Supplemental Digital Content is available in the text.
Keywords: biomarkers; gene expression; inflammation; myocardial infarction; ventricular dysfunction, left
Background:
The identification of patients with acute myocardial infarction (MI) at risk of subsequent left ventricular (LV) dysfunction remains challenging, but it is important to optimize therapies. The aim of this study was to determine the unbiased RNA profile in peripheral blood of patients with acute MI and to identify and validate new prognostic markers of LV dysfunction.
Methods:
We prospectively enrolled a discovery cohort with acute MI (n=143) and performed whole-blood RNA profiling at different time points. We then selected transcripts on admission that related to LV dysfunction at follow-up and validated them by quantitative polymerase chain reaction in the discovery cohort, in an external validation cohort (n=449), and in a representative porcine MI model with cardiac magnetic resonance–based measurements of infarct size and postmortem myocardial pathology (n=33).
Results:
RNA profiling in the discovery cohort showed upregulation of genes involved in chemotaxis, IL (interleukin)-6, and NF-κB (nuclear factor-κB) signaling in the acute phase of MI. Expression levels of the majority of these transcripts paralleled the rise in cardiac troponin T and decayed at 30 days. RNA levels of QSOX1, PLBD1, and S100A8 on admission with MI correlated with LV dysfunction at follow-up. Using quantitative polymerase chain reaction, we confirmed that QSOX1 and PLBD1 predicted LV dysfunction (odds ratio, 2.6 [95% CI, 1.1–6.1] and 3.2 [95% CI, 1.4–7.4]), whereas S100A8 did not. In the external validation cohort, we confirmed QSOX1 and PLBD1 as new independent markers of LV dysfunction (odds ratio, 1.41 [95% CI, 1.06–1.88] and 1.43 [95% CI, 1.08–1.89]). QSOX1 had an incremental predictive value in a model consisting of clinical variables and cardiac biomarkers (including NT-proBNP [N-terminal pro-B-type natriuretic peptide]). In the porcine MI model, whole-blood levels of QSOX1 and PLBD1 related to neutrophil infiltration in the ischemic myocardium in an infarct size–independent manner.
Conclusions:
Peripheral blood QSOX1 and PLBD1 in acute MI are new independent markers of LV dysfunction post-MI.
Despite marked improvements in the treatment of acute myocardial infarction (MI), the long-term outcome after MI remains poor. The rate of rehospitalization for heart failure at 1 year post-MI ranges from 5% ≤20%, despite state-of-the-art therapy.1 Up to 15% of patients with ST-segment–elevation MI have a left ventricular (LV) ejection fraction <40% at follow-up.2 Heart failure post-MI encompasses high morbidity and a high cost for rehospitalizations, drugs, and device therapies. Therefore, position papers have recently emphasized the need to identify new predictors and identify new targets for intervention.3–5
Since inflammation plays a pivotal role in the acute injury during MI and subsequent repair, a better characterization of the inflammatory drivers might improve the identification of patients at increased risk for adverse outcome.6 Conventional markers such as white blood cell count and C-reactive protein are indeed associated with outcome.7–9 However, a more comprehensive assessment of the inflammatory response upstream of these markers is lacking, but it is indispensable to identify new targets for intervention and improve the identification of high-risk patients.
Detailed assessment of the inflammatory response using whole-blood RNA in dedicated RNA tubes is a promising approach, providing detailed information on leukocyte function without the need for expedite sample handling or complex cell isolation techniques. Both coding and selected long noncoding RNAs were reported to be regulated in peripheral blood of patients with acute MI and to correlate with outcome.10–12
Therefore, the aim of this study was to characterize the unbiased circulating RNA signature during and after MI and to identify and validate new prognostic markers for LV dysfunction at follow-up.
Methods
We prospectively enrolled a discovery cohort of patients with acute MI (n=143) and performed whole-blood RNA profiling at different time points. Selected transcripts related to LV dysfunction were validated in a validation cohort (n=449) using quantitative polymerase chain reaction and in a porcine model of MI with myocardial histology and cardiac magnetic resonance (CMR) imaging. Detailed methods and data supporting the findings are available in the Data Supplement. RNA profiling data are publicly available in the Gene Expression Omnibus (GSE123342). The study protocol complies with the Declaration of Helsinki, was approved by the institution’s ethical committee, and all patients signed informed consent. Animal experiments were approved by the institution’s ethical committee and performed according to the institutional guidelines.
Results
Characteristics of the Study Cohorts
Demographic and clinical variables of patients in the discovery and validation cohorts are shown in Table 1. In the discovery cohort, the samples of 143 of 180 prospectively enrolled patients were eligible for measurement of RNA expression (study flow chart; Figure I in the Data Supplement). Forty-three percent of patients had ST-segment–elevation MI (n=61), and 13% (n=18) developed LV dysfunction at follow-up. Patients in the validation cohort had a higher risk profile: 78% of patients (n=352) had ST-segment–elevation MI, and 18% of patients (n=79) had LV dysfunction at follow-up. There were no baseline differences in the use of ACE (angiotensin-converting enzyme) inhibitors or β-blockers in patients with versus without LV dysfunction at follow-up.
Table 1.
Baseline Characteristics of the Discovery Cohort, Validation Cohort, and Stable Controls

Unbiased RNA Profiling Identifies Pathways Related to Inflammation in the Acute Phase of MI
In the discovery cohort, we first compared the unbiased RNA expression profile of 65 patients on admission with MI to 22 stable controls matched in 3:1 ratio (Figure 1A). This resulted in 1084 differentially expressed transcripts (false discovery rate–adjusted P<0.05; Figure 1B), including 469 annotated protein-coding genes and 25 long noncoding transcripts (8 intergenic RNAs, 4 antisense RNAs, and 13 pseudogenes; Figure 1C). Unbiased network analysis shows clustering related to Toll-like receptor and NF-κB (nuclear factor-κB) signaling, chemotaxis, and ubiquitination (Figure 1D). Gene ontology analysis confirms significant enrichment of biological processes related to immune response and chemotaxis in acute MI (Figure 1E, blue). The underlying pathways related to the observed immune response were identified using pathway analysis: 9 canonical pathways were significantly enriched in the acute phase of MI (Fisher exact false discovery rate–adjusted P<0.0005), shown in Figure 1E (green). The identified transcripts were predominantly expressed on the plasma membrane (Figure 1F, blue), and tissue expression profiles confirmed that they were predominantly originating from circulating leukocytes (Figure 1F, green). Taken together, the global transcriptome response highlights upregulation of different pattern recognition receptors (Toll-like receptors such as TLR4 and TLR2, C-type lectin receptors such as CLEC4E and CLEC4D, and Nod-like receptors such as NLRC4 and NLRP12), resulting in downstream NF-κB (NFKBIA and MYD88) signaling. Additional markers of leukocyte activation (IRAK3, IL1R2, and IL18R1) and mean arterial pressure kinase signaling were also upregulated. A full list of all differentially expressed transcripts, biological processes, and pathways is available in the Data Supplement.
Figure 1.
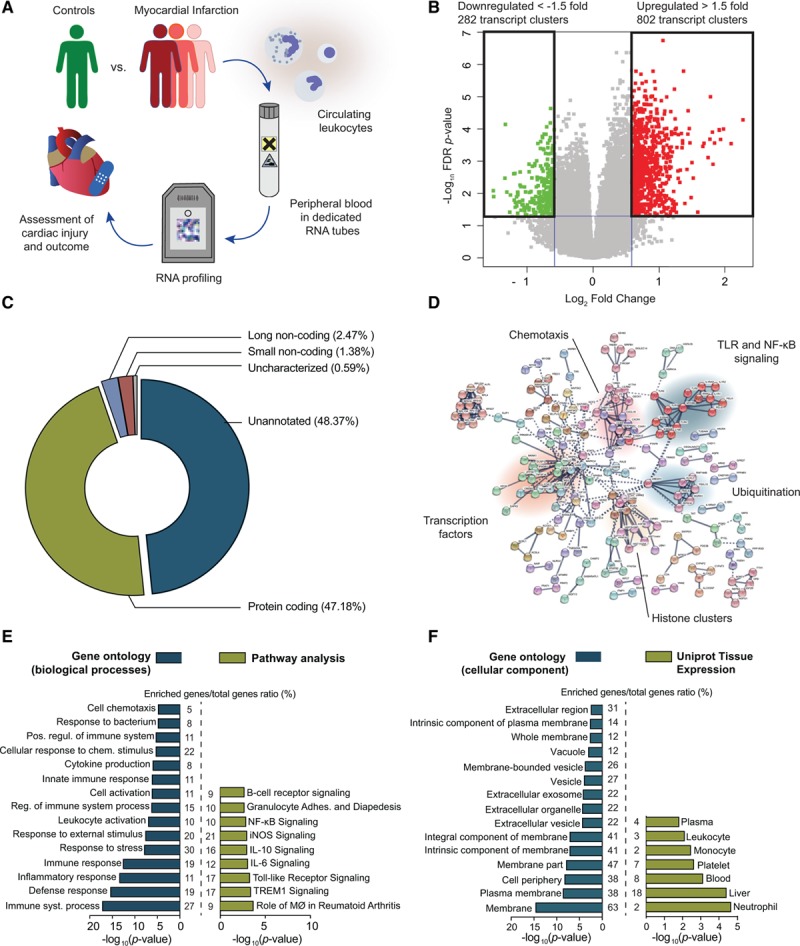
Coding and noncoding RNAs in whole blood reflect inflammatory pathways involved in the acute phase of myocardial infarction (MI). A, We performed unbiased RNA profiling in peripheral blood of patients presenting with acute MI, with the specific aim of identifying patterns that relate to cardiac injury and outcome. B, This approach revealed 802 upregulated and 282 downregulated transcript clusters in the acute phase of MI (downregulated to the left in green, upregulated to the right in red), with a fold change compared with stable controls >1.5 or <1.5 and adjusted P<0.05. C, These transcripts were predominantly protein coding (47%) but also included long noncoding transcripts (1.7%), although many remained unannotated (48.5%). D, Network analysis shows that many of these transcripts cluster together in networks associated with immune response, chemotaxis, and Toll-like receptor/NF-κB (nuclear factor-κB) signaling. E, These biological processes are confirmed by gene ontology analysis (left, blue), and additional pathways in IL (interleukin) signaling, MAPK signaling, and neutrophil activation were activated (right, green). F, The identified transcripts are predominantly expressed on the cell membrane (left, blue), and were enriched in circulating leukocytes (right, green). All P values are false discovery rate (FDR) adjusted for multiple testing. iNOS indicates inducible NO synthase; MØ, macrophages; MAPK, mitogen-activated protein kinase; TLR, toll-like receptor; and TREM1, triggering receptor expressed on myeloid cells 1.
RNA Expression in the Acute Phase Clusters With Acute Cardiac Injury and Conventional Markers of Inflammation
A large number of transcripts significantly correlated with ejection fraction in the acute phase of MI (374 transcripts with adjusted Spearman P<0.05), peak cTnT (684 transcripts), and white blood cell count (688 transcripts). RNA expression also clustered with type of presentation, with a gradient from patients with ST-segment–elevation MI over non–ST-segment–elevation MI to stable coronary artery disease (Figure II in the Data Supplement). At 30-day and 1-year follow-up, RNA expression largely normalized: only 5.8% and 3.4% of the initial 1084 transcripts remained differentially expressed compared with stable controls, with low fold changes. Further analyses, therefore, focused on RNA expression patterns in the acute phase of MI.
Identification of QSOX1, PLBD1, and S100A8 on Admission for the Prediction of LV Dysfunction at Follow-Up in the Discovery Cohort
In light of the high correlation between RNA expression and cardiac injury and inflammation, we next investigated whether RNA expression at the time of admission is related to LV function at follow-up. We selected QSOX1, PLBD1, and S100A8 as 3 top RNAs that were significantly upregulated in the acute phase of MI and related to LV function at follow-up, for further evaluation using quantitative polymerase chain reaction. We first confirmed that these 3 transcripts were significantly upregulated in MI on admission (n=138) compared with stable controls (n=20) using quantitative polymerase chain reaction (Figure 2A): 1.77-fold change for QSOX1 (P=0.013), 1.58 for PLBD1 (P<0.001), and 2.40 for S100A8 (P<0.001). Second, QSOX1 and PLBD1, but not S100A8, were significantly higher on admission in patients who developed LV dysfunction at 1-year follow-up (n=18), compared with those who did not (n=100; Figure 2B): 1.50-fold change for QSOX1 (P=0.021), 1.39-fold change for PLBD1 (P<0.001), and 1.06-fold for S100A8 (P=0.45; Figure 2B). In binary logistic regression, QSOX1 and PLBD1 were univariate predictors of LV dysfunction at follow-up (odds ratio [OR], 2.6 [95% CI, 1.1–6.1] and OR, 3.2 [95% CI, 1.4–7.4], respectively), whereas S100A8 was not (OR, 1.2 [95% CI, 0.8–1.7]).
Figure 2.

Expression levels of QSOX1, PLBD1, and S100A8 in the discovery cohort. A, In the discovery cohort, QSOX1, PLBD1, and S100A8 are upregulated in acute myocardial infarction (MI; n=138) compared with stable controls (n=20). B, QSOX1 and PLBD1 on admission were significantly higher in patients who developed left ventricular (LV) dysfunction (n=18) compared with those who did not (n=100). CAD indicates coronary artery disease; PLBD1, Phospholipase B Domain Containing 1; QSOX1, Quiescin sulfhydryl oxidase 1; S100A8, S100 calcium-binding protein A8; and SF3A1, Splicing factor 3 subunit 1.
External Validation Confirms QSOX1 and PLBD1 as Independent Predictors of LV Dysfunction at Follow-Up
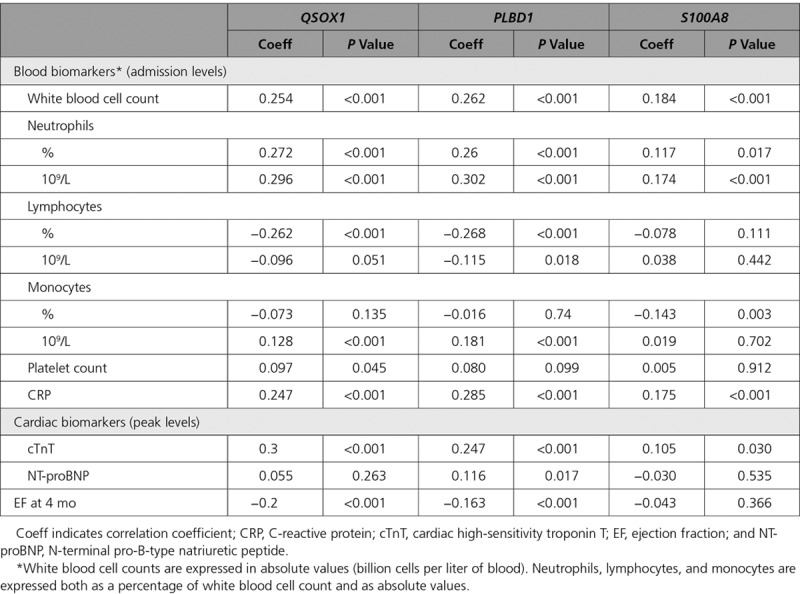
In the independent validation cohort, QSOX1 and PLBD1 were confirmed to be significantly higher expressed in the group of patients with LV dysfunction at 4 months of follow-up (n=79) compared with patients without LV dysfunction (n=370): 1.31-fold (P<0.001) and 1.32-fold (P<0.001), respectively (Figure 3A). Consistent with the discovery cohort, no difference was observed for S100A8 (1.14-fold; P=0.487). QSOX1 and PLBD1 showed statistically significant positive correlation with leukocyte and neutrophil count, peak levels of cTnT, NT-proBNP (N-terminal pro-B-type natriuretic peptide), and negative correlation with ejection fraction at 4 months of follow-up, with moderate correlation coefficients (Table 2; Table II in the Data Supplement).
Figure 3.
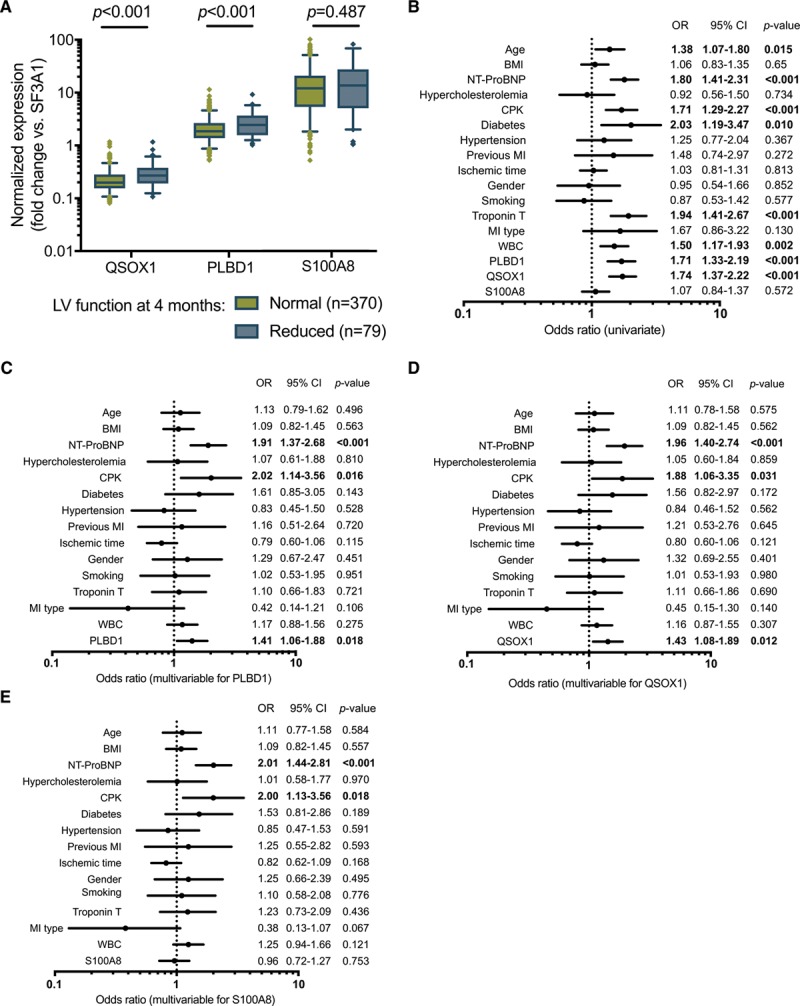
QSOX1 and PLBD1 are independent predictors of left ventricular (LV) dysfunction in the validation cohort. A, In the validation cohort (n=449), QSOX1 and PLBD1 expression at reperfusion was upregulated in patients developing LV dysfunction (n=79) compared with those who did not (n=370). B–E, The ability of QSOX1, PLBD1, and S100A8 to predict LV dysfunction was determined using univariate (B) and multivariable analyses (C–E). The parameters included in the clinical model were age, body mass index (BMI), admission level of NT-proBNP (N-terminal pro-B-type natriuretic peptide), hypercholesterolemia, peak levels of CPK (creatine phosphokinase), diabetes mellitus, hypertension, previous myocardial infarction (MI), ischemic time (ie, delay between chest pain onset and reperfusion), sex, smoking, peak levels of cardiac troponin T, MI type (ST-segment–elevation MI vs non–ST-segment–elevation MI), and white blood cell count (WBC). Odds ratios (ORs) with 95% CIs are shown, and significant associations are in bold. PLBD1 indicates Phospholipase B Domain Containing 1; QSOX1, Quiescin sulfhydryl oxidase 1; S100A8, S100 calcium-binding protein A8; and SF3A1, Splicing factor 3 subunit 1.
Table 2.
Spearman Correlation in Patients Between Expression Levels of QSOX1, PLBD1, and S100A8, Blood Biomarkers on Admission, Peak Levels of Cardiac Biomarkers, and EF at 4-mo Follow-Up in the Validation Cohort
In logistic regression, QSOX1 and PLBD1 were univariate predictors of LV dysfunction at 4 months with ORs of 1.74 (95% CI, 1.37–2.22) and 1.71 (95% CI, 1.33–2.19), respectively (Figure 3B). In multivariable analysis, QSOX1 and PLBD1 were independent predictors of LV dysfunction (ORs, 1.43 [95% CI, 1.08–1.89] and 1.41 [95% CI, 1.06–1.88], respectively), as well as admission levels of NT-proBNP and peak levels of CPK (creatine phosphokinase; Figure 3C through 3E). S100A8 was not a significant predictor of LV dysfunction in this cohort.
The incremental predictive value of these 3 new markers on top of conventional clinical and biochemical variables was calculated using the Akaike information criteria (AIC) of prediction models. The use of the AIC instead of the area under the curve allows for adjusting for the multiplication of variables entered into the model, therefore, avoiding model overfitting. A lower AIC indicates a better model fit. The AIC was calculated for the clinical model alone and after adding each combination of 1 to 3 genes (Table 3). All models were able to significantly predict LV dysfunction (Wald χ2 test P<0.001). While the addition of S100A8 to the clinical model did not provide a significant incremental predictive value, adding QSOX1 or PLBD1 to the clinical model improved prediction. The best improvement of prediction (lowest AIC) was observed for the clinical model with QSOX1. Adding PLBD1 did not provide a further incremental predictive value. In reclassification analysis, the addition of QSOX1 to the clinical model was able to reclassify a moderate but significant proportion of patients (1.6%) misclassified by the clinical model alone with a net reclassification index of 0.397 (95% CI, 0.161–0.634) and an integrated discrimination index of 0.017 (95% CI, 0.003–0.031; Table 3). Adding QSOX1 improved the sensitivity of the model from 69.6% to 73.4% and specificity from 69.5% to 70.5% (Figure III in the Data Supplement). Since the 3 identified transcripts correlated with leukocyte counts, we next adjusted the expression of QSOX1, PLBD1, and S100A8 for white blood cell count. The predictive value of the 3 genes was conserved, and we observed similar predictive values after adjustment (Figure IV in the Data Supplement).
Table 3.
Prediction and Reclassification Analyses in the Validation Cohort

Circulating QSOX1 and PLBD1 Relate to Neutrophil Count and Neutrophil Infiltration in Porcine Ischemic Myocardium
To provide direct evidence that whole-blood RNA expression as surrogate tissue is associated with myocardial injury, we correlated RNA levels from porcine blood with serial cTnT measurements, CMR-based infarct size, and myocardial tissue analysis in a representative experimental MI model. Baseline characteristics of the animals are shown in Table 4. Expression levels of QSOX1, PLBD1, and S100A8 in whole blood were significantly upregulated at 120 minutes after transient left anterior descending artery occlusion (1.6-, 2.2-, and 4.0-fold; Figure 4A).
Table 4.
Baseline Characteristics, Pathology, and CMR Imaging in Pigs Undergoing Myocardial Ischemia-Reperfusion Injury (n=33)

Figure 4.
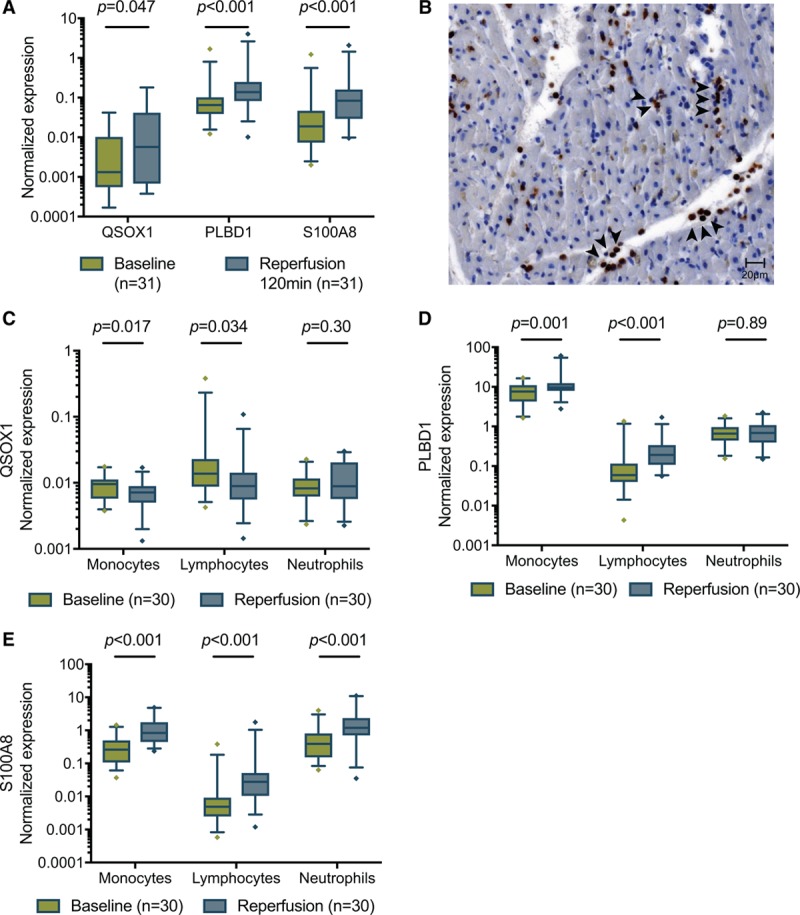
RNA expression in whole-blood and leukocyte subpopulations following acute myocardial infarction (MI) in pigs 120 min after reperfusion. The identified transcripts were validated in 33 pigs undergoing acute myocardial infarction. A, All 3 transcripts that were upregulated in acute MI in patients were also upregulated in whole blood of pigs undergoing MI. B, Representative light microscopy image of MPO (myeloperoxidase)-stained porcine ischemic myocardium, showing neutrophils (black arrowheads), cardiomyocytes, and capillaries. C–E, Expression levels of QSOX1, PLBD1, and S100A8 in isolated circulating monocytes, lymphocytes, and neutrophils. PLBD1 indicates Phospholipase B Domain Containing 1; QSOX1, Quiescin sulfhydryl oxidase 1; and S100A8, S100 calcium-binding protein A8.
Consistent with the human data, QSOX1, PLBD1, and S100A8 at 120 minutes after reperfusion correlated significantly with neutrophil count (Table 5). S100A8 and PLBD1, but not QSOX1, moderately correlated with the area under the curve of cTnT release (P=0.020 and P=0.142; n=31). However, none of the 3 transcripts correlated to CMR-based infarct size (n=18).
Table 5.
Spearman Correlation Between Porcine Peripheral Blood RNA Expression and RNA Expression in Isolated Leukocyte Subsets and Ischemic Myocardium
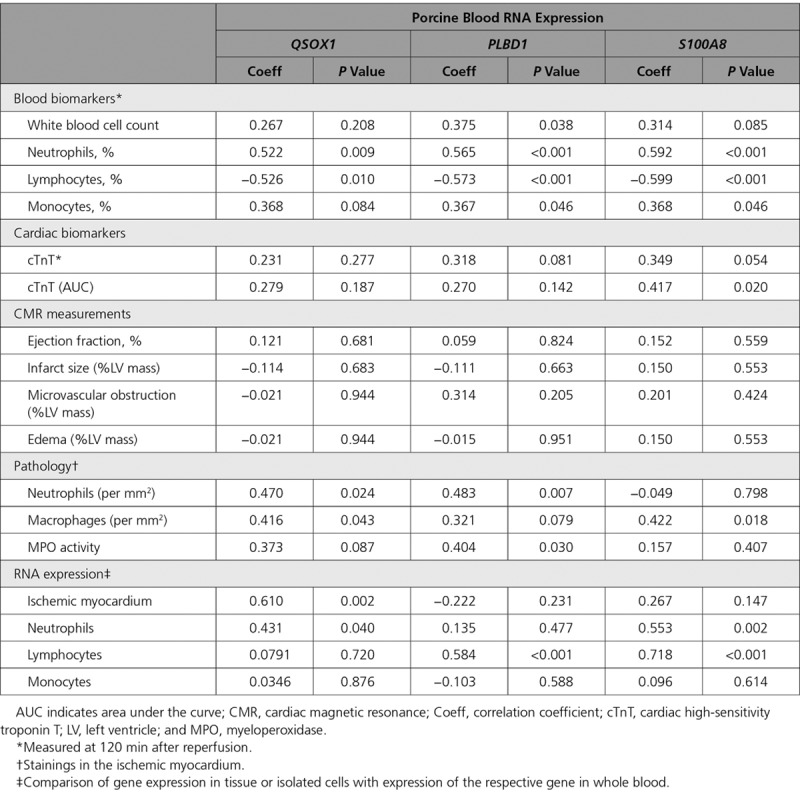
We next assessed the correlation between these transcripts and neutrophil and macrophage infiltration in the ischemic myocardium. A representative histological section, stained with a neutrophil-specific MPO (myeloperoxidase) antiserum, is shown in Figure 4B. Circulating QSOX1 and PLBD1 significantly correlated with neutrophil infiltration (r=0.47, P=0.024 and r=0.48, P=0.0007; Table 5). PLBD1 and to a lesser extent QSOX1 also correlated with neutrophil (MPO) activity in tissue (r=0.40, P=0.030 and r=0.37, P=0.087). In contrast, S100A8 correlated significantly with macrophage infiltration (r=0.42, P=0.018).
Origin of Circulating QSOX1, PLBD1, and S100A8 in Isolated Leukocytes and in Ischemic Myocardium
To identify the origin of the identified expression changes, we measured QSOX1, PLBD1, and S100A8 in leukocyte subtypes and in the ischemic myocardium. QSOX1 in whole blood only weakly correlated with expression levels in circulating neutrophils (r=0.43, P=0.04), and it did not correlate with expression levels in isolated lymphocytes or monocytes (Table 5). Moreover, QSOX1 showed only minor differential expression in these cells (Figure 4C). However, circulating QSOX1 highly correlated with expression in the ischemic zone of the myocardium (r=0.61, P=0.002). In contrast, PLBD1 levels in whole blood highly correlated with expression in circulating lymphocytes (r=0.584, P<0.001), and the latter paralleled the expression observed in whole blood after ischemic injury (Figure 4D). However, PLBD1 levels in peripheral blood did not correlate with expression levels in the myocardium. Also peripheral blood S100A8 appeared to parallel expression in neutrophils and lymphocytes, in which it was also differentially expressed (Figure 4E). In contrast to QSOX1, S100A8 was not related to the levels in the myocardium.
Discussion
New strategies to identify patients at risk for developing LV dysfunction after MI are needed to improve outcome. We here report that whole-blood transcriptome profiling at the time of hospital admission for acute MI reveals significant upregulation of pattern recognition receptors and transcripts involved in NF-κB, IL (interleukin)-6, and TLR signaling. Moreover, we identified QSOX1 and PLBD1 as new markers for the prediction of LV dysfunction at follow-up, independent of conventional risk factors, and validated these findings in a large independent MI cohort. In a large animal MI model representative of human disease, circulating QSOX1 and PLBD1 were related to neutrophil count and neutrophil infiltration in the ischemic myocardium. Circulating QSOX1 levels highly related to those measured in the ischemic myocardium, while PLBD1 levels correlated with levels in lymphocytes. Taken together, these findings indicate the diagnostic potential of whole-blood RNA to reveal disease-specific regulatory pathways in MI and to identify patients with increased risk for LV systolic dysfunction at follow-up.
The first key observation in our unbiased analysis is the identification of over 1084 coding and noncoding transcripts that are differentially expressed in whole blood of patients with ST-segment–elevation or non–ST-segment–elevation MI. These findings are in line with previous studies, which reported smaller numbers of differentially expressed transcripts in acute MI by including only selected patients with ST-segment–elevation MI or by using a targeted approach for coding RNA or selected transcripts.10,12–15 Moreover, we show that 63% of the identified transcripts relate to the extent of myocardial injury in the acute phase of MI. Many of the identified transcripts in our study relate to sterile inflammation and code for pattern recognition surface receptors such as TLR2, TLR4, and CLEC4E. These receptors respond to damage-associated molecular patterns, including HMGB1 and necrotic cell content, which are released during myocardial injury.16–18 Pathway analysis also showed activation of IL-1 signaling (IL1R2, IL18R1, IRAK3, NFKBIA, and MYD88). Several clinical trials have targeted some of these pathways in the setting of acute MI, including IL-1, CD11, and P-selectin, but failed to show marked clinical benefit.19–23 The identified transcripts in our study may point to alternative pathogenic pathways in acute MI and warrant further exploration.3,4
The second main finding is that, besides conventional markers of cardiomyocyte death such as cTnT, we identified a number of inflammation-related transcripts that also relate to LV dysfunction at follow-up. We validated QSOX1 and PLBD1. Although expression of QSOX1 and PLBD1 shows some intragroup variability and the improvement of the risk prediction by adding QSOX1 to a full model including NT-proBNP is significant but moderate, the unbiased observation that the inflammatory process measured at RNA level relates to LV function at follow-up, independent from conventional markers of cardiomyocyte death, is a key finding.
QSOX1 (quiescin sulfhydryl oxidase 1) is a sulfhydryl oxidase involved in cellular growth and extracellular matrix remodeling. Our mechanistic studies in a representative large animal model suggest that the circulating QSOX1 levels may relate to those observed in the ischemic myocardium. Previously, QSOX1 was induced in the LV of pressure-overloaded rat hearts and has been described as a marker of acute heart failure.24 QSOX1 has been shown to exhibit a cardioprotective response upon acute stress by orchestrating adequate protein folding in the endoplasmatic reticulum. Indeed, QSOX1−/− knockout mice showed a phenotype of dilated cardiomyopathy and enhanced inflammation.25 Alternatively, circulating QSOX1 also alters the redox status of soluble proteins in plasma.26 PLBD1 (Phospholipase B domain containing 1) is a phospholipase, which can generate lipid mediators of inflammation and was first identified in neutrophils.27 While our data confirm the expression of PLBD1 in monocytes and neutrophils, they also show that increased PLBD1 levels in the setting of acute MI are attributable to significant upregulation in circulating lymphocytes. Finally, we could not validate S100A8 expression as a marker of LV dysfunction. S100A8 (S100 calcium-binding protein A8) is a S100 calcium-binding protein and can act as a damage-associated molecular pattern to initiate neutrophil chemotaxis.16 Although S100A8 signaling is related to vascular inflammation and has been shown to aggravate post-MI heart failure, its expression may represent a more generic inflammatory signal detectable in many leukocyte subpopulations.28,29
Other complementary RNA-based strategies to improve risk stratification in patients with MI are currently being explored, including circular RNA in whole blood but also long noncoding RNA or microRNA in plasma.11,30 Nevertheless, whole blood seems to be a valuable source for risk stratification in MI, since neutrophils and lymphocytes contribute substantially to the expression pattern in whole blood.31,32
We recognize the limitations of our study. First, as baseline medication use was different between the acute MI patients and the control group with stable coronary artery disease, we cannot completely exclude that the identified RNA profile and pathways in the profiling phase of the study are to some extent driven by imbalances in baseline medication use. However, medication use in patients with MI who did or did not develop LV dysfunction was well balanced. Second, we lack CMR-based assessments of infarct size in our patients, and additional CMR-derived prognostic information including microvascular obstruction. However, in the subset of pigs with available CMR imaging (n=18 of 33), we did not observe associations between RNA expression and infarct size. Third, blood samples from patients (and pigs) were obtained in the acute phase of MI, at the time of reperfusion. Because the inflammatory response is time dependent and RNA profiling was performed on admission, we cannot exclude different RNA profiles at later time points. Fourth, in our bioinformatics analysis, we did not assess the possible contributions of the platelet transcriptome to the whole-blood RNA expression profile. Finally, extended follow-up preclinical studies are required to identify the exact role of QSOX1 and PLBD1 as specific markers of cardiac inflammation in acute MI and predictors of LV dysfunction.
In conclusion, whole blood as surrogate tissue provides a valuable noninvasive platform to differentiate dysregulated immune-related pathways in patients with MI at risk for LV dysfunction. Increased expression levels of QSOX1 and PLBD1 represent promising new predictors of functional impairment and targets for personalized treatment. Future studies will need to investigate whether optimizing interventions at discharge in acute MI patients, based on RNA levels measured on admission, may improve patient outcome.
Acknowledgments
The authors acknowledge all personnel of the cardiac catheterization laboratory and cardiac intensive care unit of the University Hospitals Leuven. We gratefully acknowledge the staff of the microarray facility VIB Nucleomics Core, in particular, Ruth Maes and Wout Van Delm. We furthermore thank Margaretha Van Kerrebroeck and Sofie Van Soest for the assistance during the animal experiments. We thank Christelle Nicolas, Bernadette Leners, Torkia Lalem, and Mara Luchetti for their contribution in the validation study. This article is based on collaboration supported by the European Union Cooperation in Science and Technology Action CardioRNA CA17129. Dr Vanhaverbeke, Dr Bartunek, Dr Van De Werf, Dr Janssens, and Dr Sinnaeve contributed to the study design; Dr Vanhaverbeke, Dr Wu, G. Laenen, H. Gillijns, Dr Moreau, Dr Janssens, and Dr Sinnaeve contributed to methodology; Dr Vanhaverbeke, D. Veltman, H. Gillijns, Dr Janssens, and Dr Sinnaeve enrolled patients and performed follow-up; porcine experiments were performed by Dr Vanhaverbeke, D. Veltman, Dr Wu, and H. Gillijns; data from the validation cohort was provided by M. Vausort, L. Zhang and Dr Devaux; data analysis was done by Dr Vanhaverbeke, M. Vausort, D. Veltman, L. Zhang, Dr Wu, G. Laenen, Dr Moreau, Dr Sinnaeve, and Dr Janssens; Dr Vanhaverbeke, M. Vausort, Dr Devaux, Dr Janssens, and Dr Sinnaeve contributed to the writing of the manuscript; all authors performed the revision and editing of the manuscript; and Dr Vanhaverbeke, Dr Van De Werf, Dr Devaux, Dr Janssens, and Dr Sinnaeve contributed to resources and funding acquisition.
Sources of Funding
Research was funded by Research Foundation Flanders—a score grant from the University of Leuven (PF10/014) and the Frans Van de Werf Fund for Clinical Cardiovascular Research. Dr Sinnaeve is a clinical investigator for the Research Foundation Flanders. Dr Janssens is holder of a named chair at KU Leuven, financed by AstraZeneca. The independent validation cohort was supported by the Ministry of Higher Education and Research and the Society for Research on Cardiovascular Diseases of Luxembourg.
Disclosures
None.
Supplementary Material
Footnotes
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.119.002656.
References
- 1.Cung TT, et al. Cyclosporine before PCI in patients with acute myocardial infarction. N Engl J Med. 2015;373:1021–1031. doi: 10.1056/NEJMoa1505489. doi: 10.1056/NEJMoa1505489. [DOI] [PubMed] [Google Scholar]
- 2.Janssens SP, et al. NOMI Investigators. Nitric oxide for inhalation in ST-elevation myocardial infarction (NOMI): a multicentre, double-blind, randomized controlled trial. Eur Heart J. 2018;39:2717–2725. doi: 10.1093/eurheartj/ehy232. doi: 10.1093/eurheartj/ehy232. [DOI] [PubMed] [Google Scholar]
- 3.Hausenloy DJ, et al. Novel targets and future strategies for acute cardioprotection: position paper of the European Society of cardiology working Group on cellular biology of the heart. Cardiovasc Res. 2017;113:564–585. doi: 10.1093/cvr/cvx049. doi: 10.1093/cvr/cvx049. [DOI] [PubMed] [Google Scholar]
- 4.Perrino C, et al. Epigenomic and transcriptomic approaches in the post-genomic era: path to novel targets for diagnosis and therapy of the ischaemic heart? Position paper of the European Society of cardiology working Group on cellular biology of the heart. Cardiovasc Res. 2017;113:725–736. doi: 10.1093/cvr/cvx070. doi: 10.1093/cvr/cvx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houser SR. The American Heart Association’s new institute for precision cardiovascular medicine. Circulation. 2016;134:1913–1914. doi: 10.1161/CIRCULATIONAHA.116.022138. doi: 10.1161/CIRCULATIONAHA.116.022138. [DOI] [PubMed] [Google Scholar]
- 6.Frangogiannis NG. Regulation of the inflammatory response in cardiac repair. Circ Res. 2012;110:159–173. doi: 10.1161/CIRCRESAHA.111.243162. doi: 10.1161/CIRCRESAHA.111.243162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ørn S, et al. C-reactive protein, infarct size, microvascular obstruction, and left-ventricular remodelling following acute myocardial infarction. Eur Heart J. 2009;30:1180–1186. doi: 10.1093/eurheartj/ehp070. doi: 10.1093/eurheartj/ehp070. [DOI] [PubMed] [Google Scholar]
- 8.Barron HV, et al. Association between white blood cell count, epicardial blood flow, myocardial perfusion, and clinical outcomes in the setting of acute myocardial infarction: a thrombolysis in myocardial infarction 10 substudy. Circulation. 2000;102:2329–2334. doi: 10.1161/01.cir.102.19.2329. doi: 10.1161/01.cir.102.19.2329. [DOI] [PubMed] [Google Scholar]
- 9.Arruda-Olson AM, et al. Neutrophilia predicts death and heart failure after myocardial infarction: a community-based study. Circ Cardiovasc Qual Outcomes. 2009;2:656–662. doi: 10.1161/CIRCOUTCOMES.108.831024. doi: 10.1161/CIRCOUTCOMES.108.831024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep. 2016;6:22384. doi: 10.1038/srep22384. doi: 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vausort M, et al. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 12.Kiliszek M, et al. Altered gene expression pattern in peripheral blood mononuclear cells in patients with acute myocardial infarction. PLoS One. 2012;7:e50054. doi: 10.1371/journal.pone.0050054. doi: 10.1371/journal.pone.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vausort M, et al. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115:668–677. doi: 10.1161/CIRCRESAHA.115.303836. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 14.van der Pouw Kraan TC, et al. Systemic toll-like receptor and interleukin-18 pathway activation in patients with acute ST elevation myocardial infarction. J Mol Cell Cardiol. 2014;67:94–102. doi: 10.1016/j.yjmcc.2013.12.021. doi: 10.1016/j.yjmcc.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Suresh R, et al. Transcriptome from circulating cells suggests dysregulated pathways associated with long-term recurrent events following first-time myocardial infarction. J Mol Cell Cardiol. 2014;74:13–21. doi: 10.1016/j.yjmcc.2014.04.017. doi: 10.1016/j.yjmcc.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen GY, et al. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–837. doi: 10.1038/nri2873. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Y, et al. Temporal neutrophil polarization following myocardial infarction. Cardiovasc Res. 2016;110:51–61. doi: 10.1093/cvr/cvw024. doi: 10.1093/cvr/cvw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arslan F, et al. Innate immune signaling in cardiac ischemia. Nat Rev Cardiol. 2011;8:292–300. doi: 10.1038/nrcardio.2011.38. doi: 10.1038/nrcardio.2011.38. [DOI] [PubMed] [Google Scholar]
- 19.Abbate A, et al. VCU-ART Investigators. Interleukin-1 blockade with anakinra to prevent adverse cardiac remodeling after acute myocardial infarction (Virginia Commonwealth University Anakinra Remodeling Trial [VCU-ART] Pilot study). Am J Cardiol. 2010;105:1371–1377.e1. doi: 10.1016/j.amjcard.2009.12.059. doi: 10.1016/j.amjcard.2009.12.059. [DOI] [PubMed] [Google Scholar]
- 20.Abbate A, et al. Effects of interleukin-1 blockade with anakinra on adverse cardiac remodeling and heart failure after acute myocardial infarction [from the Virginia Commonwealth University-Anakinra Remodeling Trial (2) (VCU-ART2) pilot study]. Am J Cardiol. 2013;111:1394–1400. doi: 10.1016/j.amjcard.2013.01.287. doi: 10.1016/j.amjcard.2013.01.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tardif JC, et al. Effects of the P-selectin antagonist inclacumab on myocardial damage after percutaneous coronary intervention for non-ST-segment elevation myocardial infarction: results of the SELECT-ACS trial. J Am Coll Cardiol. 2013;61:2048–2055. doi: 10.1016/j.jacc.2013.03.003. doi: 10.1016/j.jacc.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Faxon DP, et al. HALT-MI Investigators. The effect of blockade of the CD11/CD18 integrin receptor on infarct size in patients with acute myocardial infarction treated with direct angioplasty: the results of the HALT-MI study. J Am Coll Cardiol. 2002;40:1199–1204. doi: 10.1016/s0735-1097(02)02136-8. doi: 10.1016/s0735-1097(02)02136-8. [DOI] [PubMed] [Google Scholar]
- 23.Oyama J, et al. Reduced myocardial ischemia-reperfusion injury in toll-like receptor 4-deficient mice. Circulation. 2004;109:784–789. doi: 10.1161/01.CIR.0000112575.66565.84. doi: 10.1161/01.CIR.0000112575.66565.84. [DOI] [PubMed] [Google Scholar]
- 24.Mebazaa A, et al. Unbiased plasma proteomics for novel diagnostic biomarkers in cardiovascular disease: identification of quiescin Q6 as a candidate biomarker of acutely decompensated heart failure. Eur Heart J. 2012;33:2317–2324. doi: 10.1093/eurheartj/ehs162. doi: 10.1093/eurheartj/ehs162. [DOI] [PubMed] [Google Scholar]
- 25.Caillard A, et al. QSOX1, a novel actor of cardiac protection upon acute stress in mice. J Mol Cell Cardiol. 2018;119:75–86. doi: 10.1016/j.yjmcc.2018.04.014. doi: 10.1016/j.yjmcc.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 26.Israel BA, et al. Disulfide bond generation in mammalian blood serum: detection and purification of quiescin-sulfhydryl oxidase. Free Radic Biol Med. 2014;69:129–135. doi: 10.1016/j.freeradbiomed.2014.01.020. doi: 10.1016/j.freeradbiomed.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu S, et al. The identification of a phospholipase B precursor in human neutrophils. FEBS J. 2009;276:175–186. doi: 10.1111/j.1742-4658.2008.06771.x. doi: 10.1111/j.1742-4658.2008.06771.x. [DOI] [PubMed] [Google Scholar]
- 28.Du CQ, et al. The elevated serum S100A8/A9 during acute myocardial infarction is not of cardiac myocyte origin. Inflammation. 2012;35:787–796. doi: 10.1007/s10753-011-9375-8. doi: 10.1007/s10753-011-9375-8. [DOI] [PubMed] [Google Scholar]
- 29.Morrow DA, et al. Myeloid-related protein 8/14 and the risk of cardiovascular death or myocardial infarction after an acute coronary syndrome in the Pravastatin or Atorvastatin Evaluation and Infection Therapy: Thrombolysis in Myocardial Infarction (PROVE IT-TIMI 22) trial. Am Heart J. 2008;155:49–55. doi: 10.1016/j.ahj.2007.08.018. doi: 10.1016/j.ahj.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumarswamy R, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, et al. Disease-specific classification using deconvoluted whole blood gene expression. Sci Rep. 2016;6:32976. doi: 10.1038/srep32976. doi: 10.1038/srep32976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer C, et al. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]


