Abstract
The differentiation of acinar cells to ductal cells during pancreatitis and in the early development of pancreatic cancer is a key process that requires further study. To understand the mechanisms regulating acinar-to-ductal metaplasia (ADM), ex vivo 3D culture and differentiation of primary acinar cells to ductal cells offers many advantages over other systems. With the technique herein, modulation of protein expression is simple and quick, requiring only one day to isolate, stimulate or virally infect, and begin culturing primary acinar cells to investigate the ADM process. In contrast to using basement membrane matrix, the seeding of acinar cell clusters in collagen I extracellular matrix, allows acinar cells to retain their acinar identity before manipulation. This is vital when testing the contribution of various components to the induction of ADM. Not only are the effects of cytokines or other ectopically administered factors testable through this technique, but the contribution of common mutations, increased protein expression, or knockdown of protein expression is testable via viral infection of primary acinar cells, using adenoviral or lentiviral vectors. Moreover, cells can be re-isolated from collagen or basement membrane matrix at the endpoint and analyzed for protein expression.
Keywords: Primary acinar cells, acinar-to-ductal metaplasia, 3-dimensional culture, pancreatic cancer, adenoviral vectors, lentiviral vectors
SUMMARY:
Primary acinar cell isolation, protein expression or activity modulation, culture, and downstream applications are abundantly useful in the ex vivo study of acinar-to-ductal metaplasia (ADM), an early event in the development of pancreatic cancer.
INTRODUCTION:
Acinar-to-ductal metaplasia (ADM) is a protective mechanism during pancreatitis and a key process driving pancreatic cancer development1 that requires further mechanistic insight. While inflammation-induced ADM is reversible2, oncogenic KRAS mutations, which are present in 90% of pancreatic cancer cases3, prevent differentiation back to an acinar phenotype4–6. Culturing and differentiating primary acinar cells into ductal cells in 3D culture allows for study of the molecular mechanisms regulating the ADM process, which is difficult to study in vivo. Such ex vivo studies enable real-time visualization of ADM, its drivers and its regulators. Several drivers of the ADM process, as well as mechanistic insight on their downstream signaling pathways, have been identified or verified using the here described method. These include ADM induction by TGF-α (transforming growth factor alpha)-mediated expression of MMP-7 (matrix metalloproteinase-7) and activation of Notch7, as well as RANTES (regulated on activation, normal T cell expressed and secreted; also known as chemokine ligand 5 or CCL5) and TNFα (tumor necrosis factor alpha)-induced ADM through activation of NF-κB (nuclear factor-κB)8. An additional mediator of ADM is oncogenic KRas9,10, which causes a rise in oxidative stress that enhances the ADM process through increased expression of EGFR (epidermal growth factor receptor) and its ligands, EGF (epidermal growth factor) and TGF-α11.
While use of pancreatic cancer cell lines is common for in vitro studies, primary cell cultures offer many advantages. For instance, primary acinar cells from non-transgenic mice or mice harboring initiating mutations, such as KrasG12D, are the most appropriate model for studying the early event of ADM because the cells have few and controlled mutations, which are known to be present early in pancreatic cancer development. This is in contrast to many pancreatic cancer cell lines which have multiple mutations and varied expression based on passage number12. Additionally, the signaling pathways identified by such means have been verified by animal studies. One caveat of using primary acinar cells is that they cannot be transfected as typical cancer cell lines can.
The method herein details the techniques of acinar cell isolation, 3D culture that mimics ADM by adding stimulants or modulating protein expression via adenoviral or lentiviral infection, as well as re-isolation of cells at the endpoint for further analyses.
PROTOCOL:
All animal work was approved by the Mayo Clinic IACUC.
1. Preparation of Materials, Solutions, and 3-Dimensional Matrix Bases
1.1) Cut 500 μm and 105 μm polypropylene mesh into 76 mm by 76 mm squares. Fold each square in half two times to create a smaller folded square. Place one 500 μm and one 105 μm mesh square into an autoclavable pouch.
1.2) Autoclave polypropylene mesh squares, as well as two pairs of scissors and forceps.
1.3) Make 100 ml of 10X Waymouth’s solution. Stir 1 vial (14 g) of Waymouth’s powder into 50 ml of deionized (DI) water and, when dissolved, add 30 ml of 7.5% (75 g/L) sodium bicarbonate. Bring the final volume to 100 ml using DI water and filter sterilize.
1.3.1) Store 10X Waymouth’s media at 4 °C and use to prepare collagen gels and 1X Waymouth’s media. When precipitates form, discard.
1.4) Make 50 ml 1X Waymouth’s complete media per pancreas by adding 125 μl of 40 mg/ml soybean trypsin inhibitor, 12.5 μl of 4 mg/ml dexamethasone, and 500 μl fetal bovine serum (FBS). Sterilize by filtration and use within 48 hrs.
1.5) Prepare 3-dimensional matrix bases using either collagen or basement membrane matrix (see table of materials for product information). When pipetting the base, place the plate on ice, avoid bubbles, and rotate the plate immediately after pipetting each well’s volume to ensure full coverage.
Materials
| Name of Reagent/ Equipment | Company | Catalog Number |
|---|---|---|
| 37 °C shaking incubator | Thermo Scientific | SHKE4000-7 |
| 5% CO2, 37 °C Incubator | NUAIRE | NU-5500 |
| 50 ml tubes | Falcon | 352070 |
| Absorbent pad, 20” x 24” | Fisherbrand | 1420662 |
| Adenovirus, Ad-GFP | Vector Biolabs | 1060 |
| Aluminum foil | Fisherbrand | 01-213-101 |
| BD Precision Glide Needle 21G x 1/2 1 | Fisher Scientific | 305167 |
| Beaker, 600 mL | Fisherbrand | FB-101-600 |
| Bleach, 8.25% | Clorox | 31009 |
| Cell Recovery Solution 2 | Corning | 354253 |
| Centrifuge | Beckman Coulter | Allegra X-15R Centrifuge |
| Collagenase Type I, Clostridium histolyticum 3 | Sigma | C0130-1G |
| Dexamethasone 4 | Sigma | D1756 |
| Ethanol, 200 proof | Decon Laboratories | 2701 |
| Fetal Bovine Serum | Sigma | F0926-100mL |
| Forceps | Fine Science Tools | 11002-12 |
| Forceps | Fine Science Tools | 91127-12 |
| Glass slide, 8-well | Lab-Tek | 177402 |
| Hank’s Balanced Salt Solution (HBSS), No calcium, No magnesium, No phenol red | Fisher Scientific | SH3048801 |
| Ice bucket, rectangular | Fisher Scientific | 07-210-103 |
| Instant Sealing Sterilization Pouch | Fisherbrand | 01-812-51 |
| LAB GUARD specimen bags (for mouse after dissection) | Minigrip | SBL2X69S |
| Lentiviral Packaging Mix, Virapower | Invitrogen | 44-2050 |
| Matrigel 5 | Corning | 356234 |
| Parafilm 6 | Bemis | PM992 |
| PBS | Fisher Scientific | SH30028.02 |
| Penicillin-Streptomycin | ThermoFisher Scientific | 15140122 |
| Pipet tips, 10 μl | USA Scientific | 1110-3700 |
| Pipet tips, 1000 μl | Olympus Plastics | 24-165RL |
| Pipet tips, 200 μl | USA Scientific | 1111-1700 |
| Pipet-Aid | Drummond | |
| PIPETMAN Classic P10, 1-10 μl | Gilson | F144802 |
| PIPETMAN Classic P1000, 200-1000 μl | Gilson | F123602 |
| PIPETMAN Classic P20, 2-20 μl | Gilson | F123600 |
| PIPETMAN Classic P200, 20-200 μl | Gilson | F123601 |
| Pipettes, 10 ml | Falcon | 357551 |
| Pipettes, 25 ml | Falcon | 357525 |
| Pipettes, 5 ml | Falcon | 357543 |
| Plate, 12-well | Corning Costar | 3513 |
| Plate, 24-well plate | Corning Costar | 3524 |
| Plate, 35 mm | Falcon | 353001 |
| Plate, 6-well | Falcon | 353046 |
| Polybrene 7 | EMD Millipore | TR-1003-G |
| Polypropylene Mesh, 105 μm | Spectrum Labs | 146436 |
| Polypropylene Mesh, 500 μm | Spectrum Labs | 146418 |
| Scissors | Fine Science Tools | 14568-12 |
| Scissors | Fine Science Tools | 91460-11 |
| Sodium Bicarbonate (Fine White Powder) | Fisher Scientific | BP328-500 |
| Sodium Hydroxide | Fisher Scientific | S318-500 |
| Soybean Trypsin Inhibitor 8 | Gibco | 17075029 |
| Spatula | Fisherbrand | 21-401-10 |
| Steriflip 50 ml, 0.22 micron filters | Millipore | SCGP00525 |
| TGF-α | R&D Systems | 239-A-100 |
| Type I Rat Tail Collagen | Corning | 354236 |
| Waymouth MB 752/1 Medium (powder) | Sigma | W1625-10X1L |
| Weigh boat, hexagonal, medium | Fisherbrand | 02-202-101 |
Comments/Description
Use as pins for dissection
Referred to as ‘basement membrane matrix recovery solution’ in the manuscript.
Create a 100 mg/ml solution by dissolving powder in sterile molecular biology grade water, When thawing one aliquot, dilute to 10 mg/ml, filter sterilize and place 1 ml aliquots at −20 °C.
Create a 4 mg/ml solution by dissolving powder in methanol, aliquoting and storing at −20 °C.
Referred to as ‘basement membrane matrix’ in the manuscript.
Referred to as ‘plastic paraffin film’ in the manuscript.
Referred to as ‘viral infection enhancer reagent’ in the manuscript.
Create a 40 mg/ml solution by dissolving powder in sterile molecular biology grade water, aliquoting and storing at −20 °C.
1.5.1) Create collagen bases by mixing the following components on ice in a tissue culture hood: 5.5 ml of type I rat tail collagen, 550 μl of 10X Waymouth’s Media, and 366.6 μl of 0.34 M NaOH (filtered).
NOTE: This is enough solution to make collagen bases for one 24-well, 12-well, or 6-well plate. One plate is sufficient for acinar isolation from one pancreas.
1.5.2) Use the following volumes to plate the collagen base: 80 μl (8-well glass slide), 200 μl (24-well plate), 400 μl (12-well plate), or 800 μl (6-well plate).
1.5.2) For the basement membrane matrix bases, pipet the matrix without any additional components. Use the following volumes for each well: 50 μl (8-well glass slide), 120 μl (24-well plate), 240 μl (12-well plate), or 600 μl (6-well plate).
1.5.3) Let the bases solidify for at least 30 minutes in a cell culture incubator (37 °C, 5% CO2) before creating a second layer of matrix with embedded cells on top of these bases (step 4).
1.6) In preparation for acinar isolation keep one 600 ml beaker, three 50 ml tubes, an autoclaved pair of scissors, an autoclaved pair of forceps, and an ice bucket under the hood with three weigh boats. Then, make 30 ml of HBSS with 300 μl of 100X penicillin-streptomycin, putting 10 ml into each of two weigh boats and keeping the final 10 ml in a 50 ml tube (for use in steps 1.8.2 and 2.1.3).
1.7) Make 40 ml of HBSS + 5% FBS, 20 ml of HBSS + 30% FBS, and 5 ml of 2 mg/ml collagenase in HBSS. Sterilize the collagenase by filtration (0.22 μm pore) and keep at room temperature. Keep each of the HBSS solutions on ice.
1.8) Assemble a workspace for pancreas dissection, which can be done on a lab bench.
1.8.1) Place an absorbent pad on the table, along with a polystyurene lid covered in foil. Then place a paper towel with 4 pins on top of the foil.
1.8.2) Keep the following items within reach of the dissection workspace: an incineration bag, one set of autoclaved scissors and forceps, a spray bottle with 70% ethanol, and an ice bucket (with the 50 ml tube of HBSS containing penicillin-streptomycin from step 1.6).
1.9) Set a centrifuge to 4 °C and a shaker to 37 °C.
2. Acinar Cell Isolation
2.1) Sacrifice the mouse via CO2 induction, and perform cervical dislocation. Immediately dissect the pancreas. To dissect the pancreas, first pin the paws of the mouse to the polystyrene lid, orient the mouse such that the tail is facing the researcher, and spray the abdomen with 70% ethanol.
Note: The mouse used in the representative results was a 12-week old nontransgenic female with C57BL/6 background. Mouse selection is further discussed in the modifications section.
2.1.1) Using a set of autoclaved scissors and forceps, lift the fur/skin with the forceps at the midline and use the scissors to make an incision through the fur and skin from the urethral opening to the diaphragm/ribcage area.
2.1.2) Make additional incisions to the left and right such that the fur/skin is cut away to create a clear view of the abdominal cavity. Then, cut into the peritoneal lining (down the middle and to the right and left) and pull it away from the organs, as was done with the fur/skin.
2.1.3) Lift the intestines with the forceps and put it to the left side of the mouse creating space to see the pancreas, which is light pink in color and attached to the spleen which is a dark red oval. The pancreatic tissue is distinguished by its soft, spongy texture. Cut out the pancreas which will run along the stomach and intertwine with the intestines.
2.1.4) Separate the spleen from the pancreas. Then, put the pancreas in a 50 ml tube containing 10 ml of HBSS with 1X penicillin-streptomycin and bring this to the laminar flow hood.
NOTE: The remaining steps of the protocol should be done utilizing sterile technique in a laminar flow hood.
2.2) Pour the pancreas and HBSS (with penicillin-streptomycin) into an empty weigh boat and, using forceps, wash the pancreas by swirling it. Then, move the pancreas with the forceps to a second weigh boat containing HBSS (with penicillin-streptomycin). Again, wash the pancreas by swirling.
2.3) Move the pancreas to the third HBSS (with penicillin-streptomycin)-containing weigh boat and begin cutting the pancreas into small pieces of 5 mm or less. Next, pour the pancreas pieces and HBSS into an empty 50 ml tube.
2.3.1) To do this, use the forceps to move the pancreas pieces into the liquid as the weigh boat is being tipped. Pour once all the pieces are no longer attached to the weigh boat. Pick up any remaining pieces with the forceps and wash the forceps in the 50 ml tube containing the pancreas in HBSS with penicillin-streptomycin.
2.4) Centrifuge at 931 x g for 2 min at 4 °C and then remove the HBSS (and any fat that is floating) by pipetting it off with a 5 ml pipette.
2.5) Add 5 ml of collagenase (diluted in HBSS in step 1.7) to the pancreas. Ensure a sealed lid by wrapping the 50 ml tube in plastic paraffin film and then place it in an incubator shaking at 220 rpm for 20 min at 37 °C.
NOTE: The collagenase digestion time may vary, as noted in the ‘critical steps and troubleshooting’ section of the discussion. At the end of incubation, no large pieces of tissue should remain.
2.6) To stop the dissociation, place the pancreas-containing tube on ice and add 5 ml of cold HBSS + 5% FBS. Centrifuge at 931 x g for 2 min at 4 °C and then pipet off the supernatant using a 5 ml pipet.
2.7) Resuspend in 10 ml of HBSS + 5% FBS and centrifuge at 931 x g for 2 min at 4 °C. Pipet off the supernatant using a 5 ml pipet and repeat this step with another 10 ml of HBSS + 5% FBS.
2.8) Resuspend in 5 ml of HBSS + 5% FBS and, using a P1000, transfer 1 ml at a time through a 500 μm mesh into a 50 ml tube. Add an additional 5 ml of HBSS + 5% FBS through the mesh with a P1000 to wash any remaining pancreatic cells through the mesh.
2.9) Put the cell suspension from the 500 μm mesh through the 105 μm mesh by pipetting 1 ml at a time using a P1000. Next, gently pipet the cell suspension into a tube containing HBSS + 30% FBS. A layer of cell suspension will form at the top; upon centrifugation, the acinar cells will sink to form a pellet.
2.10) Centrifuge at 233 x g for 2 min at 4 °C. Remove the supernatant and resuspend the pellet in 1X Waymouth’s complete media.
2.10.1) If proceeding directly to embedment of cells in collagen or basement membrane matrix, resuspend in a volume of 7 ml per pancreas. If proceeding to adenoviral or lentiviral infection, resuspend in 4 ml per pancreas.
3. Viral Infection
3.1) One pancreas is sufficient for two viral infections (control and experimental; e.g. null and cre). Split the cell suspension between the lids of two 35 mm plates. Using the lid of the plates allows for infection in suspension and therefore easy movement onto the final substrate later.
3.1.1) When working with virus, soak all used pipette tips in 10% bleach for at least 15 minutes.
NOTE: Utilization of the 35 mm plates and the 4 ml volume works well for cells from nontransgenic mice between 8 and 16 weeks old. The plate size and volume may need to be adjusted for animals with smaller or larger pancreata.
3.2) For adenoviral infection, add the virus (with a titer of at least 107 TU/ml) at a 1:1000 dilution to the lid of each plate and swirl (Figure 2, GFP adenovirus). Place the plates in a cell culture incubator (37 °C, 5% CO2) and swirl every 15 minutes for one hour. Continue incubation for an additional 2 hours and then proceed to embedment of cells in collagen or basement membrane matrix.
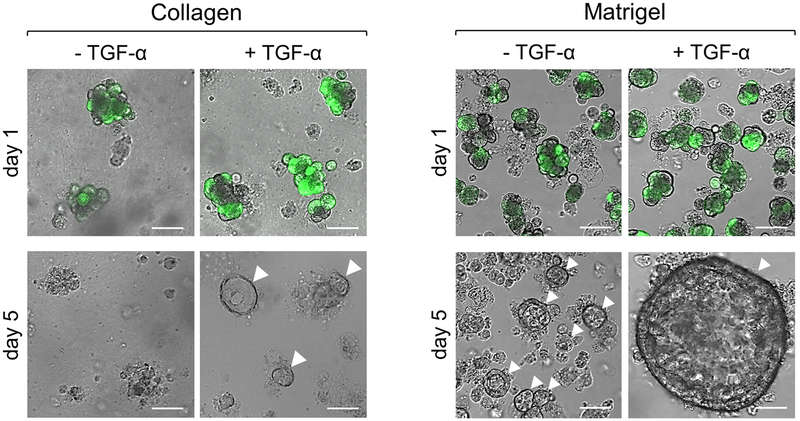
Figure 2: Representative results of primary acinar cells plated within collagen or basement membrane matrix after adenoviral infection and stimulation with TGF-α.
Primary acinar cells from a non-transgenic mouse were infected with GFP-adenovirus, embedded in collagen I or basement membrane matrix, and stimulated with or without TGF-α (50 ng/ml). Twenty-four hours post-infection with GFP adenovirus, images were captured to show infection efficiency. At five days post-infection, the images denote the differences in cells stimulated with or without TGF-α in collagen or basement membrane matrix. Duct-like structures are indicated by white arrowheads and scale bars represent 50 μm. Images were obtained using a Zeiss LSM 880 confocal microscope with the EC Plan-Neofluar 10x/0.3 M27 objective.
3.3) After completing viral work in the hood, soak all components in the hood with 10% bleach for at least 15 minutes and then thoroughly clean the bleach off using 70% ethanol.
4. Embedment of Cells in Collagen or Basement Membrane Matrix
4.1) Make collagen gel on ice by combining 7 ml type I rat tail collagen (3.3 mg/ml), 700 μl 10X Waymouth’s media, and 466.6 μl 0.34 M NaOH.
4.2) Taking equal volumes of collagen gel and cell suspension, gently mix. If adding a stimulus or inhibitor, combine this with the collagen-cell suspension. Plate one well at a time; keeping the plate on ice, swirl to evenly distribute the collagen-cell layer.
4.2.1) In each well, pipet the following volumes of collagen-cell suspension: 200 μl (8-well glass slide), 500 μl (24-well plate), 1000 μl (12-well plate), or 2000 μl (6-well plate). Note that ADM is induced via stimulation with TGF-α (50 ng/ml) in Figure 2.
4.3) For embedment of cells in basement membrane matrix, mix one part basement membrane matrix with two parts cell suspension. As with collagen, keep the matrix-cell suspension as well as the plate on ice. Pipet one well at a time using the same volumes noted in 4.2.1 and swirl for even distribution.
4.4) Place the plate in a cell culture incubator (37 °C, 5% CO2) for 30 min to solidify and then add 1X Waymouth’s complete media with any stimulus or inhibitor (Figure 2 uses TGF-α at 50 ng/ml). Change the media (with stimulus or inhibitor) the next day and then every other day. Depending on the stimulus, ADM can be observed between day 3 and day 5.
5. Harvesting Cells from Collagen or Basement Membrane Matrix
5.1) Collect the following materials needed for harvesting cells from collagen: 30 ml of 1mg/ml collagenase diluted in HBSS, 40 ml cold HBSS, 31 ml cold PBS, two spatulas, two 50 ml tubes, and ice. Adjust the number of spatulas, tubes and amount of solutions if comparing more than two culture conditions (where each condition is ½ of a plate). Set the centrifuge to 4 °C and set the shaking incubator to 37 °C.
5.2) Remove the media and use spatulas to remove the collagen disks with embedded cells. For each condition, put the disks into a separate, empty 50 ml tube.
5.3) Add 10 ml of 1 mg/ml collagenase in HBSS to each 50 ml tube. Wrap the tubes with plastic paraffin film and put in a 37 °C shaking incubator at 225 rpm for up to 45 minutes. Monitor the digestion and remove tubes when there is no more visible collagen.
5.4) Centrifuge at 233 x g at 4 °C for 2 min with minimum deceleration and then take off the supernatant and resuspend in 5 ml collagenase solution per tube. Digest in a 37 °C shaking incubator at 225 rpm for 10 min or until there is no visible collagen.
5.5) Stop the digestion by adding 5 ml cold HBSS and subsequently centrifuging at 233 x g for 2 min at 4 °C with minimum deceleration. Resuspend in 15 ml cold HBSS and again centrifuge at 233 x g for 2 min at 4 °C with minimum deceleration. Then, resuspend in 15 ml cold PBS and again centrifuge at 233 x g for 2 min at 4 °C with minimum deceleration.
5.6) Resuspend the pellet in 1 ml of PBS and move to 1.5 ml tube. Centrifuge in a swing bucket at 233 x g for 2 min at 4 °C with minimum deceleration. Remove supernatant and snap-freeze in liquid nitrogen or on dry ice.
NOTE: The cell pellet can be stored at −70°C or used immediately in down-stream applications.
5.7) To harvest basement membrane matrix-embedded cells, pipet media from each well into a 50 ml tube on ice. Then, add basement membrane matrix recovery solution to each well and scrape the cell-gel mixture into the 50 ml tube.
NOTE: The volume of basement membrane matrix recovery solution to add to each well is as follows: 150 μl (8-well glass slide), 420 μl (24-well plate), 845 μl (12-well plate), and 2000 μl (6-well plate).
5.8) Rinse each well twice with the basement membrane matrix recovery solution, adding the rinses to the 50 ml tube.
NOTE: The volume of basement membrane matrix recovery solution for each rinse is as follows: 150 μl (8-well glass slide), 420 μl (24-well plate), 845 μl (12-well plate), and 2000 μl (6-well plate).
5.9) Leave the tube on ice for 1 hour or until the basement membrane matrix dissolves and follow with centrifugation at 300 x g for 5 min at 4 °C. Remove the supernatant and resuspend the pellet in 1 ml of cold PBS (may transfer to 1.5 ml tube if cell pellet is desired).
5.10) Centrifuge at 3500 x g for 3 min at 4 °C. Remove the supernatant and either snap freeze the cell pellet or add lysis buffer and directly proceed to downstream applications.
REPRESENTATIVE RESULTS:
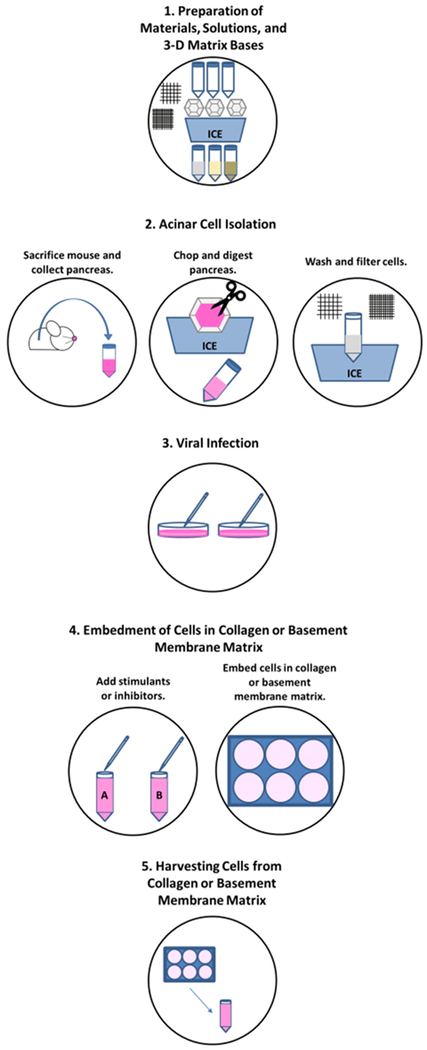
Completion of the protocol herein occurs within one day and upon stimulation, ADM is seen in 3–5 days. Figure 1 depicts the sequence of the method, whereby steps 1 through 4 are completed on the first day. This includes preparation, acinar isolation, viral infection and embedment in collagen or basement membrane matrix. While the basement membrane matrix induces ADM, acinar cells within collagen I require a stimulus, such as TGF-α, to undergo differentiation to ductal cells. On day 1, cells in collagen or basement membrane matrix have a grape-like round appearance and if utilizing viral infection with a vector expressing a fluorescent protein, infection efficiency can be checked. By day 5, cells in collagen will form ducts if given a stimulus at the time of embedment and as a supplement in the media, which is changed on days 1, 3, and 5. Cells embedded in basement membrane matrix will form ducts in the absence of a stimulus, but upon stimulation larger ducts will form, as seen in Figure 2.
Figure 1: Schematic of the procedure to isolate murine primary acinar cells, modulate protein expression and/or stimulate acinar-to-ductal metaplasia, and embed cells in collagen or basement membrane matrix.
The workflow includes isolation of acinar cells, which includes dissection of the pancreas, digestion, and filtration of the cells.
DISCUSSION:
Significance, Applications, and Limitations
The relatively short amount of time for isolation, infection, and plating primary acinar cells is an advantage of this method. In contrast, culturing acinar cells from an explant outgrowth requires little hands-on time, but it takes seven days for the outgrowth of acinar cells13. An alternative protocol for acinar isolation14 notes a very short method for obtaining acinar cells; however, EGF is an essential component in keeping acinar cells alive when isolated using that protocol. Since EGF stimulates differentiation of acinar cells to ductal cells, the method herein, which does not require ADM-stimulating components for culture, is better for studying mechanisms of induction or regulation of ADM. In this method, acinar cell identity is maintained during culturing in collagen unless stimulated to differentiate into ductal cells. Moreover, difficulties in transfecting primary cells are overcome via manipulating protein expression by viral infection.
This procedure offers direct study of ADM, by which visualization of ductal structure induction can occur via brightfield and/or immunofluorescence microscopy. For imaging applications, chamber slides (noted in materials) are best. Fixation for 15 minutes at 37 °C with 4% paraformaldehyde will fix cells embedded in basement membrane matrix without disturbing ductal structures. During this incubation, the basement membrane matrix will liquify and can be gently pipetted off with the paraformaldehyde. Immunofluorescence on collagen embedded cells is described in detail within additional protocols15,16. Further applications for analyses of involved signaling mechanisms at the endpoint of the experiment include Western blot, qPCR, and fluorescence-activated cell sorting (FACS). One sixth of a pancreas is sufficient material for one sample analysis via Western blot or qPCR.
Limitations of this protocol arise from the use of collagen or basement membrane matrix, where each has their advantages and disadvantages. While embedment of cells in collagen will only produce ducts if stimuli are given, basement membrane matrix embedment will produce ducts without additional stimuli. However, ductal size in basement membrane matrix is a measurable output. An additional limitation is the time involved in the harvesting procedures in step 5, which could affect gene expression. This limitation can be mitigated by confirming results obtained from Western blotting with immunofluorescence, in which the fixation time is considerably less than that of the harvesting time for qPCR or Western blot analysis.
Modifications
In addition to adenoviral infection, lentiviral infection can be used in primary cells. For lentiviral infection, add 6 μg/ml viral infection enhancer reagent (see table of materials) to the cells and gently swirl the plates. Subsequently, add the lentivirus (with a titer of at least 107 TU/ml) at a 1:1000 dilution and again, gently swirl. Incubate for 3 – 5 hrs (37 °C, 5% CO2) and then continue to embedment of cells in collagen or basement membrane matrix. To generate lentivirus with a plasmid of interest, use the lentiviral packaging mix noted in the table of materials.
Further modification can include isolation of cells from mice expressing KrasG12D. In these mice, which already have fibrotic areas with ADM and PanIN lesions, collagenase digestion takes longer. Careful monitoring of collagenase digestion, as described in the critical steps and troubleshooting, is required. The more abnormal tissue a mouse has, the longer the digestion will take (around an hour for a twelve-week-old p48Cre; LSL-KrasG12D mouse). Since there can be immense variation between mice of the same age, we advise to isolate acinar cells from an LSL-KrasG12D mouse and perform adenoviral or lentiviral infection utilizing cre to obtain oncogenic KRas expression. It also should be noted that our protocol is optimized for isolation of acinar cell to investigate the ADM process. The isolation of ADM and PanIN cells from KrasG12D-expressing mice for organoid culture requires altered protocols.
Critical Steps and Troubleshooting
One critical step is the amount of time that the chopped pancreas spends in collagenase. The ideal collagenase digestion time can vary from mouse to mouse and between lots of collagenase from the same company. The time noted in the protocol is appropriate for pancreata weighing about 0.250 g. Too much time can damage and kill cells, while too little time will result in incomplete digestion and therefore fewer cells that will make it through the filtering process onto the final plate. Check the dissociation visually by looking for breakdown of the chopped-up pancreas pieces; the solution should turn cloudy. Additionally, before stopping the collagenase reaction with cold HBSS, the solution can be taken up with a 5 ml pipet, where the ease with which the solution can be taken up will indicate if it is appropriate to stop the reaction. An additional consideration is the number of pancreata harvested. If more than one pancreas is isolated at the same time, the procedure works best when the pancreata are kept separate.
Another important point to ensure healthy culture of primary acinar cells is that care should be taken when pipetting the acinar cell solution. Vigorous pipetting may cause cell damage and result in a lack of duct formation, even with addition of known ADM inducers, such as TGF-α. Further, if there are viability issues, the centrifugation in steps 2.4, 2.6, and 2.7 can be taken down to 300 x g to produce a softer pellet.
ACKNOWLEDGMENTS:
This work was supported by an R01 grant (CA200572) from the NIH to PS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
DISCLOSURES:
The authors declare that they have no competing financial interests.
Contributor Information
Alicia K. Fleming Martinez, Department of Cancer Biology, Mayo Clinic Florida.
Peter Storz, Department of Cancer Biology, Mayo Clinic Florida.
REFERENCES:
- 1.Rooman I & Real FX Pancreatic ductal adenocarcinoma and acinar cells: a matter of differentiation and development? Gut. 61 (3), 449–458, (2012). [DOI] [PubMed] [Google Scholar]
- 2.Stanger BZ & Hebrok M Control of cell identity in pancreas development and regeneration. Gastroenterology. 144 (6), 1170–1179, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldas C & Kern SE K-ras mutation and pancreatic adenocarcinoma. Int J Pancreatol. 18 (1), 1–6, (1995). [DOI] [PubMed] [Google Scholar]
- 4.Kopp JL et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 22 (6), 737–750, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris J. P. t., Cano DA, Sekine S, Wang SC & Hebrok M Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. Journal of Clinical Investigation. 120 (2), 508–520, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grippo PJ, Nowlin PS, Demeure MJ, Longnecker DS & Sandgren EP Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 63 (9), 2016–2019, (2003). [PubMed] [Google Scholar]
- 7.Sawey ET, Johnson JA & Crawford HC Matrix metalloproteinase 7 controls pancreatic acinar cell transdifferentiation by activating the Notch signaling pathway. Proceedings of the National Academy of Sciences of the United States of America. 104 (49), 19327–19332, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liou GY et al. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-kappaB and MMPs. J Cell Biol. 202 (3), 563–577, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De La OJ et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proceedings of the National Academy of Sciences of the United States of America. 105 (48), 18907–18912, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habbe N et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proceedings of the National Academy of Sciences of the United States of America. 105 (48), 18913–18918, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liou GY et al. Mutant KRas-Induced Mitochondrial Oxidative Stress in Acinar Cells Upregulates EGFR Signaling to Drive Formation of Pancreatic Precancerous Lesions. Cell Rep. 14 (10), 2325–2336, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes P, Marshall D, Reid Y, Parkes H & Gelber C The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? Biotechniques. 43 (5), 575, 577,–578, 581–572 passim, (2007). [DOI] [PubMed] [Google Scholar]
- 13.Blauer M, Nordback I, Sand J & Laukkarinen J A novel explant outgrowth culture model for mouse pancreatic acinar cells with long-term maintenance of secretory phenotype. Eur J Cell Biol. 90 (12), 1052–1060, (2011). [DOI] [PubMed] [Google Scholar]
- 14.Gout J et al. Isolation and culture of mouse primary pancreatic acinar cells. J Vis Exp. 10.3791/50514 (78), (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artym VV & Matsumoto K Imaging cells in three-dimensional collagen matrix. Curr Protoc Cell Biol. Chapter 10 Unit 10 18 11–20, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geraldo S, Simon A & Vignjevic DM Revealing the cytoskeletal organization of invasive cancer cells in 3D. J Vis Exp. 10.3791/50763 (80), e50763, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]