American Type Culture Collection (ATCC) 29906 strain both in vitro [Caco-2 cells: bacterial adhesion and invasion assays, Giemsa staining, and transmission electron microscopy (TEM)] and in vivo [BALB/c mouse model: fecal character, colon injury, histological examination, immunochemistry, and western blotting (WB)]. According to the results, C02011 strain exhibited almost identical characteristics with B02005 strain in bacterial appearance and proliferation. In vitro, Caco-2 cells were infected with P. mirabilis C02011, B02005, and P. mirabilis ATCC 29906 strains. After that, Giemsa staining and TEM were used for observing the infection process of C02011 strain. Meanwhile, the adhesive abilities of different strains were rated as follows: P. mirabilis B02005 > P. mirabilis C02011 > P. mirabilis ATCC 29906 (P < 0.01). Invasive abilities of different strains were rated as follows: P. mirabilis C02011 > P. mirabilis B02005 > P. mirabilis ATCC 29906 (P < 0.01). In vivo, BALB/c mice were infected with P. mirabilis C02011 and B02005 strains. C02011 strain shows more virulence than B02005 strain in terms of the following indicators: (1) feces water content and fecal character; (2) colon length of mice; (3) histological examination on mouse intestine tissues; (4) ELISA for detecting TNF-α level in the colon; and (5) WB and immunohistochemistry (IHC) for detecting occludin protein expression in the colon. On the basis of these results, we firstly validated that the novel strain of P. mirabilis C02011 shows more gastrointestinal pathogenicity than the other strains isolated from a healthy individual. In addition, type IV secretion system (T4SS) was preliminarily confirmed to play an important role in the pathogenesis of diarrheal P. mirabilis isolated from the food poisoning incident.
Keywords: Proteus mirabilis, diarrhea, cell and mouse models, food poisoning, type IV secretion system
Introduction
Proteus mirabilis is a gram-negative bacterium with flagella around the body, which is widely found in water, soil, and human intestinal environments. Commonly, P. mirabilis is harmless to human health. Sometimes, P. mirabilis is considered as the causative pathogen in hospital cross-infection associated with urinary catheter (O’Hara et al., 2000; Hola et al., 2012). However, some novel strains have been reported to pose risks to human health and may cause serious disease in patients (Rozalski et al., 1997; Wang et al., 2010). These strains isolated from hospitals have been identified as virulent pathogens in patients suffering from human sepsis, food poisoning, peritonitis, and meningitis. In recent years, more and more food poisoning cases associated with P. mirabilis had been reported in China. In these food poisoning incidents, clinical symptoms of the patients infected with P. mirabilis included abdominal pain, diarrhea, nausea, and dizziness (Wang et al., 2010; Shi et al., 2014, 2016). For example, 256 students in a middle school were diagnosed with food poisoning of P. mirabilis in Zhejiang Province in 1998 (Liu and Lu, 2000); 34 people were diagnosed with food poisoning of P. mirabilis in Guangxi Province in 2006 (Liang et al., 2007); and four people were identified to have food poisoning of P. mirabilis in Beijing in 2013 (Huo et al., 2014). According to a surveillance report of bacterial food poisoning issued by Datong Food and Drug Inspection and Testing Center, food poisoning incidents related to P. mirabilis accounted for 3.61% in the food poisoning incidents reported in Datong from 2016 to 2017 (Shanxi Province, China).
In addition, food poisoning incidents associated with P. mirabilis have been reported worldwide for a long time, and more researchers began exploring the gastrointestinal virulence and rapid detection techniques of P. mirabilis. In the United Kingdom, a food poisoning incident related to Proteus involving eight cases was reported in Bristol Royal Infirmary; the contaminated food were brawn and tomato puree (Cooper et al., 2005). In Turkey, Reyhan and his team have explored the antimicrobial effect of garlic on foodborne pathogens including Escherichia coli, Salmonella enteritidis, and P. mirabilis AUFE 43566 (Irkin and Korukluoglu, 2009). Encyclopedia of Food Microbiology (second edition) mentioned that P. mirabilis may play a role in food spoilage and as enteropathogens (Kushwaha et al., 2014). Bad Bug Book (“Handbook of foodborne pathogenic microorganisms and natural toxins”), issued by the Food and Drug Administration [FDA] (2012), mentions that “With regard to foods, Proteus could metabolize amino acids found in meats to produce compounds that may cause putrefaction. In fish, such as tuna, Proteus is considered a histamine-producing microbe and under such circumstances could generate scombroid poisoning.” Epidemiological investigations indicated that meat products, bean products, fish, and cold dishes are commonly associated with food poisoning related to P. mirabilis (Wang et al., 2010; Cao et al., 2011; Jiang et al., 2017). Hence, P. mirabilis may pose a relatively great threat to food safety and public health.
Nowadays, the pathogenic mechanism of diarrheal P. mirabilis is still unclear. It is essential and meaningful to explore the gastrointestinal pathogenicity of the P. mirabilis, which may cause human diarrhea. Till now, few published studies have explained the pathogenic mechanism of diarrheal P. mirabilis. In our study, two strains of P. mirabilis have been isolated from clinical specimens: P. mirabilis C02011 strain was isolated from patients’ vomitus and feces specimens in a food poisoning in Shenzhen; and P. mirabilis B02005 strain was isolated from feces specimen of healthy people. Our preliminary works (Shi et al., 2014, 2016) have confirmed our speculation that several virulence genes (urease gene, hemolysin gene, and metalloproteinase gene) were detected in P. mirabilis C02011 by using whole genome sequencing and phylogenetic analysis, which were not found in P. mirabilis B02005 strain (Shi et al., 2016). Notably, type IV secretion system (T4SS), which is commonly found in pathogenic bacteria such as Klebsiella pneumoniae and Helicobacter pylori (Wang et al., 2016; Medrano and Bell, 2017), has also been identified in P. mirabilis C02011 (Shi et al., 2016).
From the above, we speculated that P. mirabilis C02011 was a novel strain with more virulence, which may possibly be associated with diarrhea in susceptible population, whereas P. mirabilis B02005 with less virulence may not cause diarrhea among the susceptible population. In order to determine the gastrointestinal pathogenicity of P. mirabilis C02011, we built an experimental model both in vitro and in vivo. In addition, T4SS was preliminarily confirmed to play an important role in the pathogenesis of diarrheal P. mirabilis C02011 in vitro in the study, so as to lay a foundation work for the following researches on exploring its mechanism.
Materials and Methods
Bacterial Strains
Ethics Statement on Bacterial Isolation
Proteus mirabilis C02011 strain was isolated, detected, and stored in Shenzhen Center for Disease Control and Prevention (SZCDC) following the standards of GB/T4789-2003 (GB/T4789.1, 17, 22, 23, and 28) and WS/T9-1996. As a reference lab and pathogenic microorganisms’ repository in local region, SZCDC is the institution that is responsible legally for investigating the pathogen contamination and food poisoning. Hence, ethics approval for the isolation of strains was not provided in the study. P. mirabilis B02005 was isolated from a healthy individual; informed consent for feces collection was agreed upon by the volunteer.
Bacterial Strains, Isolation, and Culture
On June 2002, a food poisoning occurred in Luohu District, Shenzhen, China. Five patients with clinical syndromes of nausea, vomiting, abdominal pain, watery diarrhea, and dizziness were sent to the hospital for treatment. Meanwhile, the food poisoning case was reported to SZCDC for further investigation. According to the investigation of SZCDC, P. mirabilis C02011 was isolated from the residue food (cold dishes), vomited matter, excrement, and anal swab of the patients. Results of pulsed-field gel electrophoresis (PFGE) indicated that P. mirabilis C02011 was associated with this food poisoning case (Shi et al., 2016).
In our study, food samples, stool specimens, and vomit specimens collected from the patients were sent to the laboratory for examination within 24 h. Based on the phenotypic properties of morphology and biochemical features (WS/T 9-1996: Diagnostic criterion and principles of food poisoning induced by P. mirabilis; GB/T4789-2003: National criterion for food microbiological examination), P. mirabilis was successfully identified from the food poisoning incident (Shi et al., 2016). Meanwhile, Sensititre Auto-reader (Sensititre, United Kingdom) system was used to detect the bacteria. Results consistently showed that the suspicious colonies isolated from the patients’ specimens were P. mirabilis.
Strains involved in the study were isolated from various sources in SZCDC (Shi et al., 2016). P. mirabilis C02011 strain was isolated from patients’ specimens in a food poisoning in Shenzhen, whereas P. mirabilis B02005 was isolated from the anal swab of a healthy individual with no clinical symptoms. P. mirabilis was prepared on the logarithmic phase for the experiments by inoculating in brain heart infusion (BHI) medium with a single colony grown on MacConkey (MAC) agar plates at 37°C with shaking (160 rpm) for 16 h.
Growth Curve of Bacterial Strains
Strains of P. mirabilis were inoculated on MAC agar plates at 37°C for 16 h. Then, the single colony on MAC was selected and inoculated in sterilized BHI medium. Densicheck® (Bio Mérieux, France) was used for detecting the bacteria density by measuring the turbidity at different times after inoculating at 37°C with shaking (160 rpm). Finally, growth curves of P. mirabilis were obtained based on turbidities.
Constructing the Mutant Strain With T4SS Genes Deletion
Primers were designed and synthesized according to the DNA sequences besides T4SS genes in P. mirabilis C02011. A PCR technique was used for amplifying two DNA sequences (A and B fragments) on both sides of the T4SS genes. Then, A and B fragments were combined to form pGEM-T easy vector and connected together by the same BamHI loci. After that, the vector was subcloned into suicide plasmid pCVD442. The suicide recombinant plasmid (Targeting Vector) pCVD442-ΔT4SS was constructed and electro-transformed into E. coli SMl0 λpir by conjugative transduction. The suicide plasmid pCVD442-ΔT4SS was transformed from SMl0 λpir to P. mirabilis C02011. Finally, strains with T4SS genes deletion were screened by ampicillin sensitivity test and PCR (see Supplementary Appendix 2 for technical flowchart).
Cell Assays in vitro
Cell Cultures and Reagents
Human colorectal cancer HT-29 cells (HT-29), human colon cancer LoVo cells (LoVo), and human cloning colon adenocarcinoma Caco-2 cells (Caco-2) were obtained from the Cell Bank of Chinese Academy of Sciences. Cells were cultured at 37°C in a 5% CO2-humidified incubator in 90% Dulbecco’s modified Eagle medium (DMEM) (Gibco, United States) containing 10% fetal bovine serum (FBS) (Gibco, United States), and 1% non-essential amino acid (NEAA) (Sigma, Japan). Phosphate-buffered saline (PBS, pH = 7.4) were obtained from Shanghai Double Spiral Biotech Co., Ltd. Giemsa stain solution was bought from Zhuhai Besso Biotech Co., Ltd. Other chemical reagents were of the highest grade available commercially.
Adhesion and Invasion Assays
Adhesion and invasion assays on P. mirabilis were performed following a standard method (Mathoera et al., 2002; Yamada et al., 2006). Bacteria were prepared at a concentration of 108 CFU/ml. Cells were infected with P. mirabilis for 2 h with multiplicity of infection (MOI) = 100 at 37°C in 5% CO2. After that, infected cells were washed three times with PBS. Adhesion of P. mirabilis was measured as the number of bacteria adhering to host cell. All experiments were performed in triplicate. Invasion assay (Yamada et al., 2006) on P. mirabilis was also performed in our study. As mentioned above, cells were infected with P. mirabilis for 2 h and then washed three times with PBS and covered with DMEM containing gentamicin (200 μg/ml) to kill extracellular bacteria. Afterward, invasion of P. mirabilis was measured as the number of bacteria inside the body of host cells.
Giemsa Stain
After being infected with P. mirabilis, cells were washed three times with PBS, fixed by the addition of methanol for 10 min, and stained with Giemsa stain for 15 min (Yamada et al., 2006). The stained cells on the slides were washed with tap water mildly, mounted with neutral resin after drying, and observed under an optical microscope (Axio Imager M2) (×200 magnification).
Preparation of Samples for Transmission Electron Microscopy
Caco-2 cells were prepared as described above in the invasion assay. Then, infected cells were fixed with 2.5% glutaraldehyde in 0.1 mol/L of sodium cacodylate buffer. After that, fixed cells were cut into sections and then washed with 0.1 mol/L of sodium cacodylate, dehydrated in ethanol, and finally embedded in Agar 100 resin. Sections were mounted on copper grids, stained with uranyl acetate and lead citrate, and examined using transmission electron microscopy (TEM) (Georgiev et al., 2017).
Animal Assays in vivo
Ethics Statement
Animal experiments were approved by the Animal Care Committee of Southern Medical University (Guangzhou, China). Six-week-old BALB/c mice were obtained from Animal Experimental Center of Southern Medical University. All supplements including food, water, and other nutrients were autoclaved; and animals were kept in the animal facilities. The protocol was approved by the Animal Care Committee of Southern Medical University (protocol number L2015027). The experimental process that involved infectious pathogen was conducted in biosafety level 2 laboratory. All surgeries were performed under anesthesia with ketamine and lidocaine, and utmost efforts were taken to minimize suffering.
Animals and Experimental Design
Forty-five mice were randomly divided into three groups (C02011 group, B02005 group, and control group). All mice were given sterile water containing streptomycin (SM) (5 g/L) for 3 days (Suar et al., 2006; Elder et al., 2016). One day before infection, SM water was replaced with SM-free water (Suar et al., 2006). After intestinal colonization was reduced, mice in different groups were gavaged with P. mirabilis C02011 (2.5 × 108 CFU/mouse), P. mirabilis B02005 (2.5 × 108 CFU/mouse), and saline water, respectively.
Monitoring of Animals
After infection, physiological status (body weight and fur ruffing) and feces (consistency of feces and feces water content) were observed in mice. In order to collect pathological tissues (small intestine and large intestine), animals were sacrificed for western blotting (WB), immunohistochemistry (IHC) (Xiao et al., 2015), and hematoxylin and eosin (H&E) staining (Imura et al., 2013; Zhou et al., 2018).
Pathological Evaluations on Mice
Pathological lesions were evaluated by double-blind test, based on the following aspects: inflammation, extent, regeneration, crypt damage, and percent involvement (Dieleman et al., 1998). Each tissue slice was graded by three investigators through the five indicators: (1) inflammation (0, none; 1, slight; 2, moderate; and 3, severe); (2) extent (0, none; 1, mucosa; 2, mucosa and submucosa; and 3, transmural); (3) regeneration (4, no tissue repair; 3, surface epithelium not intact; 2, regeneration with crypt depletion; 1, almost complete regeneration; and 0, complete regeneration or normal tissue); (4) crypt damage (0, none; 1, basal 1/3 damaged; 2, basal 2/3 damaged; 3, only surface epithelium intact; and 4, entire crypt and epithelium lost); and (5) percent involvement (0, 1–25%; 1, 26–50%; 2, 51–75%; and 3, 76–100%) (Dieleman et al., 1998; Cha et al., 2017; Qiu et al., 2017; Guo et al., 2018).
Immunohistochemistry
For detecting the expression of occludin protein in colon tissues, standardized method was adopted in the study (Martinez et al., 2015; Huang et al., 2018). Anti-occludin polyclonal antibody (Proteintech, United States) was mixed 1:1,000 in 5% skim milk. The sections were covered and incubated with polyclonal solutions (100 μl) at 4°C for approximately 24 h. Then the slides were washed with TBST and incubated with a secondary antibody (Dingguo Biotechnology Co., Ltd., China). The chromogenic reaction, counterstain, and dehydration were carried out; and the sections were observed by microscope (Xiao et al., 2015; Huang et al., 2016).
Western Blotting
Colon tissues were weighted and put into homogenizer to grind into tissue homogenate with mixture phenylmethylsulfonyl fluoride (PMSF) (Besbio, China) and radioimmunoprecipitation assay (RIPA) (Besbio, China). Protein concentration was measured by bicinchoninic acid assay (BCA) protein detection kit (Biyuntian, China) and micro-spectrophotometer. Proteins were electrophoresed using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (12%) and transferred to nitrocellulose membranes. Membranes were blocked with skim milk (5%) in TBST for 1 h at room temperature and incubated with a rabbit anti-mouse polyclonal occludin antibody (diluted 1:1,000) (Proteintech, United States) at 4°C overnight (Zacharek et al., 2007). After being washed thrice in TBST, the membranes were reacted with a 1:1,000 dilution of horseradish peroxidase (HRP) goat anti-rabbit IgG (Dingguo Biotechnology Co., Ltd., China) for 1 h at room temperature. After being washed thrice in TBST, membranes were reacted with the mixture of peroxide solution (Bio-Rad, United States) and Luminol enhancer solution (Bio-Rad, United States) (1:1). Finally, the results of WB were detected by Tanon 5500 chemiluminescent imager system (Tianneng, China), referencing by β-actin protein as standardization. The process was repeatedly thrice for WB experiments.
Statistical Analysis
Results were presented as mean ± SD. The data were analyzed by Student’s T-test and one-way ANOVA with SPSS 17.0 software. P < 0.05 was defined as statistically significant. ∗P < 0.05, ∗∗P < 0.01; NS, no significance.
Results
Bacterial Growth in Different Strains of Proteus mirabilis
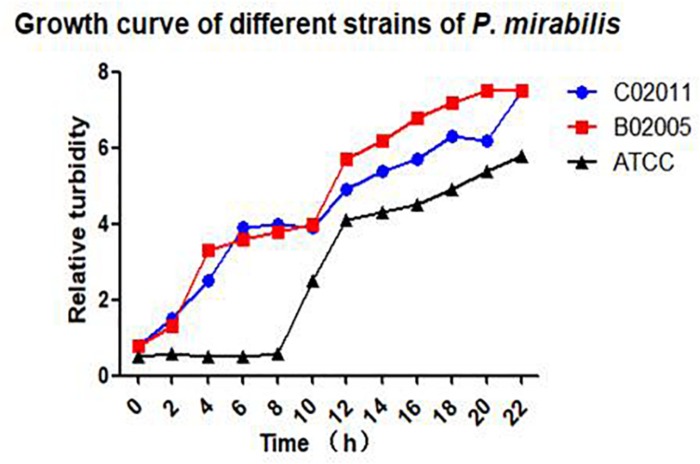
After Gram staining, morphological observations of P. mirabilis were examined under an optical microscope (×200 magnification). Short rod-like bacterial body stained with pink color could be observed in P. mirabilis C02011 and B02005 cultures. Results indicated that two isolated strains were gram-negative bacilli (see in Supplementary Appendix 1). In addition, bacterial growth was monitored by measuring optical density in cultured medium within 22 h. P. mirabilis C02011 strain exhibited an almost identical growth curve with P. mirabilis B02005 strain, whereas P. mirabilis American Type Culture Collection (ATCC) 29906 strain grew slower at 8 h (Figure 1).
FIGURE 1.
Bacterial growth in different strains of Proteus mirabilis. Growth curves of P. mirabilis C02011, B02005, and P. mirabilis American Type Culture Collection (ATCC) 29906 detected by Densimat photometer.
Caco-2 Cells May Be More Susceptible to Diarrheal Proteus mirabilis C02011
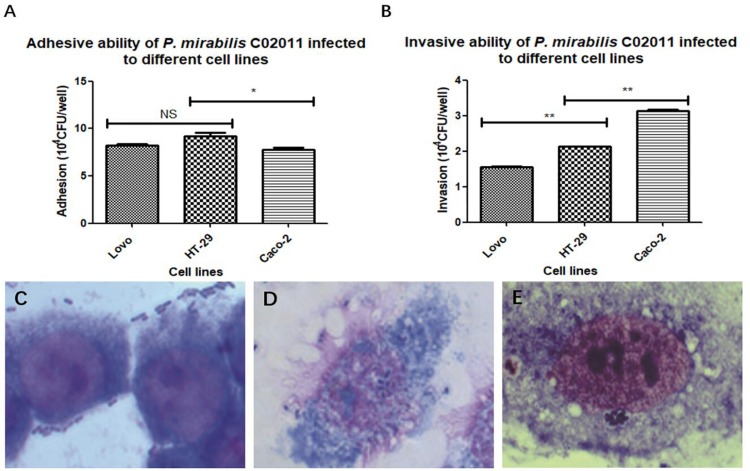
In order to screen out the most sensitive cell line to diarrheal P. mirabilis C02011, three different types of human intestinal cell lines (HT-29, Caco-2, and LoVo) were involved in our study. Adhesion and invasion assays were used to test the bacterial susceptibility to P. mirabilis C02011 in human intestinal cell. Results showed that P. mirabilis C02011 can productively infect Caco-2 cell, compared with HT-29 and LoVo cells. By using optical microscope, we could see that large numbers of bacteria efficiently adhere to human intestinal cells (Figures 2C–E). The adhesive ability of P. mirabilis C02011 to the three kinds of cells (HT-29, Caco-2, and LoVo) was examined in the study; results are listed as follows: HT-29 or LoVo > Caco-2 [P(HT–29 vs. Caco–2) = 0.014 or P(LoVo vs. Caco–2) = 0.032, ∗P < 0.05] (Figure 2A). The invasive ability of P. mirabilis C02011 to the three kinds of cells (HT-29, Caco-2, and LoVo) were also examined in the study; results are listed as follows: Caco-2 > HT-29 > LoVo [P(Caco–2 vs. HT–29/LoVo) = 0.000, ∗∗P < 0.01] (Figure 2B). Thus, the Caco-2 cell line provides excellent in vitro models for evaluating the virulence of P. mirabilis.
FIGURE 2.
HT-29, LoVo, and Caco-2 cells were infected with Proteus mirabilis C02011. (A) The adhesion ability of P. mirabilis C02011 to the three kinds of cells (HT-29, Caco-2, and LoVo) was examined. (B) The invasion ability of P. mirabilis C02011 to the three kinds of cells (HT-29, Caco-2, and LoVo) were examined. (C–E) Giemsa staining of HT-29, LoVo, and Caco-2 cells infected with P. mirabilis C02011. (C) Giemsa staining of HT-29 cells infected with P. mirabilis C02011. (D) Giemsa staining of LoVo cells infected with P. mirabilis C02011. (E) Giemsa staining of Caco-2 cells infected with P. mirabilis C02011. ∗P < 0.05, ∗∗P < 0.01; NS, no significance.
Proteus mirabilis C02011 Strain Is More Virulent Than Proteus mirabilis B02005 Strain and Proteus mirabilis American Type Culture Collection 29906 Strain in vitro
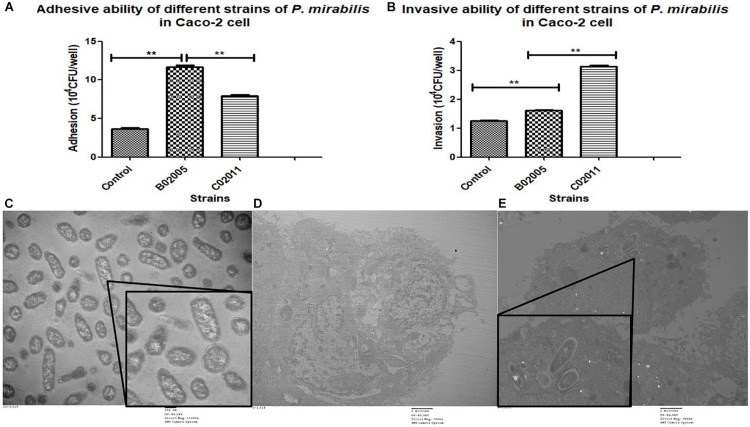
Caco-2 cells were used to evaluate the virulence of P. mirabilis C02011, P. mirabilis B02005 strain, and P. mirabilis ATCC 29906 strain. In the results, P. mirabilis B02005 strain (11.70 ± 0.36 × 104 CFU/well) is more adhesive than P. mirabilis C02011 strain (7.90 ± 0.26 × 104 CFU/well), and P. mirabilis ATCC 29906 strain (3.63 ± 0.32 × 104 CFU/well): P. mirabilis B02005 > P. mirabilis C02011 > P. mirabilis ATCC 29906 [P(B02005 vs. C02011) = 0.000, P(B02005 vs. P. mirabilisATCC 29906) = 0.000, ∗∗P < 0.01] (Figure 3A). On the other hand, P. mirabilis C02011 strain (3.13 ± 0.06 × 104 CFU/well) is more invasive than P. mirabilis B02005 strain (1.61 ± 0.04 × 104 CFU/well) and P. mirabilis ATCC 29906 strain (1.25 ± 0.03 × 104 CFU/well): P. mirabilis C02011 > P. mirabilis B02005 > P. mirabilis ATCC 29906 [P(C02011 vs. B02005) = 0.000, P(C02011 vs. P. mirabilis ATCC 29906) = 0.000, ∗∗P < 0.01] (Figure 3B). Meanwhile, TEM images were collected to observe the infection process of P. mirabilis C02011 (Figures 3C–E). In Figure 3E, well-structure bacteria of P. mirabilis C02011 could be seen clearly in the cytoplasm of Caco-2 cells. The results of adhesion and invasion assays showed that P. mirabilis C02011 strain is more virulent than P. mirabilis B02005 and P. mirabilis ATCC 29906 strains in vitro.
FIGURE 3.
Adhesion and invasion of Proteus mirabilis C02011 strain, P. mirabilis B02005, and P. mirabilis American Type Culture Collection (ATCC) 29906 strains to Caco-2 cells. (A) Caco-2 cells were infected with P. mirabilis C02011, P. mirabilis B02005, and P. mirabilis ATCC 29906 strains for 2 h. The adhesion abilities of different strains were recorded. (B) Caco-2 cells were infected with P. mirabilis C02011, P. mirabilis B02005, and P. mirabilis ATCC 29906 for 2 h, followed by treatment with gentamicin (200 μg/ml) for 2 h. The invasion abilities of different strains were recorded. (C–E) Transmission electron microscopy (TEM) images were collected for observation. (C) Morphological observation of P. mirabilis C02011 (×15,000 magnification). (D) Morphological observation of uninfected Caco-2 cell (×7,000 magnification). (E) Morphological observation of Caco-2 cell infected with P. mirabilis C02011 (×7,000 magnification). ∗∗P < 0.01.
Proteus mirabilis C02011 Causes BALB/c Mouse Diarrhea
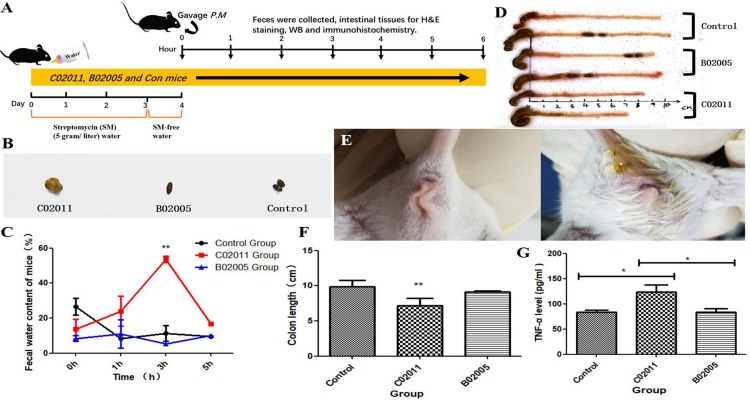
SM-treated mice demonstrate increased infection susceptibility. In our study, SM-treated mice were infected with different strains of P. mirabilis by oral administration (Figure 4A). Then anus images, feces, and colon tissues were collected to evaluate the gastrointestinal pathogenicity induced by P. mirabilis C02011. After infection for 3 h, diarrhea symptoms appeared (moist anus region and loose stools) in P. mirabilis C02011 group mice (Figures 4B,E). Figures 4C,D show the changes in feces water content of mice in different groups. Results showed that P. mirabilis C02011 may cause BALB/c mouse diarrhea, although there were no clinical changes in P. mirabilis B02005 and control groups. In order to evaluate the colonic lesion induced by P. mirabilis C02011, mice were sacrificed, and their colon length was measured. After infection for 3 h, the mean length of the colons in C02011 group was significantly shorter than that of the colons in B02005 and control groups (P = 0.044, ∗P < 0.05) (Figures 4D,F). As we have known, the expression of TNF-α could induce the increase in Caco-2 tight junction permeability in colon tissue correlated with inflammation injuries (Song et al., 2009). Therefore, it is meaningful to detect the level of TNF-α in mouse colon tissues of different groups. Results showed that TNF-α level was significantly increased in C02011 group, compared with B02005 and control groups, at 3 h after infection [P(C02011 vs. B02005) = 0.033, P(C02011 vs. Control) = 0.012, ∗P < 0.05] (Figure 4G). From the above results, it is reasonable to believe that mice in C02011 group suffered from more colonic lesions after infection.
FIGURE 4.
Diarrhea induced by Proteus mirabilis C02011 was observed in BALB/c mice. (A) Streptomycin-treated mice were gavaged with P. mirabilis C02011, P. mirabilis B02005, and normal saline. Clinical symptoms and colonic lesions were evaluated by indicators. (B) Feces images of mice in different groups. (C) Feces water content of mice in different groups was recorded at 0, 1, 3, and 5 h after infection. (D) Colon images in different groups. (E) Anal region images of mice in different groups. (F) Mouse colon length was measured in different groups. (G) The level of TNF-α in colon tissues. ∗P < 0.05, ∗∗P < 0.01.
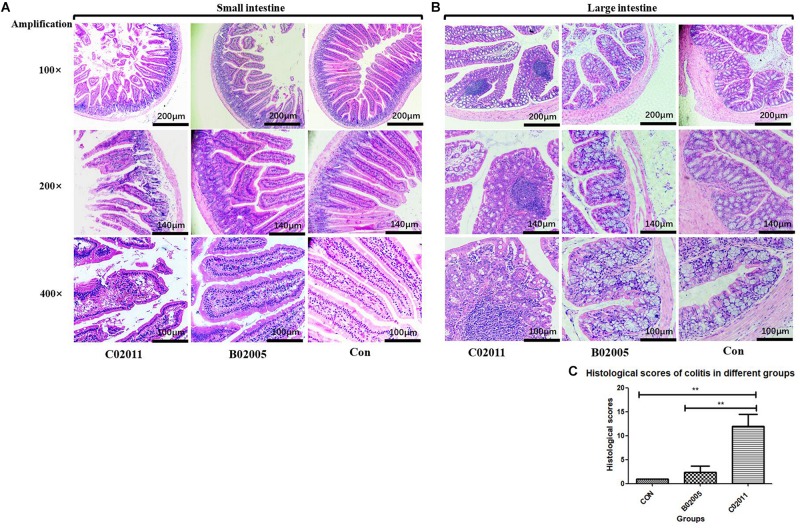
Histological Damages in the Intestine Induced by Proteus mirabilis C02011
In order to investigate the intestinal lesions induced by P. mirabilis in different groups’ mice, histological sections of the small intestine and large intestine were collected for histological observation (Figure 5). Under the optical microscope (×100, ×200, and ×400 magnification), lesions in the small intestine could be observed in C02011 group, whereas there were no changes in B02005 and control groups (Figure 5A). In B02005 and control groups, well-structured small intestine with regularly visible mucosa, submucosa, muscularis, and serosa could be observed. Intestinal epithelial cells were polarized in good order, whereas the nuclei were localized at the bottom of the epithelial cells. However, in C02011 group, histological damages were characterized by vacuolization of the enterocytes, swelling of the villus, destruction of the bases, and nuclei that were irregularly positioned within the cells. Few well-structured villi could be observed under the microscope. In addition, typical inflammatory lesions in the large intestine were also observed in Figure 5B. The colons collected from B02005 and control groups had normal architecture and cellular composition, whereas those of mice exposed to C02011 had edema and infiltration by inflammatory cells in the mucosal layers. Meanwhile, referring to histological grading of colitis by Dieleman et al. (1998), five indicators (inflammation, extent, regeneration, crypt damage, and percent involvement) were considered to evaluate the histological characters of the large intestine in different groups. The level of neutrophil infiltration in the colon tissues was evaluated. The scores in C02011 group were higher than those in B02005 and control groups [P(C02011 vs. B02005) = 0.006, P(C02011 vs. Control) = 0.002, ∗P < 0.05] (Figure 5C). Histological results showed that mice in C02011 group suffered from more colonic lesions after infection.
FIGURE 5.
Hematoxylin and eosin (H&E) staining of the small intestine and large intestine tissues in Proteus mirabilis C02011, B02005, and control groups. (A) Histopathological observations of small bowel in P. mirabilis C02011, B02005, and control groups (×100, ×200, and ×400 magnification). (B) Histopathological observations of large bowel in P. mirabilis C02011, B02005, and control groups (×100, ×200, and ×400 magnification). (C) The average for each element in the histological score (including inflammation, extent, regeneration, crypt damage, and percent involvement) for mice in different groups. ∗∗P < 0.01.
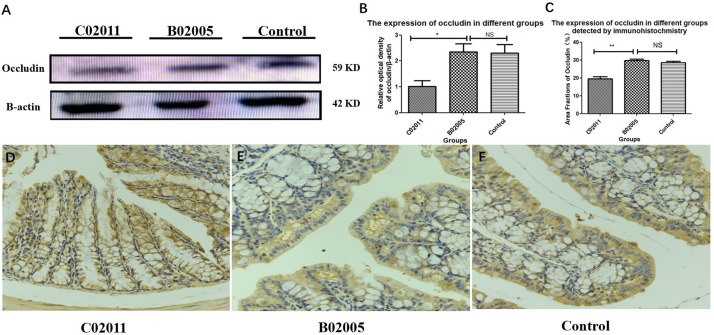
Proteus mirabilis C02011 May Induce Colonic Barrier Dysfunctions in BALB/c Mice
The expressions of tight conjunction protein (occludin) declined significantly in P. mirabilis C02011 group, compared with P. mirabilis B02005 and control groups. Results suggested that P. mirabilis C02011 may induce colonic barrier dysfunctions in BALB/c mice (Figure 6A). WB was used in our study to detect the expressions of protein (occludin), which were related to the integrity of intestinal epithelial tight conjunction. Meanwhile, ImageJ software was applied to detect the relative optical density of protein (occludin) among three groups (Figures 6B,C). In addition, IHC was also used to directly observe the expressions of protein (occludin) in colon tissues (Figures 6D–F). From the results, the expressions of protein (occludin) in three groups were listed as follows: C02011 group < control or B02005 group [P(C02011 vs. B02005) = 0.022, P(C02011 vs. Control) = 0.031, ∗P < 0.05]. These results indicated that P. mirabilis C02011 may induce colonic barrier dysfunctions in BALB/c mice.
FIGURE 6.
The expressions of protein (occludin) in colon tissues induced by Proteus mirabilis. (A) The expressions of protein (occludin) detected by western blotting (WB). (B) The occludin/β-actin intensity ratio in different groups. (C) The expressions of protein (occludin) in different groups detected by immunohistochemistry (IHC). (D–F) Large intestinal sections were stained by IHC (occludin) antibody in different groups: (D) IHC image in C02011 group; (E) IHC image in B02005 group; (F) IHC image in control group. ∗P < 0.05, ∗∗P < 0.01; NS, no significance.
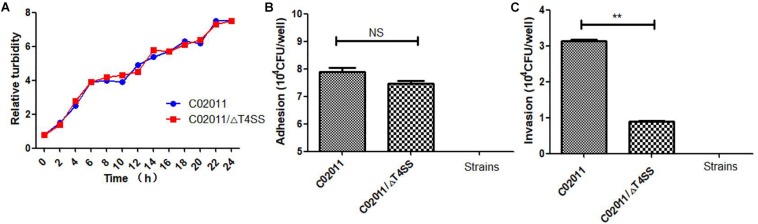
Proteus mirabilis C02011 (Wild-Type) Strain Is More Invasive Than C02011/ΔT4SS (Mutant-Type) Strain
In our recent works, we have successfully constructed the mutant strain of P. mirabilis C02011 with T4SS genes deletion. P. mirabilis C02011 wild-type (WT) strain and its mutant strain C02011/ΔT4SS (MS) were involved in the experiment to preliminarily explore the role of T4SS in the pathogenesis of diarrheal P. mirabilis. According to the bacterial growth of two strains, we found that the WT strain exhibited an identical growth curve with the MS strain during the culture period (Figure 7A). In addition, adhesion and invasion assays were used to evaluate the virulence between two strains. Results showed that the WT strain is more adhesive than the MS strain, but with no statistical significance [P(C02011 vs. C02011/Δ T4SS) = 0.070, P > 0.05] (Figure 7B). Invasion results indicated that the WT strain is more invasive than the MS strain [P(C02011 vs. C02011/Δ T4SS) = 0.000, ∗P < 0.01] (Figure 7C). Hence, we preliminarily confirmed that T4SS may play an important role in the pathogenesis of diarrheal P. mirabilis C02011 in this food poisoning case.
FIGURE 7.
Adhesion and invasion of Proteus mirabilis C02011 strain and its mutant strain to Caco-2 cells. (A) Bacterial growth of P. mirabilis C02011 and its mutant strain detected by Densimat photometer. (B) Caco-2 cells were infected with P. mirabilis C02011 and its mutant strain for 2 h. The adhesion abilities of different strains were recorded. (C) Caco-2 cells were infected with P. mirabilis C02011 and its mutant strain for 2 h, followed by treatment with gentamicin (200 μg/ml) for 2 h. The invasion abilities of different strains were recorded. ∗P < 0.05; NS, no significance.
Discussion
Proteus mirabilis is commonly associated with urinary tract infections (UTIs) in humans, especially in patients with catheter-associated UTIs or hypo-immunity (Jacobsen et al., 2008). Some researchers suggested that P. mirabilis is an opportunistic pathogen (O’Hara et al., 2000; Jacobsen et al., 2008; Hola et al., 2012). However, more and more food poisoning incidents associated with P. mirabilis were reported worldwide (Cooper et al., 2005; Wang et al., 2010). Notably, some novel strains of P. mirabilis with virulence genes including ureC, hpmA, qnrD, and T4SS have been successfully isolated from food poisoning specimens. Hence, it is meaningful to determine the gastrointestinal pathogenicity of these novel strains and explore the mechanism.
As we known, LoVo, Caco-2, and HT-29 cell lines are commonly used as in vitro models for elucidating the pathogenic mechanism in the intestine (Bu et al., 2011; Qiu et al., 2016). A previous study showed that Caco-2 cells could be used as a sensitive model for investigating the function of intestinal barrier (Bu et al., 2011). Therefore, three colon cancer cell lines were involved in our study. In order to build a cell model of infection, adhesion and invasion assays were performed to evaluate the bacterial susceptibility of diarrheal P. mirabilis to three human intestinal cell lines (Kim and Wei, 2007; Qiu et al., 2016; Yang et al., 2017). Moreover, MOI (MOI = 100) and optimal time for infection (optimal infection time = 2 h) have also been explored in our preliminary experiments. After that, qualitative assays (Giemsa staining and TEM) and quantitative assays (adhesion and invasion) were performed to better evaluate the virulence of P. mirabilis strains.
On the other hand, BALB/c mice were used to build the diarrhea animal model for P. mirabilis C02011. In order to simulate the infective route of patient in food poisoning case, mice were challenged with P. mirabilis C02011 and B02005 by oral administration. However, there is one problem: It is not easy to build a diarrheal mouse model induced by intestinal pathogen because of colonization resistance (intestinal microflora) (Kim et al., 2017). In our previous works, we have made a substantial effort to make a sensitive mouse model induced by P. mirabilis. Furthermore, we have explored oral doses of P. mirabilis (107, 108, 109, and 1010 CFU/ml), optimal culture medium of P. mirabilis (Luria broth and BHI), and culture temperature of P. mirabilis (25°C and 37°C). Nevertheless, diarrhea could not be observed in an animal model infected with P. mirabilis. In order to solve this problem, antibiotic pretreatment has been used to reduce the colonization resistance. In 2006, Suar and his team successfully built an SM-pretreated mouse model of Salmonella by oral cavity infection with typical symptoms of intestinal injury (Suar et al., 2006). Coincidently, in 2016, Elder et al. (2016) had also built a mouse model of Salmonella with inflammatory diarrhea by SM pretreatment. In 2018, Mullineaux-Sanders et al. (2018) published their work in Nature Microbiology and concluded that commensal colonization may be the reason for inhibiting enterohemorrhagic Escherichia coli (EHEC)- and Salmonella-induced colitis in a mouse model, which could be effectively improved by SM pretreatment (Kim et al., 2017; Mullineaux-Sanders et al., 2018).
In the study, the gastrointestinal pathogenicity of P. mirabilis C02011 isolated from a food poisoning case has been explored for the first time. Moreover, we successfully established the diarrheal BALB/c mouse model induced by P. mirabilis. Both in vitro and in vivo, we successfully validated that the novel strain of P. mirabilis C02011 has more diarrheagenic virulence than have other strains isolated from healthy individual. Notably, T4SS genes were identified in P. mirabilis C02011 by whole genome sequencing, which were not found in P. mirabilis B02005. On the next step, C02011/ΔT4SS, C02011/ΔVirB9 (an important structural protein in T4SS transmembrane complex) (Jakubowski et al., 2005), C02011/ΔVirB11 (a subunit of T4SS related to ATPase) (Smith et al., 2016), and their complemented strains will be built for the further explorations on the pathogenic mechanism of P. mirabilis C02011.
However, several issues need to be addressed in our study. Firstly, T4SS was considered as the reason for causing diarrhea induced by P. mirabilis C02011. But in the study, we have not yet explored the function of T4SS and its subunits (VirB9 and VirB11) completely. Secondly, P. mirabilis C02011 involved in the study was isolated from patients’ specimens in a food poisoning case. It is less convincing to completely exclude the possibility whether diarrhea disease was caused by host low immunity or bacterial virulence, so it is essential to determine the bacterial pathogenicity both in vitro and in vivo.
Data Availability Statement
The datasets analyzed in this manuscript are not publicly available. Requests to access the datasets should be directed to 1143115967@qq.com.
Ethics Statement
The animal study was reviewed and approved by the Animal Care Committee of Southern Medical University (Guangzhou, China).
Author Contributions
HC, XS, QH, and ZG designed the whole experiments. ZG and HC wrote the manuscript. ZG, FB, HZ, YuL, and YW did the animal experiments and cell experiments. XH, YiL, YQ, and QC did the technical guidance.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by the National Natural Science Foundation of China (Nos. 81773436 and 81302434) and Sanming Project of Medicine in Shenzhen (No. SZSM201811071), and the Grant from School of Public Health of Southern Medical University, China. XS is responsible for the National Natural Science Foundation of China (Nos. 81773436 and 81302434).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02810/full#supplementary-material
References
- Bu X.-D., Li N., Tian X.-Q., Huang P.-L. (2011). Caco-2 and LS174T cell lines provide different models for studying mucin expression in colon cancer. Tissue Cell 43 201–206. 10.1016/j.tice.2011.03.002 [DOI] [PubMed] [Google Scholar]
- Cao B., Li R., Xiong S., Yao F., Liu X., Wang M., et al. (2011). Use of a DNA microarray for detection and identification of bacterial pathogens associated with fishery products. Appl. Environ. Microbiol. 77 8219–8225. 10.1128/AEM.05914-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha H., Lee S., Hwan Kim S., Kim H., Lee D. S., Lee H. S., et al. (2017). Increased susceptibility of IDH2-deficient mice to dextran sodium sulfate-induced colitis. Redox Biol. 13 32–38. 10.1016/j.redox.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K., Davies J., Wiseman J. (2005). An investigation of an outbreak of food poisoning associated with organisms of the Proteus group. J. Pathol. Bacteriol. 52 91–98. 10.1002/path.1700520107 [DOI] [Google Scholar]
- Dieleman L. A., Palmen M. J., Akol H., Bloemena E., Pena A. S., Meuwissen S. G., et al. (1998). Chronic experimental colitis induced by dextran sulphate sodium (DSS) is characterized by Th1 and Th2 cytokines. Clin. Exp. Immunol. 114 385–391. 10.1046/j.1365-2249.1998.00728.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J. R., Chiok K. L., Paul N. C., Haldorson G., Guard J., Shah D. H. (2016). Erratum to: the Salmonella pathogenicity island 13 contributes to pathogenesis in streptomycin pre-treated mice but not in day-old chickens. Gut Pathog. 8:35. 10.1186/s13099-016-0114-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration [FDA] (2012). Bad Bug Book - Handbook of Foodborne Pathogenic Microorganisms and Natural Toxins. Maryland, MD: U.S. Food and Drug Administration. [Google Scholar]
- Georgiev G. P., Iliev A., Kotov G., Kinov P., Slavchev S., Landzhov B. (2017). Light and electron microscopic study of the medial collateral ligament epiligament tissue in human knees. World J. Orthoped. 8 372–378. 10.5312/wjo.v8.i5.372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Xiong J., Wang J., Wen J., Zhi F. (2018). Inhibition of Rac family protein impairs colitis and colitis-associated cancer in mice. Am. J. Cancer Res. 8 70–80. [PMC free article] [PubMed] [Google Scholar]
- Hola V., Peroutkova T., Ruzicka F. (2012). Virulence factors in Proteus bacteria from biofilm communities of catheter-associated urinary tract infections. FEMS Immunol. Med. Microbiol. 65 343–349. 10.1111/j.1574-695X.2012.00976.x [DOI] [PubMed] [Google Scholar]
- Huang W., Zhao H., Dong H., Wu Y., Yao L., Zou F., et al. (2016). High-mobility group box 1 impairs airway epithelial barrier function through the activation of the RAGE/ERK pathway. Int. J. Mol. Med. 37 1189–1198. 10.3892/ijmm.2016.2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. C., Xiao J., Leung V. Y., Lu W. W., Hu Y., Luk K. D. K. (2018). Lumbar intervertebral disc allograft transplantation: the revascularisation pattern. Eur. Spine J. 27 728–736. 10.1007/s00586-017-5419-6 [DOI] [PubMed] [Google Scholar]
- Huo Z., Bai S. Y., Gao B., San-Mei H. U. (2014). A study on the food poisoning isolated Vibrio parahaemolyticus and Proteus mirabilis. Chin. J. Health Lab. Technol. 24 3254–3256. [Google Scholar]
- Imura H., Shimada A., Naota M., Morita T., Togawa M., Hasegawa T., et al. (2013). Vanadium toxicity in mice: possible impairment of lipid metabolism and mucosal epithelial cell necrosis in the small intestine. Toxicol. Pathol. 41 842–856. 10.1177/0192623312467101 [DOI] [PubMed] [Google Scholar]
- Irkin R., Korukluoglu M. (2009). Growth inhibition of pathogenic bacteria and some yeasts by selected essential oils and survival of L. monocytogenes and C. albicans in apple-carrot juice. Foodborne Pathog. Dis. 6 387–394. 10.1089/fpd.2008.0195 [DOI] [PubMed] [Google Scholar]
- Jacobsen S. M., Stickler D. J., Mobley H. L., Shirtliff M. E. (2008). Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 21 26–59. 10.1128/CMR.00019-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski S. J., Cascales E., Krishnamoorthy V., Christie P. J. (2005). Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J. Bacteriol. 187 3486–3495. 10.1128/jb.187.10.3486-3495.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Yu T., Liu L., Li Y., Zhang K., Wang H., et al. (2017). Examination of quaternary ammonium compound resistance in Proteus mirabilis isolated from cooked meat products in China. Front. Microbiol. 8:2417. 10.3389/fmicb.2017.02417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Covington A., Pamer E. G. (2017). The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev. 279 90–105. 10.1111/imr.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Wei C. I. (2007). Antibiotic resistance and Caco-2 cell invasion of Pseudomonas aeruginosa isolates from farm environments and retail products. Int. J. Food Microbiol. 115 356–363. 10.1016/j.ijfoodmicro.2006.12.033 [DOI] [PubMed] [Google Scholar]
- Kushwaha K., Babu D., Juneja V. K. (2014). “Proteus,” in Encyclopedia of Food Microbiology (Second Edition), eds Batt C. A., Tortorello M. L. (Oxford: Academic Press; ), 238–243. [Google Scholar]
- Liang H. L., Liu X. J., Shen A. J. (2007). Investigation and analysis of a food poisoning caused by Proteus mirabilis. China Prevent. Med. 8 172–173. [Google Scholar]
- Liu Y., Lu C. N. (2000). Etiological examination of Proteus mirabilis in a food poisoning. Chin. J. Health Lab. Technol. 10 198–199. [Google Scholar]
- Martinez C. A., De Campos F. G., De Carvalho V. R., De Castro Ferreira C., Rodrigues M. R., Sato D. T., et al. (2015). Claudin-3 and occludin tissue content in the glands of colonic mucosa with and without a fecal stream. J. Mol. Histol. 46 183–194. 10.1007/s10735-015-9610-y [DOI] [PubMed] [Google Scholar]
- Mathoera R. B., Kok D. J., Verduin C. M., Nijman R. J. (2002). Pathological and therapeutic significance of cellular invasion by Proteus mirabilis in an enterocystoplasty infection stone model. Infect. Immun. 70 7022–7032. 10.1128/iai.70.12.7022-7032.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano E. G., Bell A. A. (2017). Demonstration that a Klebsiella pneumoniae subsp. pneumoniae isolated from an insect (Nezara viridula) harbors a plasmid-borne type IV secretion system. Curr. Microbiol. 74 1033–1042. 10.1007/s00284-017-1277-z [DOI] [PubMed] [Google Scholar]
- Mullineaux-Sanders C., Suez J., Elinav E., Frankel G. (2018). Sieving through gut models of colonization resistance. Nat. Microbiol. 3 132–140. 10.1038/s41564-017-0095-1 [DOI] [PubMed] [Google Scholar]
- O’Hara C. M., Brenner F. W., Miller J. M. (2000). Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin. Microbiol. Rev. 13 534–546. 10.1128/cmr.13.4.534-546.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu E. Q., Guo W., Cheng T. M., Yao Y. L., Zhu W., Liu S. D., et al. (2017). Diagnostic classification of endosonography for differentiating colorectal ulcerative diseases: a new statistical method. World J. Gastroenterol. 23 8207–8216. 10.3748/wjg.v23.i46.8207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. W., He X. L., Zhang B., Du L., Zeng Q., Li S., et al. (2016). [Effect on Muc2 gene knockdown in Ht29 cells by CRISPR/Cas9 on probiotics-mediated inhibition of E.coli K1 adhesion and invasion]. Nan Fang Yi Ke Da Xue Xue Bao 36 819–823. [PubMed] [Google Scholar]
- Rozalski A., Sidorczyk Z., Kotelko K. (1997). Potential virulence factors of Proteus bacilli. Microbiol. Mol. Biol. Rev. 61 65–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Hu Q., Lin Y., Qiu Y., Li Y., Jiang M., et al. (2014). [Etiological and molecular characteristics of diarrhea caused Proteus mirabilis]. Zhonghua Liu Xing Bing Xue Za Zhi 35 724–728. [PubMed] [Google Scholar]
- Shi X., Lin Y., Qiu Y., Li Y., Jiang M., Chen Q., et al. (2016). Comparative screening of digestion tract toxic genes in Proteus mirabilis. PLoS One 11:e0151873. 10.1371/journal.pone.0151873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. P., Miller C. N., Child R., Cundiff J. A., Celli J. (2016). Postreplication roles of the Brucella VirB Type IV secretion system uncovered via conditional expression of the VirB11 ATPase. mBio 7:e01730-16. 10.1128/mBio.01730-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H. L., Lv S., Liu P. (2009). The roles of tumor necrosis factor-alpha in colon tight junction protein expression and intestinal mucosa structure in a mouse model of acute liver failure. BMC Gastroenterol. 9:70. 10.1186/1471-230X-9-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suar M., Jantsch J., Hapfelmeier S., Kremer M., Stallmach T., Barrow P. A., et al. (2006). Virulence of broad- and narrow-host-range Salmonella enterica serovars in the streptomycin-pretreated mouse model. Infect. Immun. 74 632–644. 10.1128/iai.74.1.632-644.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ling F., Wang H., Yu M., Zhu H., Chen C., et al. (2016). The Helicobacter pylori Cag pathogenicity island protein Cag1 is associated with the Function of T4SS. Curr. Microbiol. 73 22–30. 10.1007/s00284-016-1016-x [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang S., Yu J., Zhang H., Yuan Z., Sun Y., et al. (2010). An outbreak of Proteus mirabilis food poisoning associated with eating stewed pork balls in brown sauce, Beijing. Food Control 21 302–305. 10.1016/j.foodcont.2009.06.009 [DOI] [Google Scholar]
- Xiao G., Yuan F., Geng Y., Qiu X., Liu Z., Lu J., et al. (2015). Eicosapentaenoic acid enhances heatstroke-impaired intestinal epithelial barrier function in rats. Shock 44 348–356. 10.1097/SHK.0000000000000417 [DOI] [PubMed] [Google Scholar]
- Yamada F., Ueda F., Ochiai Y., Mochizuki M., Shoji H., Ogawa-Goto K., et al. (2006). Invasion assay of Listeria monocytogenes using Vero and Caco-2 cells. J. Microbiol. Methods 66 96–103. 10.1016/j.mimet.2005.10.017 [DOI] [PubMed] [Google Scholar]
- Yang Y., Weng W., Peng J., Hong L., Yang L., Toiyama Y., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of MicroRNA-21. Gastroenterology 152 851.e24–866.e24. 10.1053/j.gastro.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharek A., Chen J., Cui X., Li A., Li Y., Roberts C., et al. (2007). Angiopoietin1/Tie2 and VEGF/Flk1 induced by MSC treatment amplifies angiogenesis and vascular stabilization after stroke. J. Cereb Blood Flow Metab. 27 1684–1691. 10.1038/sj.jcbfm.9600475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Hamza T., Fleur A. S., Zhang Y., Yu H., Chen K., et al. (2018). Mice with inflammatory bowel disease are susceptible to clostridium difficile infection with severe disease outcomes. Inflamm. Bowel Dis. 24 573–582. 10.1093/ibd/izx059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed in this manuscript are not publicly available. Requests to access the datasets should be directed to 1143115967@qq.com.