Abstract
The present study defines RNA-dependent amplification of βAPP mRNA as a molecular basis of beta-amyloid overproduction in Alzheimer’s disease. In this process, βAPP mRNA serves as a template for RNA-dependent RNA polymerase, RdRp complex. The resulting antisense RNA self-primes its extension utilizing two complementary elements: 3’-terminal and internal, located within an antisense segment corresponding to the coding portion of βAPP mRNA. The extension produces 3’-terminal fragment of βAPP mRNA, a part of a hairpin-structured antisense/sense RNA molecule. Cleavage at the 3’ end of the hairpin loop produces RNA end product encoding a C-terminal fragment of βAPP. Since each conventional βAPP mRNA can be used repeatedly as a template, the process constitutes an asymmetric mRNA amplification. The 5’-most translation initiation codon of the amplified mRNA is the AUG preceding immediately and in-frame the Aβ-coding segment. Translation from this codon overproduces Aβ independently of βAPP. Such process can occur in humans but not in mice and other animals where segments of βAPP antisense RNA required for self-priming have little, if any, complementarity. This explains why Alzheimer’s disease occurs exclusively in humans and implies that βAPP mRNA amplification is requisite in AD. In AD, therefore, there are two pathways of beta-amyloid production: βAPP proteolytic pathway and βAPP mRNA amplification pathway independent of βAPP and insensitive to beta-secretase inhibition. This implies that in healthy humans, where only the proteolytic pathway is in operation, Aβ production should be suppressed by the BACE inhibition, and indeed it is. However, since βAPP-independent pathway operating in AD is by far the predominant one, BACE inhibition has no effect in Alzheimer’s disease. It appears that, physiologically, the extent of beta-amyloid overproduction sufficient to trigger amyloid cascade culminating in AD requires asymmetric RNA-dependent amplification of βAPP mRNA and cannot be reached without it. In turn, the occurrence of mRNA amplification process depends on the activation of inducible components of RdRp complex by certain stresses, for example the ER stress in case of amplification of mRNA encoding extracellular matrix proteins. In case of Alzheimer’s disease, such an induction appears to be triggered by stresses associated with mitochondrial dysfunction, a phenomenon closely linked to AD. The cause-and-effect relationships between mitochondrial dysfunction and AD appear to be very different in familial, FAD, and sporadic, SAD cases. In FAD, increased levels or more toxic species of Aβ resulting from the abnormal proteolysis of βAPP trigger mitochondrial dysfunction, activate mRNA amplification and increase the production of Aβ, reinforcing the cycle. Thus in FAD, mitochondrial dysfunction is an intrinsic component of the amyloid cascade. The reverse sequence is true in SAD where aging-related mitochondrial dysfunction activates amplification of βAPP mRNA and enhances the production of Aβ. This causes further mitochondrial dysfunction, the cycle repeats and degeneration increases. Thus in SAD, the initial mitochondrial dysfunction arises prior to the disease, independently of and upstream from the increased Aβ production, i.e. in SAD, mitochondrial pathology hierarchically supersedes Aβ pathology. This is the primary reason for the formulation of the Mitochondrial Cascade Hypothesis. But even in terms of the MCH, the core of the disease is the amyloid cascade as defined in the amyloid cascade hypothesis, ACH. The role of mitochondrial dysfunction in relation to this core is causative in SAD and auxiliary in FAD. In FAD, the initial increase in the production of Aβ is mutations-based and occurs relatively early in life, whereas in SAD it is coerced by an aging-contingent component, but both lead to mechanistically identical self-perpetuating mutual Aβ/mitochondrial dysfunction feedback cycles, an engine that drives, via RNA-dependent βAPP mRNA amplification, overproduction of beta-amyloid and, consequently, AD; hence drastic difference in the age of onset, yet profound pathological and symptomatic similarity in the progression, of familial and sporadic forms of Alzheimer’s disease. Interestingly, the recent findings that mitochondrial microprotein PIGBOS interacts with the ER in mitigating the unfolded protein response indicate a possible connection between mitochondrial dysfunction and ER stress, implicated in activation of RNA-dependent mRNA amplification pathway. The possible involvement of mitochondrial dysfunction in βAPP mRNA amplification makes it a promising therapeutic target. Recent successes in mitigating, and even reversing, Aβ-induced metabolic defects with anti-diabetes drug metformin are encouraging in this respect.
Keywords: Alzheimer’s disease, Mitochondrial dysfunction, RNA-dependent βAPP mRNA amplification, Therapeutic strategies
Conventional Pathway of Beta-Amyloid Generation
Beta-amyloid, Aβ, the peptide associated with and widely believed to have a pivotal early role in etiology of Alzheimer’s Disease (AD), is generated ostensibly by proteolytic cleavages of a much larger molecule, beta-amyloid precursor protein, βAPP. In the nonamyloidogenic proteolytic pathway, a cleavage by the alpha-secretase enzyme occurs within the Aβ-containing segment of βAPP, thus preventing the generation of beta-amyloid. In the amyloidogenic proteolytic pathway, two sequential cleavages of βAPP are involved in the production of Aβ. The first is a cleavage of βAPP by the beta-secretase enzyme. It occurs between residues 671 and 672 of the βAPP molecule (isoform 770 numbering), generating the N-terminus of Aβ, yielding the 12kDa membrane-bound C-terminal fragment, C99 (residues 672–770), releasing a large ectodomain of βAPP, soluble sAPPβ (residues 1–671), and precluding activity of alpha-secretase which cleaves βAPP within its Aβ-containing segment but cannot cut within C99 or Aβ [1–3]. The second cleavage, by gamma-secretase activity, occurs at one of multiple sites within C99 and generates the C-terminus of Aβ. Thus released, Aβ is secreted from the cell. The size of Aβ ranges from 36 to 43 amino acids, with Aβ40 being the most abundant species normally formed. Studies of the inherited forms of the disease, FAD (Familial Alzheimer’s Disease), strongly indicated that cerebral Aβ accumulation is essential for and underlies the etiology of the disease [4–6]. This notion, formalized in a theory of AD known as “Amyloid Cascade Hypothesis”, ACH [7–12], has become the dominant model of AD pathogenesis and has guided the development of potential treatments. Most therapeutic strategies attempted to date have been based on this model and virtually all preclinical tests and clinical trials discussed below have been designed within the framework of ACH. Several alternative theories of AD have also been proposed. They include Mitochondrial Cascade Hypothesis, MCH, reviewed in [13] and discussed below, Presenilin Hypothesis, PSH, and βAPP Matrix Approach, AMA, both reviewed in [14]. Over two hundred autosomal dominant mutations associated with FAD have been identified in genes for βAPP and presenilins, the components of gamma-secretase complex [6]. In βAPP gene most of the mutations cluster around alpha-, beta-, and gamma-secretases cleavage sites and increase either the production of total Aβ or the relative proportion of a more neurotoxic 42-residue form of Aβ, Aβ42. In terms of the ACH, there is little doubt that abnormal processing of βAPP and increased production of total Aβ or its 42-amino acid isoform are pivotal events in the pathogenesis of FAD. Although the number of individuals affected by FAD is substantial, in relative terms this form of the disease is quite rare, representing less than 5%, in fact less than 1% by some estimates, of the total Alzheimer’s disease burden [5,14,15]. Since the pathological lesions and symptoms in the non-hereditary form of the disease, SAD (Sporadic Alzheimer’s Disease), are analogous to those seen in the familial forms, it has been assumed that abnormal amyloidogenic proteolytic processing of βAPP of a type(s) seen in FAD also underlies the pathogenesis of SAD [4,5]. The assumption that ACH applies to both forms of AD implied that any therapeutic approach effective in FAD would also be successful in treatment of SAD.
Inhibition of Beta-Secretase is Very Efficient in Preclinical Studies and Animal Models but Completely Ineffective in Alzheimer’s Disease. Can Aβ be Produced Independently of βAPP in AD?
Success of BACE (Beta-site APP-Cleaving Enzyme) inhibitors in preclinical tests
In the framework of ACH, it was understood early on that inhibition of Aβ production might benefit affected individuals. In light of the above discussion, beta-secretase activity was viewed as a strategic target of choice: Inhibit beta-secretase cleavage and there is no beta-amyloid. Moreover, such inhibition would shift the equilibrium between alpha- and beta-secretase cleavages toward the former, thus augmenting its efficacy. Therefore, since the identification of BACE as beta-secretase [16–18], it became the primary therapeutic target for treatment of AD. Designing BACE-inhibiting agents presented major challenges of cell penetration, oral bioavailability, metabolic clearance, and brain access, but intense efforts, mainly by the pharmaceutical industry, led to development of a number of brain-penetrant small-molecule BACE inhibitors that have been vigorously investigated. The results obtained in the early investigations of BACE inhibition, first appearing around 2007 [19–27], are truly striking. As an example, Merck researchers reported in 2012 the discovery of “compound 16”, which robustly reduced cortex and CSF levels of Aβ when administered orally to rats [28]. Continuous efforts to improve upon “compound 16” culminated in the development of verubecestat [MK-8931]. Preclinical tests of this agent achieved dramatic results [29]. Levels of Aβ and sAPPβ were reduced by up to 90% in plasma, brain, and CSF after even a single administration of verubecestat to healthy subjects including rats, monkeys, and humans [29]. The acute reduction of over 80% in CSF and cortical Aβ and sAPPβ produced by verubecestat was maintained after chronic administration for 9 months in monkeys [29]. Because of its favorable initial safety profile and its ability to markedly reduce cerebral and CSF Aβ and sAPPβ concentrations, verubecestat was the first BACE inhibitor to progress to phase III clinical trials. Preclinical evaluation of a number of independently developed BACE inhibitors, such as BI 1181181, LY2811376, LY 2886721, AZD3293 (lanabacestat, LY3314814), CNP520, E2609 (elenbacestat), JNJ-54861911, CTS-21166, HPP854, PF-05297909, RG7129, TAK-070, VTP-37948 yielded similarly impressive results in animals and healthy volunteers and all these agents have entered clinical trials.
Inhibition of Beta-Secretase Activity Rescues Functional Impairments in Animal Models of Alzheimer’s Disease
With the ability to significantly reduce the production and lower the levels of Aβ thus established, the question remained whether such a reduction would translate into a “treatment” of the disease. This question was answered resolutely and convincingly, with animal models bioengineered to mimic FAD, in two recent studies using different approaches to inhibit beta-secretase activity. One study utilized BACE inhibitor NB-360 [30]. It was based on a previous study [31] showing NB-360 to be a potent, brain penetrable BACE inhibitor capable of completely blocking Aβ deposition in the brains of βAPP transgenic mice, as well as of rats and dogs. Moreover, this inhibitor blocked accumulation of activated inflammatory cells in the brains of βAPP transgenic mice. The more recent study with NB-360 [30] further assessed the notion that suppression of Aβ production can have beneficial downstream effects on the progression of Alzheimer’s disease. Using histochemistry, in vivo Ca2+ imaging, and behavioral analyses in a mouse model of AD, the authors demonstrated that along with reducing prefibrillary Aβ surrounding plaques, the inhibition of BACE activity rescued neuronal hyperactivity, impaired long-range circuit function and memory defects. That all these effects were due to inhibition of Aβ production was strongly indicated by the observation that functional neuronal impairments reappeared after infusion of soluble Aβ [30].
In the second study [32], mimicking BACE1 inhibition in adult organisms, the authors generated BACE1 conditional knockout (BACE1fl/fl) mice and bred them with ubiquitin-Cre mice to induce deletion of BACE1 after passing early developmental stages. Strikingly, sequential deletions of BACE1 in an adult AD mouse model were capable of reversing amyloid deposition and resulted in significant improvement in gliosis and neuritic dystrophy. Moreover, in correlation with amyloid plaque reversal, it also significantly improved synaptic functions, as was determined by long-term potentiation and contextual fear conditioning experiments. These studies offered great hope that sustained inhibition of BACE1 activity can constitute a treatment, or at least be beneficial, for AD patients. This assumption was tested in several clinical trials.
BACE Inhibition is Completely Ineffective in Alzheimer’s Disease
The results of clinical trials of BACE inhibitors in humans, however, do not support this assumption. All BACE inhibitor clinical trials that ended to date, ended in failure. Some trials, such as that of BI 1181181, LY2811376, LY2886721 and RG7129, were terminated because of technical and safety issues. On the other hand, there were no such issues in the trials of the verubecestat (MK-8931). This agent was shown to be very efficient in suppressing Aβ production in preclinical tests and was proven safe in clinical trials. Yet, its Phase III, 2000 patient-strong “EPOCH” trial in mild to moderate AD patients was terminated prematurely in February 2017 for the lack of efficacy, with an interim analysis by an external data-monitoring committee giving the trial “virtually no chance of finding a positive effect”. At that time, a separate large Phase III clinical trial of verubecestat in prodromal AD patients, the “APECS”, set to run until 2019, was continued as investigators found no signs of safety issues. In February 2018, however, this trial, too, was terminated prematurely and for the same reason; lack of efficacy. The clinical trials of several other BACE inhibitors are still in progress but the verubecestat results do not inspire confidence in their successful outcome.
Results of Clinical Trials in Humans can be Explained by βAPP-Independent and therefore BACE Inhibition-Insensitive Generation of Beta-Amyloid in Alzheimer’s Disease
One rational explanation for the strikingly different effects of BACE inhibition in animal models and healthy human subjects and in Alzheimer’s disease is that in the majority of AD cases, in addition to conventional βAPP/beta-secretase-dependent component of Aβ production that operates in AD models (and in healthy human subjects), there is another, unconventional, Aβ-generating component in operation, possibly facilitated or enabled by epigenetic changes associated with the disease [33], which is βAPP–independent and thus bypasses the requirement for beta-secretase activity. In these AD cases, administration of safe and effective BACE inhibitors would suppress the βAPP-dependent component, but would have no effects on the second, βAPP- and beta-secretase-independent, component. The extent of suppression of total Aβ production by BACE inhibitors would depend on the relative input of two components in the generation of Aβ, and if the input of the second significantly exceeds that of the first component, BACE inhibitors would be ineffective both in lowering Aβ levels and in the treatment of Alzheimer’s disease.
The AUG Encoding Met671 in βAPP mRNA is a Key to βAPP-Independent Generation of Aβ in Alzheimer’s Disease
The proposed mechanism for a βAPP-independent overproduction of Aβ in Alzheimer’s disease [34–36] is based on two foundations. The first is an intriguing possibility suggested by the primary structure of the human βAPP mRNA. In this molecule, the Aβ-encoding segment is preceded immediately and in-frame by the AUG codon normally encoding methionine in position 671 of the βAPP (isoform 770 numbering). If translation were initiated at this position, it would produce 12kDa C-terminal βAPP fragment (C99, after the removal of methionine by the N-terminal methionine aminopeptidase) independently of βAPP. Interestingly, the AUG in question is situated within a nucleotide context optimal for the initiation of translation (an “A” in position −3 and a “G” in position +4 relative to the “A” of the AUG codon). In fact, of the twenty AUG codons encoding methionine residues in the βAPP mRNA, only the AUG encoding Met671 (not even the AUG encoding Met1) is located within an optimal translation initiation context. Such favorable positioning of the AUG encoding Met671 of βAPP was the basis for a proposal by Breimer and Denny that in Alzheimer’s disease C99 βAPP fragment may be generated independently from βAPP by the internal initiation of translation of the intact βAPP mRNA [37]. Such precursor-independent generation of C99 would be an efficient way to overproduce Aβ. This is because (a) C99 is not susceptible to the alpha-secretase cleavage [1–3], (b) cleavage by gamma-secretase was shown to be not the rate-limiting step in the production of Aβ [1–3], and (c) Aβ produced from C99 and either containing or lacking signal sequence were shown to be secreted with similar efficiency [2]. The possibility of internal initiation of translation, proposed by Breimer and Denny, has been subsequently ruled out by experiments of Citron and co-investigators [38]. There is, however, another possible utilization of the AUG in question as a translation initiation codon, namely, the generation of a severely 5’-truncated βAPP mRNA in which the AUG encoding Met671in the intact mRNA becomes the first AUG codon. As described below, such molecule can be generated by a process known as “chimeric RNA-dependent mRNA amplification” [47–49], a process that constitutes the second foundation of βAPP-independent overproduction of Aβ in Alzheimer’s disease.
Chimeric RNA-Dependent mRNA Amplification Pathway
De novo production of RNA on an RNA template, a process known as RNA-dependent RNA synthesis (RdRs) and the enzymatic activity conducting it, RNA-dependent RNA polymerase (RdRp), were initially considered to be exclusively virus-specific. Eventually, however, the occurrence of RdRs and the ubiquitous presence of conventional RdRp were demonstrated in numerous eukaryotic organisms [39]. The evidence that the enzymatic machinery capable of RdRs is present in mammalian cells was derived from studies of viruses, such as hepatitis delta virus, HDV, that do not encode RdRp, yet undergo a robust RNA replication once inside the mammalian host [39–41]; thus firmly establishing its occurrence and functionality. Moreover, it became clear that RdRp activity, apparently in a non-conventional form [42–44], is constitutively present in most, if not in all, mammalian cells. Because such activity was shown to produce short transcripts, because of its apparent involvement in RNA interference phenomena, and because double-stranded RNA is known to trigger cellular responses leading to its degradation, it was generally assumed that its role in mammalian cells is restricted to a regulatory function. However, at the same time, an enzymatic activity capable of generating complete antisense RNA complements of mRNAs was discovered in mammalian cells undergoing terminal differentiation [45]. Moreover, observations of widespread synthesis of antisense RNA initiating at the 3’poly(A) of mRNAs in human cells [46] suggested an extensive cellular utilization of mammalian RdRp activity. These results led to the development of a model of RdRp-facilitated and antisense RNA-mediated amplification of mammalian mRNA [47–49]. Recent detection of the major model-predicted identifiers, chimeric RNA intermediates containing both sense and antisense RNA strands covalently joined in a rigorously predicted and uniquely defined manner [49,50], as well as the identification of a putative chimeric RNA end product of this process [49], validated the proposed model.
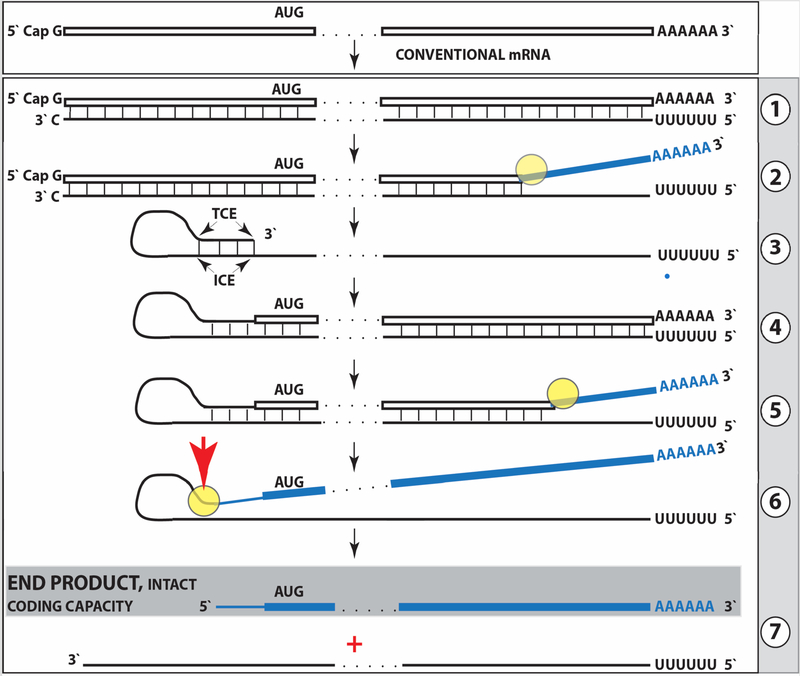
The process of RNA-dependent amplification of mammalian mRNA is diagrammed in Figure 1 below and can be briefly summarized as follows. The amplification process occurs in the cytoplasm and starts with transcription of the antisense complement from a conventional spliced mRNA template, initiating at the 3’poly(A), possibly with the help of a uridilated protein, as seen in viral RdRs [51] (Figure 1, Step 1), and terminating at the 3’end with the “C”, a transcript of the capG of mRNA [48–50]. Generation of a complete antisense transcript requires the presence of an eligible RNA template and a compatible polymerase activity. The only major prerequisite for a potential RNA template appears to be the presence of the poly(A) segment at its 3’ terminus [46–49]. The compatible polymerase activity is RdRp. The RdRp activity in mammalian cells appears to be non-conventional; two possible candidates for this role are the RNA polymerase II complex or its components [42,43] and RdRp activity of the TERT complex [44], both ubiquitously present in all cells.
Figure 1: Projected stages of the chimeric pathway of RdRp-facilitated, antisense RNA-mediated amplification of mammalian mRNA.
Top panel: Conventional, genome-originated mRNA molecule. Bottom panel: Projected stages of antisense RNA-mediated mRNA amplification. Boxed line – sense strand RNA. Single line – antisense strand RNA. “AUG” – functional translation initiation codon (could be other than “AUG”). “TCE”– 3’-terminal complementary element; “ICE”– internal complementary element, both on the antisense RNA strand. Yellow circle – helicase/modifying activity complex. Blue lines (both single and boxed) – RNA strand modified and separated from its complement by a helicase complex. Red arrowhead – position of cleavage of the chimeric intermediate. Step 1: Synthesis of antisense strand; step 2: Strand separation; step 3: Folding of antisense strand into self-priming configuration; step 4: Extension of self-primed antisense RNA; step 5: Strand separation; step 6: Cleavage of the chimeric intermediate; stage 7: End-products of amplification. Note that chimeric RNA end product retains the intact coding capacity of conventional mRNA.
Under regular circumstances the RdRp activity in mammalian cells produces only short antisense RNA transcripts. For example, a widespread synthesis of diverse short antisense RNA transcripts initiating at the 3’poly(A) of mRNA was observed in human cells [46]. On the other hand, RdRp activity isolated from rabbit reticulocytes [45] was able to produce, in assays, long antisense RNA transcripts. Subsequent studies identified full-length antisense transcripts of globin mRNA in erythroid cells [47–49]. It could be argued that the component responsible for the production of long antisense transcripts in mammalian cells is a processivity conferring co-factor of RdRp activity that is induced under special circumstances when overproduction of specific proteins is required [48–50]. The notion of a processivity co-factor is strongly supported by studies of HDV replication in “normal” (i.e. apparently lacking processivity co-factor) mammalian cells [39–41]. Within the framework of the above considerations, the ability of RdRp-deficient viruses to use RdRp activity of mammalian cell for their replication implies that they should encode a processivity co-factor of cellular RdRp. In case of HDV, it appears to be hepatitis delta antigen HDAg, the only protein encoded by HDV. HDAg is essential both for production of long transcripts by cellular RdRp, and for viral replication [41]. In its absence only short transcripts are generated [41]. These observations provide a proof of concept for the notion of RdRp processivity co-factor, central for our understanding of mammalian mRNA amplification. Identification of a cellular homolog of HDAg, DIPA [52,53] suggests directions for a search for the cellular RdRp processivity co-factor.
The resulting double-stranded sense/antisense structure is then separated into single-stranded molecules by a helicase activity that mounts the poly(A) segment of the 3’poly(A)-containing strand (the sense-oriented strand) of the double-helical structure and proceeds along this strand modifying, on average, every fifth nucleotide in the process [48,49] (Figure 1, Step 2). Only purines, the “A” and the “G” appear to be modified in the separation/modification process [48,49]. The 5’ poly(U)-containing antisense strand remains unmodified during and after the separation [47–49]; this being essential, as described below, for the production of a new sense strand since modifications were shown to interfere with and would preclude complementary interactions required in this process [48,49].
The vast majority of mammalian mRNA species contains 3’ -terminal poly(A) segments. The notion that many, or possibly most, of them could be eligible templates for RdRp was suggested in our previous studies [47–49]. Subsequent observations by Kapranov et al. [46] showed a widespread synthesis of antisense RNA initiating, apparently indiscriminately, at the 3’ poly(A) of mRNA in human cells. This, seemingly undiscerning, RdRp template eligibility of the bulk of mammalian mRNA species raises questions with regard to mechanisms underlying the manifestly stringent specificity of the mRNA amplification process [47–49]. The specificity of mRNA amplification appears to be determined at the 3’ terminus of an antisense transcript by its ability or inability to support production of a complementary sense strand RNA molecule; the end product of the amplification process.
The generation of a sense strand on an antisense template occurs via the extension of the 3’ terminus of a self-primed antisense template and requires the presence within the antisense transcript of two spatially independent complementary elements [48,49,54]. One of these is the strictly 3’-Terminal Complementary Element (TCE), the other is the Internal Complementary Element (ICE). These elements (Figure 1, Step 3) must be complementary to a sufficient extent to form a priming structure but may contain mismatches and utilize unconventional G/U pairings. The generation of a sense strand also requires the thermodynamic feasibility, enhanced/enabled by the occurrence of two complementary and topologically compatible elements, of the antisense strand folding into a self-priming configuration.
Provided that a self-priming structure is formed, the 3’ end of the folded antisense strand is extended by RdRp into a sense-oriented molecule terminating with the poly(A) at the 3’end (Figure 1, Step 4), thus generating a hairpin-structured chimeric intermediate consisting of covalently joined sense and antisense strands. The double-stranded portion of the resulting structure is separated by a helicase activity invoked above, which mounts the 3’poly(A) of a newly synthesized sense strand component of the chimeric intermediate and proceeds along this strand in the 5’ direction modifying the molecule as it advances (Figure 1, Step 5). When the helicase activity reaches a single-stranded portion of the hairpin structure, it, or associated activities, cleave the molecule either within the TCE, at a TCE/ICE mismatch, or immediately upstream of the TCE; the cleavage occurs between the 5’ hydroxyl group and the 3’ phosphate [48,49] (red arrowhead, Figure 1, Step 6).
Strand separation, in conjunction with the cleavage, produces two single-stranded molecules (Figure 1, Step 7) one of which is a chimeric mRNA, the functional mRNA end product of amplification and the basis for defining this pathway as the “chimeric”. The chimeric nature of this end product is due to the presence at its 5’ end of a 3’-terminal segment of the antisense strand consisting, depending on the site of cleavage of the chimeric intermediate, of either the entire TCE or a portion thereof covalently attached, in a 5’ to 3’ orientation, to the 5’-truncated sense strand. This chimeric molecule is modified and 3’ polyadenylated. In contrast to conventional mRNA that can be repeatedly used as RdRp template [48,49], it cannot be further amplified because its antisense complement would be lacking the TCE but can be translated into conventional mRNA-encoded polypeptide [48,49]. In the chimeric pathway of mRNA amplification, the cleavage of the chimeric intermediate following the strand separation and the associated modification of its poly(A)-containing component of the double-stranded structure is the ultimate act in the generation of the chimeric mRNA end product. Consequently, it is formed already modified and is never present in the unmodified form. Therefore, because the modified amplified RNA is resistant to reverse transcription [48,49], it cannot be detected by conventional reverse transcription-based sequencing methods.
The chimeric RNA-dependent mRNA amplification process illustrated in Figure 1 above results in an mRNA molecule containing the entire protein-coding region of a conventional, genome-transcribed, mRNA and can be translated into the original, conventional mRNA-encoded, polypeptide as was described elsewhere [48,49]. If this process were to apply to βAPP mRNA, it would result in the complete βAPP polypeptide. However, in the proposed scenario of βAPP-independent generation of beta-amyloid in Alzheimer’s disease, the expected translational outcome is a segment of amyloid precursor protein containing Aβ at its N-end. Such an outcome can indeed be achieved in an asymmetric RNA-dependent mRNA amplification pathway.
Asymmetric RNA-Dependent mRNA Amplification Pathway
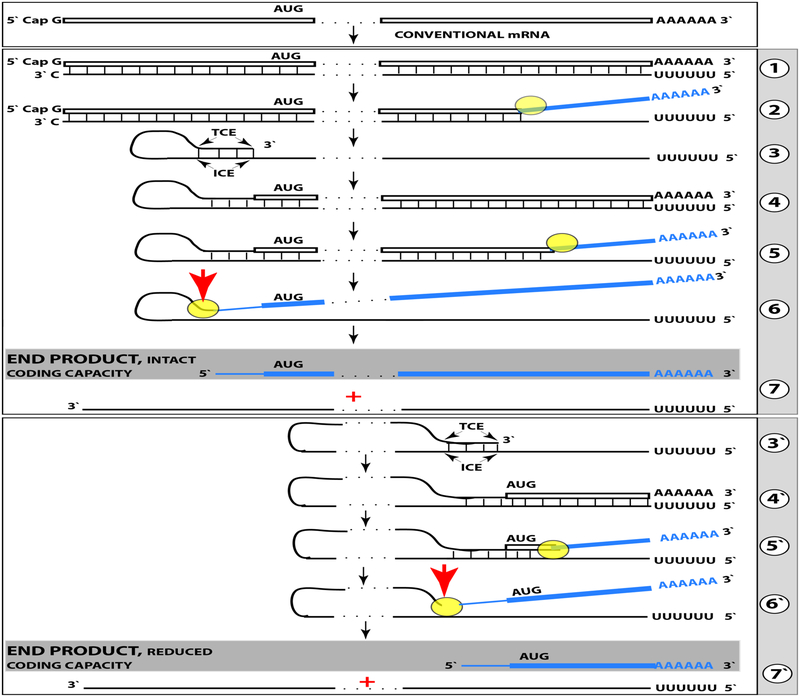
Previous section discussed a situation where both complementary elements required for an appropriate folding and self-priming of the antisense strand, TCE and ICE, are located within its segment corresponding to the 5’UTR of a conventional genome-encoded mRNA. In such a situation, depicted in steps 3 trough 7 of Figure 2, the chimeric RNA end product contains the entire protein coding region of a conventional mRNA and can be translated into the original, conventional mRNA-encoded, polypeptide. In the chimeric mRNA amplification pathway, the position of the TCE within the antisense molecule is fixed; it is always strictly 3’-terminal. In contrast, the intramolecular location of the ICE is variable, and potentially it can be positioned within a segment of the antisense strand corresponding to the coding portion of an mRNA, a scenario diagrammed in steps 3’ trough 7’ of Figure 2. In this scenario, the chimeric RNA end product would consist of a 3’-terminal segment of the antisense strand (the TCE or its fraction) and a 3’ portion of a conventional mRNA progenitor with a 5’-truncated coding region. In such a case, the translational outcome would be decided by the position of the first functional (capable of initiation of translation) AUG or other initiation-competent codon. If it were in-frame with the protein-encoding information content of conventional mRNA, translation would result in the C-terminal fragment, CTF, of a conventionally encoded polypeptide. This variant of RNA-dependent mRNA amplification pathway would be asymmetric. Indeed, only one end, a 3-terminal portion, of conventional mRNA coding region would be amplified, and its translation would produce only one end of conventional genome-encoded polypeptide, its C-terminal fragment. The implications for βAPP mRNA amplifications by such a pathway are clear: If in the resulting 5’-truncated chimeric mRNA the first, 5’-most, functional translation initiation codon were the AUG encoding Met671 in the conventional βAPP mRNA, the translational outcome would be the C99 fragment of amyloid precursor protein produced independently of βAPP and containing Aβ at its N-terminus. Is such an outcome feasible?
Figure 2: RNA-dependent mRNA amplification can result in a 5’-truncated molecule encoding C-terminal fragment of a conventionally encoded polypeptide.
Boxed line-sense strand RNA. Single line-antisense strand RNA. “AUG”-functional translation initiation codon (could be other than AUG). “TCE”– 3’-terminal complementary element; “ICE”– internal complementary element, both on the antisense RNA strand. Yellow circle – helicase/modifying activity complex. Blue lines (both single and boxed) – RNA strand modified and separated from its complement by a helicase complex. Red arrow – position of cleavage of the chimeric intermediate. Step 1: Synthesis of antisense strand; step 2: Strand separation; step 3: Folding of antisense strand into self-priming configuration; step 4: Extension of self-primed antisense RNA; step 5: Strand separation; step 6: Cleavage of the chimeric intermediate; stage 7: End-products of RNA amplification. Steps 3’−7’ correspond to steps 3–7. Top panel: Conventional, genome-transcribed mRNA molecule. Middle panel: Projected stages of RNA-dependent mRNA amplification. “ICE” is located within a segment of antisense RNA corresponding to the 5’UTR of conventional mRNA; the chimeric RNA end product contains the entire coding content of conventional mRNA. Bottom panel: “ICE” is located within a segment of antisense RNA corresponding to the coding region of conventional mRNA. The amplified chimeric end product contains a 5’-truncated coding region of conventional mRNA. The translational outcome is decided by position of the first functional translation initiation codon; if in-frame, a CTF of conventional polypeptide is produced.
Projected Pathway of Asymmetric Amplification of βAPP mRNA Resulting in Chimeric mRNA Encoding C99 Fragment of Beta-Amyloid Precursor Protein
To determine if an mRNA species of interest can potentially be a subject of RNA-dependent mRNA amplification (provided that cellular RdRs machinery is activated), one needs to assess whether its antisense complement contains both TCE and ICE and is capable of folding into a self-priming configuration. If it were, the position of the ICE would indicate the possible translational outcome. Such an assessment can be conducted in a model experiment where an mRNA of interest serves as a template for synthesis of cDNA, initiating at the 3’-terminal poly(A), and is subsequently removed by RNAse H activity present in a preparation of reverse transcriptase used. If an mRNA were fully transcribed, if complementary elements were present within the antisense strand (cDNA), if one of them were 3’-terminal, and if they were topologically compatible, self-priming and the extension synthesis of a segment of the sense strand would occur. The junction between the antisense and sense components would define the site of self-priming and facilitate identification of the TCE and ICE. Just such an experiment was inadvertently carried out with human βAPP mRNA [55]. The results of this experiment, misinterpreted and eventually dismissed by the authors as an artifact [56], indicated the occurrence of topologically compatible TCE and ICE within the antisense strand of βAPP mRNA and defined their sequence as well as the position of self-priming. Based on these results, the TCE/ICE-guided folding of the antisense strand of βAPP mRNA [34–36,57] can be depicted as shown in Figure 3.
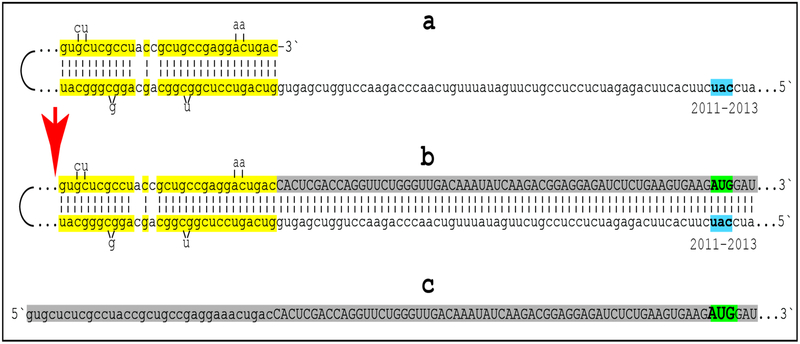
Figure 3: Projected topology of RNA-dependent generation of 5’-truncated mRNA encoding Aβ-containing C-terminal fragment of human beta-amyloid precursor protein.
Lowercase letters -- nucleotide sequence of the antisense RNA; uppercase letters -- nucleotide sequence of the sense RNA. Double-stranded portions highlighted in yellow: TCE (top) and ICE (bottom) elements of the antisense RNA. Note that the TCE and the ICE are separated by about 2000 nucleotides. “2011–2013”: nucleotide positions on the antisense RNA (starting from the complement of the AUG encoding Met1 of the βAPP) of the “uac” (highlighted in blue) corresponding to the “AUG” (highlighted in green) encoding Met671 in the βAPP mRNA. a: TCE/ICE-guided folding of the antisense βAPP RNA. 3’-terminal “c” corresponds to one of multiple transcription start sites of βAPP mRNA 149 nucleotides upstream from its AUG initiation codon [27]; note that such folding configuration would accommodate an additional 3’C, a transcript of the capG of βAPP mRNA (not shown). b: Extension of self-primed antisense RNA into sense RNA and cleavage (red arrow; may also occur at one of the TCE/ICE mismatches), after strand separation, of the chimeric intermediate. c: Chimeric RNA end product contains 5’terminal antisense segment extending into severely 5’-truncated βAPP mRNA. Its translation initiates from the “AUG” (highlighted in green and encoding Met671 in conventional βAPP mRNA) immediately preceding the beta amyloid-encoding region.
Approximately 30 nucleotide-long 3’-terminal segment of the antisense strand of βAPP mRNA constitutes the TCE. Its counterpart, the ICE, is separated by nearly 2000 nucleotides, yet these elements are topologically compatible and the folding of the antisense molecule results in a self-priming configuration capable to accommodate an additional 3’C if the capG of βAPP mRNA is transcribed [48–50] (Figure 3a). The TCE serves as a primer and is extended; thus, generating the sense strand as shown in the Figure 3b. Strands are then separated as illustrated in Steps 5’ and 6’ of Figure 2, and cleavage occurs either at the mismatches within the TCE or immediately upstream from it as indicated by the arrow in Figure 3b. The resulting chimeric RNA end product, shown in Figure 3c, consists of an antisense segment (TCE or its portion) continued into a sense-orientation molecule. The translational outcome is decided by the first, 5’-most, initiation-competent AUG codon. As shown in Figure 3b and c, the first AUG codon is located 58 nucleotides downstream from the TCE portion of the chimeric RNA end-product and it is, in fact, the AUG encoding Met671 in the intact βAPP mRNA! Translation from this position would produce beta amyloid-containing CTF, the C99 fragment of amyloid precursor protein, in the βAPP-independent manner. The major prediction of such a mechanism is a complete inefficiency of beta-secretase inhibition in Alzheimer’s disease. This prediction was, in fact, borne out in several massive stage III clinical trials [57].
Metabolic Mitochondrial Dysfunction: Potential Mediator/Initiator of Asymmetric Amplification of βAPP mRNA in Alzheimer’s Disease
As was argued above, the core enzymatic machinery required for RNA-dependent mRNA amplification appears to be constitutively present, albeit in a non-conventional form [42–44], in all mammalian cells. Under regular circumstances, this RdRp activity produces only short antisense transcripts due to the lack of a processivity co-factor [48,49]. Our current understanding indicates that mammalian core RdRp activity is constitutively expressed and that its processivity co-factor is inducible [48,49]. It doesn’t inform us on the status of other RdRs components involved such as helicase/modifying activity and single-strand-cleaving activity [48,49]. Inducible components of mammalian RdRp complex appear to be expressed under special circumstances requiring a substantial overproduction of specific proteins. Their induction is likely triggered by a cellular stress. One possible example of such regulation is RNA-dependent amplification of mRNA encoding secreted extracellular matrix proteins.
Previously, we demonstrated the occurrence of RNA-dependent amplification of mRNA encoding all three chains of laminin in mouse tissue producing very large quantities of this protein [50]. It was suggested [48–50] that the initial ER stress resulting from increased transcription and subsequent translation of conventional mRNA encoding a secreted protein could be one of potentially multiple cellular events which may trigger mRNA amplification. In this case, one can envision that conventional overproduction of a secreted protein induces ER stress and activates multiple transcription factors [58,59] that complement cellular core RdRp activity and enable RNA-dependent mRNA amplification process. It could be argued that this can further exacerbate the ER stress and trigger cell death. However, the amplified and heavily modified mRNA could behave in ways that are different spatially, qualitatively and quantitatively from those of conventional mRNA. One cellular response to ER stress appears to be a shift of translation and secretion outside the ER [60]. Therefore, nucleotide modifications of amplified mRNA may direct its translation and secretion of the resulting protein via pathways bypassing the ER, despite the presence of a signal peptide sequence. In such a case, mRNA amplification triggered by ER stress would eventually relieve the stress because not only the modified chimeric RNA end product but also conventional mRNA molecules, used as templates for the production of antisense RNA and modified during strand separation (Figure 1, step 2), would be translationally processed outside the ER.
In the case of Alzheimer’s disease, a probable trigger of asymmetric RNA-dependent βAPP mRNA amplification, resulting in 5’truncated mRNA encoding Aβ-containing C99 fragment is stress of metabolic mitochondrial dysfunction. Mitochondria are dynamic ATP-generating organelles which contribute to many cellular functions, including bioenergetic processes, intracellular calcium regulation, alteration of reduction-oxidation potential of cells, free radical scavenging and activation of caspase-mediated cell death. There is mounting evidence showing that mitochondrial damage plays an important role in Alzheimer disease. Increased oxygen species generation and deficient mitochondrial dynamic balance have been suggested to be the reason as well as the consequence of Alzheimer-related pathology. In AD, mitochondrial functions can be negatively affected by beta-amyloid, which can interact with mitochondria and cause mitochondrial dysfunction and consequent oxidative stress. This, in turn, mediates an increased production of Aβ and thus drives the disease.
Mitochondrial dysfunction is a prominent and early feature of AD [61], with reduced energy metabolism as one of the best documented early abnormalities [62]. Key mitochondrial enzymes of oxidative metabolism (i.e., cytochrome C oxidase, KGDHC, and PDHC) are deficient in AD [63,64]. Early deficits in synaptic mitochondria were also detected in an AD mouse model [65]. In addition, damaged mitochondrial DNA (mtDNA), including DNA mutation and DNA defects are also found to be involved in AD [66]. Both beta-amyloid precursor protein and Aβ were localized in mitochondria [67–71], and Aβ not only contributes to significant oxidative damage of mtDNA, but leads also to impaired mtDNA gene expression [72,73]. More recent studies suggest abnormal mitochondrial dynamics, including excessive mitochondrial fragmentation and abnormal mitochondrial distribution, plays a critical role in mitochondrial dysfunction in AD [74–81]. It appears that beta-amyloid is taken up by the organelle from elsewhere inside the cell. In fact, it was demonstrated that Aβ is internalized by cells from an extracellular source and then imported into the mitochondria via the translocase of the outer membrane complex before accumulating in the mitochondrial cristae [71].
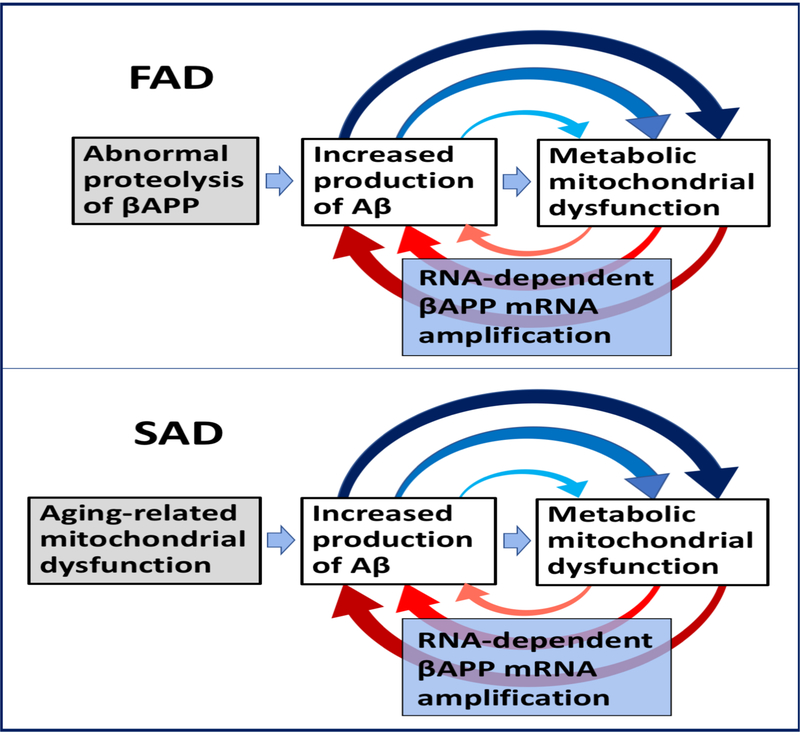
The cause-and-effect relationships between mitochondrial dysfunction and Alzheimer’s disease appear to be very different, in fact diametrically opposite, in FAD and SAD. The primary basis of FAD is the abnormal proteolysis of beta-amyloid precursor protein, which, in turn, is due to the presence of specific mutations in the genes for βAPP and/or presenilins 1 and 2. The increase in levels of Aβ, or a shift to its more neurotoxic species, result in mitochondrial dysfunction and augmented Reactive Oxygen Species (ROS) levels. This, in turn, causes the additional increase in production of Aβ and reinforces the cycle. In SAD, the reverse sequence appears to occur. Aging-related mitochondria dysfunction and mitochondria-derived reactive oxygen species cause an enhanced beta-amyloid formation [78], and the Aβ increase leads to further mitochondrial dysfunction, resulting in even higher ROS levels [79]. Then, the cycle repeats and degeneration increases. In FAD, therefore, Aβ-induced mitochondrial dysfunction is an intrinsic part of the amyloid cascade. In contrast, in SAD the initial mitochondrial dysfunction arises prior to the disease, exists independently of Aβ and lies upstream of the increased Aβ production. In other words, in SAD mitochondrial pathology hierarchically supersedes Aβ pathology. This is the primary reason for the formulation of the Mitochondrial Cascade Hypothesis [13]. But even in terms of the MCH, the core of the disease is the amyloid cascade as defined in the ACH [7–12]. The role of mitochondrial dysfunction in relation to this core is causative in the case of SAD and auxiliary in FAD.
How stresses associated with mitochondrial dysfunction enhance the production of Aβ in Alzheimer’s disease is currently less than clear. We propose that they activate the expression of RdRp processivity co-factor and of other inducible components of the RdRp complex and thus enable asymmetric RNA-dependent amplification of βAPP mRNA, the overproduction of the C99 fragment, and, consequently, the overproduction of Aβ. This implies that a substantial overproduction of Aβ, to the extent sufficient to activate beta-amyloid cascade and trigger Alzheimer’s disease, cannot occur without βAPP mRNA amplification. This notion is supported by the observation that in mice where, as described below, RNA-dependent amplification of its βAPP mRNA cannot take place, mitochondrial dysfunction, even in long terms, results in neither a substantial Aβ accumulation, nor in beta-amyloid plaque formation and neurodegeneration [82]. One of mitochondrial components, a microprotein PIGBOS, was shown to interact with the ER in mitigating the Unfolded Protein Response (UPR) [84]. It is feasible that mitochondrial dysfunction suppresses the occurrence and/or functionality of PIGBOS; this, in turn, would lead to suppression of UPR and, consequently, to ER stress. It is possible, therefore that mitochondrial dysfunction triggers the expression of inducible components of RdRp complex by initiating ER stress, implicated in activation of RNA-dependent mRNA amplification pathway [50].
Of Mice and Men: Why, Unlike Men, Mice (and Elephants) never Develop Alzheimer’s Disease and why BACE Inhibition Works in Mice (and in Healthy Men) but not in AD
Humans are not alone in accumulating Aβ in their brain. Some other animals, notably non-human primates, develop copious beta-amyloid plaques. However, most animals accumulate very little beta-amyloid as they age, and even those that do are spared neurodegeneration and outright dementia that consistently follows the onset of Aβ pathology in men. For example, mice accumulate little beta-amyloid and never develop Alzheimer’s disease. It could be argued that this is due to their size and a short life span. However, this is also true at the other end of the spectrum on both counts: There is no buildup of amyloid plaques and no AD in aging elephants. Does only Homo sapiens get Alzheimer’s disease? And if so, why? The answer is indicated by nucleotide sequence analysis of βAPP in different species that shows that mice and other animals lack the crucial component of RNA-dependent mRNA amplification process, namely the ability of the βAPP antisense RNA to self-prime the synthesis of a new sense strand. Indeed, in non-human species, segments of βAPP antisense RNA corresponding to 3’-terminal and internal complementary elements, TCE and ICE, of human βAPP antisense RNA show very little, if any, complementarity [36]. In fact, it was proposed that mice could be rendered, under certain stress conditions, susceptible to AD by mutating its βAPP gene so as to confer self-priming ability on the antisense strand, thus creating an experimental model that would more faithfully reflect the human condition [36].
The above considerations imply that in non-human species beta-amyloid can be generated only by the proteolysis of beta-amyloid precursor protein. In such a case, its production in these species has to be sensitive to the inhibition of beta-secretase, and it is [29]. Moreover, the same considerations also imply that in healthy humans Aβ is also produced solely via proteolysis of βAPP and therefore must be suppressed by the BACE inhibition. This prediction has been borne out experimentally. Even a single administration of verubecestat to healthy human controls results in the 90% reduction in levels of Aβ and sAPPβ in plasma and CSF [29]. A nearly complete inefficiency of the BACE inhibition in suppression of beta-amyloid production in Alzheimer’s disease indicates that in AD the bulk of Aβ is produced via asymmetric RNA-dependent amplification of βAPP mRNA in an amyloid precursor protein-independent manner. In other words, it appears that the physiological emergence and progression of AD requires the occurrence of RNA-dependent amplification of βAPP mRNA. The mRNA amplification process, in turn, depends on the activation of its inducible components by stresses apparently associated with mitochondrial dysfunction.
In mouse and rat models of AD the desired effect is achieved non-physiologically by expression of multiple inserted copies of human βAPP-encoding DNA. In many cases, the inserted DNA encodes only a 5’-truncated portion of the βAPP gene. In such a case, even if RdRs is enabled by Aβ-induced mitochondrial dysfunction, beta-amyloid-encoding mRNA would not be eligible for amplification because its antisense complement would be lacking the 3’-terminal complementary element, TCE. Even when a full copy of βAPP gene is inserted, it is in a non-physiological chromosomal location, under a non-physiological promoter, and would likely result in a non-physiological position of Transcription Start Site (TSS). In such a case, TCE of the antisense strand would either be absent or be non-terminal; in both cases the amplification would not be feasible. In other words, in most animal models of AD, beta-amyloid production occurs solely by the proteolysis of a precursor protein and is, therefore, susceptible to the BACE inhibition.
βAPP Transcription Start Site Selection: Second Regulatory Tier of βAPP mRNA Amplification
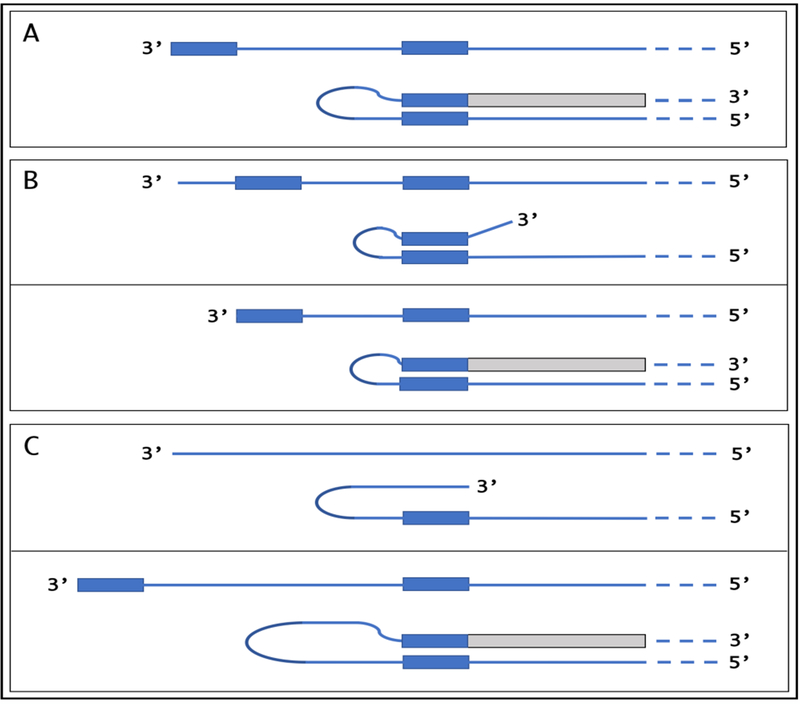
The presence of a regulatory element known as a “TATA-box” is characteristic for a large class of mammalian genes. Usually, it occurs about 30 nucleotides upstream from the TSS and rigidly defines its position. βAPP gene belongs to a class of TATA-less genes that are characterized by multiple transcription start sites. There are at least five and possibly more positions where transcription of human βAPP mRNA can be initiated [34,83]. Of those, only one, 149 nucleotides upstream from the AUG translation initiation codon, results in mRNA molecule eligible for RNA-dependent mRNA amplification because only for this transcript the position of TCE on its antisense strand would be strictly 3’-terminal [34], and an additional 3’C, a transcript of the capG of βAPP mRNA [48–50], could be accommodated in the antisense self-priming structure (Figure 3). It could be argued that with mRNA transcripts of TATA-less genes in general and of βAPP gene in particular, where transcription can be initiated from multiple positions, the eligibility for RdRp-mediated amplification could be in part regulated by a shift in the TSS position. The concepts of such a regulation are summarized in Figure 4.
Figure 4: TSS shift as potential regulator of the eligibility of an mRNA for the amplification process.
Single line: 3’terminus of the antisense strand. Filled grey boxes: Sense strand. Filled blue boxes: Topologically compatible complementary elements on the antisense strand. A: One of the complementary elements is 3’-terminal; folding results in a self-priming structure that is extended into the sense strand. B: Both complementary elements are internal, no self-priming is possible; TSS shift in the downstream direction makes one of the elements 3’-terminal and allows self-priming and extension into the sense strand. C: There are no complementary elements/no self-priming; TSS shift in the upstream direction generates 3’-terminal complementary element and thus enables self-priming and extension. Note that processes depicted in panels B and C can occur in reverse, resulting in a loss, rather than the acquisition, of the eligibility.
If the 3’-distant complementary element of the antisense strand is terminal (Figure 4, panel A), it can form a self-priming structure. If, however, both elements are internal (Figure 4, panel B), the downstream shift of the TSS position can make one of them a 3’-terminal and thus enable self-priming. When, on the other hand, the 3’ segment of the antisense strand has no topologically compatible complementary sequences, an upstream shift of the TSS position (Figure 4, panel C) can generate such elements and make the transcript eligible for amplification. The events diagrammed in panels B and C of Figure 4 can also occur in reverse, making previously eligible mRNAs ineligible for the amplification process. In the previous section, an argument was made that stress-induced activation of inducible components of RNA-dependent mRNA amplification process is necessary but not sufficient for it to occur for a particular mRNA species; proper TCE and ICE must also be present in the antisense strand. Here, we extend this argument to include shifts in positions of complementary elements required for self-priming of antisense RNA as a secondary regulatory level.
Conclusion
The present study posits RNA-dependent amplification of βAPP mRNA as a molecular basis of beta-amyloid overproduction in Alzheimer’s disease [34–36,47–49]. In this process, βAPP mRNA serves as a transcription template for RdRp complex. The resulting antisense folds into a self-priming configuration utilizing two complementary elements, one 3’-terminal and another internal. The internal complementary element is located within an antisense segment corresponding to the coding portion of human βAPP mRNA. The extension of self-primed antisense RNA produces a 3’-terminal fragment of βAPP mRNA, a part of a hairpin-structured antisense/sense RNA molecule. Cleavage at the 3’ end of the hairpin structure’s loop produces RNA end product encoding a C-terminal fragment of βAPP polypeptide. Since each conventional βAPP mRNA can be used repeatedly as a template, the process constitutes an asymmetric mRNA amplification. In the amplified mRNA, the first, 5’-most translation initiation codon is the AUG encoding Met671 in the intact βAPP mRNA. Translation from this codon, located immediately upstream from and contiguous to βAPP mRNA segment encoding beta-amyloid, would produce Aβ-containing C99 fragment of βAPP. Since this process is completely independent of amyloid precursor protein and does not require beta-secretase cleavage, it is insensitive to the BACE inhibition. Such process can occur in humans but not in mice and other animals where segments of βAPP antisense RNA required for self-priming have little, if any, complementarity [36]. This explains why Alzheimer’s disease, defined ultimately by Aβ overproduction-initiated neurodegeneration and outright dementia, occurs exclusively in humans, and implies that asymmetric RNA-dependent βAPP mRNA amplification is requisite in AD. In AD, therefore, there are two pathways of beta-amyloid production; one, βAPP proteolytic pathway and another, RNA-dependent βAPP mRNA amplification pathway independent of βAPP and insensitive to beta-secretase inhibition. In healthy humans only the proteolytic pathway is in operation. This implies that in healthy humans, unlike in AD patients, Aβ production should be suppressed by the BACE inhibition, and indeed it is [29]. However, since βAPP-independent pathway operating in AD is by far the predominant one, BACE inhibition has no effect in Alzheimer’s disease.
It appears that, physiologically, the extent of beta-amyloid overproduction sufficient to trigger amyloid cascade culminating in AD requires asymmetric RNA-dependent amplification of βAPP mRNA and cannot be reached without it. In turn, the occurrence of mRNA amplification process depends on the activation of inducible components of RdRp complex. Such an induction can be a consequence of certain stresses, for example the ER stress in case of RNA-dependent amplification of mRNA encoding extracellular matrix proteins [50]. In case of Alzheimer’s disease, such an induction appears to be triggered by stresses associated with mitochondrial dysfunction, a phenomenon closely linked to AD and shown to be stimulated by increased levels of beta-amyloid. The cause-and-effect relationships between mitochondrial dysfunction and Alzheimer’s disease appear to be very different, in fact diametrically opposite, in FAD and SAD. In FAD, increased levels or more toxic species of Aβ, resulting from the mutations-mediated abnormal proteolysis of βAPP, trigger metabolic mitochondrial dysfunction and augmented ROS levels. This, in turn, activates βAPP mRNA amplification, causes the additional increase in production of Aβ, and reinforces the cycle. Thus in FAD, mitochondrial dysfunction is an intrinsic component of the amyloid cascade. The reverse sequence is true in SAD where aging-related mitochondrial dysfunction and mitochondria-derived reactive oxygen species activate RNA-dependent amplification of βAPP mRNA and enhanced production of Aβ. This causes further mitochondrial dysfunction, the cycle repeats and degeneration increases. Thus in SAD, the initial mitochondrial dysfunction arises prior to the disease, independently of and upstream from the increased Aβ production. In other words, in SAD, in contrast to FAD, mitochondrial pathology hierarchically supersedes Aβ pathology. This is the primary reason for the formulation of the Mitochondrial Cascade Hypothesis [13]. But even in terms of the MCH, the core of the disease is the amyloid cascade as defined in the ACH [7–12]. The role of mitochondrial dysfunction in relation to this core is causative in the case of SAD and auxiliary in FAD.
The initial increases in levels of Aβ are attained differently in different forms of AD. In FAD, it is caused by mutations in and, consequently, abnormal proteolysis of βAPP and occurs relatively early in life. In SAD, it is stimulated by an aging-dependent component and takes place, accordingly, late in life. Regardless of their origin, however, upon reaching a certain threshold, elevated levels of beta-amyloid instigate, in both forms of AD, mechanistically identical self-perpetuating mutual Aβ/mitochondrial dysfunction feedback cycles that drive, via RNA-dependent βAPP mRNA amplification, Aβ overproduction and are an essential element of the amyloid cascade. This explains drastic differences in the age of onset, yet profound pathological and symptomatic similarities in the progression, of familial and sporadic forms of Alzheimer’s disease. These relationships are diagrammatically summarized in Figure 5; it depicts the mutual feedback cycles as a two-stroke engine, an engine that drives beta-amyloid overproduction and, consequently, Alzheimer’s disease. Interestingly, the recent findings that mitochondrial microprotein PIGBOS interacts with the ER (via the ER protein CLCC1) in mitigating the unfolded protein response [84] indicate possible connection between mitochondrial dysfunction and ER stress, implicated in activation of RNA-dependent mRNA amplification pathway [50].
Figure 5: The engine that drives AD: Mitochondrial dysfunction-mediated overproduction of beta-amyloid (and vice versa) in Alzheimer’s disease.
FAD: Familial Alzheimer’s disease; SAD: Sporadic Alzheimer’s disease; Highlighted in grey: Initial stimuli of the increased production of Aβ (different in FAD and SAD); Highlighted in blue: asymmetric RNA-dependent βAPP mRNA amplification, a molecular basis of beta-amyloid overproduction in Alzheimer’s disease (note a requirement for βAPP TSS utilization discussed in main text above); Horizontal arrows: the initial Aβ overproduction cycle; Arched arrows: Mutual feedback cycles; Blue arrows: Aβ-mediated induction of mitochondrial dysfunction and, possibly, ER stress; Red arrows: Mitochondrial dysfunction (and, possibly, ER stress)-mediated asymmetric RNA-dependent amplification of βAPP mRNA resulting in overproduction of Aβ. Note that in FAD, mitochondrial dysfunction is an intrinsic component of the amyloid cascade whereas in SAD, the initial mitochondrial pathology hierarchically supersedes and triggers Aβ pathology, where self-perpetuating mutual Aβ/mitochondrial dysfunction feedback cycles are, as in FAD, a central component of the amyloid cascade. In FAD, the initial increased production of Aβ results from mutations-driven abnormal proteolysis of βAPP and occurs relatively early in life, whereas in SAD, it is compelled by an aging-dependent component; hence drastic temporal difference in the age of onset yet profound pathological and symptomatic similarity in the progression of familial and sporadic Alzheimer’s disease, reflecting mechanistically identical nature of feedback cycles in both forms of AD.
Regardless of the initial cause-and-effect relationships between mitochondrial dysfunction and different forms of AD, neither SAD nor FAD can occur without stresses of mitochondrial dysfunction that enable RNA-dependent amplification of βAPP mRNA. The RdRs-enabling potential denotes mitochondrial dysfunction as possible therapeutic target; this is especially important in view of the fact that success with targeting other presumptive targets associated with the production of beta-amyloid has proved to be elusive. Moreover, if the aim is to pursue therapeutically RNA-dependent mRNA amplification process as the plausible molecular basis of beta-amyloid overproduction in Alzheimer’s disease, mitochondrial dysfunction, an ostensive trigger of this process, is the target of choice. Indeed, systemically targeting any component of the enzymatic machinery of RNA-dependent RNA synthesis would interfere with normal physiological functions of this process, such as, for example, specific mRNAs amplification in both, erythroid differentiation [47–49] and the deposition of extracellular matrix proteins [50], and would have highly deleterious consequences. On the other hand, successfully targeting mitochondrial dysfunction would potentially result in two highly beneficial outcomes, namely, switching off an abnormal RNA-dependent βAPP mRNA amplification process and repairing metabolic defects. In this respect, a recent study with C. elegans [85] is very encouraging. In this study beta-amyloid-induced mitochondrial dysfunction was modeled and achieved by expressing human Aβ specifically in neurons (GRU102). Importantly, treatment with an anti-diabetes drug, metformin, reversed Aβ-induced metabolic defects, reduced protein aggregation and normalized lifespan of GRU102 thus establishing metabolic mitochondrial dysfunction as a promising and feasible therapeutic intervention target.
It should be emphasized that the above considerations radically change the outlook on the dynamics of Alzheimer’s disease. Until now, it was presumed, at least in terms of the ACH, that the development of AD occurs as one prolonged process where damages slowly accumulate and culminate in manifestation of the disease. This viewpoint implied that preemptive therapeutic intervention late in life in case of potential SAD, or mid-age in case of FAD, is largely futile since the damage has already occurred over the preceding decades. To be effective, according to this view, it should start early in life. The outlook suggested by the present study is drastically different. According to it, the disease evolves in two stages. The first stage is slow process of beta-amyloid accumulation. It occurs in humans and animals and results neither in significant damage nor in the manifestation of the disease. The second stage, which occurs only in humans and produces significant damages, is fast. It commences with the activation of βAPP mRNA amplification and results in rapid accumulation of Aβ and in AD. This means that preventive therapeutic intervention, an AD-”statin”, c an be initiated and be effective at any time prior to the second stage, i.e. late in life in case of potential SAD. As for possible treatment strategies, they should address not only mitochondrial dysfunction and/or ER stress but also the regulation of utilization of βAPP transcription start sites, with the aim of preventing the production of βAPP mRNA eligible for RNA-dependent amplification process. Moreover, βAPP TSSs profile may serve as a prognostic indicator of susceptibility to AD.
Funding
NIH R21 GM056179; NIH RO1 AR036819.
Footnotes
Conflict of Interest
Authors declare no conflict of interest.
References
- 1.Haass C, Lemere C, Capell A, Citron M, Seubert P, Schenk D, et al. The Swedish mutation causes early-onset Alzheimer’s disease by beta-secretase cleavage within the secretory pathway. Nat Med. 1995;1(12):1291–6. [DOI] [PubMed] [Google Scholar]
- 2.Dyrks T, Dyrks E, Monning U, Urmoneit B, Turner J, Beyreuther, K. Generation of beta A4 from the amyloid protein precursor and fragments thereof. FEBS Lett. 1993;335(1):89–93. [DOI] [PubMed] [Google Scholar]
- 3.Iizuka T, Shoji M, Kawarabayashi T, Sato M, Kobayashi T, Tada N, et al. Intracellular generation of amyloid beta-protein from amyloid beta-protein precursor fragment by direct cleavage with beta- and gamma-secretase. Biochem. Biophys. Res Commun. 1996;218(1):238–42. [DOI] [PubMed] [Google Scholar]
- 4.De Strooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci. 2000;113 (11):1857–70. [DOI] [PubMed] [Google Scholar]
- 5.Barber RC. The genetics of Alzheimer’s disease. Scientifica (Cairo). 2012;2012:246210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vassar R. BACE inhibitor drugs in clinical trials for Alzheimer’s disease. Alzheimers Res Ther. 2014;6(9):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyreuther K, Masters C. Amyloid precursor protein (APP) and beta A4 amyloid in the etiology of Alzheimer’s disease: precursor-product relationships in the derangement of neuronal function. Brain Pathol. 1991;1(4):241–51. [DOI] [PubMed] [Google Scholar]
- 8.Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol Sci. 1991;12(10):383–8. [DOI] [PubMed] [Google Scholar]
- 9.Selkoe DJ. The molecular pathology of Alzheimer’s disease. Neuron. 1991;6(4):487–98. [DOI] [PubMed] [Google Scholar]
- 10.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256(5054):184–5. [DOI] [PubMed] [Google Scholar]
- 11.Hardy J, Selkoe D. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–6. [DOI] [PubMed] [Google Scholar]
- 12.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8(6):595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swerdlow RH. Mitochondria and Mitochondrial Cascades in Alzheimer’s Disease. J Alzheimers Dis. 2018;62(3):1403–16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter S, Brayne C. Understanding the roles of mutations in the amyloid precursor protein in Alzheimer disease. Mol Psychiatry. 2018;23(1):81–93. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S, Wang Z, Cai F, Zhang M, Wu Y, Zhang J, et al. BACE1 Cleavage Site Selection Critical for Amyloidogenesis and Alzheimer’s Pathogenesis. J Neurosci. 2017;37(29):6915–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vassar R, Bennett B, Babu-Khan S, Kahn S, Mendiaz E, Denis P, Teplow D, et al. Beta-secretase cleavage of Alheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–41. [DOI] [PubMed] [Google Scholar]
- 17.Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, et al. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol Cell Neurosci. 1999;14(6):419–27. [DOI] [PubMed] [Google Scholar]
- 18.Sinha S, Anderson J, Barbour R, Basl G, Caccavello R, Davis D, et al. Purification and cloning of amyloid precursor protein beta secretase from human brain. Nature. 1999;402(6761):537–40. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Strickland C, Voigt J, Kennedy M, Beyer B, Senior MM, et al. Application of fragment-based NMR screening, X-ray crystallography, structure-based design, and focused chemical library design to identify novel μM leads for the development of nM BACE-1 inhibitors. J Med Chem. 2010;53(3):942–50. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Z, Sun Z, Ye Y, Voigt J, Strickland C, Smith E, et al. Discovery of cyclic acylguanidines as highly potent and selective ß-site amyloid cleaving enzyme (BACE) inhibitors: Part I - inhibitor design and validation. J Med Chem. 2010;53(3):951–65. [DOI] [PubMed] [Google Scholar]
- 21.Cumming J, Smith E, Wang L, Misiaszek J, Durkin J, Pan J, et al. Structure based design of available, centrally active BACE1 iminohydantoin BACE1 inhibitors: identification of an orally inhibitor. Bioorg Med Chem Lett. 2012;22(7):2444–9. [DOI] [PubMed] [Google Scholar]
- 22.Edwards P, Albert J, Sylvester M, Aharony D, Andisik D, et al. Application of fragment-based lead generation to the discovery of novel, cyclic amidine ß -secretase inhibitors with nanomolar potency, cellular activity, and high ligand efficiency. J Med Chem. 2007;50:5912–25. [DOI] [PubMed] [Google Scholar]
- 23.Barrow J, Stauffer S, Rittle K, Ngo P, Yang Z, Selnick H, et al. Discovery and X-ray crystallographic analysis of a S-piropiperidine iminohydantoin inhibitor of ß -secretase. J Med Chem. 2008;51:6259–62. [DOI] [PubMed] [Google Scholar]
- 24.Malamas M, Erdei J, Gunawan I, Turner J, Hu Y, Wagner E, et al. Design and synthesis of 5,5’-disubstituted aminohydantoins as potent and selective human ß -secretase (BACE1) inhibitors. J Med Chem. 2010;53:1146–58. [DOI] [PubMed] [Google Scholar]
- 25.Rueeger H, Rondeau J, McCarthy C, Moebitz H, Tintelnot-Blomley M, Neumann U, et al. Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors. Bioorg Med Chem Lett. 2011;21(7):1942–7. [DOI] [PubMed] [Google Scholar]
- 26.Probst G, Xu Y. Small-Molecule BACE1 Inhibitors: a patent literature review (2006–2011). Expert Opin Ther Pat. 2012;22(5):511–40. [DOI] [PubMed] [Google Scholar]
- 27.May P, Dean R, Lowe S, Martenyi F, Sheehan S, Boggs L, et al. Robust central reduction of amyloid-ß in humans with an orally available, non-peptidic ß -secretase inhibitor. J Neurosci. 2011;31:6507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamford A, Scott J, Li S, Babu S, Tadesse D, Hunter R, et al. Discovery of an orally available, brain penetrant BACE1 inhibitor that affords robust CNS Aß reduction. ACS Med. ACS Med Chem Lett. 2012;3(11):897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy M, Stamford A, Chen X, Cox K, Cumming J, Dockendorf M, et al. The BACE1 inhibitor verubecestat (MK-8931) reduces CNS b-amyloid in animal models and in Alzheimer’s disease patients. Sci Transl Med. 2016;8(363):363ra150. [DOI] [PubMed] [Google Scholar]
- 30.Keskin AD, KekuÅ¡ M, Adelsberger H, Neumann U, Shimshek DR, Song B, et al. BACE inhibition-dependent repair of Alzheimer’s pathophysiology. Proc Natl Acad Sci U S A. 2017;114(32):8631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neumann U, Rueeger H, Machauer R, Veenstra S, Lueoend R, Tintelnot-Blomley M, et al. A novel BACE inhibitor NB-360 shows a superior pharmacological profile and robust reduction of amyloid-ß and neuroinflammation in APP transgenic mice. Mol Neurodegener. 2015;10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu X, Das B, Hou H, He W, Yan R. BACE1 deletion in the adult mouse reverses preformed amyloid deposition and improves cognitive functions. J Exp Med. 2018;215(3):927–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nativio R, Donahue G, Berson A, Lan Y, Amlie-Wolf A, Tuzer F, et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nature Neurosci. 2018;10:1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volloch V. A mechanism for ß-amyloid overproduction in Alzheimer’s disease: Precursor-independent generation of ß-amyloid via antisense RNA-primed mRNA synthesis. FEBS Lett. 1996;390:124–8. [DOI] [PubMed] [Google Scholar]
- 35.Volloch V. Mechanism for ß-amyloid overproduction in sporadic Alzheimer’s Disease: Possible antisense RNA-mediated generation of a 5’-truncated ßAPP mRNA encoding 12 kDa C-terminal fragment of ßAPP, the immediate precursor of Aß In: Molecular Mechanisms of Dementia. 1997, Wasco W and Tanzi R, Eds. [Google Scholar]
- 36.Volloch V. Possible mechanism for resistance to Alzheimer’s disease (AD) in mice suggests new approach to generate a mouse model for sporadic AD and may explain familial resistance to AD in man. Exp Neurol. 1997;144(1):214–8. [DOI] [PubMed] [Google Scholar]
- 37.Breimer LH, Denny P. Alzheimer amyloid aspects. Nature. 1987;326(6115):749–50. [DOI] [PubMed] [Google Scholar]
- 38.Citron M, Haass C, Selkoe DJ. Production of amyloid-beta-peptide by cultured cells: no evidence for internal initiation of translation at Met596. Neurobiol Aging. 1993;14(6):571–3. [DOI] [PubMed] [Google Scholar]
- 39.Lai M. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol 2005;79:7951–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor J. Replication of human hepatitis delta virus: recent developments. Trends Microbiol. 2003;11:185–90. [DOI] [PubMed] [Google Scholar]
- 41.Tseng CH, Lai MM. Hepatitis delta virus RNA replication. Viruses. 2009;1(3):818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehmann E, Brueckner F, Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450(7168):445–9. [DOI] [PubMed] [Google Scholar]
- 43.Wagner SD, Yakovchuk P, Gilman B, Ponicsan SL, Drullinger LF, Kugel JF, et al. RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J. 2013;32(6):781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maida Y, Yasukawa M, Masutomi K. De Novo RNA Synthesis by RNA-Dependent RNA Polymerase Activity of Telomerase Reverse Transcriptase. Mol Cell Biol. 2016;36(8):1248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Downey KM, Byrnes JJ, Jurmark BS, So AG. Reticulocyte RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 1973;70(12):3400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapranov P, Ozsolak F, Kim S, Foissac S, Lipson D, Hart C, et al. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature. 2010;466(7306):642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volloch V, Schweitzer B, Rits S. Antisense Globin RNA in Murine Erythroid Tissues: Structure, Origin and Possible Function. Proc Natl Acad Sci USA. 1996;93(6):2476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volloch V. Protein-encoding RNA to RNA transfer in mammatian cells: principles of RNA-dependent mRNA amplification. Ann Integr Mol Med. 2019;1(1):1002. [PMC free article] [PubMed] [Google Scholar]
- 49.Rits S, Olsen BR, Volloch V. Protein-encoding RNA to RNA information transfer in mammalian cells:RNA-dependent mRNA ampli cation. Identi cation of chimeric RNA intermediates and putative RNA end products. Ann Integr Mol Med. 2019;1(1):1003. [PMC free article] [PubMed] [Google Scholar]
- 50.Volloch V, Rits S, Olsen BR. RNA-dependent Amplification of Mammalian mRNA Encoding Extracellullar Matrix Proteins: Identification of Chimeric RNA Intermediates for a1, ß1, and ?1 Chains of Laminin. Ann Integr Mol Med. 2019;1(1):1004. [PMC free article] [PubMed] [Google Scholar]
- 51.Richards OC, Ehrenfeld E. Poliovirus RNA replication. Curr Top Microbiol Immunol. 1990;161:89–119. [DOI] [PubMed] [Google Scholar]
- 52.Brazas R, Ganem D. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science. 1996;5284:90–4. [DOI] [PubMed] [Google Scholar]
- 53.Huang CR, Lo SJ. Evolution and diversity of the human hepatitis d virus genome. Adv Bioinformatics. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volloch V, Schwetizer B, Rits S. Evolutionarily Conserved Elements in the 5’-untranslated Region of beta-Globin mRNA Mediate Site-specific Priming of a Unique Hairpin Structure during cDNA Synthesis. Nucleic Acids Res. 1994;22(24):5302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mita S, Sadlock J, Herbert J, Schon E. A cDNA specifying the human amyloid beta precursor protein encodes a 95-kDa polypeptide. Nucleic Acids Res. 1988;16:9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mita S, Sadlock J, Herbert J, Schon E. A cDNA specifying the human amyloid beta precursor protein encodes a 95-kDa polypeptide. Nucleic Acids Res. 1988;16(19):9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Volloch V, Rits S. Results of beta secretase-inhibitor clinical trials support amyloid precursor protein-independent generation of beta amyloid in sporadic Alzheimer’s disease. Med Sci (Basel). 2018;6(2);pii: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–91. [DOI] [PubMed] [Google Scholar]
- 59.Jiang S, Zhang E, Zhang R, Li X. Altered activity patterns of transcription factors induced by endoplasmic reticulum stress. BMC Biochem. 2016;17:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27(3):230–40. [DOI] [PubMed] [Google Scholar]
- 61.Zhu X, Perry G, Moreira PI, Aliev G, Cash AD, Hirai K, et al. Mitochondrial abnormalities and oxidative imbalance in Alzheimer disease. J Alzheimers Dis. 2006;9:147–53. [DOI] [PubMed] [Google Scholar]
- 62.Blass JP. The mitochondrial spiral. An adequate cause of dementia in the Alzheimer’s syndrome. Ann N Y Acad Sci. 2000;924:170–83. [DOI] [PubMed] [Google Scholar]
- 63.Manczak M, Park BS, Jung Y, Reddy PH. Differential expression of oxidative phosphorylation genes in patients with Alzheimer’s disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med. 2004;5:147–62. [DOI] [PubMed] [Google Scholar]
- 64.Qin W, Haroutunian V, Katsel P, Cardozo CP, Ho L, Buxbaum JD, et al. PGC-1alpha expression decreases in the Alzheimer disease brain as a function of dementia. Arch Neurol. 2009;66:352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS. Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci U S A. 2010;107:18670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin MT, Simon DK, Ahn CH, Kim LM, Beal MF. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum Mol Genet. 2002;11:133–45. [DOI] [PubMed] [Google Scholar]
- 67.Calkins M, Manczak M, Mao P, Shirendeb U, Reddy PH. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer’s disease. Hum Mol Genet. 2011;20(23):4515–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anandatheerthavarada HK, Biswas G, Robin MA, Avadhani NG. Mitochondrial targeting and a novel transmembrane arrest of Alzheimer’s amyloid precursor protein impairs mitochondrial function in neuronal cells. J Cell Biol. 2003;161:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caspersen C, Wang N, Yao J, Sosunov A, Chen X, Lustbader JW, et al. Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer’s disease. FASEB J. 2005;19(14):2040–1. [DOI] [PubMed] [Google Scholar]
- 70.Chen JX, Yan SS. Role of mitochondrial amyloid-beta in Alzheimer’s disease. J Alzheimers Dis. 2010;20 Suppl 2:S569–78. [DOI] [PubMed] [Google Scholar]
- 71.Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, et al. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc Natl Acad Sci U S A. 2008;105(35):13145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de la Monte SM, Luong T, Neely TR, Robinson D, Wands JR. Mitochondrial DNA damage as a mechanism of cell loss in Alzheimer’s disease. Lab Invest. 2000;80(8):1323–35. [DOI] [PubMed] [Google Scholar]
- 73.Brooks WM, Lynch PJ, Ingle CC, Hatton A, Emson PC, Faull RL, Starkey MP. Gene expression profiles of metabolic enzyme transcripts in Alzheimer’s disease. Brain Res. 2007;1127(1):127–35. [DOI] [PubMed] [Google Scholar]
- 74.Wang X, Perry G, Smith MA, Zhu X. Amyloid-beta-derived diffusible ligands cause impaired axonal transport of mitochondria in neurons. Neurodegener Dis. 2010;7(1–3):56–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X, Su B, Fujioka H, Zhu X. Dynamin-like protein 1 reduction underlies mitochondrial morphology and distribution abnormalities in fibroblasts from sporadic Alzheimer’s disease patients. Am J Pathol. 2008;173:470–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, et al. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29(28):9090–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, et al. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci U S A. 2008;105:19318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Su B, Wang X, Bonda D, Perry G, Smith M, Zhu X. Abnormal mitochondrial dynamics--a novel therapeutic target for Alzheimer’s disease? Mol Neurobiol. 2010;41:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manczak M, Calkins MJ, Reddy PH. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage. Hum Mol Genet. 2011;20:2495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leuner K, Schütt T, Kurz C, Eckert SH, Schiller C, Occhipinti A, et al. Mitochondrion-derived reactive oxygen species lead to enhanced amyloid beta formation. Antioxidants and Redox Signaling. 2012;16(12):1421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.LaFerla FM, Green KN, Oddo S. Intracellular amyloid-ß in Alzheimer’s disease. Nat Rev Neurosci. 2007;8(7):499–509. [DOI] [PubMed] [Google Scholar]
- 82.Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/− mice. J Biol Chem. 2008;283(38):26217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salbaum JM, Weidemann A, Lemaire HG, Masters CL, Beyreuther K. The promoter of Alzheimer’s disease amyloid A4 precursor gene. EMBO J. 1988;7(9):2807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chu Q, Martinez T, Novak S, Donaldson C, Tan D, Joan MV, et al. Regulation of the ER stress response by a mitochondrial microprotein. Nat Commu. 2019;10:4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teo E, Ravi S, Barardo D, Kim H, Fong S, Tan TY, et al. Metabolic stress is a primary pathogenic event in transgenic Caenorhabditis elegans expressing pan-neuronal human amyloid beta. Elife. 2019;8 pii: e50069. [DOI] [PMC free article] [PubMed] [Google Scholar]