Abstract
The identification of medical concepts, their attributes and the relations between concepts in a large corpus of Electroencephalography (EEG) reports is a crucial step in the development of an EEG-specific patient cohort retrieval system. However, the recognition of multiple types of medical concepts, along with the many attributes characterizing them is challenging, and so is the recognition of the possible relations between them, especially when desiring to make use of active learning. To address these challenges, in this paper we present the Self-Attention Concept, Attribute and Relation (SACAR) identifier, which relies on a powerful encoding mechanism based on the recently introduced Transformer neural architecture [1]. The SACAR identifier enabled us to consider a recently introduced framework for active learning which uses deep imitation learning for its selection policy. Our experimental results show that SACAR was able to identify medical concepts more precisely and exhibited enhanced recall, compared with previous methods. Moreover, SACAR achieves superior performance in attribute classification for attribute categories of interest, while identifying the relations between concepts with performance competitive with our previous techniques. As a multi-task network, SACAR achieves this performance on the three prediction tasks simultaneously, with a single, complex neural network. The learning curves obtained in the active learning process when using the novel Active Learning Policy Neural Network (ALPNN) show a significant increase in performance as the active learning progresses. These promising results enable the extraction of clinical knowledge available in a large collection of EEG reports.
Keywords: deep learning, electroencephalography, active learning, long-distance relation identification, concept detection, attribute classification
1. Introduction
Clinical electroencephalography (EEG) is the main investigation tool used for the diagnosis and management of epilepsy. It is also used to evaluate other types of brain disorders [2], including encephalopathies, neurological infections, Creutzfeldt-Jacob disease, and even in the evaluation of the progression of Alzheimers disease. An EEG records electrical activity along the scalp and measures spontaneous electrical activity of the brain. The signals measured along the scalp can be correlated with brain activity, which makes it a primary tool for diagnosis of brain-related illnesses [3]. But, as noted in [4], the complexity of the EEG signal, interpreted and documented in EEG reports, produces inter-observer agreement known to be moderate. As more clinical EEG signals and reports become available, the interpretation of EEG signals can be improved by providing neurologists with results of search for patients that exhibit similar EEG characteristics.
Recently, in Goodwin et al. [5] we have described the MERCuRY (Multi-modal EncephalogRam patient Cohort discoveRY) system that uses deep learning to represent the EEG signal and operates on a multi-modal EEG index resulting from the automatic processing of both the EEG signal and the EEG reports that document and interpret them. The MERCuRY system allows neurologist to search a vast data archive of clinical electroencephalography (EEG) signals and EEG reports, enabling them to discover patient populations relevant to queries like Q: Patients with triphasic waves suspected of encephalopathy. The discovery of a relevant patient cohort satisfying the characteristics expressed in queries such as Q relies on the ability to automatically and accurately recognize various medical concepts and their attributes, both in the queries and throughout the EEG reports. In Q we could recognize that triphasic waves represents an EEG activity, while encephalopathy is a medical problem. To find relevant patients for Q based on their EEG reports, the relevance models and the index implemented in the MERCuRY system consider the concepts identified in the query as well as the concepts identified in the EEG reports. For example, a patient from the cohort relevant to Q has the EEG report illustrated in Figure 1, where identified concepts have annotations indicating medical problems [PROB], treatments [TR], tests [TEST], EEG activities [ACT], and EEG events [EV]. This EEG report is relevant to Q because, as indicated in its impression section, the patient’s triphasic waves represent one of the explanations for the abnormal EEG, while in the report’s clinical correlation section, it is mentioned that the EEG activity identified by triphasic waves is a manifestation of the medical problem encephalopathy. Thus the EEG report illustrated in Figure 1 is relevant to the query Q because of the identified medical concepts and their attributes.
Figure 1:

Synthetic Example EEG Report.
In previous work [6] we reported a multi-task neural active learning method, capable of recognizing medical concepts and their attributes in queries and EEG reports. However, the identification of the medical concepts from the query and in the EEG reports is not sufficient, as many false positives can be produced. For example, the query Q does not only ask about the concepts it mentions, but it also implicitly asks about the relation between the concepts [triphasic waves]ACT and [encephalopathy]PROB. Because the memory-augmented relation detection technique reported in [7] identifies in the EEG report illustrated in Figure 1 two relations between the medical concepts mentioned in Q, namely: (R1):[triphasic waves]ACT –Evidences→ [metabolic_encephalopathy]PROB; (R2):[triphasic waves]ACT –Evidences→ [hepatic encephalopathy]PROB; the EEG report is judged relevant to the query Q. However, if only the query concepts would be considered to infer relevance of an EEG report, the EEG report illustrated in Figure 1 would be deemed relevant also for the query Q’: Patients with theta waves suspected of encephalopathy. Both concepts from Q’ are mentioned in the EEG report, but no relation between those concepts can be automatically detected in the EEG report, which correctly indicates that it should not be judged as relevant to Q’.
Relation detection methods, such as the one reported in [7], assume that medical concepts and their attributes are already identified in EEG reports. This entails that a first round of active learning is applied to obtain the desired performance of concept and attribute recognition, as we have done with the method reported in [6]. Then, a second cycle of active learning is required to guarantee the desired performance for relation extraction as well, as made possible by the framework presented in [7]. However, two separate cycles of active learning are burdensome. Thus we asked ourselves if it would be possible to perform active learning only once, while still taking advantage of the deep learning methods that perform quite well on a large dataset of EEG reports. The answer to this research question led to the main contributions of this paper.
This paper presents two contributions to the problem of knowledge extraction from large datasets of clinical narratives. First, it details a neural learning framework that enables the joint learning of medical concepts, their attributes and relations between concepts. This framework benefits from a self-attention mechanism that produces a representation of the clinical narratives that is shared by the concept recognizer, the attribute recognizer and the relation recognizer neural architectures. Although we have employed this neural learning framework to EEG reports, it can be used for any type of clinical narratives on which annotation schemas for concept, attribute and relation identification are defined. Our second contribution consists of a neural active learning methodology that learns how to select the examples in the active learning loop, instead of using heuristics, as we have done in [6] and [7]. We are releasing the implementations of the neural architecture for joint learning of medical concepts, attributes and relations as well as for the neural active learning at: https://github.com/r-mal/sacar.
2. Background
Prior work on clinical narratives related to EEG tests consider either clinical discharge summaries or EEG reports. The Epilepsy Data Extraction and Annotation (EpiDEA) [8] system operates on discharge summaries. These discharge summaries originated from the Epilepsy Monitoring Unit (EMU) which collects clinical data about patients with potential of Sudden Unexpected Death in Epilepsy (SUDEP) at four centers: University Hospitals Case Medical Center (UH CMC Cleveland), Ronald Reagan University of California Los Angeles Medical Center (RRUMC-Los Angeles), The National Hospital for Neurology and Neurosurgery (NHNN, London, UK), and the Northwestern Memorial Hospital (NMH Chicago). The EMU patient discharge summaries had four sections: (Section 1) Epilepsy Classification, describing the etiology, seizure semiology, epileptogenic zone and co-morbidities of the patient; (Section 2) History and Exam, describing the seizure types, evolution and frequency of the seizure, risk factors and family history, medications and results of the physical and neurological examination as well as the phycological history of the patient; (Section 3) Evaluation, which presents the results of the EEG, Magnetic Resonance Imagining (MRI) and sleep study, and (Section 4) Conclusions and Recommendations, recorded by the attending physician. These four sections have interleaving unstructured free text and semi-structured “attribute-value” text. The EpiDEA system operated on 662 EMU discharge summaries by applying regular expressions, concept identification provided by cTakes [9] and negation detection delivered by the Negex algorithm [10] to map the clinical narratives into the concept classes provided by the Epilepsy and Seizure Ontology (EpSO) [11]. EpSO has more than 1000 classes modeling the etiology of epilepsy.
Cui et al [12] describes natural language processing (NLP) methods aiming at extracting epilepsy-related phenotypes from the same discharge summaries as those reported in Cui et al [8]. The epilepsy phenotypes were related to (a) anatomical locations for the identification Epileptogenic Zone; (b) the Seizure Semiology; and (c) Lateralizing Signs, Interictal EEG Patterns, and Ictal EEG Patterns, which is a sub-set of the EEG activities we targeted in the research presented in this paper. The NLP methodology for phenotype extraction was also centered on the semantics of the EpSO ontology, but it incorporated the detection of anatomical locations available from MetaMap [13] into the rule-based concept detector described in Cui et al [8].
To our knowledge, the only NLP techniques operating on EEG reports have been published in our previous work [6, 7, 14-17]. All these publications showcased NLP methodologies operating on the 25,000 reports EEG reports available from the Temple University Hospital (TUH) 1. A deep learning method, augmented by active learning for identifying EEG-specific concepts and their attributes was presented in [6]. The automatic discovery of relations between such concepts was described first in [17] and then in [7]. Taylor & Harabagiu [14] have presented the need for processing negation in EEG reports for patient cohort retrieval and compared several neural NLP methods for tackling negation. In addition, since often in EEG reports some of the mandatory sections are not present [15] has presented novel NLP methods for inferring the under-specified Interpretation section while [16] has presented a novel NLP neural methodology for recovering the clinical correlation section in EEG reports.
This paper benefits from the NLP framework initiated in [6] and [7] and explores a joint neural method for discovering concepts, their attributes and the relations between them, while also considering a novel active learning paradigm that leverages the benefits of imitation learning in a neural setting. It operates on a clinical narrative dataset which is two orders of magnitude larger than the dataset of discharge summaries on which the methods from Cui et al. [8, 12] operate. Furthermore, it incorporates a rich set of attributes, including spatial (which consider anatomical locations) and temporal, and it covers the full set of EEG activities and events (including seizures) that are collected and interpreted during the EEG test. The NLP methods described in [6, 7, 14-17] as well as in this paper, provide an information extraction framework operating on EEG-specific clinical narratives that takes advantage of Big Data technologies (including Deep Learning) in contrast with the methods described in [8, 12], which rely on the EpSO ontology [11].
Active learning (AL) allows an NLP model operating on EEG reports to be improved by (a) querying for the most informative instances of annotations; (b) presenting the results of NLP to an expert for acceptance or correction; and then (c) retraining the model; — a cycle that is repeated until the desired performance is obtained. The performance of AL exceeds that of random learners in most cases. But more importantly, the active learning approach requires significantly fewer manual annotations while maintaining comparable performance to traditional supervised learning, or even improving it. AL has been used to enhance supervised NLP methods in health narratives in the past. For example, AL was used to annotate pathological phenomena in Medline abstracts in the PathoJen system [18] by relying on a set of classifiers and computing the disagreement between them to inform the selection of the annotation that needs to be validated or edited. AL has also been used for high-throughput phenotyping algorithms. By integrating AL with SVM-based classifiers, the research published by Chen et al. [19] showed that AL can reduce the number of sampled annotations required for achieving an area under the curve (AUC) of 0.95. Dligash et al. [20] used AL with Naive Bayes classification for extracting phenotypes and observed that AL generated a significant reduction in annotation efforts: only one third of annotations were required. More recently, a new study targeting a cost-sensitive AL for clinical phenotyping was published by Ji et al. [21], using the identification of breast cancer patients as a use case. The cost model was generated based on linear regression using some heuristic features, and used to maximize the informativeness/cost ratio when selecting samples for validation/editing.
The neural AL framework that we present in this paper is enhancing our previous multi-task AL systems that operate on neural learning architectures. In [6] and [7] we have reported on the first AL systems operating on EEG reports and using Deep Learning methods for identifying concepts and their attributes or long-distance relations between concepts. As more Neural NLP methods are developed to operate on clinical narratives, we expect that an AL framework in which the selection of samples is informed by a policy that is learned through a neural method to be adopted and used in other clinical NLP tasks, that are in need of AL or have considered AL in the past, e.g. supervised word sense disambiguation in Medline [22] or classifying the results of imaging examinations into reportable or non-reportable cancer cases [23].
3. Deep Learning of Concepts, Attributes and Relations from EEG Reports
In this work, medical concepts, their attributes and the relations between concepts are identified in the TUH corpus of EEG reports comprised of over 25,000 reports from over 15,000 patients collected over 12 years. EEG reports are designed to convey a written impression of the visual analysis of the EEG along with an interpretation of its clinical significance. In accordance with the American Clinical Neurophysiology Society Guidelines for writing EEG reports, the reports from the TUH EEG Corpus start with a clinical history of the patient including information about the patients age, gender, current medical conditions (e.g. “right leg swelling”), and relevant past medical conditions (e.g. “history of seizures”) followed by a list of medications the patient is currently taking (e.g. “Dilantin”), described in a separate section. Together, these two initial sections depict the clinical picture and therapy of the patient, containing a wealth of medical concepts including medical problems (e.g. “stroke”), symptoms (e.g. “facial droop”), signs (e.g. “twitching”) and treatments (e.g. “Zofran”, “gastrocnemius surgery”). After the clinical picture and therapy of the patient is established, the introduction section of the EEG report describes the techniques used for the current EEG (e.g. “digital video EEG using standard 10-20 system of electrode placement with one channel of EKG), the patients condition at the time of the record (e.g. drowsy), and possible activating procedures carried out (e.g. “stimulation of the patient”).
The description section is the mandatory part of the report, meant to provide a complete and objective description of the EEG, noting all observed EEG activities (e.g. “sharp waves”, “PLEDs”), and EEG events (e.g. “stimulation”, “hyperventilation”). The impression section indicates whether or not the EEG test is abnormal and, if so, lists the abnormalities in decreasing order of importance. These abnormalities are usually describing EEG activities (e.g. “triphasic waves”), but can also be EEG Events (e.g. “myoclonic seizures”). Finally, the clinical correlation section explains what the EEG findings mean in terms of clinical interpretation, (e.g. “findings are supportive of bihemispheric disturbance of cerebral function”).
From the narratives of each section from every EEG report, we decided to extract only five types of concepts: (1) EEG activities, (2) EEG events, (3) medical problems, (4) medical treatments, and (5) medical tests, because they represent the predominant types of concepts in the EEG reports (as illustrated in Figure 1). We were able to take advantage of the definitions of three types of medical concepts, which were used in the 2010 i2b2 challenge [24], namely medical problems (e.g., disease, injury), tests (e.g., diagnostic procedure, lab test), and treatments (e.g., drug, preventive procedure, medical device). For the EEG-specific medical concepts (i.e. EEG activities and EEG events) we created our own definitions, which are presented in Section 3.1. When deciding on the attributes associated with the five types of medical concepts from EEG reports, as illustrated in Table 1, we distinguished between attributes that apply to all types of concepts, e.g. polarity and modality, and attributes that are specific only to EEG activities. For identifying the polarity of medical concepts in EEG reports, we relied on the definition used in the 2012 i2b2 challenge[25], considering that each concept can have either a “positive or a “negative polarity, depending on the absence or presence of negation of its finding. When we considered the recognition of the modality of concepts, we took advantage of the definitions used in the same i2b2 challenge, where modality was used to capture whether a medical event discerned from a medical record actually happens, is merely proposed, mentioned as conditional, or described as possible. We extended this definition such that the possible modality values of “factual, “possible, and “proposed indicate that medical concepts mentioned in the EEGs are actual findings, possible findings and findings that may be true at some point in the future, respectively. Through the identification of polarity and modality of the medical concepts, we aimed to capture the neurologists beliefs about the medical concepts mentioned in the EEG report. For example, in the EEG report illustrated in Figure 1, we identified the medical problem “right leg swelling” with a “factual” modality and a “positive” polarity in the clinical history section, whereas the medical problem “structural brain disease” in the clinical correlation section was found to have the modality “possible” and the polarity “positive”. In the same section, the medical problem “seizures” had the modality “factual” and the polarity “negative”.
Table 1:
Medical concept types and their attribute types.
| Concept Type |
Polarity |
Modality |
EEG Activity-Specific |
|---|---|---|---|
| EEG Activity | ✓ | ✓ | ✓ |
| EEG Event | ✓ | ✓ | |
| Problem | ✓ | ✓ | |
| Test | ✓ | ✓ | |
| Treatment | ✓ | ✓ |
3.1. EEG-specific Medical Concepts and Attributes
An EEG activity is defined as an EEG wave or sequence of waves, while an EEG event is defined as a stimulus that activates the EEG by the International Federation of Clinical Neurophysiology [26]. Although previous efforts of identifying medical concepts in clinical narratives assumed that it is sufficient to automatically discover (a) the boundary of each mention of a concept; (b) the concept type; (c) its modality and (d) its polarity, the EEG activities could not be recognized in the same way. First, as reported in [6], we noticed that EEG activities are not mentioned in a continuous expression. For example, in the narrative fragment: “there are also bursts of irregular, frontally predominant [sharply contoured delta activity]ACT, some of which seem to have an underlying [spike complex]ACT from the left mid-temporal region” we can recognize one EEG activity that is mentioned by distant expressions in the narrative. To address this problem, we considered (a) the anchors of EEG activities and (b) their attributes. This allows us to automatically identify the anchors of EEG activities, annotate them in EEG reports while also recognizing the attributes of EEG activities and attaching them to the anchors, without needing to annotate all text spans referring to EEG activities in the reports. For this purpose, we defined 16 attributes which are specific to EEG activities, listed and defined in Table 2.
Table 2:
Attributes specific to EEG activities.
Attribute 1: Morphology ::= represents the type or “form” of EEG waves.
|
Attribute 2: Frequency Band
|
Attribute 3: Background
| |
Attribute 4: Magnitude ::= describes the amplitude of the EEG activity if it is emphasized in the EEG report
| |
Attribute 5: Recurrence ::= describes how often the EEG activity occurs
| |
Attribute 6: Dispersal ::= describes the spread of the activity over regions of the brain
| |
Attribute 7: Hemisphere ::= describes which hemisphere of the brain the activity occurs in
| |
Location Attributes: Brain Location ::= describes the region of the brain in which the EEG activity occurs. The BRAIN LOCATION attribute of the EEG Activity indicates the location/area of the activity (corresponding to the electrode placement under the standard 10-20 system).
| |
Because Morphology best defines the EEG activities, we decided to use it as the anchor of each EEG activity, but its values also expressed the attributes of the EEG activities. When considering the Morphology of EEG activities, we relied on a hierarchy of values, distinguishing first two types: (1) Rhythm and (2) Transient. In addition, the Transient type contains three subtypes: Single Wave, Complex and Pattern. Each of these sub-types can take multiple possible values, illustrated in Table 2. In addition to Morphology, we considered three classes of attributes for EEG activities, namely (a) general attributes of the waves, e.g. the Frequency Band , the Background - which asserts whether the EEG activity occurs in the background or not; and Magnitude; (b) temporal attributes and (c) spatial attributes. The only temporal attribute considered is Recurrence, which describes how often the EEG activity occurs. As spacial attributes, we considered the Dispersal, the Hemisphere and eight additional attributes for the Brain Location where the EEG activity is observed, since an activity can simultaneously occur in more than one brain location. All attributes specific to EEG activities have multiple possible values associated with them. Table 2 defines each of the 16 attributes of EEG activities and illustrates the possible values each of these attributes.
In contrast, EEG events, which are frequently mentioned in EEG reports as well, can be recognized only by identifying the text span where they are mentioned.
3.2. Relations between Medical Concepts in EEG Reports
First, we decided to consider only binary relations between the five types of medical concepts. Second, we decided to consider only four types of relations: (1) Evidences; (2) Evokes; (3) Clinical-Correlation and (4) Treatment-For. The decision to focus on these four relations was motivated by discussions with practicing neurologists, as the relations represent implicit knowledge gleaned from the EEG reports which informs their reading of the Impression and Clinical Correlation sections of the EEG report. This relation-annotation schema is adapted from the schema reported in previous work [7, 17]. The Evidences relation considers EEG activities or medical problems as providing evidence for medical problems mentioned in the EEG report. The Evokes relation represents the relationship where a medical concept evokes an EEG activity. EEG events, medical problems and treatments can all evoke EEG activities. The Clinical-Correlation relation connects the EEG activities and medical problems mentioned in the Clinical Correlation section of the EEG report if the activity clinically correlates with the medical problem. The Treatment-For relation links treatments to the medical problems for which they are prescribed.
We also made the decision to annotate relations between medical concepts, and not between their mentions in the EEG report. Because the same concept can be mentioned multiple times in the same EEG report, the representation of concepts is achieved by (i) normalized their mentions and (ii) identifying their attributes. This made it possible to recognize coreferring mentions of the same concept by simply grouping concepts with the same normalized mention name and attribute values. Therefore, a relation annotated between any two concept mentions is applied to the normalized concepts associated with the two mentions. We normalized each concept mention into a canonical form using the (i) the morphology attribute for EEG activities and (ii) the United Medical Language System (UMLS) [27] preferred name of the concepts of other types.
3.3. Joint Learning of Concepts, Attributes and Relations Using Transformers
Inspired by the Bi-affine Relation Attention Networks (BRANs) presented in [28], we have designed a neural network architecture capable of extracting (a) medical concepts; (b) their attributes and (c) the relations between them simultaneously. The architecture uses self-attention to learn a representation of all the words in the EEG report. Self-attention is an attention mechanism relating different positions of a single sequence (of words) with one another in order to compute a representation of the sequence. Self-attention has been used successfully before in abstractive summarization [29], textual entailment [30] and learning task-independent sentence representations [31].
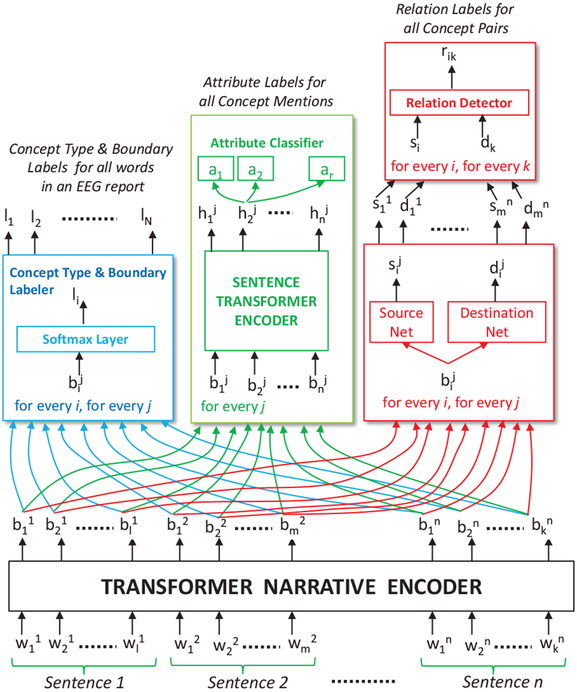
We designed the Self-Attention Concept, Attribute and Relation (SACAR) identifier for automatically recognizing concepts, their attributes, and relations relations between concept (even long-distance relations across multiple sentences from the EEG report). As shown in Figure 2, SACAR first uses a transformer narrative encoder to generate an encoding, , for each word, , in each sentence Sentencej in the narrative of the EEG report2. These encodings serve as input to: (1) the concept type and boundary labeler that detects the type and the boundaries of each concept mentioned in an EEG report; (2) the attribute classifier that recognizes the attributes of each concept mentioned in the EEG report; and (c) the relation detector that identifies relations between the concepts in EEG reports. As shown in Figure 2, all these three modules operate jointly because they share the encoding of the words and sentences produced by the transformer narrative encoder. Moreover, each of these three modules has an associated loss function, namely for the concept type and boundary labeler, for the attribute classifier and for the relation detector, respectively. We shall define the loss functions later in the Section, when we detail the functionality of each module. The parameters of each module as well as the transformer narrative encoder are learned jointly by minimizing the combined loss:
| (1) |
where , is the Adaptive Computation Time loss used in the encoder for learning the encodings of words from the EEG report, defined later in the Section, while γC, γA, γR, and γT are hyperparameters (set to 0.85, 0.65, 1.0, and 0.1 in this work, respectively). The combined loss is minimized using Adam [33], a widely-used stochastic optimization algorithm.
Figure 2:
Architecture for Self-Attention Concept, Attribute and Relation (SACAR) Identification.
A. The Transformer Narrative Encoder.
The transformer narrative encoder (TNE) learns a contextualized encoding, , for each word, , in an EEG report given the full context of the EEG report using self-attention. The TNE consists of B recurrent blocks, denoted as TNEk, with k ∈ {1, . . . , B}. Each block TNEk shares parameters and is made up of two sub-components: (a) a multi-head attention and (b) a series of convolutions, as in Verga at al. [28]. The output of the kth block for the ith word, denoted as , is connected to its input, through a residual connection [34]:
| (2) |
The ith input to the first block of the TNE, , is the word embedding of the ith word in the EEG report. It should be noted that word embeddings are learned jointly along with the other parameters of this model in this work.
Each block TNEk uses the multi-headed self-attention mechanism introduced in Vaswani et al. [35], which allows our learning model to jointly attend to information from different representation subspaces at different positions in the sequence of words from each sentence of the EEG report. This amounts to using multiple, parallel self-attention functions, one for each head. The self-attention function maps a sequence of input vectors to a sequence of output vectors, where each output vector is a weighted sum of the input vectors and the weights are computed using a compatibility function that compares the sequence of input vectors to itself (hence the name self-attention). In this way, for head h using an input vector , the self-attention function computes the output vector , as:
| (3) |
where is a learned weight matrix, do is the dimension of the vector oih, ☉ denotes the element-wide multiplication, and αijh is the attention weight between inputs i and j of attention head h computed by:
| (4) |
where are learned weight matrices.
The outputs of each attention-head, oih, are concatenated and projected back into d dimensions using a linear layer:
| (5) |
where H is the number of attention heads, is a learned weight matrix and [·; ·] is the concatenation operation. A residual connection combined with Layer Normalization [36], denoted as LN(·), is then applied to the output vectors:
| (6) |
In addition to the multi-head attention, as in Verga et al. [28], for each block TNEk we have used a feed-forward network of three convolutional layers cl followed by a final residual connection and Layer Norm:
| (7) |
| (8) |
| (9) |
| (10) |
where CL(·) represents a convolution operator of kernel width L and is the output of block k for word i. It is to be noted that in EEG reports, certain words tend to be more ambiguous than others, suggesting that computing the encodings of these words requires additional processing to correctly capture their meaning from the contexts in which they appear. Therefore, some words will require a larger number of TNE blocks than others. Inspired by Dehgani et al. [1], we dynamically adjusted the number of blocks used to produce the encoding for each word in the narrative using Adaptive Computation Time (ACT) [37]. ACT allows the model to learn how many processing steps (blocks) are necessary to encode each word and to dynamically halt processing for one word, but to continue processing for other words whose encodings still require further refinement. Once ACT has halted for a word in the EEG report narrative, its encoding is simply copied to the next block until processing has halted for all other words in the narrative or until a maximum number of blocks has been reached (12 in this work).
To determine if processing should halt for word i at block k, we used a sigmoidal halting unit with weights and bias [37]:
| (11) |
The halting score, , for word i at block k is used to calculate (i) the number of recurrent updates for word i, N(i); (ii) the halting probability ; and (iii) the remainder, R(i), defined as:
| (12) |
| (13) |
| (14) |
where epsilon is a small constant (0.01 in this work). Equation 12 bounds the number of recurrent updates for word i by the total halting score for all blocks such that processing halts when . The remainder, R(i), represents the amount of halting score remaining before processing halts. The final encoding for word i produced by the transformer narrative encoder is the sum of the output at each block weighted by the halting probability for that block:
| (15) |
In order to allow the neural network to learn when to halt or continue processing, we add the differentiable loss term described in Graves (2016) [37] which bounds the total recurrent computation performed by the TNE:
| (16) |
B. Concept Type and Boundary Annotator.
The concept type and boundary annotator first identifies spans of text that correspond to medical concept mentions by assigning a label to each word in the narrative of the EEG report indicating that the word begins a medical concept mention (B), is inside of a medical concept mention but not at the beginning (I), or is outside of a medical concept mention (O). In this way, medical concept mentions can be identified by continuous sequences of words starting with a word labeled B optionally followed by words labeled I.
In order to also identify the type of the medical concepts from the EEG reports and to distinguish EEG activities, EEG events, medical problems, treatments, and tests, we extended the IOB labeling system to include separate B and I labels for each concept type, yielding 11 total medical concept boundary and type labels: LC ={B-ACT, I-ACT, B-EV, I-EV, B-PR, I-PR, B-TR, I-TR, B-TE, I-TE, O}. The concept type and boundary labeler assigns a label li ∈ LC to each word wi in the EEG report by passing each word’s encoding produced by the TNE through a fully connected softmax layer to produce a distribution over the labels:
| (17) |
where is a weight matrix and is a bias vector. Then, the predicted label for word wi is the label with the highest probability .
We trained the concept type and boundary labeler to maximize the likelihood of labeling each word correctly, considering that the labels are conditionally independent, given the word encoding produced by the transformer narrative encoder, namely bi. This allowed us to define:
| (18) |
where the probability is the probability assigned to the label in .
C. Attribute Classifier.
Each medical concept automatically identified in an EEG report is associated with several attributes including its modality, and polarity. In addition, EEG activities have 16 specific attributes defined in Table 2. After each medical concept is identified in the EEG report, the attribute classifier determines the values of each attribute type for each medical concept using another transformer encoder – this time at the sentence level – and a series of linear classifiers, one for each attribute type.
In each EEG report, the textual cues that signal attribute values for a medical concept are typically found in the same sentence as the concept mention. Therefore, the attribute classifier needs to further refine the encoding for each word using the sentence transformer encoder (STE), which is a transformer module similar to the TNE, operating at the sentence level instead of the full narrative. By operating at the sentence level, the STE allows the model to attend to each word in the surrounding sentence, determining which context words are most informative for attribute classification, while ignoring irrelevant out-of-sentence words. The STE is defined exactly the same as the TNE, consisting of a set of Bs identical blocks (8 in this work) where the number of blocks used is dynamically determined for each word using Adaptive Computation Time. For each sentence S j in the EEG report, the STE produces an encoding for each word in S j using the sequence of encodings for the words in sentence S j produced by the TNE, as illustrated in Figure 2. We also took into account the annotations produced by the concept type and boundary annotator on S j such that for each concept identified in S j we considered only the encoding of the sentence words found within the boundaries of . In addition we took into account the type of concept to decide which of the 18 attributes should be identified for that concept. If = EegActivity each of the 18 attributes will be considered but if ∈ {EegEvent, MedicalProblem, Treatment, Test}, we shall only consider the attributes polarity and modality. In this way, given any attribute a selected for concept , we computed the distribution over the values for attribute a as follows:
| (19) |
where , are weight matrices, d is the dimension of the encodings produced by the STE, dA is the dimension of the hidden state, Δ(a) is the number of distinct attribute values of attribute a, and , are bias vectors.
Similarly to the concept type and boundary annotator, the attribute classifier is trained to maximize the likelihood of correctly classifying each attribute of every concept identified in an EEG report, where the attribute values, are conditionally independent given the concept encodings, :
| (20) |
where Φ is the set of (sentence, concept) indices (j, k) denoting concept mentions and the probability is given by the probability assigned to attribute value in the predicted distribution, , for attribute a.
D. Relation Detector.
The relation detector automatically identifies several types of relations between pairs of concepts recognized in EEG reports (the definition of relations types was provided in Section 3.2). Because in the same EEG report, the same entity corresponding to a unique medical concept may be mentioned several times, we distinguish between concept mentions and concept entities. Figure 3 illustrates two concept entities and their corresponding concept mentions. As it can be seen, the concept entities are illustrated through (1) their normalized name; (2) their type; and (3) all their identified attributes. It should be noted that the automatic identification of medical concept type and boundaries performed by the SACAR neural system recognizes only concept mentions. To identify the concept entities, we assumed that concept mentions from the same EEG report that (1) have the same normalized name (described in Section 3.2), (2) type, and (3) the same values of their attributes are coreferring to the same concept entity. Moreover, as illustrated in Figure 3, we considered for each pair of concept entities two possible cases, namely (case 1) in which the first concept entity is the the source concept of the potential relation and the second concept entity is the destination of the relation; or (case 2) in which the first concept entity is the destination whereas the second concept entity is the source.
Figure 3:
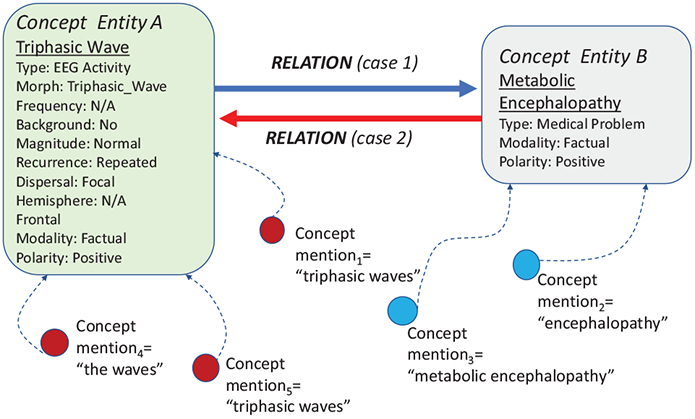
Concept entities, their mentions and possible relations between them.
In this framework relation discovery was cast as a prediction of the most likely relation type between two concept entities. The two cases (above described) of selecting in each pair of concepts the relation source and destination were addressed simultaneously, considering that the first concept of the pair is the source while the second concept is the destination for one possible relation, and conversely for a second possible relation. To enable the prediction of the most likely relation, we have generated for each concept mention of every concept entity both a source encoding, and a destination encoding using the Source Net, and the Destination Net, respectively, as illustrated in Figure 2, implemented as two-layer neural networks:
| (21) |
| (22) |
where , , , are weight matrices. These encodings enabled us to represent all possible relations between each pair of concept entities from an EEG report in terms of their mentions using an N × R × N tensor, L, where N is the number of concept mentions discovered in the EEG report and R is the number of possible relation types. For each source concept entity S and each destination concept entity D, we consider (a) the source encodings of all the mentions of S in the EEG report, denoted as sm; and (b) the destination encodings we produced for all the mentions of D, denoted as dn. In order to compute each value of L, we used a bi-affine function between a source encoding, sm, and a destination encoding, dn, for each relation type r ∈ R:
| (23) |
where is a learned tensor representing a set of d×d embedding matrices (one for each relation type r ∈ R). This enabled us to compute the scores of all relation types between a pair of concept entities, in which the source is S and the destination is D using the LogSumExp function:
| (24) |
where M(S) are all the mentions of concept entity S and M(D) are all the mentions of the concept entity D in the same EEG report. Note that scores is a vector of scalar scores for each relation type between S and D and the LogSumExp function is a smooth approximation to the max function [38]. The probability distribution over the possible relation types between S and D can then be calculated using the softmax function:
| (25) |
To train the relation detector, we maximized the likelihood of classifying each relation between each pair of concepts entities correctly:
| (26) |
where is the type of the relation from concept entity S to concept entity D and is the probability assigned to relation type in the distribution .
4. Multi-Task Active Deep Learning
As in our previous work on recognizing medical concepts in EEG reports [6] or identifying distant relations between such concepts [7], we considered a multi-task active deep learning (MTADL) framework that combines the strength of Active Learning with the advantages of deep learning. The MTADL framework allowed us to rely on the active learning loop to enhance the quality of the concepts, attributes and relations we have discovered automatically in the EEG report. Not only does the SACAR identifier, presented in the previous section, produce on its own superior results than those obtained using MTADL in our previous work [6, 7], but it also enables us to consider a novel framework for learning how to actively learn, using a deep imitation learning approach as introduced in Liu et al. [39]. In our previous work employing the MTADL framework, we used uncertainty sampling to select the most beneficial EEG reports for expert validation, where uncertainty was calculated using the Shannon Entropy. However, combining the uncertainty of the medical concepts and their attributes in an EEG report with the uncertainty of the relations between medical concepts in the same EEG report is not trivial, as it may favor sampling based on concepts over relations in some instances, and vice-versa in other instances. Therefore we chose to learn the EEG report selection strategy best suited for the identification of concepts, attributes and relations in our corpus of EEG reports. As in Liu et al. [39] we cast the problem of learning the example selection policy as a imitation learning problem. We were able to train a deep imitation learning network capable of learning a policy for selecting EEG reports which was later applied in each active learning loop. Our MTADL framework using deep imitation learning is illustrated in Figure 4. As shown in the Figure, MTADL uses the following six steps:
Figure 4:
Multi-Task Active Deep Learning of Concepts, Attributes and Relations from EEG Reports.
STEP 1: The initial manual annotation of medical concepts, attributes and relations between medical concepts in EEG reports. In addition to the expert annotations, that we have used before in previous work [6, 7], we also made use of silver annotations, produced on the entire corpus of EEG reports, using our previous methods for concept and attribute recognition as well as relation identification;
STEP 2: Learn to recognize concepts, their attributes and relations between concepts by training SACAR on the current manually annotated training data along with the silver annotations;
STEP 3: Automatically annotate all concepts, their attributes and relations between concepts given the current SACAR model;
STEP 4: Deep imitation learning of the active learning policy is performed if a selection policy is not yet learned. The learned policy is then applied on the entire set of un-annotated EEG reports, given the current state of the SACAR model. This step is performed only once, while the resulting, learned selection of the annotated EEG reports will be used repeatedly, inside the AL loop, as illustrated in Figure 4;
STEP 5: Accept/Edit annotations of concepts, attributes and relations in sampled EEG reports, made available by the learned selection of annotated EEG reports;
STEP 6: Re-training SACAR with the new training data, containing the validated annotations. Go to Step 3 until the desired performance is obtained or the time for active learning is exhausted.
As shown in Figure 4, the active learning loop is comprised of Steps 2, 3, 5 and 6. Central to MTADL is the active learning policy which, once learned in Step 4, it is applied in the AL loop in a static fashion.
Deep Imitation Learning of the Active Learning Policy
To select unlabeled examples for manual annotation in the active learning loop, we used a neural network, namely the the Active Learning Policy Neural Network (ALPNN), illustrated in Figure 5, which we trained with the deep imitation learning algorithm developed by Liu et al. [39]. The ALPNN represents an active learning problem in terms of (1) a model (in this case the SACAR model); (2) a set of labeled data for training, DT; (3) a set of labeled data for evaluation, DE; and (4) a set of unlabeled data DU. Given an AL problem, the Policy Network determines a score for each unlabeled data example and returns the example with the highest score for manual annotation.
Figure 5:

The Active Learning Policy Neural Network (ALPNN).
In order to train the ALPNN, we use the initial manually annotated dataset, D, to develop a series of simulated active learning problems. Each simulated active learning problem consists of (1) three random partition of D to form (a) the training data, DT, (b) the evaluation data DE, and (c) the unlabeled data, DU; along with (2) the SACAR model. The ALPNN is trained to select the optimal unlabeled example for each simulated problem. To determine the optimal selection, K examples X1, X2, . . . XK are randomly selected from DU and K different SACAR models are trained using DT augmented with one of the examples, e.g. SACARi is trained using DT augmented with Xi, for each i ∈ {1 . . . K}. When evaluating each of the K SACAR models using the evaluation data DE, we were able to determine the SACAR model with the best performance, e.g. SACARj and thus conclude that example Xj is the the optimal selection. In this way, the ALPNN is trained to imitate an expert selection policy that selects the unlabeled example that will most improve the performance of SACAR.
The ALPNN is a two-layer feed-forward neural network that calculates a score, s(dU), for an unlabeled example, dU, given an active learning problem. The inputs to the ALPNN, as shown in Figure 5 consist of 3 fixed-size vector representations: (1) v(DU) representing the entire unlabeled data set, DU; (2) v(DT) representing the entire labeled data set, DT; and (3) v(dU) representing the unlabeled example, dU, along with the predicted labels for it, , generated by SACAR. Specifically, v(DU), the vector representation of the unlabeled data set DU, is produced by aggregating the encodings for each example, d ∈ DU:
| (27) |
where bi is the encoding produced by SACAR’s TNE for word wi in the EEG report d; and |d| is the number of words in d. Similarly, the vector representation, v(DT), for the labeled data set consists of an aggregation of (i) the encodings for each example d ∈ DT concatenated with vectors for the empirical distributions of class labels for (ii) the concept type & boundary detection task; (iii) the 18 attribute classification tasks, and (iv) the relation prediction task in the labeled data set:
| (28) |
| (29) |
| (30) |
where x is the sum of the average TNE encoding for each word in each example d ∈ DT; is the probability of task t having class l under the empirical distribution of DT; and vT is the concatenation of x and all of the distributions, yt for each task t. The vector representation v(dU) of the unlabeled example, dU, along with its predicted labels, , is derived as follows:
| (31) |
| (32) |
| (33) |
where vx is the average TNE encoding for each word in the example dU, is the sum of the predicted distributions (from Equations 17, 19, and 25) for each instance of task t in dU, and v(dU) is the concatenation of vx and all of the combined predicted distributions for each task t.
The score s(dU), illustrated in Figure 5, is then calculated as:
| (34) |
where and are weight matrices, dp is the dimension of the hidden state, d0 is the dimension of the three input vectors concatenated together, and , are bias vectors.
The ALPNN is trained using the imitation learning algorithm described in Liu at al. [39] using the following loss function which maximizes the probability of selecting the optimal unlabeled example, , from DU for each simulated problem:
| (35) |
where the tuple (, DU, DT, DE) represents a simulated active learning problem, and SIM is the set of simulated examples. The set SIM is dynamically generated during training of ALPNN using the algorithm described in Liu et al. [39], which uses Dataset Aggregation method [40] which is meant to increase the generalization of the Policy Network by exposing it to problems similar to those it is likely to encounter during the active learning loop.
5. Experimental Results
We evaluate SACAR using a subset of 140 EEG reports from the TUH EEG corpus in which concepts, attributes, and relations were manually annotated. This subset was constructed using the ALPNN system starting from a seed set of 40 manually annotated reports using ten rounds of active learning in which 10 reports were sampled each round. For training purposes, another subset of 1,000 EEG reports with silver-standard annotation produced by the systems in [6] and [7] are used to augment the training data. Statistics for concept and relation types for the evaluation dataset are provided in Table 3, while fine-grained attribute type statistics are presented later in the section. The evaluations were performed using 7-fold cross-validation on the gold subset of 140 EEG reports while all of the silver-standard data was used for training in each round of cross validation. Manual annotation was performed by three graduate students after extensive consultation with practicing neurologists. Average inter-annotator agreement, measured using Jaccard Score [41], was 0.9658, 0.9518, and 0.8843 for concept boundary, attribute, and relation identification, respectively.
Table 3:
Data statistics for the evaluation dataset.
| Concept Types | Relation Types | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Annotations | Activity | Event | Problem | Test | Treatment | Evidences | Evokes | Treatment-For | Clinical-Correlation |
| Gold | 1438 | 452 | 798 | 716 | 539 | 397 | 342 | 356 | 195 |
| Silver | 8820 | 3033 | 5367 | 5049 | 3416 | 2850 | 2326 | 2065 | 1228 |
In our experiments we evaluated (1) the results of the SACAR identifier for automatically recognizing concepts, their attributes, and relations spanning them; as well as (2) the impact of the ALPNN as a selection policy during active learning. First, we compared the results of SACAR to three neural baselines that were trained to perform (a) concept type and boundary detection, (b) attribute classification, or (c) relation identification, respectively. Each of the these neural baselines was trained to perform one of the three tasks discussed above, while SACAR is trained to perform all tasks, jointly. As baseline for the concept type and boundary detection, we use the stacked LSTM models presented in our previous work [6], consisting of a stacked LSTM operating at sentence level. In fact, we used two separate LSTM-based neural networks: one for detecting only the boundaries of EEG activities, and the other for detecting the types and boundaries of all other medical concepts. The results of this baseline, denoted as LSTM, are presented in Table 4. The results for concept type and boundary detection are presented in Table 4 in terms of precision, recall, and F1 score, where predicted concept boundaries are considered correct if they exactly match a manually annotated boundary. When evaluating the results of SACAR, we took also into account the fact that SACAR uses two transformer encoders – the TNE and the STE – to produce internal representations of each EEG report. We were interested to evaluate two important properties of SACAR’s transformer encoders: (1) recurrence and (2) Adaptive Computation Time.
Table 4:
Evaluation Results for Concept Type and Boundary Recognition.
| EEG Activity | EEG Event | Medical Problem | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 |
| LSTM | 0.8949 | 0.8125 | 0.8517 | 0.8842 | 0.8301 | 0.8563 | 0.8391 | 0.7863 | 0.8118 |
| SACAR-V | 0.8352 | 0.8336 | 0.8344 | 0.8068 | 0.8170 | 0.8119 | 0.6992 | 0.6159 | 0.6549 |
| SACAR-U | 0.8494 | 0.9153 | 0.8811 | 0.8335 | 0.8674 | 0.8501 | 0.7658 | 0.7516 | 0.7586 |
| SACAR-A | 0.9460 | 0.9080 | 0.9266 | 0.9072 | 0.8299 | 0.8668 | 0.8866 | 0.8358 | 0.8605 |
| Treatment | Test | All Types (Macro Average) | |||||||
| Model | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 |
| LSTM | 0.9257 | 0.8687 | 0.8963 | 0.8904 | 0.9250 | 0.9074 | 0.8869 | 0.8645 | 0.8756 |
| SACAR-V | 0.8072 | 0.7540 | 0.7797 | 0.8207 | 0.8654 | 0.8425 | 0.7937 | 0.7772 | 0.7854 |
| SACAR-U | 0.9332 | 0.9048 | 0.9188 | 0.8171 | 0.9470 | 0.8773 | 0.8398 | 0.8772 | 0.8581 |
| SACAR-A | 0.9511 | 0.8814 | 0.9149 | 0.9050 | 0.9280 | 0.9164 | 0.9192 | 0.8766 | 0.8974 |
Inspired by Dehghani et al. [1] we hypothesized that introducing recurrence to the transformer encoders could help SACAR learn better from our small amount of labeled data and that ACT could further boost performance by allowing SACAR to dynamically allocate more resources to more complicated encodings In this section, we will refer to the full SACAR model described in Section 3.3 that uses both recurrence and ACT based on the Adaptive Universal Transformer [1] as SACAR-A. We evaluate SACAR-A against two alternative configurations: (1) SACAR-V which uses a simple vanilla transformer encoder without recurrence or ACT as described in Vaswani et al. [35]; and (2) SACAR-U based on the Universal Transformer described in Dehgani et al. [1] which uses recurrence, but not ACT. Formally, SACAR-V is equivalent to SACAR-A where each TNE and STE block has its own parameters and Equation 15 is replaced by given B blocks. Similarly, SACAR-U is equivalent to SACAR-V where parameters are shared between TNE blocks and between STE blocks (i.e. the transformer encoders are recurrent). Results of all implementations of SACAR for concept type and boundary recognition are listed in Table 4. As we can see from the Table, the SACAR-A model outperforms the baseline as well as the other alternative implementations of the transformers for all concept types, except for Treatment, where it is slightly outperformed by SACAR-U. Interestingly, the only model that does not make use of recurrence, SACAR-T, performs worst – obtaining results that are outperformed even by the LSTM baseline.
When evaluating the results for attribute classification, we used as baseline the Deep ReLU Network (DRNet) reported before in [6]. The DRNet consists of five fully-connected ReLU layers and a series of softmax layers, one for each attribute type. Moreover, we used two DRNet models: one for EEG activity attributes, and one for the attributes of all other medical concepts. It should be noted that both baselines used for concept type and boundary detection and for attribute classification, namely the LSTM and DRNet baselines, rely on hand-crafted features while the SACAR models are trained end-to-end with no feature extraction needed. The results of the baseline used for attribute classification as well as the results of all three SACAR model are listed in Table 5, evaluated with Precision, Recall, and F1 score. Table 5 presents the results for each value of each attribute along with their prevalence in the gold annotated data (indicated by the ‘#’ symbol). Attribute values with no examples in the training data are omitted. Aggregated metrics are presented for each attribute type using micro-average, however precision and recall are omitted for multi-class classification tasks since they are equivalent to F1. All three SACAR models tend to outperform the DRNet baseline in micro-averaged F1 score.
Table 5:
Evaluation Results for Attribute Classification.
| Precision | Recall | F1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Attribute | # | DRNet | SACAR-V | SACAR-U | SACAR-A | DRNet | SACAR-V | SACAR-U | SACAR-A | DRNet | SACAR-V | SACAR-U | SACAR-A |
| Morphology | 1438 | – | – | – | – | – | – | – | – | 0.7716 | 0.8548 | 0.8263 | 0.8868 |
| Rhythm | 388 | 0.6087 | 0.8941 | 0.8254 | 0.9549 | 0.8660 | 0.9965 | 0.9820 | 0.9961 | 0.7149 | 0.9425 | 0.8969 | 0.9751 |
| V wave | 37 | 0.6977 | 0.6849 | 0.5217 | 0.7143 | 0.8108 | 0.8228 | 0.8000 | 1.0000 | 0.7500 | 0.7475 | 0.6316 | 0.8333 |
| Wicket spikes | 11 | 0.5000 | 0.7000 | 0.5000 | 1.0000 | 0.1818 | 0.6363 | 0.1818 | 1.0000 | 0.2667 | 0.6667 | 0.2677 | 1.0000 |
| Spike | 43 | 0.8000 | 0.5319 | 0.7083 | 0.7500 | 0.5581 | 0.5814 | 0.3953 | 0.3023 | 0.6575 | 0.5556 | 0.5075 | 0.4309 |
| Sharp wave | 107 | 0.9195 | 0.7091 | 0.6696 | 0.6522 | 0.7577 | 0.7290 | 0.7009 | 0.8333 | 0.8247 | 0.7189 | 0.6849 | 0.7317 |
| Slow wave | 64 | 0.8929 | 0.8305 | 0.8276 | 0.9048 | 0.7813 | 0.7656 | 0.7500 | 0.8261 | 0.8333 | 0.7967 | 0.7869 | 0.8636 |
| K-complex | 11 | 0.8889 | 0.9000 | 0.6667 | 1.0000 | 0.7273 | 0.8181 | 0.5454 | 1.0000 | 0.8000 | 0.8571 | 0.6000 | 1.0000 |
| Sleep spindles | 28 | 0.5526 | 0.7248 | 0.6875 | 0.8333 | 0.7500 | 0.8081 | 0.7917 | 1.0000 | 0.6364 | 0.7642 | 0.7360 | 0.9091 |
| Spike-and-slow | 106 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.6132 | 0.5934 | 0.4615 | 0.6250 | 0.7602 | 0.7448 | 0.6316 | 0.7692 |
| Triphasic wave | 16 | 0.8333 | 0.7143 | 0.8000 | 1.0000 | 0.6250 | 0.3125 | 0.2500 | 0.8000 | 0.7143 | 0.4348 | 0.3810 | 0.8889 |
| Polyspike complex | 29 | 0.7500 | 0.9286 | 0.8750 | 0.4000 | 0.5172 | 0.4483 | 0.2414 | 1.0000 | 0.6122 | 0.6047 | 0.3784 | 0.5714 |
| Suppression | 48 | 0.5814 | 1.0000 | 0.6923 | 1.0000 | 0.5208 | 0.1667 | 0.3103 | 0.2857 | 0.5495 | 0.2857 | 0.4286 | 0.4444 |
| Slowing | 174 | 0.9371 | 0.7910 | 0.7453 | 0.8710 | 0.7701 | 0.9138 | 0.9080 | 0.9000 | 0.8454 | 0.84480 | 0.8187 | 0.8852 |
| Breach Rhythm | 12 | 0.8000 | 1.0000 | 0.8889 | 1.0000 | 0.6667 | 1.0000 | 0.6667 | 1.0000 | 0.7273 | 1.0000 | 0.7619 | 1.0000 |
| Photic Driving | 51 | 0.8750 | 1.0000 | 0.9329 | 1.0000 | 0.6863 | 0.8294 | 0.7414 | 1.0000 | 0.7692 | 0.9067 | 0.8262 | 1.0000 |
| PLEDs | 20 | 0.5625 | 0.4167 | 0.2222 | 0.3333 | 0.4500 | 0.2500 | 0.2000 | 0.2500 | 0.5000 | 0.3125 | 0.2105 | 0.2857 |
| Epileptiform discharge | 136 | 0.7934 | 0.6519 | 0.6173 | 0.7000 | 0.7059 | 0.7574 | 0.7353 | 0.6364 | 0.7471 | 0.7007 | 0.6711 | 0.6667 |
| Disorganization | 98 | 0.8000 | 0.7416 | 0.6574 | 0.7895 | 0.6122 | 0.6735 | 0.7245 | 0.6250 | 0.6936 | 0.7059 | 0.6893 | 0.6977 |
| Unspecified | 59 | 0.3855 | 0.8158 | 0.7941 | 1.0000 | 0.5424 | 0.5254 | 0.4576 | 0.4286 | 0.4507 | 0.6392 | 0.5806 | 0.6000 |
| Frequecy Band | 1438 | – | – | – | – | – | – | – | – | 0.8754 | 0.8911 | 0.8862 | 0.9356 |
| Alpha | 128 | 0.8632 | 0.8710 | 0.8779 | 0.9286 | 0.7891 | 0.8438 | 0.8984 | 0.8387 | 0.8245 | 0.8571 | 0.8880 | 0.8814 |
| Beta | 91 | 0.6881 | 0.3929 | 0.3704 | 0.8750 | 0.8242 | 0.2418 | 0.2198 | 0.6364 | 0.7500 | 0.2993 | 0.2759 | 0.7368 |
| Delta | 129 | 0.8416 | 0.7381 | 0.7949 | 0.9524 | 0.6589 | 0.4806 | 0.4806 | 0.7143 | 0.7391 | 0.5822 | 0.5990 | 0.8163 |
| Theta | 118 | 0.7755 | 0.7471 | 0.7944 | 0.9000 | 0.6441 | 0.5508 | 0.7203 | 0.6923 | 0.7037 | 0.6341 | 0.7556 | 0.7826 |
| N/A | 972 | 0.9067 | 0.9683 | 0.9593 | 0.9744 | 0.9410 | 0.9889 | 0.9797 | 0.9954 | 0.9235 | 0.9785 | 0.9694 | 0.9848 |
| Background | 1438 | 0.8904 | 0.9506 | 0.9657 | 0.9704 | 0.8197 | 0.9110 | 0.9506 | 0.9428 | 0.8543 | 0.9340 | 0.9581 | 0.9564 |
| Magnitude | 1438 | – | – | – | – | – | – | – | – | 0.8148 | 0.9175 | 0.9461 | 0.9273 |
| Low | 184 | 0.7548 | 0.7669 | 0.7669 | 0.7273 | 0.5939 | 0.6757 | 0.6757 | 0.4444 | 0.6648 | 0.7184 | 0.7184 | 0.5517 |
| High | 175 | 0.6273 | 0.5119 | 0.5119 | 0.8421 | 0.6900 | 0.5890 | 0.4674 | 0.4848 | 0.6571 | 0.5478 | 0.7184 | 0.6154 |
| Normal | 1079 | 0.9210 | 0.9627 | 0.9909 | 0.9636 | 0.9396 | 1.0000 | 0.9992 | 0.9934 | 0.9302 | 0.9810 | 0.9851 | 0.9783 |
| Recurrence | 1438 | – | – | – | – | – | – | – | – | 0.7174 | 0.8746 | 0.8802 | 0.9045 |
| Continuous | 224 | 0.7244 | 0.6459 | 0.6286 | 0.6750 | 0.6546 | 0.7411 | 0.6185 | 0.8438 | 0.6878 | 0.6902 | 0.6235 | 0.7500 |
| Repeated | 262 | 0.7974 | 0.6809 | 0.7333 | 0.7179 | 0.6461 | 0.7328 | 0.6740 | 0.8740 | 0.7138 | 0.7059 | 0.7024 | 0.7883 |
| None | 952 | 0.6607 | 0.9626 | 0.9677 | 0.9761 | 0.8163 | 0.9563 | 0.9603 | 0.9574 | 0.7303 | 0.9594 | 0.9640 | 0.9667 |
| Dispersal | 1438 | – | – | – | – | – | – | – | – | 0.7283 | 0.8812 | 0.9222 | 0.8951 |
| Localized | 330 | 0.5955 | 0.8144 | 0.8174 | 0.7714 | 0.6424 | 0.8061 | 0.9091 | 0.5094 | 0.6181 | 0.8509 | 0.8608 | 0.6136 |
| Generalized | 246 | 0.5162 | 0.8061 | 0.8132 | 0.8400 | 0.5813 | 0.8618 | 0.9024 | 0.4118 | 0.5468 | 0.8330 | 0.8555 | 0.5526 |
| N/A | 862 | 0.8441 | 0.8935 | 0.9713 | 0.9464 | 0.7947 | 0.9153 | 0.9408 | 0.9943 | 0.8186 | 0.9043 | 0.9558 | 0.9698 |
| Hemisphere | 1438 | – | – | – | – | – | – | – | – | 0.8148 | 0.8878 | 0.9042 | 0.9117 |
| Right | 96 | 0.7634 | 0.8346 | 0.8571 | 0.8182 | 0.7396 | 0.5362 | 0.5556 | 0.6429 | 0.7513 | 0.6529 | 0.6742 | 0.7200 |
| Left | 159 | 0.8257 | 0.8519 | 0.8810 | 0.7647 | 0.5660 | 0.7233 | 0.6981 | 0.5200 | 0.6716 | 0.7823 | 0.7789 | 0.6190 |
| Both | 246 | 0.6027 | 0.9052 | 0.8832 | 0.8182 | 0.5488 | 0.7764 | 0.7683 | 0.4737 | 0.5745 | 0.8359 | 0.8217 | 0.6000 |
| N/A | 937 | 0.8603 | 0.9149 | 0.9248 | 0.9540 | 0.9218 | 0.9755 | 0.9840 | 0.9920 | 0.8900 | 0.9442 | 0.9535 | 0.9727 |
| Modality | 1438 | – | – | – | – | – | – | – | – | 0.9337 | 0.9613 | 0.9690 | 0.9550 |
| Factual | 1366 | 0.9696 | 0.9753 | 0.9754 | 0.9682 | 0.9939 | 0.9846 | 0.9883 | 0.9682 | 0.9816 | 0.9800 | 0.9818 | 0.9682 |
| Possible | 70 | 0.4308 | 0.8600 | 0.8627 | 0.7824 | 0.4000 | 0.6143 | 0.6286 | 0.5027 | 0.4148 | 0.7167 | 0.7273 | 0.6121 |
| Proposed | 76 | 0.2318 | 0.8776 | 0.8800 | 0.7429 | 0.4605 | 0.5658 | 0.5789 | 0.5098 | 0.3084 | 0.6880 | 0.6984 | 0.6049 |
| Polarity | 1438 | 0.9100 | 0.8864 | 0.9245 | 0.9165 | 0.7389 | 0.8121 | 0.8857 | 0.9071 | 0.8156 | 0.8476 | 0.9047 | 0.9118 |
| Location | 692 | 0.7450 | 0.6982 | 0.7285 | 0.7506 | 0.5958 | 0.6145 | 0.5753 | 0.6062 | 0.6618 | 0.6537 | 0.6429 | 0.6707 |
| Frontal | 189 | 0.7302 | 0.7091 | 0.7258 | 0.7522 | 0.4868 | 0.5044 | 0.4990 | 0.5951 | 0.5841 | 0.5895 | 0.5914 | 0.6645 |
| Occipital | 268 | 0.7871 | 0.7088 | 0.7500 | 0.7944 | 0.7313 | 0.8055 | 0.7216 | 0.7401 | 0.7582 | 0.7541 | 0.7355 | 0.7663 |
| Temporal | 128 | 0.6786 | 0.7338 | 0.7348 | 0.7561 | 0.5938 | 0.6418 | 0.6289 | 0.6257 | 0.6333 | 0.6847 | 0.6777 | 0.6847 |
| Central | 30 | 0.7500 | 0.6724 | 0.6827 | 0.6201 | 0.4109 | 0.5006 | 0.4548 | 0.3195 | 0.5309 | 0.5739 | 0.5459 | 0.4217 |
| Parietal | 10 | 0.5714 | 0.2500 | 0.6000 | 0.5018 | 0.4000 | 0.4000 | 0.5000 | 0.3520 | 0.4706 | 0.3077 | 0.5455 | 0.4138 |
| Frontocentral | 50 | 0.8824 | 0.8247 | 0.8422 | 0.8563 | 0.6000 | 0.5818 | 0.5981 | 0.6244 | 0.7143 | 0.6823 | 0.6995 | 0.7222 |
| Frontotemporal | 17 | 0.5618 | 0.4086 | 0.5374 | 0.4218 | 0.4963 | 0.3238 | 0.3877 | 0.3380 | 0.5270 | 0.3613 | 0.4504 | 0.3753 |
For relation identification, we compare against the EEG-RelNet model presented in our previous work [7]. Like SACAR, EEG-RelNet processes the entire EEG report, detecting relations between distant medical concept entities and requires no feature extraction. The results for relation identification are presented in Table 6. In general both SACAR-A and SACAR-U are able to produce similar – yet slightly worse – results as the baseline, with SACAR-V consistently performing worst. However, it should be noted that the SACAR models do not only identify relations, but also recognize concept types and boundaries and classify concept attributes as well.
Table 6:
Evaluation Results for Relation Identification.
| Evokes | Evidences | Treatment-For | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 |
| EEG-RelNet | 0.8563 | 0.8187 | 0.8371 | 0.6592 | 0.6205 | 0.6392 | 0.5905 | 0.8953 | 0.7116 |
| SACAR-V | 0.7521 | 0.7231 | 0.7373 | 0.5648 | 0.5801 | 0.5723 | 0.5029 | 0.9219 | 0.65079 |
| SACAR-U | 0.8021 | 0.8418 | 0.8215 | 0.5980 | 0.5854 | 0.5916 | 0.5824 | 0.9093 | 0.7100 |
| SACAR-A | 0.8402 | 0.8569 | 0.8485 | 0.5955 | 0.6109 | 0.6031 | 0.5806 | 0.9042 | 0.7071 |
| Clinical-Correlation | All Relations (Macro Average) | ||||||||
| Model | Precision | Recall | F1 | Precision | Recall | F1 | |||
| EEG-RelNet | 0.8380 | 0.8124 | 0.8250 | 0.7360 | 0.7777 | 0.7563 | |||
| SACAR-V | 0.8204 | 0.8259 | 0.8231 | 0.6601 | 0.7627 | 0.7077 | |||
| SACAR-U | 0.8018 | 0.8370 | 0.8190 | 0.6961 | 0.7934 | 0.7415 | |||
| SACAR-A | 0.8287 | 0.8006 | 0.8144 | 0.7112 | 0.7932 | 0.7499 | |||
The impact of the ALPNN as a selection policy is evaluated by measuring the change in performance after each additional round of active learning, as in our previous work [6, 7]. The active learning systems are evaluated for 10 rounds of active learning where 10 unlabeled EEG reports are selected from the entire unlabeled pool for manual annotation, starting with a seed set of 40 labeled documents and the 1000 silver-annotated documents. The ALPNN policy is learned using the seed set of 40 labeled documents. Table 7 illustrates the increase in Micro-F1 of ALPNN as learning progresses for relation identification, concept type and boundary recognition, and attribute classification respectively evaluated using 7-fold cross-validation. We compare the learning curves produced by ALPNN with the MTADL system [6] for concept type and boundary detection as well as attribute classification and MAADL system [7] for relation identification. We also compare against random sampling using the SACAR learner (RAND). Table 7 clearly illustrates a significant increase in performance as active learning progresses using ALPNN, resulting in 55%, 13%, and 47% increases for relation identification, concept type and boundary detection, and attribute classification, respectively. The baselines produce 21%, 8% and 33% increases in the same number of active learning rounds while random sampling produces 44%, 10%, and 36% increases. The improved relative increases of ALPNN for each task compared to random sampling illustrates the effectiveness of the approach. Moreover, we found that ALPNN selects documents with slightly fewer concepts and a similar number of relations compared to random sampling. Specifically, the average document sampled by ALPNN contained 24.54 concepts and 7.57 relations while the average randomly sampled document contained 27.20 concepts with 7.89 relations. This indicates that the policy learned by ALPNN doesn’t simply bias towards longer documents with more annotations to trivially outperform random sampling.
Table 7:
Performance of the identification of concepts, their attributes, and relations between them as active learning progresses for ten rounds, measured by F1.
| Task | System | Round 1 | Round 2 | Round 3 | Round 4 | Round 5 | Round 6 | Round 7 | Round 8 | Round 9 | Round 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Concepts | RAND | 0.7949 | 0.8120 | 0.8118 | 0.8149 | 0.8268 | 0.8347 | 0.8452 | 0.8601 | 0.8597 | 0.8714 |
| MTADL | 0.8108 | 0.8188 | 0.8076 | 0.8123 | 0.8230 | 0.8305 | 0.8468 | 0.8502 | 0.8617 | 0.8756 | |
| ALPNN | 0.7949 | 0.8144 | 0.8119 | 0.8314 | 0.8449 | 0.8544 | 0.8478 | 0.8733 | 0.8718 | 0.8998 | |
| Attributes | RAND | 0.6074 | 0.6426 | 0.6619 | 0.6870 | 0.7004 | 0.7274 | 0.7607 | 0.7819 | 0.8000 | 0.8241 |
| MTADL | 0.6017 | 0.6481 | 0.6565 | 0.6528 | 0.6812 | 0.7085 | 0.7128 | 0.7420 | 0.7784 | 0.7988 | |
| ALPNN | 0.6074 | 0.6292 | 0.6670 | 0.7008 | 0.7126 | 0.7615 | 0.7803 | 0.8191 | 0.8469 | 0.8955 | |
| Relations | RAND | 0.4910 | 0.4957 | 0.5355 | 0.5498 | 0.5506 | 0.5800 | 0.6269 | 0.6500 | 0.6712 | 0.7095 |
| MAADL | 0.6340 | 0.6386 | 0.6392 | 0.6571 | 0.6843 | 0.7081 | 0.7198 | 0.7321 | 0.7640 | 0.7729 | |
| ALPNN | 0.4910 | 0.5162 | 0.5514 | 0.5685 | 0.6057 | 0.6369 | 0.6561 | 0.6833 | 0.7044 | 0.7628 |
Compared to MAADL, ALPNN starts with much lower initial performance for relation identification. This could be due to the fact that the underlying model of ALPNN, SACAR, is performing concept boundary detection and attribute classification in addition to relation identification while MAADL’s model is able to focus on relation identification alone. While the SACAR model used by ALPNN is just starting active learning, it focuses on concept boundaries and attributes as there are more concepts and attributes than relations to be identified, dominating the loss function. However, as the performance for concepts and attributes increases, so to does the performance for relation identification - to the point where the performance is comparable to that of MAADL. In this way, ALPNN is able to match the performance of the dedicated relation identification system while also out-performing another dedicated system for concept boundary and attribute identification at the same time.
6. Discussion
The experimental results indicate that the SACAR identifier is able to jointly extract medical concepts from EEG reports, classify their attributes, and detect relations between them. While SACAR is able to out-perform existing methods for concept type and boundary detection and attribute classification in EEG reports, it does not out-perform EEG-RelNet [7] for relation detection, achieving similar results. This could be due to the way EEG-RelNet represents medical concept entities, updating a series of concept- and relation-memory cells. In future work, we plan on experimenting with integrating EEG-RelNet-style concept- and relation-memory cells into SACAR.
Recall from Section 3.3 that the word embeddings used in SACAR are learned jointly along with the other parameters. The decision to learn word embeddings from scratch was made empirically. While the use of pre-trained word embeddings [42] has proved to be effective, more recent work [43, 44] has shown that pre-training entire representation layers that learn to contextualize word embeddings can be more effective. In order to determine if such a pre-training paradigm would improve SACAR, we adopted the BERT [44] pre-training procedure to pre-train the Transformer Narrative Encoder. We pre-trained the TNE on the text of the entire TUH EEG corpus for 256 epochs before incorporating it into the full SACAR system. Surprisingly, this resulted in slight performance decreases across the board. We believe this is likely due to the comparatively small size of the TUH EEG corpus – just 25,000 reports – allowing the TNE to overfit. How to best pre-train SACAR remains a question for future work with possible areas of investigation including: (a) incorporating large amounts of auxiliary clinical text; (b) the use of word-piece tokenization; and (c) fine tuning a massively pre-trained model on the EEG report domain.
The results show that the SACAR-V model which did not make use of recurrence or Adaptive Computation Time consistently performed worst among the SACAR and baseline models in concept type and boundary detection, as well as relation identification. Interestingly, SACAR-V is the only model evaluated that did not make use of recurrence. However, it should be noted that the baseline models apply recurrence sequentially, using the same recurrent cells for each input, while the SACAR-U and SACAR-A models apply recurrence in a parallel manner, using separate recurrent blocks for each input.
In order to analyze the properties of Adaptive Computation Time, we graphed the number of Transformer Narrative Encoder blocks used per word in an example sentence along with the concept type and boundary labels of each word in Figure 6. The sentence we considered is: “Male with, spells, childhood epilepsy and chronic right hemispheric infarct.” We can see that ACT tends to allocate more computation time to words at the boundaries of concept mentions. ACT allocated the maximum of twelve TNE blocks to each of words 4-10 in the sentence as those words are constantly changing boundary labels, corresponding to two concept mentions, “childhood epilepsy” and “chronic right hemispheric infarct”, separated by a single word, “and”. This supports the suggestion made in Graves et al. [37] that ACT can learn to ‘ponder’ longer on inputs that indicate implicit boundaries or transitions in sequential data.
Figure 6:

Example of the number of blocks used per word in a sentence in the Transformer Narrative Encoder, determined with Adaptive Computation Time.
To better understand the impact of multi-headed self-attention in SACAR, we visualized the attention weights for two attention heads produced by the last block of the Sentence Transformer Encoder for the example sentence: “Hyperventilation was performed and results in symmetric slowing of the background activity.” This visualization is presented in Figure 7. Each column in the Figure shows the attention distribution for each word in the vertical axis over the words in the horizontal axis. For the word “slowing”, which corresponds to a mention of an EEG Activity, we notice that the attention head 1 places high attention weights on the words: “symmetric” – indicating Attribute 6: Dispersal = Generalized; and “background” – indicating Attribute 3: Background = Yes. However, for the same word “slowing” the attention head 2 does not place a high weight on “symmetric”, attending to the attribute “background”, but it places high attention weights for the word “results”, indicating that the two heads tend to focus on different parts of the context.
Figure 7:
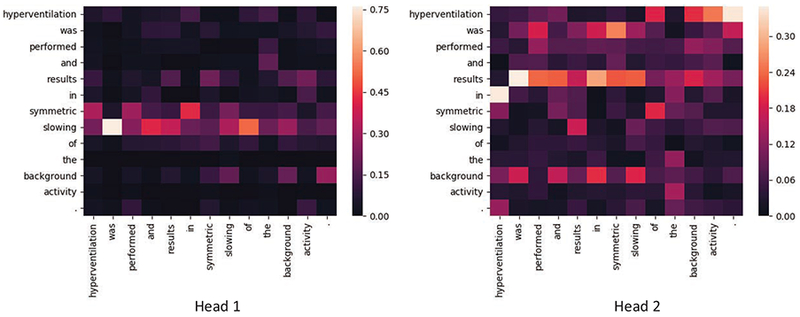
Self-Attention weights generated by the Transformer Sentence Encoder for an example sentence.
In order to better understand the SACAR model, we conducted a brief ad hoc error analysis on a few synthetic excerpts illustrating common errors. Consider the following sentence indicative of typical text found in the description section of an EEG report: “In addition, there is asymmetric anterior predominant small amplitude polyspikes and spike and waves seen generalized that at times have an EMG correlate noted in the arms and in the video on the face.” SACAR is unable to correctly identify that two EEG activity anchors are present (“polyspikes” and “spike and wave”), instead annotating a single erroneous anchor (“polyspikes and spike and waves”). This incorrect boundary identification causes a cascade of failures in both attribute classification and relation identification. Since there is only one anchor identified, the anchor is determined to have the Polyspike-complex morphology, leaving the spike-and-slow-wave complex un-annotated. Since the spike-and-slow-wave complex is not identified, the Evokes relation it has with the EEG Event “EMG correlate” is also not identified. Another source of errors for SACAR stems from the complicated – and sometimes ungrammatical – way in which EEG activities are described. For instance, we ran SACAR on the example sentence from [6]: “There are rare sharp transients noted in the record but without after going slow waves as would be expected in epileptiform sharp waves.” This sentence is meant to convey the fact that sharp waves have occurred, but not sharp-and-slow-wave-complexes which would indicate epileptiform activity. However, three anchors are identified by SACAR, “sharp transients”, “slow waves”, and “epileptiform sharp waves” with morphologies Sharp-wave, Slow-wave, and Epileptiform-discharge(unspecified), respectively. SACAR is unable to associate the text ‘slow waves” with “sharp transients” to identify that the morphology of the “slow waves” anchor should be sharp-and-slow-wave-complex. Moreover, the polarity of the epileptiform activity is not correctly identified as being negative.
As can be seen in Table 6, SACAR exhibits moderate performance in identifying Treatment-For relations due to low precision. A typical erroneous Treatment-For relation is exemplified in the following synthetic excerpt: “CLINICAL HISTORY: 50 year old with history of seizures status post code. MEDICATIONS: Dilantin, Trileptal, Keppra, Hydralazine, Ativan.” In this excerpt, all of the treatments (medications) listed other than Hydralazine are treatments for the medical problem, “seizures”. Since they share the same context, SACAR tends to link all of the treatments in Treatment-For relations with “seizures”. We believe that incorporating medical knowledge from biomedical ontologies can address this shortcoming since medications and the possible medical problems they can treat are represented in such ontologies. To that end we plan to investigate the incorporation of medical knowledge embeddings generated from both biomedical ontologies and free text into SACAR.
We believe that the automatic and simultaneous identification of medical concepts and relations in a large collection of EEG reports will enable the generation of EEG-specific knowledge embeddings of high accuracy. High-quality embeddings have been shown recently [45] to be crucial in designing relevance models that rely on deep learning, and thus produce excellent results. We intend in future work to learn knowledge embeddings from the large collection of EEG reports and use them in an enhanced MERCuRY system.
The system presented in this work has two important limitations that should be noted. The first is that both ALPNN and SACAR require substantial resources to train. ALPNN requires training S(1 + B(K + 1)) SACAR models in total, where S is the number of simulated active learning problems, B is the number of times each simulated problem is used, and K is the number examples sampled from DU at each step. In this work, learning the ALPNN policy required training 820 SACAR models (with S = 20, B = 10, K = 4) over 1 week on a single 8GB GTX 1070. Likewise, the final SACAR model itself is comprised of 12,346,589 parameters - which is relatively small for a Transformer-based model. Recent work [44, 46, 47] indicates that, given enough data, the performance of Transformer-based models, like SACAR, can be further improved by scaling the size of the model well beyond the 12 million parameters of this work. Second, document-level training results in fewer training examples than sentence- or word- level training from the same amount of data. Each EEG report represents a single training example for SACAR, while the same report would yield more than ten times as many examples for sentence level training, on average. In this way, document-level training necessitates the use of silver-annotated data to adequately train SACAR, requiring pre-trained models capable of producing such silver annotations.
7. Conclusion
In this paper we have presented the Self-Attention Concept, Attribute and Relation (SACAR) Identification system, capable to jointly identify EEG-specific medical concepts, their attributes and several forms of relations between concepts. We have shown how the SACAR system can be used successfully in a neural active learning framework, which enables the distillation of clinical knowledge from a large collection of EEG reports. This clinical knowledge consists of medical concepts characterized by a rich set of attributes, while concepts participating in relations are often identified at long distance in the same EEG report. The experimental results show great promise, indicating that it is possible to learn EEG-specific knowledge that could be assembled in a knowledge graph. Such a knowledge graph may inform the deep learning of EEG-specific knowledge embeddings, which can be used to enhance patient cohort retrieval systems operating on EEG clinical data.
Transformer encoders enable end-to-end, accurate knowledge extraction from EEG reports
Medical concepts, their attributes and relations between them can be extracted jointly
Joint learning enables active learning policy that selects based on all 3 tasks
Active learning policy itself can be learned with imitation learning and a seed dataset
Acknowledgments
Research reported in this publication was supported by the National Human Genome Research Institute (NHGRI) of the National Institutes of Health under award number U01HG008468, respectively. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
There are no conflicts of interest.
Public access to this large corpus of EEG reports is granted at: https://www.isip.piconepress.com/projects/tuh_eeg/
Tokenization was performed using the GENIA tagger [32] and sentence splitting was performed using OpenNLP (opennlp.apache.org).
References
- [1].Dehghani M, Gouws S, Vinyals O, Uszkoreit J, Kaiser Ł, Universal transformers, arXiv preprint arXiv:1807.03819. [Google Scholar]
- [2].Smith SJM, Eeg in the diagnosis, classification, and management of patients with epilepsy, Journal of Neurology, Neurosurgery & Psychiatry 76 (suppl 2) (2005) ii2–ii7. arXiv:https://jnnp.bmj.com/content/76/suppl_2/ii2.full.pdf, doi: 10.1136/jnnp.2005.069245. URL https://jnnp.bmj.com/content/76/suppl_2/ii2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tatum W, AM H, Benbadis S, Kaplan P, Handbook of EEG interpretation., 2007. [Google Scholar]
- [4].Beniczky S, Hirsch L, Kaplan P, Pressler R, Bauer G, Aurlien H, Brogger J, Trinka E, Unified eeg terminology and criteria for nonconvulsive status epilepticus 54 Suppl 6(2013) 28–9. [DOI] [PubMed] [Google Scholar]
- [5].Goodwin T, Harabagiu SM, Multi-modal patient cohort identification from eeg report and signal data, in: AMIA Annual Symposium Proceedings, Vol. 2016, 2016, pp. 1794–1803. [PMC free article] [PubMed] [Google Scholar]
- [6].Maldonado R, Goodwin TR, Harabagiu SM, Active deep learning-based annotation of electroencephalography reports for cohort identification, in: AMIA Summits on Translational Science Proceedings, Vol. 2017, American Medical Informatics Association, 2017, pp. 229–238. [PMC free article] [PubMed] [Google Scholar]
- [7].Maldonado R, Goodwin T, Harabagiu SM, Memory-augmented active deep learning for identifying relations between distant medical concepts in electroencephalography reports, in: AMIA Joint Summits on Translational Science proceedings. AMIA Joint Summits on Translational Science, 2018. [PMC free article] [PubMed] [Google Scholar]
- [8].Cui L, Bozorgi A, Lhatoo S, Zhang G-Q, S Sahoo S, Epidea: Extracting structured epilepsy and seizure information from patient discharge summaries for cohort identification, AMIA … Annual Symposium proceedings / AMIA Symposium. AMIA Symposium 2012 (2012) 1191–200. [PMC free article] [PubMed] [Google Scholar]
- [9].Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG, Mayo clinical text analysis and knowledge extraction system (ctakes): architecture, component evaluation and applications, Journal of the American Medical Informatics Association 17 (5) (2010) 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG, A simple algorithm for identifying negated findings and diseases in discharge summaries, Journal of biomedical informatics 34 (5) (2001) 301–310. [DOI] [PubMed] [Google Scholar]
- [11].Sahoo SS, Lhatoo SD, Gupta DK, Cui L, Zhao M, Jayapandian C, Bozorgi A, Zhang G-Q, Epilepsy and seizure ontology: towards an epilepsy informatics infrastructure for clinical research and patient care, Journal of the American Medical Informatics Association 21 (1) (2013) 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cui L, Sahoo SS, Lhatoo SD, Garg G, Rai P, Bozorgi A, Zhang G-Q, Complex epilepsy phenotype extraction from narrative clinical discharge summaries, Journal of biomedical informatics 51 (2014) 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Aronson AR, Effective mapping of biomedical text to the umls metathesaurus: the metamap program, in: Proceedings of the AMIA Symposium, American Medical Informatics Association, 2001, p. 17. [PMC free article] [PubMed] [Google Scholar]
- [14].Taylor SJ, Harabagiu SM, The role of a deep-learning method for negation detection in patient cohort identification from electroencephalography reports, in: AMIA Annual Symposium Proceedings, Vol. 2018, American Medical Informatics Association, 2018, p. 1018. [PMC free article] [PubMed] [Google Scholar]
- [15].Goodwin TR, Harabagiu SM, Deep learning from eeg reports for inferring underspecified information, AMIA Summits on Translational Science Proceedings 2017 (2017) 112. [PMC free article] [PubMed] [Google Scholar]
- [16].Goodwin TR, Harabagiu SM, Inferring clinical correlations from eeg reports with deep neural learning, in: AMIA Annual Symposium Proceedings, Vol. 2017, American Medical Informatics Association, 2017, p. 770. [PMC free article] [PubMed] [Google Scholar]
- [17].Maldonado R, Goodwin TR, Skinner MA, Harabagiu SM, Deep learning meets biomedical ontologies: knowledge embeddings for epilepsy, in: AMIA Annual Symposium Proceedings, Vol. 2017, American Medical Informatics Association, 2017, p. 1233. [PMC free article] [PubMed] [Google Scholar]
- [18].Hahn U, Beisswanger E, Buyko E, Faessler E, Active learning-based corpus annotation–the pathojen experience, in: AMIA Annual Symposium Proceedings, Vol. 2012, American Medical Informatics Association, 310M, p. 301. [PMC free article] [PubMed] [Google Scholar]
- [19].Chen Y, Carroll RJ, McPeek Hinz ER, Shah A, Eyler AE, Denny J, Xu H, Applying active learning to high-throughput phenotyping algorithms for electronic health records data, Journal of the American Medical Informatics Association : JAMIA 20. doi: 10.1136/amiajnl-2013-001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dligach D, Miller T, Savova G, Active learning for phenotyping tasks, in: Proceedings of the Workshop on NLP for Medicine and Biology associated with RANLP 2013, INCOMA Ltd. Shoumen, BULGARIA, Hissar, Bulgaria, 2013, pp. 1–8. URL https://www.aclweb.org/anthology/W13-5101 [Google Scholar]
- [21].Ji Z, Wei Q, Franklin A, Cohen T, Xu H, Cost-sensitive active learning for phenotyping of electronic health records, in: 2019 AMIA Informatics Summit Proceedings, Vol. 2019, American Medical Informatics Association, 838, p. 829. [PMC free article] [PubMed] [Google Scholar]
- [22].Chen Y, Cao H, Mei Q, KAi Z, Xu H, Applying active learning to supervised word sense disambiguation in medline, Journal of the American Medical Informatics Association 20 (5) (2013) 1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nguyen DHM, Patrick J, Supervised machine learning and active learning in classification of radiology reports, Journal of the American Medical Informatics Association 21 (2014) 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Uzuner Ö, South BR, Shen S, DuVall SL, 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text, Journal of the American Medical Informatics Association 18 (5) (2011) 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun W, Rumshisky A, Uzuner O, Evaluating temporal relations in clinical text: 2012 i2b2 challenge, Journal of the American Medical Informatics Association 20 (5) (2013) 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Noachtar S, Binnie CA, Ebersole JS, Mauguière F, Sakamoto A, Westmoreland BF, A glossary of terms most commonly used by clinical electroencephalographers and proposal for the report form for the eeg findings. the international federation of clinical neurophysiology., Electroencephalogr Clin Neurophysiol Suppl 52. [PubMed] [Google Scholar]
- [27].Bodenreider O, The unified medical language system (umls): integrating biomedical terminology, Nucleic acids research 32 (suppl 1) (2004) D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Verga P, Strubell E, McCallum A, Simultaneously self-attending to all mentions for full-abstract biological relation extraction, in: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), Association for Computational Linguistics, 2018, pp. 872–884. [Google Scholar]
- [29].Paulus R, Xiong C, Socher R, A deep reinforced model for abstractive summarization, in: International Conference on Learning Representations, 2018. URL https://openreview.net/forum?id=HkAClQgA- [Google Scholar]
- [30].Parikh A, Täckström O, Das D, Uszkoreit J, A decomposable attention model for natural language inference, in: Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Association for Computational Linguistics, Austin, Texas, 2016, pp. 2249–2255. URL https://aclweb.org/anthology/D16-1244 [Google Scholar]
- [31].Lin Z, Feng M, dos Santos CN, Yu M, Xiang B, Zhou B, Bengio Y, A structured self-attentive sentence embedding, CoRR. [Google Scholar]
- [32].Tsuruoka Y, Tateishi Y, Kim J-D, Ohta T, McNaught J, Ananiadou S, Tsujii J, Developing a robust part-of-speech tagger for biomedical text, in: Panhellenic Conference on Informatics, Springer, 2005, pp. 382–392. [Google Scholar]
- [33].Kingma DP, Ba J, Adam: A method for stochastic optimization, arXiv preprint arXiv:1412.6980. [Google Scholar]
- [34].He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition, in: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016, pp. 770–778. [Google Scholar]
- [35].Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I, Attention is all you need, in: NIPS, 2017. [Google Scholar]
- [36].Ba JL, Kiros JR, Hinton GE, Layer normalization, arXiv preprint arXiv:1607.06450. [Google Scholar]
- [37].Graves A, Adaptive computation time for recurrent neural networks, arXiv preprint arXiv:1603.08983. [Google Scholar]
- [38].Das R, Neelakantan A, Belanger D, McCallum A, Chains of reasoning over entities, relations, and text using recurrent neural networks, in: Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics: Volume 1, Long Papers, Vol. 1, 2017, pp. 132–141. [Google Scholar]
- [39].Liu M, Buntine W, Haffari G, Learning how to actively learn: A deep imitation learning approach, in: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Association for Computational Linguistics, 2018, pp. 1874–1883. [Google Scholar]
- [40].Ross S, Gordon G, Bagnell D, A reduction of imitation learning and structured prediction to no-regret online learning, in: Proceedings of the fourteenth international conference on artificial intelligence and statistics, 2011, pp. 627–635. [Google Scholar]
- [41].Levandowsky M, Winter D, Distance between sets, Nature 234 (5323) (1971) 34. [Google Scholar]
- [42].Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J, Distributed representations of words and phrases and their compositionality, in: Advances in neural information processing systems, 2013, pp. 3111–3119. [Google Scholar]
- [43].Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, Zettlemoyer L, Deep contextualized word representations, arXiv preprint arXiv:1802.05365. [Google Scholar]
- [44].Devlin J, Chang M-W, Lee K, Toutanova K, Bert: Pre-training of deep bidirectional transformers for language understanding, arXiv preprint arXiv:1810.04805. [Google Scholar]
- [45].Jameel S, Bouraoui Z, Schockaert S, Member: Max-margin based embeddings for entity retrieval, in: Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval, SIGIR ‘17, ACM, New York, NY, USA, 2017, pp. 783–792. [Google Scholar]
- [46].Radford A, Wu J, Child R, Luan D, Amodei D, Sutskever I, Language models are unsupervised multitask learners.
- [47].Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov R, Le QV, Xlnet: Generalized autoregressive pretraining for language understanding, arXiv preprint arXiv:1906.08237. [Google Scholar]