Abstract
Objective
Automatic segmentation for biochemical cartilage evaluation holds promise for an efficient and reader-independent analysis. This pilot study aims to investigate the feasibility and to compare delayed gadolinium-enhanced magnetic resonance imaging of cartilage (dGEMRIC) hip joint assessment with manual segmentation of acetabular and femoral head cartilage and dGEMRIC hip joint assessment using automatic surface and volume processing software at 3 Tesla.
Design
Three-dimensional (3D) dGEMRIC data sets of 6 patients with hip-related pathology were assessed (1) manually including multiplanar image reformatting and regions of interest (ROI) analysis and (2) automated by using a combined surface and volume processing software. For both techniques, T1Gd values were obtained in acetabular and femoral head cartilage at 7 regions (anterior, anterior-superior, superior-anterior, superior, superior-posterior, posterior-superior, and posterior) in central and peripheral portions. Correlation between both techniques was calculated utilizing Spearman’s rank correlation coefficient.
Results
A high correlation between both techniques was observed for acetabular (ρ = 0.897; P < 0.001) and femoral head (ρ = 0.894; P < 0.001) cartilage in all analyzed regions of the hip joint (ρ between 0.755 and 0.955; P < 0.001).
Conclusions
Automatic cartilage segmentation with dGEMRIC assessment for hip joint cartilage evaluation seems feasible providing high to excellent correlation with manually performed ROI analysis. This technique is feasible for an objective, reader-independant and reliable assessment of biochemical cartilage status.
Keywords: hip, cartilage, MRI, dGEMRIC, automatic segmentation
Introduction
Osteoarthritis (OA) is a multifactorial degenerative joint disease that usually progressively leads to pain and functional impairment. Management strategies include symptomatic treatment, joint preservation surgeries in cases of underlying pre-arthritic conditions (e.g., hip dysplasia or femoroacetabular impingement [FAI]), and joint replacement.
Magnetic resonance imaging (MRI) is the modality of choice to evaluate the cartilage status. Although technical developments (e.g., the introduction of higher field strengths and design of cartilage-specific sequences) have improved morphological cartilage assessment with standard MRI, the ability to detect cartilage matrix alterations that occur early on a molecular level remains limited. Various biochemical-sensitive MRI techniques have evolved and proven to be reliable tools that may add valuable information concerning cartilage composition. Some of these methods are sensitive to the cartilage water content and collagen fiber orientation (e.g., T21- and T2*-mapping2), the cartilage glycosaminoglycan (GAG) content (gagCEST3), or to the cartilage GAG and water content (T1rho imaging4).
The technique of delayed gadolinium-enhanced magnetic resonance imaging of cartilage (dGEMRIC)5 is noted to be sensitive to the charge density of hyaline cartilage. Notably, following administration of a negatively charged gadolinium-based contrast agent, which penetrates the cartilage and reduces T1 relaxation, higher uptake of contrast media in GAG-depleted cartilage areas (resulting in lower T1 values) indicates cartilage degeneration and vice versa.
Major challenges for MR imaging of hip joint include its deep position in the body and the thin, spherical, and crescent-shaped cartilage layers of the femoral head and the acetabulum. However, with sufficient image resolution and contrast-to-noise-ratio, it is possible to distinguish between acetabular and femoral cartilage layers.
As for dGEMRIC analysis, T1Gd relaxation is commonly evaluated by region of interest (ROI) analysis in selected slices throughout the joint, where different marker points are manually placed within cartilage boundaries,6-8 usually leading to a fairly time-consuming process (depending on the number of cartilage areas being investigated). Understandably, it is prone to intra- and inter-reader variability as well. Both are quite relevant factors, why this technique—and probably other ROI-based (semi-) quantitative techniques as well—have not yet found their way into clinical routine and follow-up studies.
While implementing an automatic surface and volume processing software (Clinical Graphics) for a reader-independent and fast cartilage analysis, the study question was as follows: Is there is a good correlation between manual and automatic cartilage segmentation for the evaluation of hip joint cartilage with dGEMRIC?
Methods
Study Population
This study was approved by the local ethics committee. We identified 6 patients (5 females, 1 male, mean age: 27.0 ± 8.6 years, range 19-43 years, mean body mass index: 23.5 ± 3.0 kg/m2, 3 left hips) who underwent 3D dGEMRIC MR imaging in the clinical setting of hip-related pain due to symptomatic FAI (4 patients) or suspected labral tear without typical radiographic evidence of FAI (2 patients). No patient had any notable/advanced OA of the hip (Tönnis grade ⩽1) on previously obtained radiographic films. Before participation, contraindications related to MRI or intravenous contrast media administration were excluded. Written informed consent was obtained in all cases.
MRI Protocol
FDA-approved Gd-DOTA− (0.4 mL/kg, 0.2 mmol Gd/kg, Dotarem; Guerbet GmbH, Sulzbach, Germany) was administered intravenously, and patients were instructed to walk around the MRI facility for 30 minutes to allow for contrast media uptake before MRI acquisition was started. All MR images were acquired in the supine position with the investigated hip in neutral rotation on a 3 Tesla (T) scanner (Magnetom Trio, Siemens Medical Solutions, Erlangen, Germany) using a flexible body-matrix phased-array coil. The MRI protocol comprised localizer images, 2D T1- and T2-weighted sequences (data not shown), a 3D double-echo steady-state (DESS) sequence with water excitation for morphological cartilage assessment, and a dual flip angle 3D gradient echo sequence with volumetric interpolated breath-hold examination (VIBE) for T1Gd evaluation ( Table 1 ).
Table 1.
MRI Settings for the 3D Double-Echo Steady-State (DESS) Sequence and 3D Volumetric Interpolated Breath-Hold Examination (VIBE).
| 3D DESS, Water Excitation | 3D VIBE, T1Gd Mapping | |
|---|---|---|
| TR (repetition time, ms) | 14.75 | 15 |
| TE (echo time, ms) | 5.00 | 2.24 |
| FA (flip angle) | 25 | 5,26 |
| NEX (number of excitation) | 1 | 1 |
| FOV (field of view, mm2) | 192 | 192 |
| Slice thickness (mm) | 0.6 | 0.6 |
| In-plane resolution (mm) | 0.6 × 0.6 | 0.6 × 0.6 |
| Slice gap (mm) | 0.2 | 0.2 |
| Bandwidth (Hz/pixel) | 260 | 260 |
| TA (acquisition time, minutes) | 13.17 | 14.31 |
Manual Cartilage Assessment
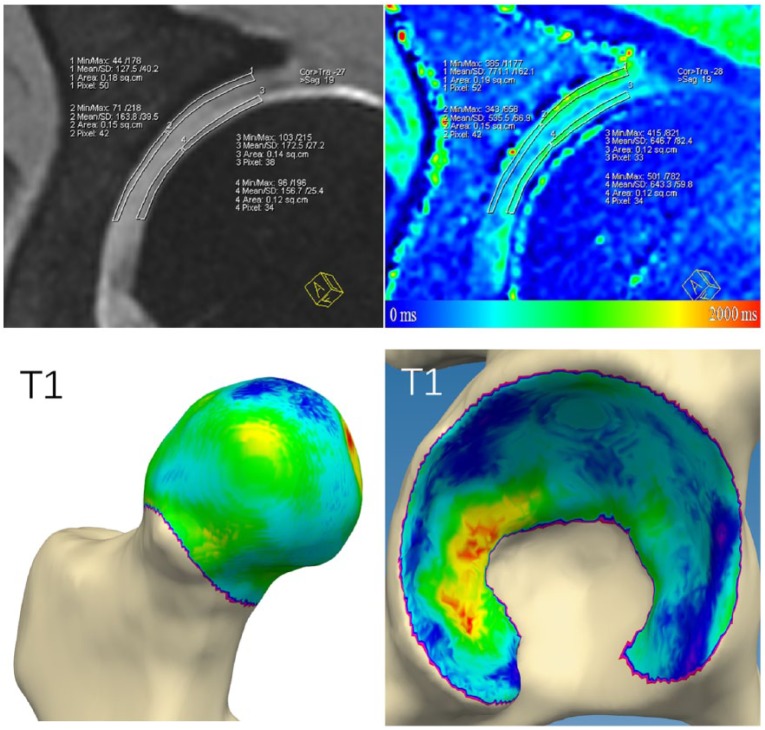
Manual cartilage assessment was conducted in accordance with previously published methodology.9 Initially, the MR data sets of DESS and VIBE, which included the inline 3D T1Gd maps, were transferred to a Leonardo workstation (Siemens Medical Solutions, Erlangen, Germany). Using multiplanar reconstruction software, 7 radial reformats (anterior, anterior-superior, superior-anterior, superior, superior-posterior, posterior-superior, and posterior) with a slice thickness of 2 mm were then created around the femoral neck with the femoral head as the center of rotation. In each reformat ROI analysis was conducted in acetabular and femoral cartilage. Both acetabular and femoral cartilages were further divided into a peripheral zone and a central zone. Peripheral acetabular (and corresponding femoral cartilage) was defined as the lateral half of acetabular cartilage between the fossa and the cartilage-labral junction. Each ROI was delineated by precise placement of multiple marker points. The corresponding DESS reformats served as a morphological reference to warrant ROI placement within cartilage boundaries.
Computerized Cartilage Assessment Using Proprietary Software
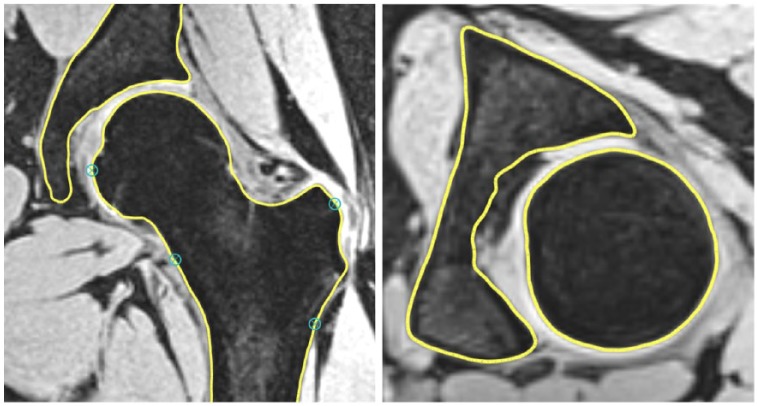
Automated cartilage assessment was based on bone segmentations of the acetabulum and proximal femur extracted from the VIBE sequence. The bone segmentations were created by fitting a statistical shape model.10 Two research engineers placed a dense cloud of fitting points on the volumetric image data (Fig. 1). Verification and local corrections (where deemed necessary) were applied manually by 2 research engineers. Following the segmentation, the center of the femoral head was determined using regression analysis of the surface of the segmented 3D proximal femur model. Based on this femoral head center, and similar to the manual cartilage assessment, 7 radial areas were defined on the femoral head and the acetabular lunate surface (anterior, anterior-superior, superior-anterior, superior, superior-posterior, posterior-superior, and posterior). Each of these radial areas was subdivided into a peripheral zone and a central zone. The software then automatically determined the 3D T1Gd image values by sampling 500 points for each zone, taking the mean value as the final result for that area (Fig. 2).
Figure 1.
The bone segmentations were created by fitting a statistical shape model to the volumetric image data. Verification and—where needed—local corrections were applied manually by 2 research engineers.
Figure 2.
dGEMRIC analysis of hip joint cartilage was conducted in central and peripheral acetabular and femoral head cartilage by region of interest analysis and by automated cartilage assessment based on bone segmentations of the acetabulum and proximal femur.
Statistical Analysis
Spearman’s correlation coefficient (ρ) was calculated for correlation between manual and automatic cartilage segmentation. P values below 0.05 were considered statistically significant.
Results
We evaluated a total of 168 ROIs (6 patients, acetabular and femoral cartilage; 7 regions, peripheral and central zones). A total of 28 ROIs had to be excluded from analysis due to artifacts (and consecutive inability to reliably delineate acetabular and femoral cartilage) or notable cartilage loss in the investigated regions. Therefore, a total of 140 ROIs underwent statistical analysis. Between both techniques we observed excellent correlation for overall acetabular (ρ = 0.897; P < 0.001) and femoral (ρ = 0.894; P < 0.001) cartilage assessment. Correlation was also noted high for regional (acetabular: ρ ranging from 0.829 to 0.944, P < 0.001; femoral: ρ ranging from 0.755 to 0.938, P < 0.001) and zonal (acetabular central: 0.862, acetabular peripheral: 0.889, P < 0.001; femoral central: 0.909, femoral peripheral: 0.868, P < 0.001) cartilage evaluation ( Table 2 ).
Table 2.
Spearman’s Correlation Coefficient (ρ) for Correlation between Manual and Automatic Cartilage Segmentation in Global, Zonal, and Regional Evaluation of Acetabular and Femoral Cartilage.
| Acetabular | Femoral | P | |
|---|---|---|---|
| Global | 0.897 | 0.894 | <0.001 |
| Zone | |||
| Peripheral | 0.889 | 0.868 | <0.001 |
| Central | 0.862 | 0.909 | <0.001 |
| Region | |||
| Anterior Anterior-superior |
0.944 | 0.755 | <0.001 |
| Superior-anterior Superior Superior-posterior |
0.829 | 0.881 | <0.001 |
| Posterior-superior Posterior |
0.844 | 0.938 | <0.001 |
Discussion
In this pilot study, we sought to determine the feasibility of a newly developed technique of fully automatic cartilage segmentation for biochemical hip joint cartilage assessment with dGEMRIC. Although it was a limited cohort of 6 patients, we could demonstrate an excellent correlation to manual cartilage evaluation by ROI analysis.
In pathologies of the hip joint, particularly for guiding treatment decisions ranging from different cartilage repair techniques to joint replacement and for monitoring treatment, a reliable modality of hip joint cartilage evaluation is critical. Notably, quantitative cartilage analysis (e.g., volume/thickness measurements, biochemical assessment) can be achieved in a reproducible manner. In recent studies, quantitative hip joint cartilage evaluation mostly relies on manual ROI analysis. However, postprocessing the 3D MR data sets and manual ROI placement requires adequately trained users. Furthermore, it is time-consuming and inherently prone to intra- and inter-reader variability. Automatic software approaches to evaluate acetabular and femoral cartilage layers could promote the clinical application of biochemical cartilage evaluation, especially in settings with a high patient turnover or on clinical studies that embrace a large number of participants.
Approaches for automatic cartilage segmentation have been published previously. However, most of the reported data focus on knee joint11-13 and glenohumeral cartilage assessment14 and only a few studies are available for automatic segmentation of hip joint cartilage. In 2004, Nishii et al.15 reported a fully automated segmentation algorithm for acetabular cartilage thickness measurements in 45 patients with hip dysplasia. An edge detection approach was utilized, and for a sharp distinction between femoral and acetabular cartilage, continuous leg traction during MRI was applied in that study. However, to the best of our understanding, validation of their technique was not reported as no comparative manual analysis was undertaken.
With differences in methodology, Xia et al.16 demonstrated an automatic hip joint cartilage segmentation in 52 asymptomatic volunteers without the need for joint distraction. With the extraction of the bone-cartilage interfaces (BCI) and arc weighted graph searching, cartilage volume analysis revealed Dice’s similarity coefficients between 0.72 ± 0.05 and 0.81 ± 0.03 for automatic and manual segmentation. Notably, the study was conducted in 3D true fast imaging with steady-state precession (TrueFISP) images, which are not routinely used in clinical workflows. Therefore, application of their software technique to MRI sequences with other contrast characteristics or biochemical imaging needs further validation.
As an advancement of the work of Xia et al., utilizing different MRI sequences and improvements in BCI extraction, Chandra et al.17 reported an approach for extracting biochemical information from hip joint cartilage with automated T2 mapping analysis. In their study of 24 asymptomatic volunteers, a mean voxel overlap between manual and automated analysis was reported to be 73% with little difference error in median T2 average signals in the investigated cartilage areas.
For biochemical MRI in an OA cohort, Pedoia et al.18 published a fully automatic voxel-based relaxometry method for T1rho imaging. In that study, overall (16 patients with radiographic hip OA and 37 asymptomatic controls) Pearson correlation coefficients between fully automatic and manual cartilage segmentation were noted: R = 0.79 (P < 0.001) for acetabular, and R = 0.90 (P < 0.001) for femoral assessment. These results are somewhat comparable to ours. However, it has to be noted that correlation for acetabular cartilage in patients with OA (Kellgren-Lawrence 2 or 3) was only noted to be moderate (R = 0.57; P = 0.022), most likely due to thinner/absent cartilage layers in these patients. Given that our population was not diagnosed with higher grades of OA and cartilage analysis was only conducted in regions with no advanced morphological cartilage damage, the value of our proposed technique has to be further investigated in populations with ongoing progressive or advanced cartilage degeneration.
With an approach to examine the cartilage status across the entire hip joint, 2 previously published studies19,20 reported a methodology that uses multi-template based label fusion21 to automatically segment morphological TrueFISP and dGEMRIC data sets. These segmentations were further processed and “unfolded” to planar T1Gd and morphological 2D maps for cartilage analysis. Introduced by Sieversson et al., in a study in 15 symptomatic patients with mild or no OA, average Dice coefficients for cartilage segmentation between 0.76 and 0.82 were reported for 2 different algorithms, depending on the number of templates that were used (4 templates vs. 14 templates). However, some limitations have to be considered: Previously acquired manual segmentations are required to serve as a reference set to guide the automated segmentation algorithm, and the whole processing time was reported around 3 hours, which somewhat questions the clinical applicability. Furthermore, utilizing a 1.5-T MR scanner, femoral and acetabular cartilage could not be distinguished, and both cartilage layers were segmented together and subsequently divided. An accurate assessment of acetabular and femoral changes may be affected/biased in this case as joint fluid between both layers might have been taken into analysis. Additionally, the physiological discrepancy between femoral and acetabular thickness is not considered with this approach.
Our study certainly has limitations. With only 6 patients included, our study lacks statistical power. However, it is important to note that the purpose of this pilot study was not to validate this technique but to prove its feasibility for biochemical cartilage evaluation in future applications. A limitation of our technical approach is that the zone definitions may cause relevant defects to be missed, for example, when these are located on the border of 2 zones. The software takes the average value on a per-zone basis, and this could potentially lead to a misleading quantification of cartilage quality. For the automated cartilage assessment, no reproducibility analysis was carried out. However, since the automatic algorithm is deterministic in its outcome, rerunning the algorithm with the same data as input would yield identical results. Variability may be expected as a result of the segmentation quality. However, since a total of 500 points are sampled for each zone, taking the mean value of these samples, the resulting variability is negligible. Only if the segmentation would be significantly off for 500 of the bone surface points in a specific zone we would expect a difference, but in that case the segmentation would be incorrect. In this feasibility study, we chose to have 2 observers agree on the correctness of the segmentation, in which some degree of variation—albeit negligible—among observers during the semi-automated segmentation process of bone surfaces need to be considered.
In summary, this preliminary study sought to investigate the feasibility of a new approach for automatic hip joint cartilage segmentation and biochemical cartilage analysis with dGEMRIC. Notably, with lately reported concerns regarding gadolinium deposition and nephrogenic systemic fibrosis,22 it is of interest that the described computerized cartilage assessment technique is applicable to other non–contrast-based biochemical-sensitive, quantitative MRI techniques provided that there is a high-resolution volumetric image data set available. With promising results in this cohort of patients, future studies embracing larger groups with ongoing cartilage degeneration should be encouraged.
Footnotes
Acknowledgments and Funding: We would like to thank Sebastian Ullrich, who helped with conducting the statistical analysis in this study. The authors received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Ethical approval for this study was obtained from the local ethics committee (Study-ID 5574R).
Informed Consent: Written informed consent was obtained from all subjects before the study.
Trial Registration: Not applicable.
References
- 1. Ho CP, Surowiec RK, Ferro FP, Lucas EP, Saroki AJ, Dornan GJ, et al. Subregional anatomical distribution of T2 values of articular cartilage in asymptomatic hips. Cartilage. 2014;5(3):154-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hesper T, Hosalkar HS, Bittersohl D, Welsch GH, Krauspe R, Zilkens C, et al. T2* mapping for articular cartilage assessment: principles, current applications, and future prospects. Skeletal Radiol. 2014;43(10):1429-45. [DOI] [PubMed] [Google Scholar]
- 3. Schleich C, Bittersohl B, Miese F, Schmitt B, Muller-Lutz A, Sondern M, et al. Glycosaminoglycan chemical exchange saturation transfer at 3T MRI in asymptomatic knee joints. Acta Radiol. 2016;57(5):627-32. [DOI] [PubMed] [Google Scholar]
- 4. Le J, Peng Q, Sperling K. Biochemical magnetic resonance imaging of knee articular cartilage: T1rho and T2 mapping as cartilage degeneration biomarkers. Ann N Y Acad Sci. 2016;1383(1):34-42. [DOI] [PubMed] [Google Scholar]
- 5. Zilkens C, Miese F, Bittersohl B, Jäger M, Schultz J, Holstein A, et al. Delayed gadolinium-enhanced magnetic resonance imaging of cartilage (dGEMRIC), after slipped capital femoral epiphysis. Eur J Radiol. 2011;79(3):400-6. [DOI] [PubMed] [Google Scholar]
- 6. Hesper T, Bulat E, Bixby S, Akhondi-Asl A, Afacan O, Miller P, et al. Both 3-T dGEMRIC and acetabular-femoral T2 difference may detect cartilage damage at the chondrolabral junction. Clin Orthop Relat Res. 2017;475(4):1058-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tiderius CJ, Jessel R, Kim YJ, Burstein D. Hip dGEMRIC in asymptomatic volunteers and patients with early osteoarthritis: the influence of timing after contrast injection. Magn Reson Med. 2007;57(4):803-5. [DOI] [PubMed] [Google Scholar]
- 8. Bittersohl B, Hosalkar HS, Sondern M, Miese FR, Antoch G, Krauspe R, et al. Spectrum of T2* values in knee joint cartilage at 3 T: a cross-sectional analysis in asymptomatic young adult volunteers. Skeletal Radiol. 2014;43(4):443-52. [DOI] [PubMed] [Google Scholar]
- 9. Bittersohl B, Miese FR, Hosalkar HS, Mamisch TC, Antoch G, Krauspe R, et al. T2* mapping of acetabular and femoral hip joint cartilage at 3 T: a prospective controlled study. Invest Radiol. 2012;47(7):392-7. [DOI] [PubMed] [Google Scholar]
- 10. Khanduja V, Baelde N, Dobbelaere A, Van Houcke J, Li H, Pattyn C, et al. Patient-specific assessment of dysmorphism of the femoral head-neck junction: a statistical shape model approach. Int J Med Robot. 2016;12(4):765-72. [DOI] [PubMed] [Google Scholar]
- 11. Öztürk CN, Albayrak S. Automatic segmentation of cartilage in high-field magnetic resonance images of the knee joint with an improved voxel-classification-driven region-growing algorithm using vicinity-correlated subsampling. Comput Biol Med. 2016;72:90-107. [DOI] [PubMed] [Google Scholar]
- 12. Pang J, Li P, Qiu M, Chen W, Qiao L. Automatic articular cartilage segmentation based on pattern recognition from knee MRI images. J Digit Imaging. 2015;28(6):695-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahn C, Bui TD, Lee YW, Shin J, Park H. Fully automated, level set-based segmentation for knee MRIs using an adaptive force function and template: data from the osteoarthritis initiative. Biomed Eng Online. 2016;15:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neubert A, Yang Z, Engstrom C, Xia Y, Strudwick MW, Chandra SS, et al. Automatic segmentation of the glenohumeral cartilages from magnetic resonance images. Med Phys. 2016;43:5370. [DOI] [PubMed] [Google Scholar]
- 15. Nishii T, Sugano N, Sato Y, Tanaka H, Miki H, Yoshikawa H. Three-dimensional distribution of acetabular cartilage thickness in patients with hip dysplasia: a fully automated computational analysis of MR imaging. Osteoarthritis Cartilage. 2004;12(8):650-7. [DOI] [PubMed] [Google Scholar]
- 16. Xia Y, Chandra SS, Engstrom C, Strudwick MW, Crozier S, Fripp J. Automatic hip cartilage segmentation from 3D MR images using arc-weighted graph searching. Phys Med Biol. 2014;59(23):7245-66. [DOI] [PubMed] [Google Scholar]
- 17. Chandra SS, Surowiec R, Ho C, Xia Y, Engstrom C, Crozier S, et al. Automated analysis of hip joint cartilage combining MR T2 and three-dimensional fast-spin-echo images. Magn Reson Med. 2016;75(1):403-13. [DOI] [PubMed] [Google Scholar]
- 18. Pedoia V, Gallo MC, Souza RB, Majumdar S. Longitudinal study using voxel-based relaxometry: association between cartilage T1ρ and T2 and patient reported outcome changes in hip osteoarthritis. J Magn Reson Imaging. 2017;45(5):1523-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siversson C, Akhondi-Asl A, Bixby S, Kim YJ, Warfield SK. Three-dimensional hip cartilage quality assessment of morphology and dGEMRIC by planar maps and automated segmentation. Osteoarthritis Cartilage. 2014;22(10):1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bulat E, Bixby SD, Siversson C, Kalish LA, Warfield SK, Kim YJ. Planar dGEMRIC maps may aid imaging assessment of cartilage damage in femoroacetabular impingement. Clin Orthop Relat Res. 2016;474(2):467-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage. 2009;46(3):726-38. [DOI] [PubMed] [Google Scholar]
- 22. Fraum TJ, Ludwig DR, Bashir MR, Fowler KJ. Gadolinium-based contrast agents: a comprehensive risk assessment. J Magn Res Imaging. 2017;46(2):338-53. [DOI] [PubMed] [Google Scholar]