Abstract
Background:
Several studies have explored the associations between interleukin-6 (IL-6) gene polymorphisms and the susceptibility to liver diseases, however, results remain ambiguous. The goal of this study was to conduct a meta-analysis to provide more credible evidence.
Methods:
Studies identified in the PubMed, Cochrane Library, and EMBASE databases were used to perform a meta-analysis via the STATA software. Pooled odds ratios (OR) were calculated under fixed- and random-effects models to estimate the potential genetic associations.
Results:
Twenty-five case-control studies involving 5813 cases and 5298 controls were included in this meta-analysis. Overall, the pooled results suggested that rs1800795 polymorphism was significantly associated with the risk of liver diseases in heterozygote (GC vs CC; OR = 1.57) and dominant (GG+GC vs CC: OR = 1.47) models; rs1800796 polymorphism was significantly associated with the susceptibility to liver diseases in heterozygote (GG vs GC; OR = 0.58) and recessive (GG vs GC+CC: OR = 0.68) models; rs1800797 polymorphism was significantly associated with genetic predisposition to liver diseases in homozygote (GG vs AA: OR = 1.63), heterozygote (GA vs AA; OR = 1.53) and dominant (GG + GA vs AA: OR = 1.61) models. A similar conclusion was found in the HBV, HCV, HCC, NASH and alcoholic liver disease of all ethnic populations for rs1800795; HBV and Asian subgroups for rs1800796; HCV and non-Asian subgroups for rs1800797. However, IL-6 rs2069837 and rs2066992 polymorphisms did not exhibit significant associations with the risk of liver diseases under any genetic models.
Conclusion:
This meta-analysis suggests that patients carrying G (rs1800795), C (rs1800796) or G (rs1800797) allele or genotypes of IL-6 may be more likely to suffer from liver diseases, which was ethnic-dependent.
Keywords: genetic variation, hepatitis B virus, hepatitis C virus, interleukin-6, liver diseases, transformation
1. Introduction
The liver is one of the key organs of the body, which performs many pivotal functions essential for human life, including carbohydrate, protein and fat metabolism,[1] immune response against pathogens[2] as well as detoxification of xenobiotic.[3] The consequence of hepatic impairments, including viral hepatitis, alcoholic or nonalcoholic steatohepatitis (NASH), drug-induced liver injury, autoimmune hepatitis, fatty liver, liver cirrhosis (LC) and liver cancer, may be serious and even lethal.[4] Thus, it is vital to understand the etiology of liver diseases for developing efficiently predictive, preventive and therapeutic strategies.
Despite the pathogenesis remains unclear, increasing evidence has suggested liver diseases are of an inflammatory nature.[5] Interleukin-6 (IL-6) is an important inflammatory cytokine and may play a central role for the development and progression of liver diseases. Serum IL-6 concentration was detected to be significantly higher in alcoholic or non-alcoholic cirrhosis and toxic hepatitis when compared to controls.[6] Higher level of IL-6 was observed to be produced in CD4(+) T cells from acute-on-chronic hepatitis B virus (HBV) liver failure patients.[7] Higher level of IL-6 was significantly associated with advanced liver fibrosis in human immunodeficiency virus-type 1 (HIV)-infected patients [adjusted odds ratio (OR) = 11.78, 95% confidence interval (CI): 1.17–118.19, P = .036].[8] High plasma IL-6 was also suggested as a biomarker for poor prognosis of patients with hepatocellular carcinoma (HCC).[9] IL-6 promoted HCC cell proliferation and migration by activating signal transducer and activator of transcription 3 (STAT3) signaling pathway.[10] These findings imply any factor that influences the expression of IL-6 may be an underlying contributor for the development of liver diseases.
Recently, some scholars have found genetic mutations in the IL-6 gene could alter its expression, with genotype CC carriers of rs1800796 showing higher level of IL-6 mRNA compared with genotype CG/GG carriers.[11,12] Therefore, this IL-6 single nucleotide polymorphism (SNP) may be a possible risk factor to contribute to the susceptibility to liver diseases. This hypothesis has been validated as follows: genotyping of IL-6 rs1800796 SNP showed a significant increase in GC genotypes, but reduction in GG genotype in HBV infection group compared with controls. A direct positive correlation was also detected between HBV and the presence of GC genotype and C allele.[13] Riazalhosseini et al also observed the frequency of allele G of rs1800796 was higher among healthy controls than that among chronic HBV patients (0.303 vs 0.258) and GC+CC genotype was associated with a protection mechanism against HBV infection (OR = 0.40, 95% CI: 0.34–0.48).[14] However, inconsistent conclusions were also reported, with no significant associations of rs1800796 polymorphism with HBV infection,[15] hepatitis C virus (HCV) infection,[16] LC and HCC.[15,17] Furthermore, there were also studies to investigate the associations between the risk of liver diseases and other polymorphisms in IL-6, including rs1800795, rs1800797,[13,16] rs2066992,[18,19] and rs2069837[14,18,20] and the controversial outcomes were also present in them. These equivocal results may be attributed to small sample size and limited statistical power of each individual study.
The goal of this study was to conduct a meta-analysis to comprehensively estimate the associations of IL-6 polymorphisms and genetic predisposition to all liver diseases.
2. Materials and methods
2.1. Literature search
Our study was performed according to the Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) standard.[21] PubMed, the Cochrane Library and EMBASE databases were searched for papers published before February, 2019 using the keywords: interleukin-6 (OR IL-6) AND polymorphism (OR SNP OR variant OR mutation) AND liver diseases (OR hepatitis OR liver cirrhosis OR hepatocellular carcinoma OR liver injury OR fatty liver). The publication language was restricted to English. Furthermore, potentially eligible literatures were supplemented through manually mining bibliographies of relevant studies.
2.2. Inclusion and exclusion criteria
Studies were included if they satisfied the following criteria:
-
(1)
human genotyping;
-
(2)
case-control design;
-
(3)
healthy (HC) or infection resolved (IF) controls;
-
(4)
evaluation of the associations between IL-6 polymorphisms and liver diseases in more than 2 articles; and
-
(5)
providing adequate data to calculate the OR and its corresponding 95%CI.
Studies having the following characteristics were excluded:
-
(1)
repeated studies;
-
(2)
animal studies, reviews, case reports, series, meeting abstracts, as well as comment;
-
(3)
the data of genotype frequency were unavailable;
-
(4)
studies that investigated the therapy response; and
-
(5)
some controls showing HBV positive or having other liver diseases.
2.3. Data extraction and quality assessment
Two investigators independently extracted the data from each eligible study, including first author's name, year of publication, country, ethnicity, liver disease type, genotyping method, number of cases and controls, source of control, and frequency of genotypes. If articles included more than 1 disease type, each group was considered as an independent dataset. The quality of individual studies was also assessed independently by two authors using the New-castle–Ottawa Scale (NOS) system[22] that includes 3 aspects: selection (0–4 points), comparability (0–2 points) and exposure (0–3 points). The NOS ranges from zero (worst) to 9 stars (best). Studies scored more than 7 stars were considered to be of high quality. Any disagreements in data extraction and quality assessment were resolved by the involvement of a third part.
2.4. Statistical analysis
STATA software (version 13.0; STATA Corporation, USA) was used for this meta-analysis. The associations between IL-6 polymorphisms (rs1800795, rs1800796, rs1800797, rs2066992, and rs2069837) and the risk of liver diseases were estimated based on pooled ORs and 95%CI under various genetic models. P value of Cochran's Q-statistic >0.1 or I2 value <50% indicated the absence of heterogeneity among studies and thus of a fixed-effect model was utilized in the association test; otherwise (P < .10 or I2 > 50), a random-effect model was chosen. The significance of the pooled ORs was determined by the Z test, and P < .05 was considered statistically significant. Potential publication bias was evaluated using the Egger linear regression test. If there was evidence of publication bias (P < .05), trim and fill method was used to adjust for the effect of publication bias.[23] Sensitivity analysis was performed to evaluate the stability of the results by omitting each study at a time.
3. Results
3.1. Study characteristics
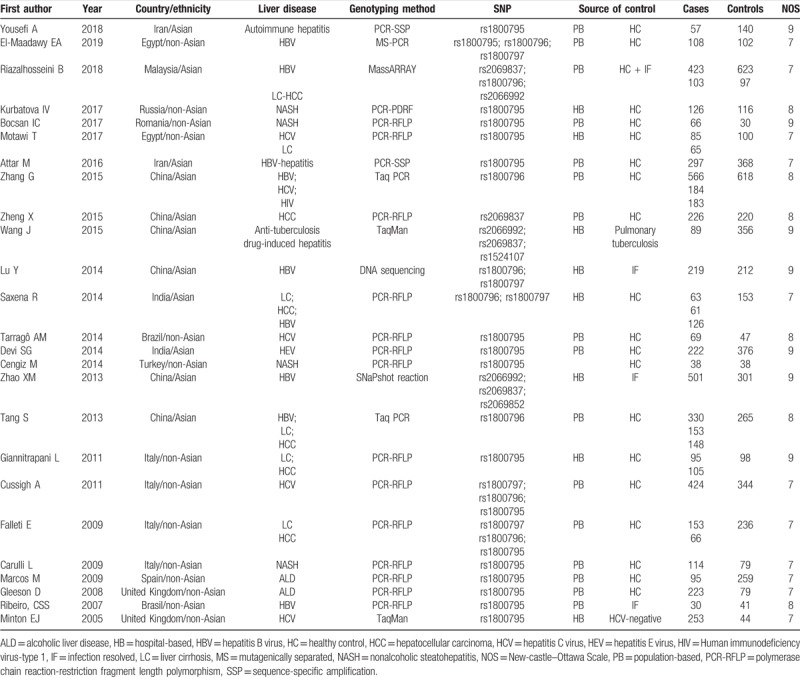
The search strategy retrieved 1035 relevant papers. Based on the inclusion and exclusion criteria (Fig. 1), 25 case-control studies including 5813 cases and 5298 controls were finally included for this meta-analysis.[12–16,18–20,24–40] Among these 25 studies published between 2005 and 2018, 17 of them with 20 datasets investigated the associations between rs1800795 polymorphism of IL-6 gene and liver diseases (including 1 for autoimmune hepatitis, 3 for HBV infection, 4 for NASH, 4 for HCV infection, 3 for LC, 1 for HEV infection, 2 for HCC and 2 for alcoholic liver disease), 8 studies with 16 datasets involved rs1800796 (including 6 for HBV infection, 2 for HCV infection, 1 for HIV infection, 3 for LC, 3 for HCC and 1 for LC/HCC), 5 studies with 8 datasets analyzed rs1800797 (including 3 for HBV infection, 1 for HCV infection, 2 for LC and 2 for HCC), 4 studies with 5 datasets explored rs2069837 (including 2 for HBV infection, 1 for anti-tuberculosis drug-induced hepatitis, 1 for LC-HCC and 1 for HCC) and 3 studies with 4 datasets surveyed rs2066992 (including 2 for HBV infection, 1 for anti-tuberculosis drug-induced hepatitis and 1 for LC-HCC). According to the NOS, all the included studies were of high quality. The detailed characteristics of included studies are listed in Table 1.
Figure 1.
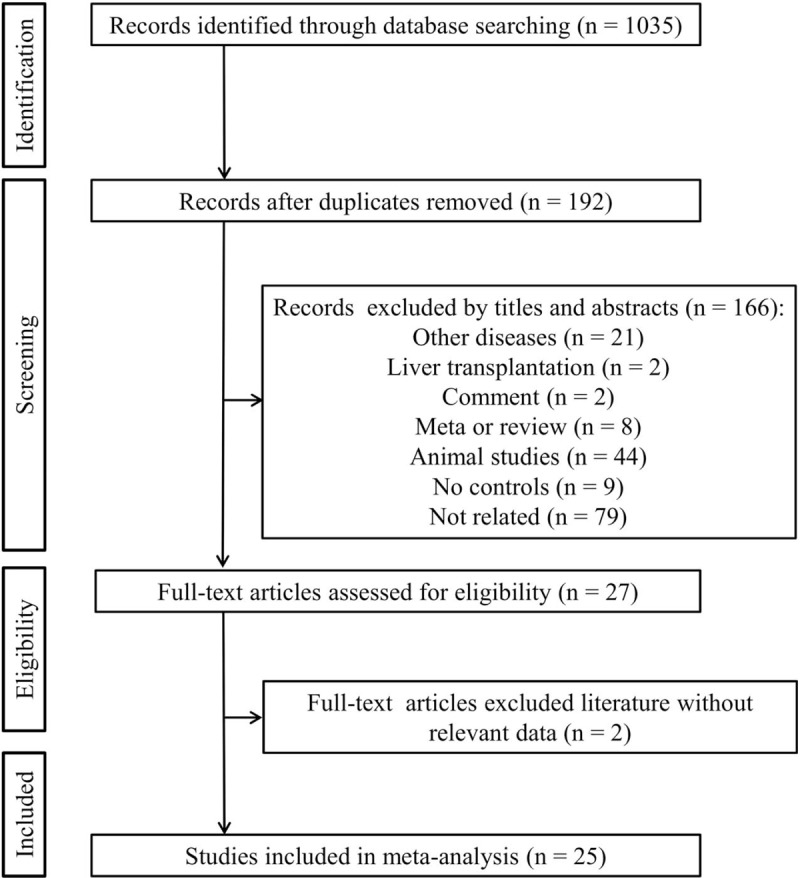
Flow diagram of the study selection process.
Table 1.
Characteristics of studies included in this meta-analysis.
3.2. Meta-analysis
The meta-analysis results of the correlations between five IL-6 polymorphisms and vulnerability to liver diseases in all genetic models are shown in Table 2. The pooled results suggested that rs1800795 polymorphism was significantly associated with the risk of liver diseases in heterozygote (GC vs CC: OR = 1.57, 95% CI = 1.32–1.88, P < .0001) (Fig. 2A) and dominant (GG + GC vs CC: OR = 1.47, 95% CI = 1.14–1.91, P < .0001) (Fig. 2B) models; rs1800796 polymorphism was significantly associated with the susceptibility to liver diseases in heterozygote (GG vs GC: OR = 0.58, 95% CI = 0.39–0.85, P = .006) (Fig. 3A) and recessive (GG vs GC+CC: OR = 0.68, 95% CI = 0.50–0.91, P = .009) (Fig. 3B) models; rs1800797 polymorphism was significantly associated with genetic predisposition to liver diseases in homozygote (GG vs AA: OR = 1.63, 95% CI = 1.17–2.27, P = .004) (Fig. 4A), heterozygote (GA vs AA: OR = 1.53, 95% CI = 1.09–2.14, P = .013) (Fig. 4B) and dominant (GG + GA vs AA: OR = 1.61, 95% CI = 1.17–2.22, P = .003) (Fig. 4C) models. However, IL-6 rs2069837 and rs2066992 polymorphisms did not exhibit significant associations with liver disease risk in any genetic model.
Table 2.
Overall meta-analysis results.
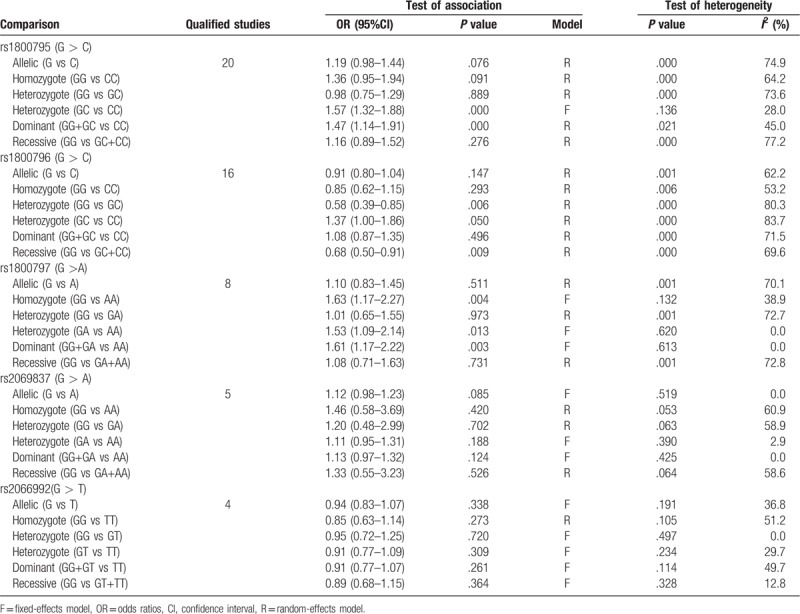
Figure 2.
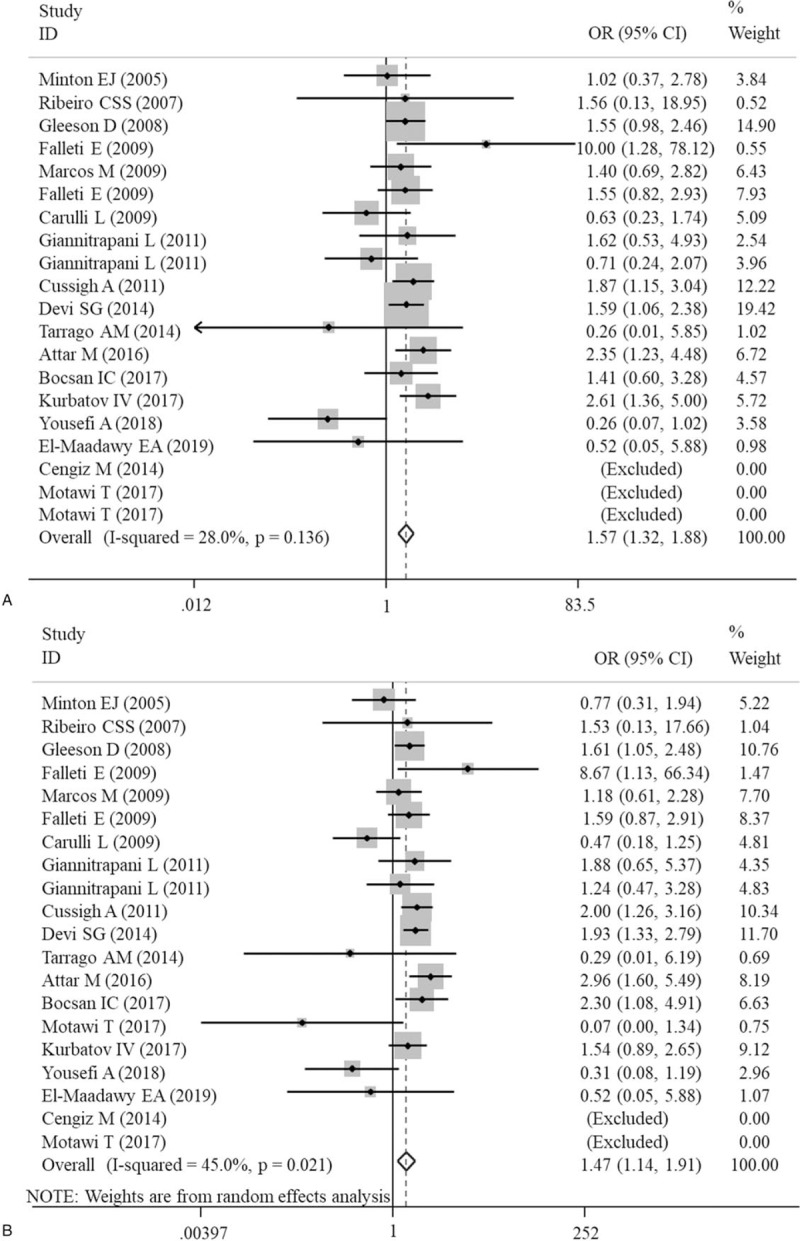
Forest plots of the association of IL-6 gene rs1800795 polymorphism with an increased risk of liver diseases under heterozygote (GC vs CC) and dominant (GG+GC vs CC) models. A, heterozygote; B, dominant. CI = confidence intervals; OR = odds ratio.
Figure 3.
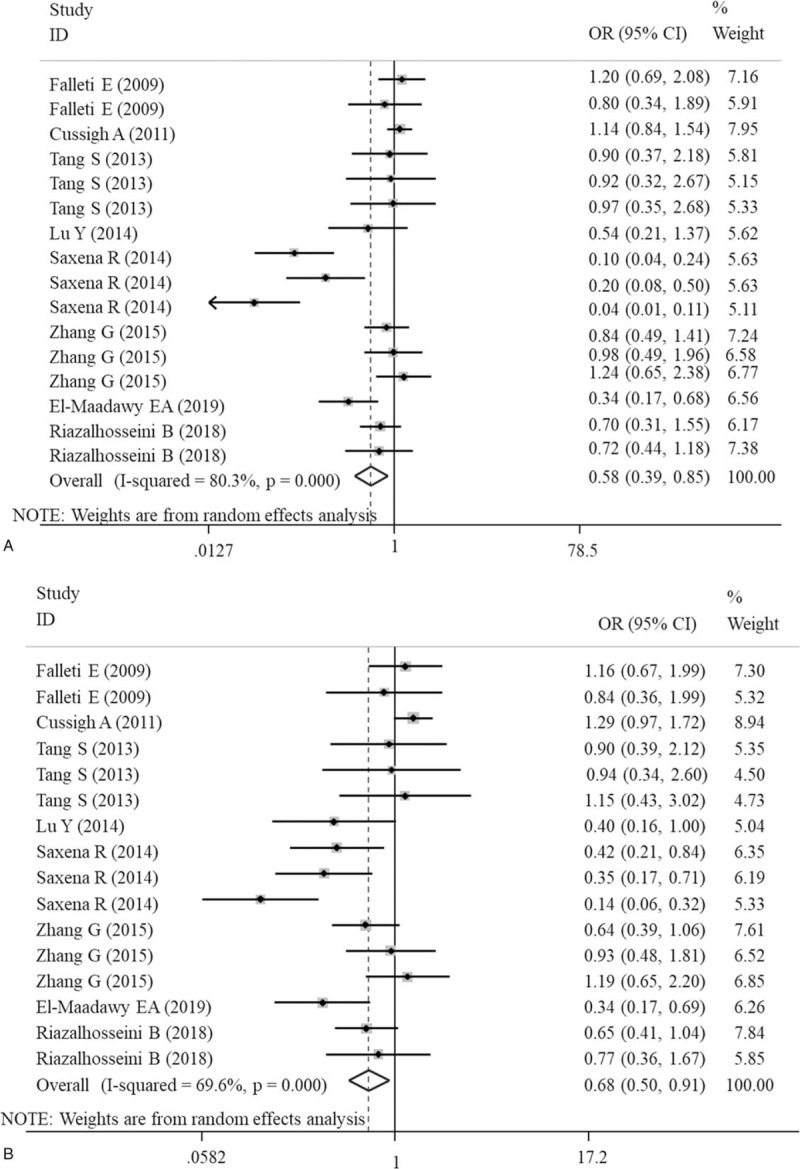
Forest plots of the association of IL-6 gene rs1800796 polymorphism with an increased risk of liver diseases under heterozygote (GC vs CC) and recessive (GG vs GC+CC) models. A, heterozygote; B, recessive. CI = confidence intervals; OR = odds ratio.
Figure 4.
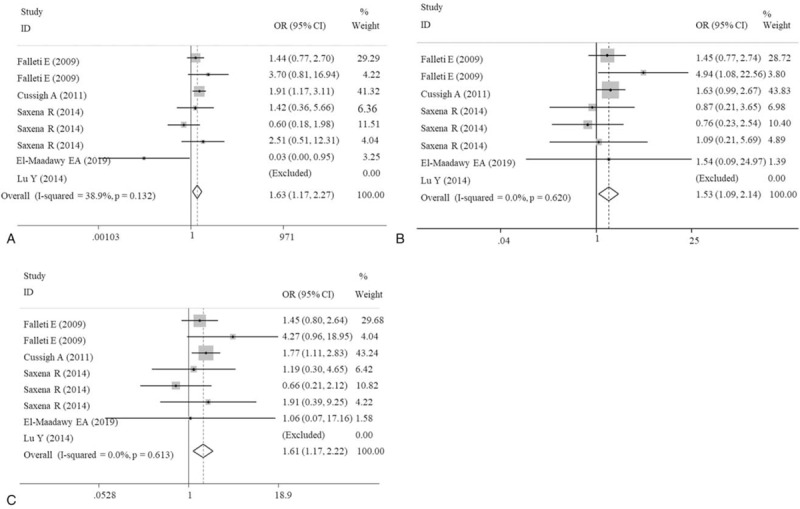
Forest plots of the association of IL-6 gene rs1800797 polymorphism with an increased risk of liver diseases under homozygote (GG vs AA), heterozygote (GA vs AA) and dominant (GG+GA vs AA) models. A, homozygote; B, heterozygote; C, dominant. CI = confidence intervals; OR = odds ratio.
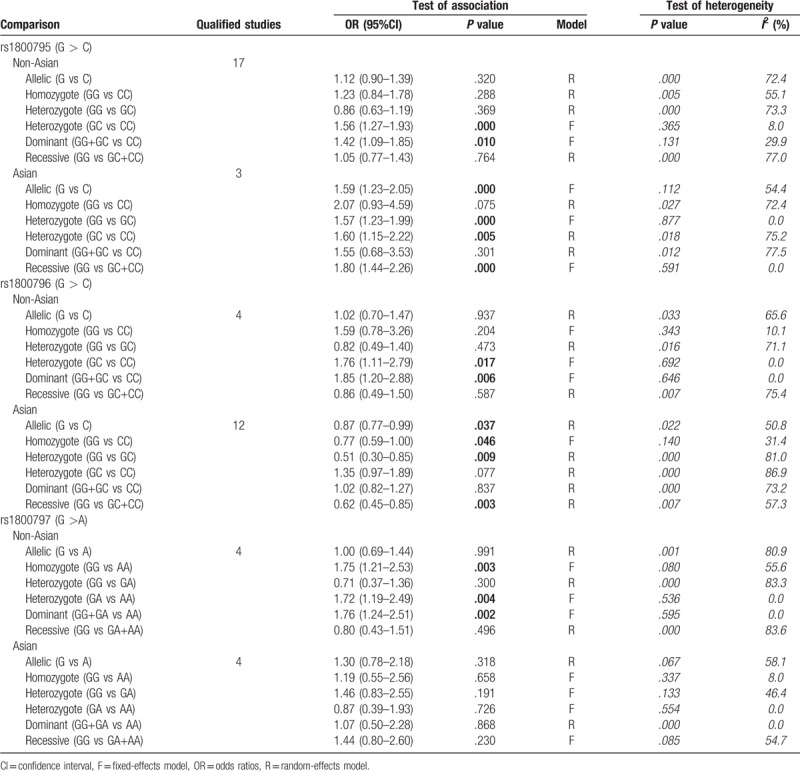
Due to the presence of significant heterogeneity in some overall analysis (Table 2), subgroup analyses were conducted based on liver disease type and ethnicity. For rs1800795 polymorphism, only a significant association was observed for patients with HBV [G vs C: OR = 1.75, 95% CI = 1.13–2.72, P = .012; GG vs CC: OR = 2.98, 95% CI = 1.63–5.45, P < .001; GC vs CC: OR = 2.08, 95% CI = 1.15–3.77, P = .016; GG+GC vs CC: OR = 2.54, 95% CI = 1.37–4.71, P = .003; GG vs GC+CC: OR = 1.91, 95% CI = 1.03–3.56, P = .041], NASH (GC vs CC: OR = 1.60, 95% CI = 1.03–2.49, P = .038), HCV (GC vs CC: OR = 1.58, 95% CI = 1.04–2.42, P = .034), HCC (GC vs CC: OR = 3.11, 95% CI = 1.24–7.79, P = .015) and alcoholic liver disease [GC vs CC: OR = 1.51, 95% CI = 1.03–2.21, P = .036; GG vs GC+CC: OR = 1.47, 95% CI = 1.03–2.10, P = .036] (Table 3). For rs1800796 polymorphism, only a significant association was detected for patients with HBV [G vs C: OR = 0.74, 95% CI = 0.65–0.85, P < .001; GG vs CC: OR = 0.56, 95% CI = 0.42–0.74, P < .001; GC vs GC: OR = 0.42, 95% CI = 0.20–0.87, P = .020; GG vs GC + CC: OR = 0.46, 95% CI = 0.29–0.73, P = .001] (Table 4). For rs1800797 polymorphism, no significant association was detected for most of liver disease patients other than HCV (Table 5), but only 1 literature was included for HCV and this result remained inconclusive. In both of Asian and non-Asian population, a significant increased risk to develop liver diseases can be observed in G allelic carriers of rs1800795 polymorphism; G allele (OR = 0.87, 95% CI = 0.77–0.99, P = .037) or GG genotype (GG vs CC: OR = 0.77, 95% CI = 0.59–1.00, P = .046; GG vs GC: OR = 0.51, 95% CI = 0.30–0.85, P = .009; GG vs GC+CC: OR = 0.62, 95% CI = 0.45–0.85, P = .003) of rs1800796 polymorphism was related with the lower risk of liver diseases only in Asian population, but contrast results for the non-Asian (GC vs CC: OR = 1.76, 95% CI = 1.11–2.79, P = .017; GG vs GC+CC: OR = 1.85, 95% CI = 1.20–2.88, P = .006); rs1800797 polymorphism was significantly associated with the susceptibility to liver diseases only in non-Asian population (GG vs AA: OR = 1.75, 95% CI = 1.21–2.53, P = .003; GA vs AA: OR = 1.72, 95% CI = 1.19–2.49, P = .004; GG+GA vs AA: OR = 1.76, 95% CI = 1.24–2.51, P = .002) (Table 6).
Table 3.
Subgroup analysis for rs1800795.
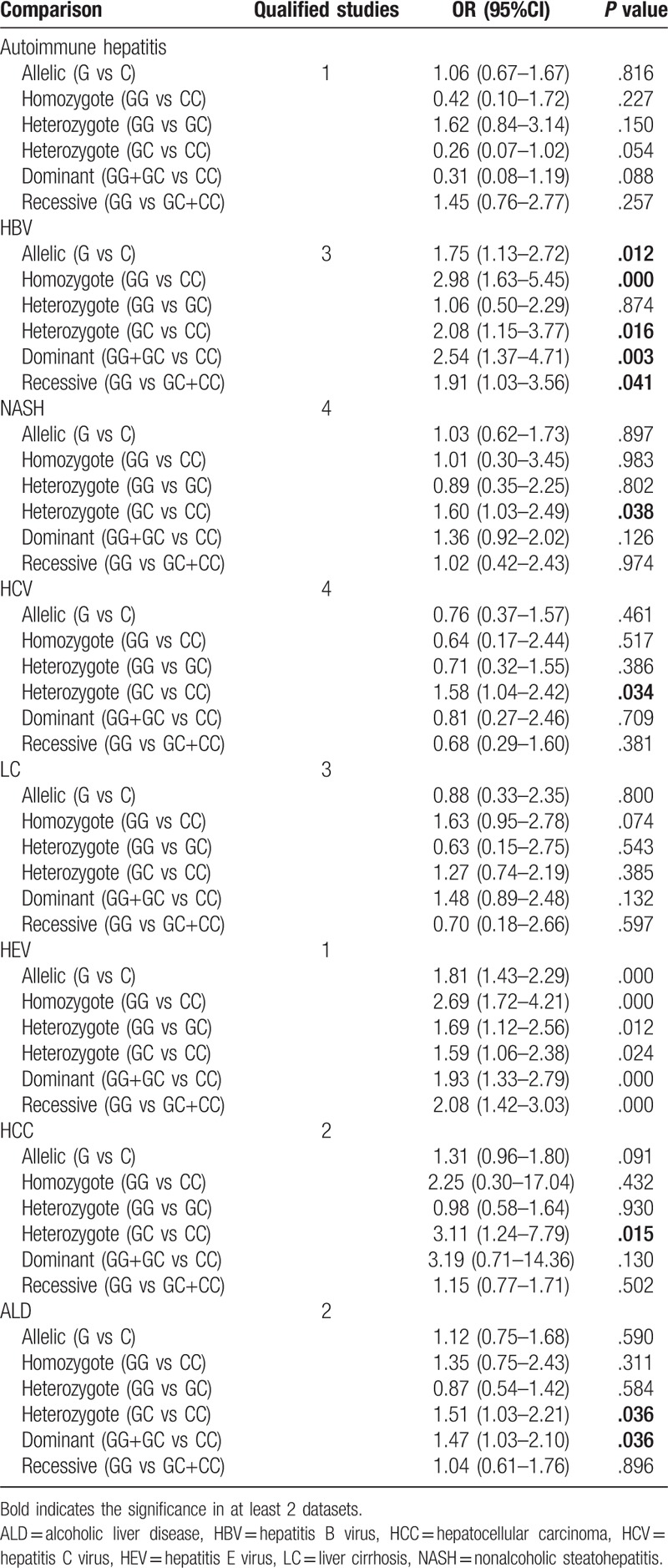
Table 4.
Subgroup analysis for rs1800796.
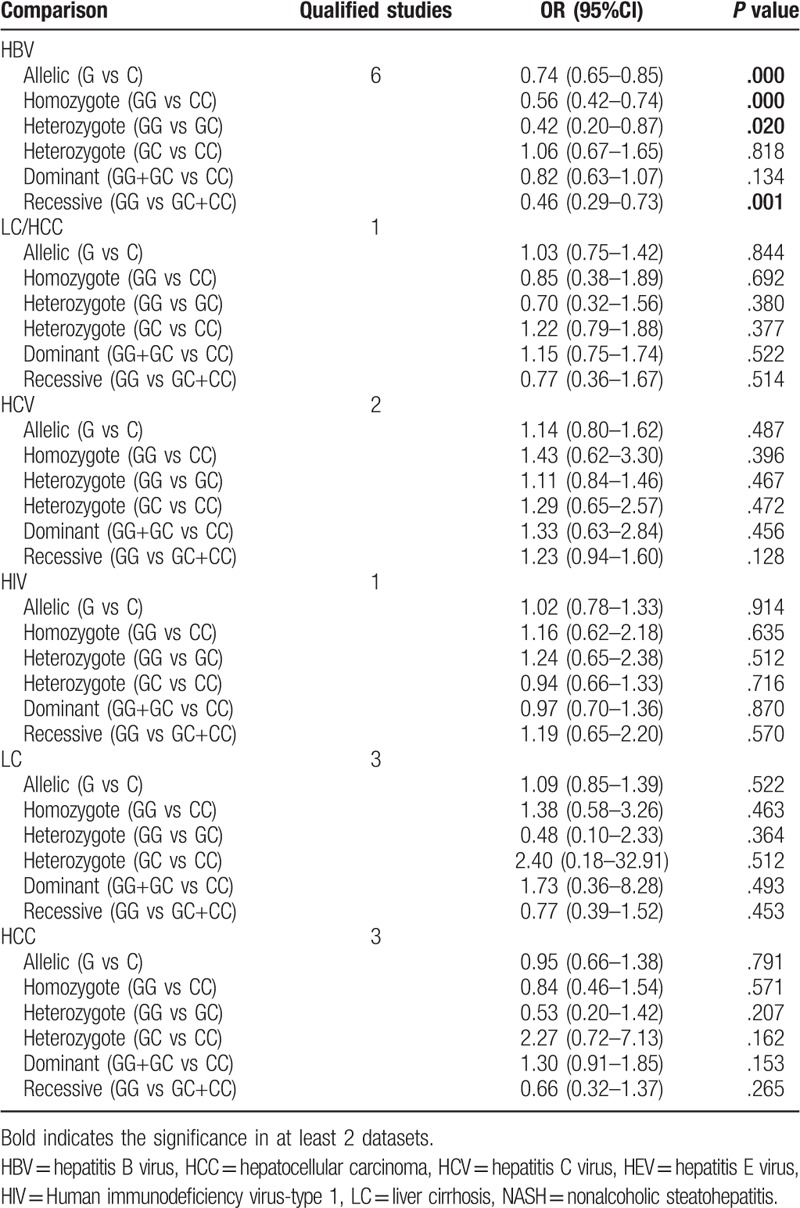
Table 5.
Subgroup analysis for rs1800797.
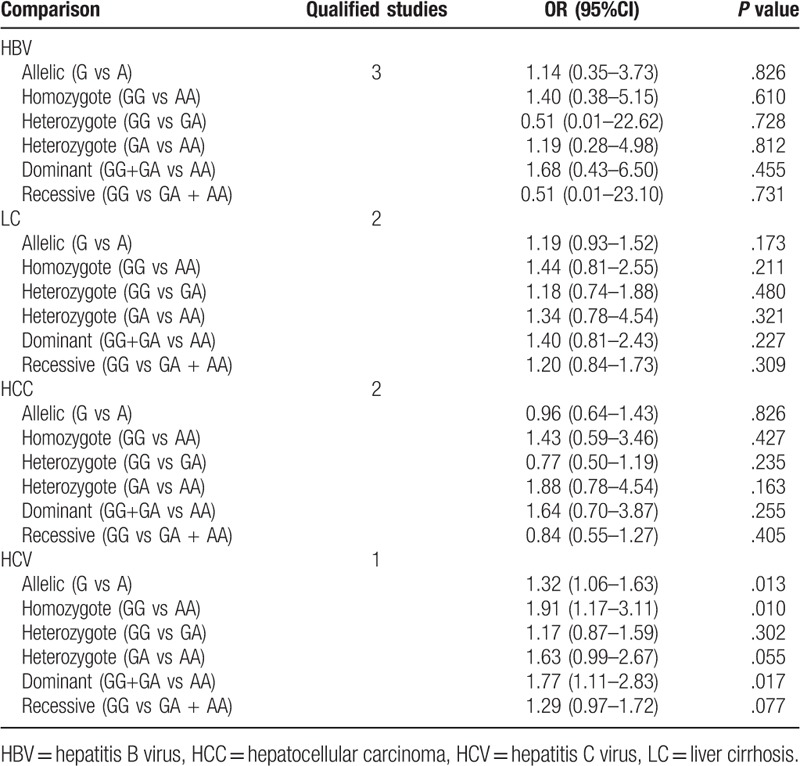
Table 6.
Ethnicity-based subgroup meta-analysis.
3.3. Publication bias and sensitivity analysis
Egger linear regression test was performed to investigate the potential publication bias for significant results in overall meta-analysis. The results showed the intercept did not pass through the origin (that is, asymmetry) in association analysis of rs1800796 under heterozygote model (GG vs GC) (Fig. 5A), indicating the presence of publication bias (P = .017). Subsequently, trim and fill method was used to further adjust for the publication bias (Fig. 5B). The results showed the association remained significant after correcting the publication bias (OR = 0.76, 95% CI = 0.64–0.90, P = .001), implying our results were statistically robust. No obvious asymmetry was observed in the evaluation of publication bias for rs1800795 (GC vs CC: P = 0.072; GG+GC vs CC: P = .182) and rs1800797 (GG vs AA: P = .242; GA vs AA: OR = 1.53, P = .316; GG + GA vs AA: P = .321), suggesting no evidence of publication bias.
Figure 5.
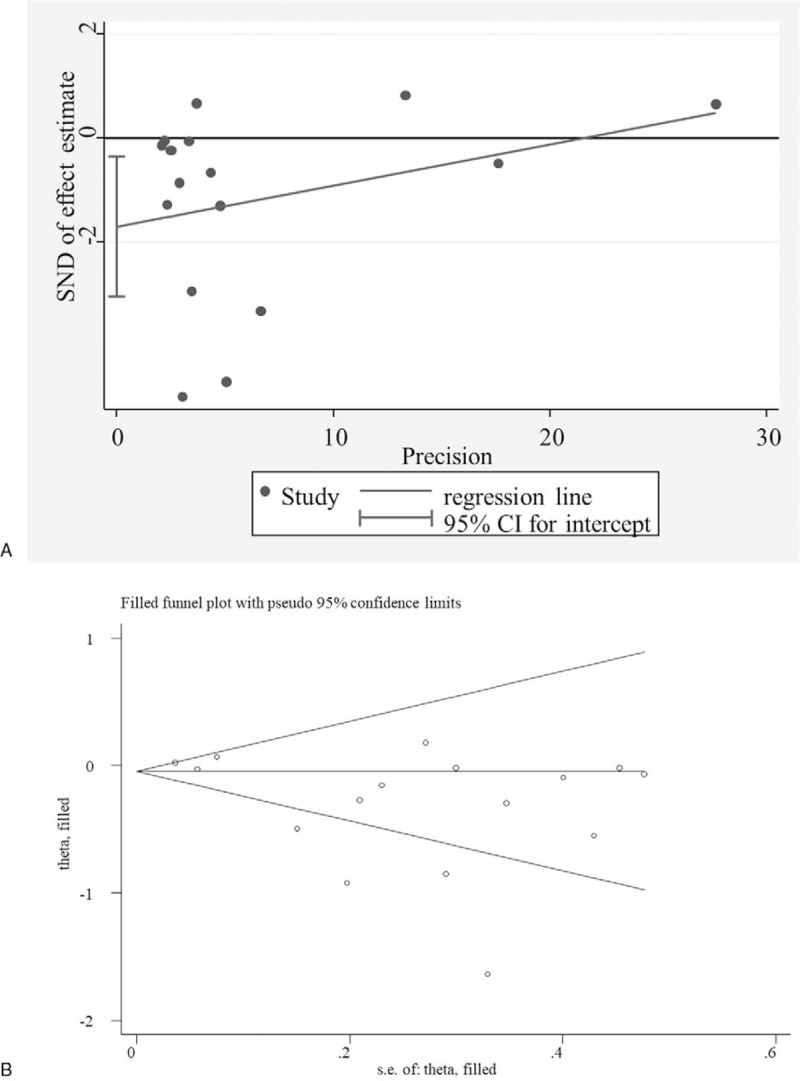
Potential publication bias. A, Egger funnel plot for the assessment of rs1800795 polymorphism (GG+GC vs CC); Trim and fill for adjusting publication bias. CI = confidence intervals, SND = standard normal deviation.
As shown in Figure 6, the omission of any single study did not significantly affect the pooled ORs or 95% CIs, indicating our results may be reliable.
Figure 6.
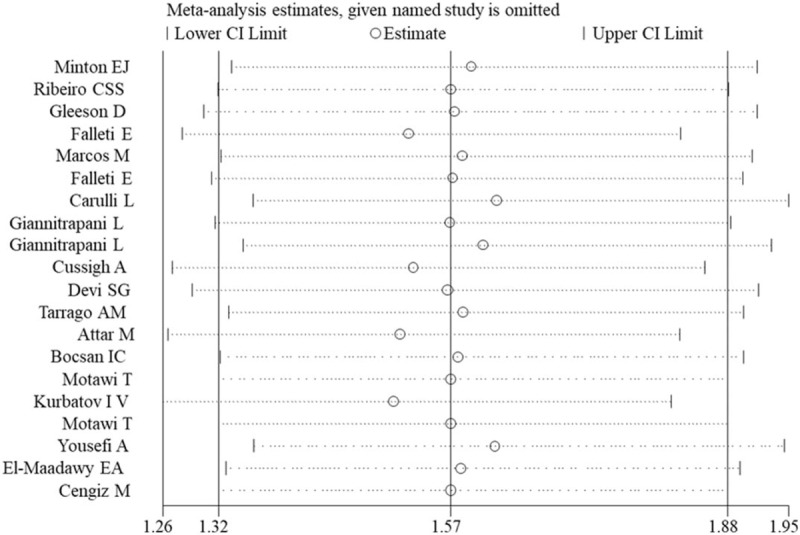
Sensitivity analysis for the assessment of result stability for rs1800795 polymorphism (GC vs CC). CI = confidence intervals.
4. Discussion
In this study, we performed a meta-analysis to investigate the associations of IL-6 SNPs with liver diseases. Our findings showed that IL-6 rs1800795 and rs1800796 polymorphisms may be potential genetic factors for the development of liver diseases. Patients with G allele or GG, GC and GG+GC genotypes of rs1800795 had significantly increased risks for developing liver diseases in all ethnic populations, especially HBV, HCV, HCC, NASH and alcoholic liver disease subgroups. On the contrary, G allele, GG or GC genotypes of rs1800796 may be significant protective factors for the development of liver diseases, especially HBV and Asian population. Although the overall and ethnic meta-analysis showed people carrying the GG, GA or GG+GA genotypes of rs1800797 had a higher risk of suffering liver diseases in non-Asian population, subgroup analysis seemed to show no significant association between this polymorphism and various subtypes of liver diseases except HCV identified in one article. For IL-6 rs2069837 and rs2066992 polymorphisms, we did not find any association with liver disease risk, although this was the first meta-analysis study to investigate them in liver diseases.
Previously, there have 2 meta studies to explore the associations between IL-6 polymorphisms and liver diseases,[41,42] but they were obviously different from our study:
-
(1)
only the HBV-related liver diseases or HCC were analyzed in the study of Chang et al[42] and Liu et al,[41] but not all types of liver diseases as reported in our study;
-
(2)
Chinese papers were included in these studies, but not in our study;
-
(3)
articles with HBV carriers as controls were included, which were excluded in our study; and
-
(4)
these 2 meta-analyses only searched the published papers up to 2015.
The differences in these 4 aspects may contribute to the slight deviation of our results from them. For example, the significant associations between IL-6 rs1800795 polymorphism and risk of HCC under homozygote model (CC vs GG: OR = 0.36; 95% CI = 0.16–0.85) and recessive model (GG+CG vs CC: OR = 2.82; 95% CI = 1.26–6.28) identified by Liu et al[41] were not observed in our study, but only significant under heterozygote model (GC vs CC: OR = 3.11, 95% CI = 1.24–7.79); significant associations between IL-6 rs1800797 polymorphism and the risk of HBV under allelic (G vs A: OR = 1.89; 95% CI = 1.11–3.20), heterozygote (GG vs GA: OR = 2.21; 95% CI = 1.12–3.92) and recessive (GA + AA vs GG: OR = 0.47; 95% CI = 0.26–0.86) models identified by Chang et al,[42] were not shown in our study, but we found some novel conclusions, including significant associations with HBV, HCV, NASH, and alcoholic liver disease of rs1800795.
rs1800795 polymorphism is located at the 174 base pair upstream of IL-6 gene promoter and variation from G to C at this region was reported to reduce this gene's transcription rate and lead to the lower production of IL-6.[43,44] A recent study even found IL-6 mRNA expression was especially higher in the GC than in the GG and CC cases.[45] It had been demonstrated hepatitis B core antigen transfection increased the expression and secretion of IL-6 through activating extracellular signal-related kinase, p38 mitogen-activated protein kinase and nuclear factor-kappa B in hepatocytes.[46] Subsequently, HBV-IL-6 activated the transcription and translation of angiogenin and vascular endothelial growth factor genes via the STAT3 pathway and ultimately promoted HCC cell proliferation.[10,47] Furthermore, activation of IL6/STAT3 pathway also could support HBV replication to further deteriorate HBV-related carcinogenesis.[48] HCV infection was also proved to play important roles in the development of liver diseases by IL-6/STAT3 pathway.[49] IL-6 level, which activated downstream immune and oxidative stress signaling to exacerbate inflammation infiltration, was also found to be increased in patients with NASH[50] and alcoholic liver injury.[51] Accordingly, we believe patients carrying GC genotype of rs1800795 may have higher risks to suffer HBV, HCV infection, HCC, NASH and alcoholic liver disease, which was confirmed in our study.
rs1800796 polymorphism (-572 G/C) is also located within the promoter region of IL-6 gene. The individuals harboring -572GG or GC genotype was observed to have significantly lower IL-6 levels than those harboring the -572CC genotype.[52] Also, CD14(+) monocytes from subjects carrying the rs1800796C allele were shown to produce more IL-6 in response to in vitro HBV core antigen stimulation than those carrying G allele.[12] Thus, rs1800796 C allelic or genotype (CC or GC) polymorphism may be associated with an increased risk to HBV infection, which was confirmed in both of our study involving 1772 cases and 1973 controls and the study of Chang et al[42] involving 426 cases and 777 controls. However, this conclusion seemed to be only suitable to the Asian population. In the non-Asian group, GC and GG+GC genotype were risk factors for the development of liver diseases, which was in line with some studies showing the mRNA expressions of IL-6 was higher in the rs1800796 GG genotype compare with others.[45,53]
rs1800797 (-579 G/A) is also another polymorphism located within the promoter region of IL-6 gene. Our overall, non-Asian subgroup analysis and the study of Chang et al,[42] showed GG and GA genotype may be risk factors for liver diseases, indicating patients with these genotypes may have higher IL-6 levels. However, recent studies on lung cancer or obesity revealed IL-6 expression level was increased in an A allelic dose-dependent manner (that is, the highest for AA),[54,55] which may be attributed to the dual-function of IL-6[56] or disease difference.
There are several limitations in this meta-analysis. First, the number of studies in some liver disease subtypes was relatively small and thus statistical power may be still sufficient to estimate the correlation between the IL-6 gene polymorphisms with them. Second, articles in languages other than English were not included in this meta-analysis. Third, although the meta-analysis only included case-control designed studies, several studies did not report whether they were age and sex matched, which may influence the creditability of conclusions. Fourth, although there were studies to indicate a linkage disequilibrium between some SNPs of IL-6 (such as rs1800796-rs1800797,[13] rs1800796-rs2066992,[14] rs2069837-rs17147230,[20] rs2069837-rs1524107-rs2066992,[19] rs17147230-rs2066992-rs2069837-rs2069852,[18] rs1800796-rs1800797,[13] and rs1800795-rs1800797[17]) and haplotypes were calculated for more effective markers for prediction the risk of liver diseases, no meta-analysis was conducted for these haplotypes because no same haplotypes were reported. Fifth, the association between IL-6 level and IL-6 gene polymorphisms could not be evaluated to reveal the function mechanisms due to the lack of the related data.
In conclusion, our meta-analysis of 25 studies revealed that IL-6 rs1800795 (all ethnic populations) and rs1800797 (non-Asian) polymorphisms may be associated with an increased risk of liver diseases, while rs1800796 polymorphism was associated with a decreased susceptibility factor for liver diseases in Asian population. The absence of a relationship between IL-6 rs2069837 and rs2066992 polymorphisms and the risk of liver diseases was demonstrated. A similar conclusion was found in the HBV, HCV, HCC, NASH and alcoholic liver disease population for rs1800795; HBV subgroup for rs1800796; and HCV subgroup for rs1800797.
Author contributions
Conceptualization: Xuehan Wang.
Data curation: Xuehan Wang, Zhenghui Yan.
Formal analysis: Xuehan Wang, Zhenghui Yan, Qingjian Ye.
Investigation: Zhenghui Yan.
Methodology: Zhenghui Yan.
Writing – original draft: Xuehan Wang, Qingjian Ye.
Writing – review & editing: Xuehan Wang, Qingjian Ye.
Footnotes
Abbreviations: CI = confidence interval, HBV = hepatitis B virus, HC = healthy, HCC = hepatocellular carcinoma, HCV = hepatitis C virus, HIV = human immunodeficiency virus-type 1, IF = infection resolved, IL-6 = interleukin-6, LC = liver cirrhosis, NASH = nonalcoholic steatohepatitis, NOS = New-castle–Ottawa Scale, OR = odds ratio, PRISMA = Preferred Reporting Items for Systematic Review and Meta-analysis, SNP = single nucleotide polymorphism, STAT3 = signal transducer and activator of transcription 3.
How to cite this article: Wang X, Yan Z, Ye Q. Interleukin-6 gene polymorphisms and susceptibility to liver diseases: A meta-analysis. Medicine. 2019;98:50(e18408).
The authors have no funding and conflicts of interests to disclose.
References
- [1].Vikramjit M, Jane M. Metabolic functions of the liver. Anaesth Intensive Care Med 2009;10:334–5. [Google Scholar]
- [2].Knolle PA, Wohlleber D. Immunological functions of liver sinusoidal endothelial cells. Cell Mol Immunol 2016;13:347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Grant DM. Detoxification pathways in the liver. J Inherit Metab Dis 1991;14:421–30. [DOI] [PubMed] [Google Scholar]
- [4].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Ca Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [5].Tacke F. Targeting hepatic macrophages to treat liver diseases. J Hepatol 2017;66:1300–12. [DOI] [PubMed] [Google Scholar]
- [6].Gudowska-Sawczuk M, Wrona A, Gruszewska E, et al. Serum level of interleukin-6 (IL-6) and N-terminal propeptide of procollagen type I (PINP) in patients with liver diseases. Scand J Clin Lab Invest 2018;78:125–30. [DOI] [PubMed] [Google Scholar]
- [7].Kim HY, Jhun JY, Cho ML, et al. Interleukin-6 upregulates Th17 response via mTOR/STAT3 pathway in acute-on-chronic hepatitis B liver failure. J Gastroenterol Hepatol 2014;49:1264–73. [DOI] [PubMed] [Google Scholar]
- [8].Fuster D, Tsui JI, Cheng DM, et al. Interleukin-6 is associated with noninvasive markers of liver fibrosis in HIV-infected patients with alcohol problems. AIDS Res Hum Retroviruses 2013;29:1110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shao YY, Lin H, Li YS, et al. High plasma interleukin-6 levels associated with poor prognosis of patients with advanced hepatocellular carcinoma. Jpn J Clin Oncol 2017;47:949–53. [DOI] [PubMed] [Google Scholar]
- [10].Hu Z, Luo D, Wang D, et al. IL-17 activates the IL-6/STAT3 signal pathway in the proliferation of hepatitis B virus-related hepatocellular carcinoma. Cell Physiol Biochem 2017;43:2379–90. [DOI] [PubMed] [Google Scholar]
- [11].Wang Z, Wu S, Liao J, et al. Interleukin-6 and rs1800796 locus single nucleotide polymorphisms in response to hypoxia/reoxygenation in hepatocytes. Int J Mol Med 2013;38:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang G, Wang W, Li S, et al. IL6 gene allele-specific C/EBP(-binding activity affects the development of HBV infection through modulation of Th17/Treg balance. Genes Immun 2015;16:528–35. [DOI] [PubMed] [Google Scholar]
- [13].El-Maadawy EA, Talaat RM, Ahmed MM, et al. Interleukin-6 promotor gene polymorphisms and susceptibility to chronic hepatitis B virus in Egyptians. Hum Immunol 2019;80:208–14. [DOI] [PubMed] [Google Scholar]
- [14].Riazalhosseini B, Mohamed Z, Apalasamy YD, et al. Interleukin-6 gene variants are associated with reduced risk of chronicity in hepatitis B virus infection in a Malaysian population. Biomed Rep 2018;9:213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tang S, Liu Z, Zhang Y, et al. Rather than Rs1800796 polymorphism, expression of interleukin-6 is associated with disease progression of chronic HBV infection in a Chinese Han population. Dis Markers 2013;35:799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cussigh A, Falleti E, Fabris C, et al. Interleukin 6 promoter polymorphisms influence the outcome of chronic hepatitis C. Immunogenetics 2015;63:33–41. [DOI] [PubMed] [Google Scholar]
- [17].Falleti E, Fabris C, Toniutto P, et al. Interleukin-6 polymorphisms and gender: relationship with the occurrence of hepatocellular carcinoma in patients with end-stage liver disease. Oncology 2015;77:304–13. [DOI] [PubMed] [Google Scholar]
- [18].Zhao XM, Gao YF, Zhou Q, et al. Relationship between interleukin-6 polymorphism and susceptibility to chronic hepatitis B virus infection. World J Gastroenterol 2013;19:6888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang J, Chen R, Tang S, et al. Analysis of IL-6, STAT3 and HSPA1L gene polymorphisms in anti-tuberculosis drug-induced hepatitis in a nested case-control study. PLoS One 2015;10:e0118862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zheng X, Han C, Shan R, et al. Association of interleukin-6 polymorphisms with susceptibility to hepatocellular carcinoma. Int J Clin Exp Med 2015;8:6252–6. [PMC free article] [PubMed] [Google Scholar]
- [21].Shamseer L, Moher D, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ 2015;349:g7647–17647. [DOI] [PubMed] [Google Scholar]
- [22].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [23].Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000;56:455–63. [DOI] [PubMed] [Google Scholar]
- [24].Yousefi A, Najafi M, Motamed F, et al. Association of Interleukin-6 and Interleukin-1 Family Gene Polymorphisms in Autoimmune Hepatitis. Ann Hepatol 2018;17:1021–5. [DOI] [PubMed] [Google Scholar]
- [25].Kurbatova IV, Topchieva LV, Dudanova OP. Caspase 3, 6, 8, and 9 gene expression in peripheral blood leukocytes and plasma concentrations of il-6 and TNF-( in carriers of different polymorphic marker -174G>C genotypes of IL6 gene associated with the risk of nonalcoholic steatohepatitis. Bull Exp Biol Med 2017;162:370–4. [DOI] [PubMed] [Google Scholar]
- [26].Bocsan IC, Milaciu MV, Pop RM, et al. Cytokines genotype-phenotype correlation in nonalcoholic steatohepatitis. Oxid Med Cell Longev 2017;2017:4297206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Motawi T, Shaker OG, Hussein RM, et al. Polymorphisms of (1-antitrypsin and Interleukin-6 genes and the progression of hepatic cirrhosis in patients with a hepatitis C virus infection. Balkan J Med Genet 2017;19:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Attar M, Azar SS, Shahbazi M. Interleukin-6-174 promoter polymorphism and susceptibility to hepatitis B virus infection as a risk factor for hepatocellular carcinoma in Iran. Asian Pac J Cancer Prev 2017;17:2395–9. [PubMed] [Google Scholar]
- [29].Lu Y, Peng J, Wang C, et al. IL-6 promoter functional polymorphism -572C/G affects spontaneous clearance of hepatitis B virus infection. Clin Lab 2014;60:1903–7. [DOI] [PubMed] [Google Scholar]
- [30].Saxena R, Chawla YK, Verma I, et al. IL-6(2572/2597) polymorphism and expression in HBV disease chronicity in an Indian population. Am J Hum Biol 2017;26:549–55. [DOI] [PubMed] [Google Scholar]
- [31].Tarragô AM, da Costa AG, Pimentel JP, et al. Combined impact of hepatitis C virus genotype 1 and interleukin-6 and tumor necrosis factor-( polymorphisms on serum levels of pro-inflammatory cytokines in Brazilian HCV-infected patients. Hum Immunol 2014;75:1075–83. [DOI] [PubMed] [Google Scholar]
- [32].Devi SG, Kumar A, Kar P, et al. Association of pregnancy outcome with cytokine gene polymorphisms in HEV infection during pregnancy. J Med Virol 2017;86:1366–76. [DOI] [PubMed] [Google Scholar]
- [33].Giannitrapani L, Soresi M, Giacalone A, et al. IL-6 -174G/C polymorphism and IL-6 serum levels in patients with liver cirrhosis and hepatocellular carcinoma. OMICS 2011;15:183–6. [DOI] [PubMed] [Google Scholar]
- [34].Falleti E, Fabris C, Toniutto P, et al. Interleukin-6 polymorphisms and gender: relationship with the occurrence of hepatocellular carcinoma in patients with end-stage liver disease. Oncology 2009;77:304–13. [DOI] [PubMed] [Google Scholar]
- [35].Carulli L, Canedi I, Rondinella S, et al. Genetic polymorphisms in non-alcoholic fatty liver disease: interleukin-6-174G/C polymorphism is associated with non-alcoholic steatohepatitis. Dig Liver Dis 2009;41:823–8. [DOI] [PubMed] [Google Scholar]
- [36].Marcos M, Pastor I, González-Sarmiento R, et al. Common polymorphisms in interleukin genes (IL4, IL6, IL8 and IL12) are not associated with alcoholic liver disease or alcoholism in Spanish men. Cytokine 2009;45:158–61. [DOI] [PubMed] [Google Scholar]
- [37].Gleeson D, Bradley MP, Jones J, et al. Cytokine gene polymorphisms in heavy drinkers with and without decompensated liver disease: a case-control study. Am J Gastroenterol 2008;103:3039–46. [DOI] [PubMed] [Google Scholar]
- [38].Ribeiro CS, Visentainer JE, Moliterno RA. Association of cytokine genetic polymorphism with hepatitis B infection evolution in adult patients. Mem Inst Oswaldo Cruz 2007;102:435–40. [DOI] [PubMed] [Google Scholar]
- [39].Minton EJ, Smillie D, Smith P, et al. Clearance of hepatitis C virus is not associated with single nucleotide polymorphisms in the IL-1, -6, or -10 genes. Hum Immunol 2005;66:127–32. [DOI] [PubMed] [Google Scholar]
- [40].Mustafa C, Demet Gokalp Y, Mehmet Ali E, et al. The role of interleukin-6 and interleukin-8 gene polymorphisms in non-alcoholic steatohepatitis. Hepat Mon 2014;14:e24635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu Y, Gao SJ, Du BX, et al. Association of IL-6 polymorphisms with hepatocellular carcinoma risk: evidences from a meta-analysis. Tumour Biol 2014;35:3551–61. [DOI] [PubMed] [Google Scholar]
- [42].Chang L, Lan T, Wu L, et al. The association between three IL-6 polymorphisms and HBV-related liver diseases: a meta-analysis. Int J Clin Exp Med 2016;8:17036–45. [PMC free article] [PubMed] [Google Scholar]
- [43].Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest 1998;102:1369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Belluco C, Olivieri F, Bonafè M, et al. -174 G>C polymorphism of interleukin 6 gene promoter affects interleukin 6 serum level in patients with colorectal cancer. Clin Cancer Res 2003;9:2173–6. [PubMed] [Google Scholar]
- [45].Lu QK, Zhang JT, Zhao N, et al. Association of IL-6 gene (-174 and -572 G/C) polymorphisms with proliferative diabetic retinopathy of type 2 diabetes in a Chinese population. Ophthalmic Res 2017;58:162–7. [DOI] [PubMed] [Google Scholar]
- [46].Chen Z, Li YX, Fu HJ, et al. Hepatitis B virus core antigen stimulates IL-6 expression via p38, ERK and NF -κB pathways in hepatocytes. Cell Physiol Biochem 2002;41:91–100. [DOI] [PubMed] [Google Scholar]
- [47].Zhou X, Yang F, Yang Y, et al. HBV facilitated hepatocellular carcinoma cells proliferation by up-regulating angiogenin expression through IL-6. Cell Physiol Biochem 2018;46:461–70. [DOI] [PubMed] [Google Scholar]
- [48].Hösel M, Quasdorff M, Ringelhan M, et al. Hepatitis B virus activates signal transducer and activator of transcription 3 supporting hepatocyte survival and virus replication. Cell Mol Gastroenterol Hepatol 2002;4:339–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dai CY, Tsai YS, Chou WW, et al. The IL-6/STAT3 pathway upregulates microRNA-125b expression in hepatitis C virus infection. Oncotarget 2018;9:11291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Min HK, Mirshahi F, Verdianelli A, et al. Activation of the GP130-STAT3 axis and its potential implications in nonalcoholic fatty liver disease. Am J Physiol Gastrointest Liver Physiol 2015;308:G794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Li M, He Y, Zhou Z, et al. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47phox-oxidative stress pathway in neutrophils. Gut 2017;66:705–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fang M, Huang Y, Zhang Y, et al. Interleukin-6 -572C/G polymorphism is associated with serum interleukin-6 levels and risk of idiopathic pulmonary arterial hypertension. J Am Soc Hypertens 2017;11:171–7. [DOI] [PubMed] [Google Scholar]
- [53].Ding Y, Feng Q, Chen J, et al. TLR4/NF-κB signaling pathway gene single nucleotide polymorphisms alter gene expression levels and affect ARDS occurrence and prognosis outcomes. Medicine 2019;98:e16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Leng S, Thomas CL, Snider AM, et al. Radon exposure, IL-6 promoter variants, and lung squamous cell carcinoma in former uranium miners. Environ Health Perspect 2016;124:445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Boeta-Lopez K, Duran J, Elizondo D, et al. Association of interleukin-6 polymorphisms with obesity or metabolic traits in young Mexican-Americans. Obes Sci Pract 2017;4:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Scheller J, Chalaris A, Schmidt-Arras D, et al. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta 2011;1813:878–88. [DOI] [PubMed] [Google Scholar]


