Abstract
The purpose of our research was to evaluate diagnostic performance of serum microRNA-135a (miR-135a) in non-small cell lung cancer (NSCLC).
Quantitative real time-polymerase chain reaction was employed to detect the expression serum of miR-135a in NSCLC patients and controls. The influence of serum miR-135a level on clinical characteristics of NSCLC patients was explored through the Chi-square test. Serum carcinoembryonic antigen (CEA) level was estimated via enzyme-linked immunosorbent assay. Receiver operating characteristic (ROC) curve was plotted to elucidate diagnostic roles of serum miR-135a and CEA in NSCLC.
The expression level of serum miR-135a was significantly lower in NSCLC patients than in healthy controls (0.40 ± 0.29 vs 1.00 ± 0.40, P < .001). Moreover, miR-135a expression was related to lymph node metastasis (P = .021), tumor differentiation (P = .020), and tumor node metastasis stage (P = .031). ROC curve showed serum miR-135a level could discriminate NSCLC patients from healthy controls (P < .0001) with a corresponding cutoff value of 0.665, and a sensitivity and specificity of 81.3% and 83.1%, respectively. The area under the curve was 0.888. In diagnosis analysis on the combination of miR-135a and CEA, when its specificity was maintained at 90%, diagnosis cut-off point reached 0.678.
Serum miR-135a level is significantly downregulated in NSCLC and serves as a potential diagnostic biomarker for the disease.
Keywords: diagnosis, miR-135a, non-small cell lung cancer, ROC
1. Introduction
Lung cancer is one of the most frequent and aggressive malignancies with high morbidity all over the world.[1,2] Besides, lung cancer is also reported to be the leading cause of cancer-related deaths.[3,4] According to the World Health Organization classification, lung cancer could be divided into small cell lung cancer and non-small cell lung cancer (NSCLC), with the later accounting for approximately 85% of total lung cancer cases.[5–7] Currently, complete surgical resection is the most effective treatment for early NSCLC.[8] However, it has been reported that the majority of NSCLC cases have already entered into advanced stages when diagnosed due to the lack of specific symptoms in early stages, leading to dismal survival.[9,10] Early screening and treatment are key to improve the prognosis of NSCLC patients. Serum carcinoembryonic antigen (CEA) is widely accepted in early diagnosis of NSCLC. Diagnostic specificity of serum CEA for NSCLC may be more than 90%, but its sensitivity is relatively low, leading to missed diagnosis and delay in treatments.[11,12] Thus, noninvasive and sensitive biological biomarkers are needed for early detection of NSCLC.
MicroRNAs (miRNAs) are endogenous, conserved short noncoding RNAs containing about 22 nucleotides.[13,14] Up to date, thousands of miRNAs have been identified as tumor suppressors or oncogenes. They could regulate most of human genes at post-transcription level via binding to the 3′-untranslated region of target genes.[15,16] A growing number of researches have demonstrated that miRNAs play important roles in various biological activities, such as the proliferation, invasion, metastasis, differentiation, apoptosis, and survival of cells or organisms.[17,18] Expression patterns of miRNAs show specific to cell types, tissues, and development stages. Dysregulated miRNAs can influence several signaling pathways through their target genes, thus leading to human diseases, like cancer.[19] MiRNAs are stable and abound in human fluids, such as blood, urine, and saliva, and thus can be detected through easy and noninvasion methods.[20] Therefore, miRNAs are considered as promising biomarkers in cancer diagnosis. MiR-135a is a common member of miRNA family, and its dysregulation has been reported in NSCLC by Shi et al.[21] Accordingly, the expression of miR-135a was significantly downregulated in NSCLC tissues, while its restored expression might decrease the migration and invasion of lung cancer cells. MiR-135a might act as a tumor suppressor in NSCLC. Additionally, the study performed by Zhang et al reported that serum levels of miR-135a were significantly decreased in NSCLC patients compared to healthy individuals and showed close association with the disease progression.[22] Serum miR-135a might be a potential biomarker for NSCLC. Based on published articles, we speculated that serum levels of miR-135a might be correlated with NSCLC inhibition, development and progression. Serum miR-135a might be used for early diagnosis of NSCLC. However, whether serum miR-135a could be employed as diagnostic biomarker for NSCLC was still unclear.
The present study was conducted to determine diagnostic role of serum miR-135a in NSCLC using receiver operating characteristic (ROC) curve. The present study might develop a quick and convenient way for NSCLC diagnosis.
2. Materials and methods
2.1. Patients and specimens
The enrolled 117 patients, who were pathologically diagnosed with NSCLC, were from Ankang Central Hospital. Other 83 blood donors in the same hospital were collected as healthy controls. No radio- or chemotherapeutic modality was conducted on these patients before sampling. And their clinical information was recorded in a database. An aliquote of 5 mL peripheral blood was obtained from every participant. And serum specimens were centrifuged and stored at −80°C for later RNA isolation. This study was permitted by the Ethics Committee of Ankang Central Hospital. All participants had signed an informed consent in advance.
2.2. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from previously prepared serum samples using miRNA isolation Kit (Applied Biosystems, Foster City, CA) following the manufacture's instruction. The first strand of complementary DNA was reversely transcribed from total RNA using the Taqman MicroRNA Reverse Transcription Kit (Applied Biosystem). U6 was used as an internal reference. The primers for miR-135a and U6 were as follows: miR-135a (Forward: 5′-ACACTCCAGCTGGGTATGGCTTTTTATTCCT-3′; Reverse: 5′-GGTGTCGTGGAGTCGGCAA-3′); U6 (Forward: 5′-CTCGCTTCGGCAGCACA-3′; Reverse: 5′-AACGCTTCACGAATTTGCGT-3′). PCR amplification for genes was carried out through the following thermal conditions: 95°C for 30 seconds followed by 40 cycles of 95°C for 5 seconds and 60°C for 30 seconds. PCR conditions for miR-135a amplification were: 95°C for 20 seconds, followed by 40 cycles of 95°C for 10 seconds and 60°C for 20 seconds, with a final incubation at 70°C for 5 seconds. qRT-PCR was carried out under optimal conditions. Expression level of miR-135a was normalized to U6, and determined using the 2−ΔΔCt method.
2.3. Enzyme-linked immunosorbent assay (ELISA)
Serum levels of CEA were detected using ELISA method. The assay was performed using the third generation ELISA kits (Can Ag, Canada). The procedures were carried out according to the introduction of the manufacturer. Each sample was repeated at least 3 times.
2.4. Statistical analysis
All data were analyzed by SPSS 18.0 software. Expression values of serum miR-135a and CEA were expressed as mean ± standard deviation, and different expression of miR-35a between NSCLC and healthy controls were analyzed using the Student t test. The associations between serum miR-135a expression and patients’ clinical parameters were detected via the Pearson Chi-square test. Diagnostic value of serum miR-135a in NSCLC was estimated adopting the ROC analysis. Moreover, we explored diagnostic value of miR-35a combined with CEA in NSCLC through a method of a biomarker combined with a standard screening test marker reported by Shaw et al.[23] Relative ROC curve was drawn to express relative true positive rate versus relative false positive rate as biomarker threshold for positivity varies. P < .05 was considered to be of statistical significance.
3. Results
3.1. Baseline characteristics of the study subjects
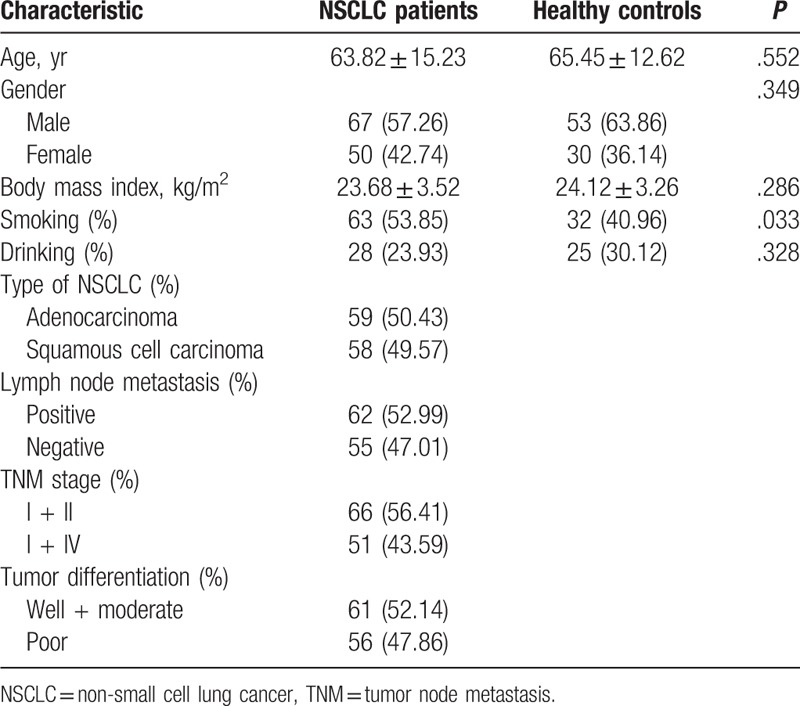
A total of 117 NSCLC patients and 83 healthy individuals were included in our study. There were 67 males and 50 females in NSCLC group, while the control group included 53 men and 30 women. Gender distributions did not show significant differences between case and control groups (P = .349). The average age of NSCLC patients was 63.82 ± 15.23 years, and the mean age of the included healthy individuals was 65.45 ± 12.62 years. The 2 study groups did not show obvious difference in age (P = .552). The NSCLC and control groups were matched in age and gender (P > .05 for both). Among NSCLC patients, 63 had smoking history, while there were 32 smokers in the healthy control group. The percentage of smokers was significantly higher in the NSCLC group than in the control group (P = .033). We found no obvious difference between the 2 groups in either body mass index or drinking status (P > .05). Detailed data were listed in Table 1.
Table 1.
The clinical characteristics of subjects in NSCLC patients and healthy controls.
According to pathological examinations, 59 patients were diagnosed with adenocarcinoma, while 58 with squamous cell carcinoma. Lymph node metastasis was observed in 62 patients. Additionally, 61 patients exhibited well and moderate differentiation, while 56 showed poor differentiation. According to tumor node metastasis (TNM) staging system, 66 patients were classified into stages I + II, and 51 into III + IV. Clinical characteristics of the patients were summarized in Table 2.
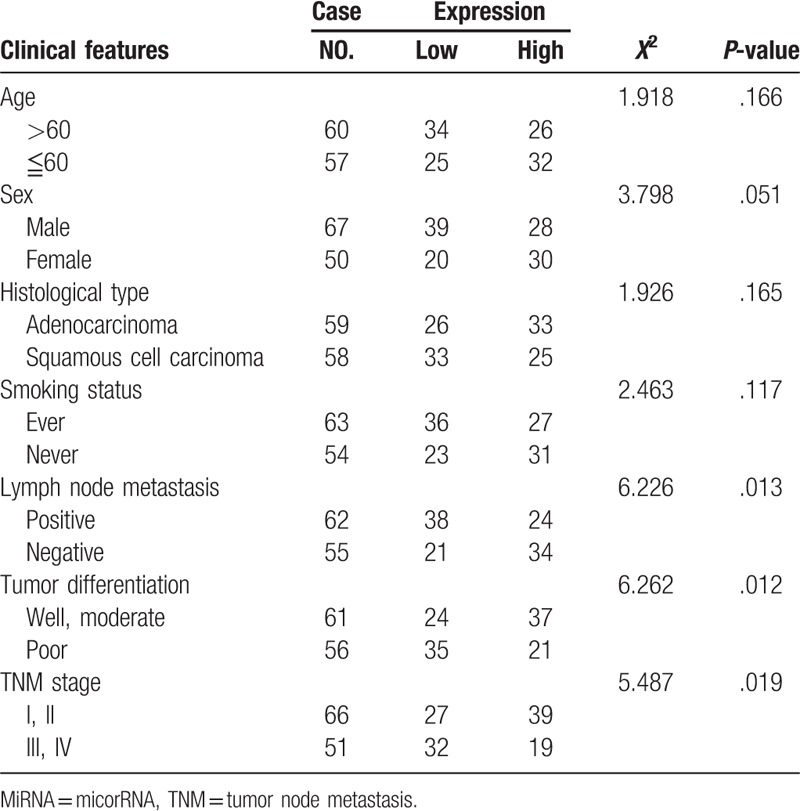
Table 2.
Relationship between miR-135a expression and clinical characteristics.
3.2. Decreased expression of serum miR-135a in NSCLC patients
Expression levels of serum miR-135a in NSCLC patients and healthy controls were detected using qRT-PCR. As shown in Figure 1, the level of serum miR-135a was significantly decreased in NSCLC patients (0.40 ± 0.29) compared with healthy controls (1.00 ± 0.40) (P < .001).
Figure 1.
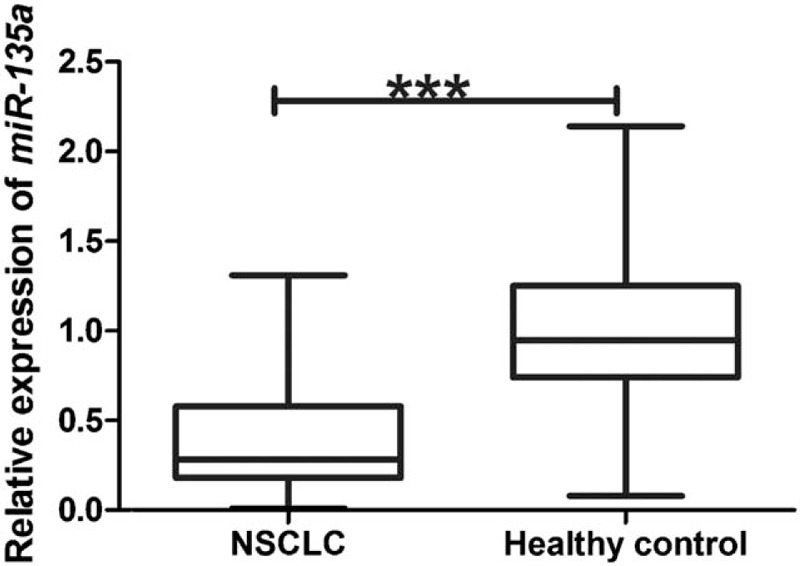
The expression of serum miR-135a in NSCLC patients and healthy controls. Serum miR-135a was weakly expressed in NSCLC patients (mean ± SD: 0.40 ± 0.29) compared with healthy individuals (1.00 ± 0.40). The differences in serum miR-135a levels between NSCLC and healthy controls were significant, according to the Student t test. ∗∗∗P < .001. NSCLC = non-small cell lung cancer, SD = standard deviation.
3.3. Association of serum miR-135a expression with clinicopathological characteristics of NSCLC patients
To determine the correlation of miR-135a expression with clinical characteristics, NSCLC patients were divided into high (n = 58) and low (n = 59) level groups based on their median value of serum miR-135a expression. As listed in Table 1, low miR-135a expression was positively associated with lymph node metastasis (P = .013), poor tumor differentiation (P = .012), and advanced TNM stage (P = .019). However, no significant relationship was found between miR-135a expression and other clinicopathological parameters, including age (P = .166), sex (P = .051), histological type (P = .165), and smoking status (P = .117).
3.4. The expression and diagnostic value of CEA in NSCLC
CEA is a widely accepted blood biomarker for early detection of NSCLC. In our study, we estimated serum CEA levels of the subjects. Mean CEA level was 14.82 ± 17.24 ng/mL in NSCLC patients, and such figure in healthy control was 3.37 ± 2.74 ng/mL. Compared with healthy controls, NSCLC patients showed significantly increased CEA levels (P < .001) (Fig. 2A). ROC curve was plotted to estimate diagnostic value of serum CEA in NSCLC. As displayed in Figure 2B, serum CEA level could distinguish NSCLC patients from the healthy individuals with an area under the curve (AUC) of 0.811 (95% confidence interval [CI]: 0.751–0.872). The cut-off value of serum CEA for NSCLC diagnosis was 6.50 ng/mL, and the corresponding sensitivity and specificity were 60.7% and 97.6%, respectively. From the curve, we also found that when the specificity remained at 90%, diagnostic sensitivity was about 64%.
Figure 2.
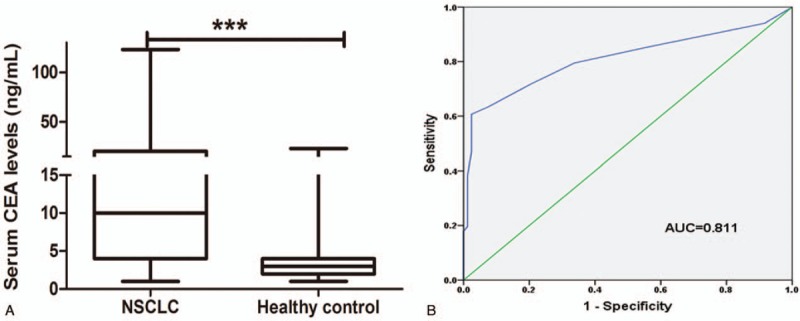
Expression patterns and diagnostic value of serum CEA in NSCLC. Compared to healthy controls (3.37 ± 2.74 ng/mL), NSCLC patients exhibited elevated levels of serum CEA (14.82 ± 17.24 ng/mL). Student t test revealed significant differences (A). ROC curve demonstrated that serum CEA could discriminate between NSCLC patients and healthy individuals (B). CEA = carcinoembryonic antigen, NSCLC = non-small cell lung cancer, ROC = receiver operating characteristic.
3.5. Diagnostic significance of serum miR-135a in NSCLC
To estimate diagnostic utility of serum miR-135a in NSCLC, we performed ROC curve analysis. As indicated in Figure 3, the curve identified serum miR-135a as a promising diagnostic biomarker for NSCLC, with an AUC of 0.888 (95% CI: 0.834–0.943). In addition, corresponding optimal cut-off point was 0.665, providing a sensitivity and specificity of 81.2% and 83.1%, respectively. The results showed that miR-135a could distinguish NSCLC cases from healthy controls. Serum miR-135a might improve the sensitivity of serum CEA in NSCLC diagnosis.
Figure 3.
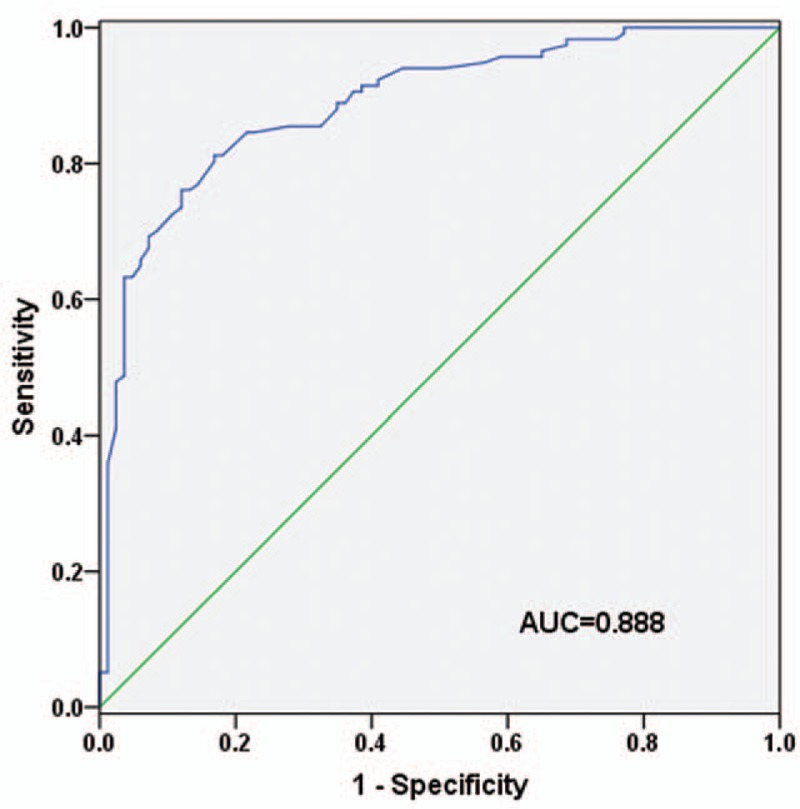
Diagnostic accuracy of serum miR-135a in NSCLC. The result showed that serum miR-135a possessed high diagnostic value in NSCLC (P < .001). NSCLC = non-small cell lung cancer.
Moreover, based on the method reported by Shaw et al,[23] we estimated diagnostic performance for the combination of serum miR-135a and CEA in NSCLC. As displayed in Figure 4, relative AUC was 0.919 (95% CI: 0.879–0.960), with a relative sensitivity of 91.5% and a relative specificity of 83.1%. When relative specificity was maintained at 90%, the diagnosis cut-off point was 0.678. When prediction probability among individuals equal or was greater than 0.678, they were deemed as positive.
Figure 4.
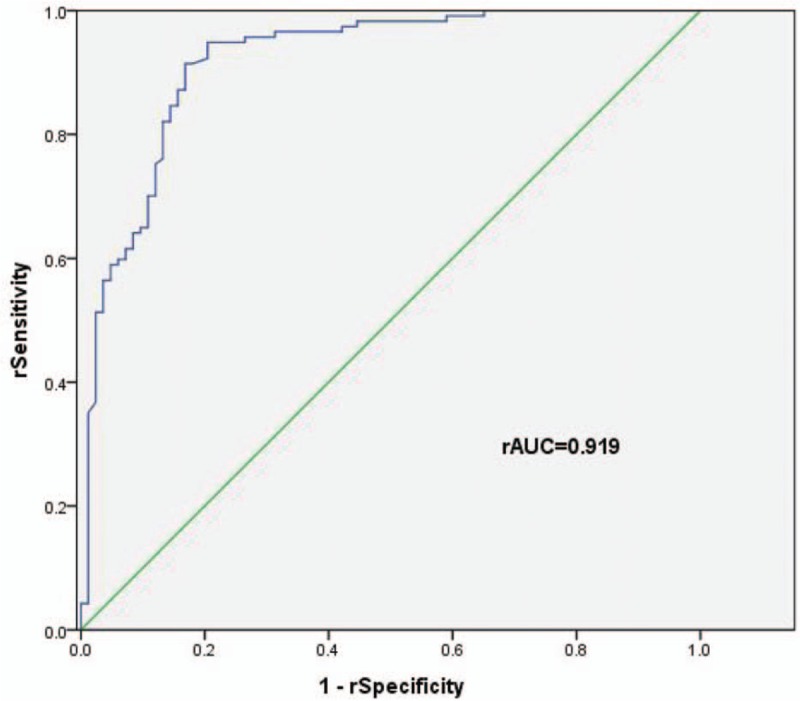
Relative ROC curve plotted for the combination CEA and miR-135a in NSCLC cases and healthy individuals. Serum miR-135a might increase diagnostic sensitivity of CEA in NSCLC by 3%. CEA = carcinoembryonic antigen, NSCLC = non-small cell lung cancer, ROC = receiver operating characteristic.
4. Discussion
Lung cancer is one of the tumors with highest incidence and mortality, and its patients are frequently diagnosed after entering advanced stages, resulting in relatively low 5-year survival rate. Thus, early detection and intervention of NSCLC are particularly important to improve patients’ outcomes. At present, screening methods for NSCLC mainly contain sputum cytology, computed tomography, positron emission tomography, and magnetic resonance imaging, all of which are expensive, invasive, and radioactive.[24] Moreover, to our knowledge, the abnormality of tumor markers in peripheral blood often occurs earlier than that of imaging detection, which might be of significant meaning for cancers with no clinical symptoms at early stages. Therefore, we would like to find a low-cost and noninvasive way for early NSCLC diagnosis.
In recent years, the discovery of miRNAs as disease markers has attracted attentions from numerous scientists, especially for cancer diagnosis and prognosis. MiRNAs are stable in serum and plasma, and they are under little effects from repeated freezing and thawing or pH change. More importantly, they might offer a minimally invasive method in clinical to exhibit health status for people only through blood testing. These advantages made it possible to use miRNAs detection as a screening or auxiliary diagnosis method. So far, considerable studies have explored the role of serum miRNAs in diverse cancers. Toiyama et al demonstrated that serum miR-21 could act as a diagnostic candidate for colorectal cancer.[25] In the study of Sun et al, serum miR-367 displayed great diagnostic significance in esophageal squamous cell carcinoma.[26] However, diagnostic performance of miR-135a in NSCLC has not been explored.
Based on existent documents, we discussed diagnostic role of serum miR-135a in NSCLC. Serum miR-135a was weakly expressed in NSCLC patients compared to healthy controls, which agreed with former findings.[21,22] The downregulation of miR-135a in tumorigenesis might be mediated by DNA methylation.[27] Such conclusion needed further investigation. Besides, abnormal expression of serum miR-135a was significantly related to lymph node metastasis, tumor differentiation, and TNM stage, indicating serum miR-135a might be involved in the development and progression of NSCLC. On the basis of previous findings, we further plotted ROC curve to calculate diagnostic value for serum miR-135a in NSCLC. The results demonstrated that serum miR-135a could distinguish NSCLC patients from the healthy individuals. Moreover, diagnostic accuracy and sensitivity of serum miR-135a were superior to CEA among our study subjects. Besides, we explored diagnostic performance for the combination of miR-135a with CEA in NSCLC. As a result, when the specificity was maintained at 90%, the diagnosis cut-off was 0.678. That is, when prediction probability for patients was equal to or greater than 0.678, the patients were diagnosed with NSCLC. The combination of biomarker with a standard screening test marker improved diagnostic accuracy, compared with 1 single biomarker.
In the present study, we confirmed diagnostic value of serum miR-135a in NSCLC. However, several limitations should be stated. First, concrete mechanisms how serum miR-135a functions in NSCLC tumorigenesis and development are still unclear. As mentioned above, miRNAs could affect tumor progression through regulating target genes. Published studies have explained that miR-135a plays an important role in tumor proliferation, invasion and migration through targeting FOXO1, TRPC1, and JAK2.[28–30] Further investigations should be carried out based on these findings. Second, published article reported that miR-135a downregulation in tumorigenesis was mediated by DNA methylation.[27] Whether the conclusion was valid in NSCLC needs further exploration. Thirdly, due to short study period, follow-up investigation was not performed among the study population. Only 1 cohort of patients from the Chinese Han population was used in this study, and no public available dataset was used to evaluate the diagnostic value of miR-135a. The study sample size was also relative small. Clinical significance of miR-135a in prognosis evaluation among NSCLC patients was not explored in our study. In addition, the sample size was relatively small that might influence statistical power of our results. So the application of serum miR-135a in NSCLC detection might cause diagnostic errors. The combination of serum miR-135a with other biomarkers might be an effective way to improve early screening of NSCLC. Therefore, further studies with extended sample size are required to improve serum miR-135a-involved measures for NSCLC diagnosis.
In conclusion, downregulated expression of miR-135a was observed in NSCLC, which might be associated with the development of the disease. Serum miR-135a may be employed as an auxiliary biomarker for early diagnosis of NSCLC. Moreover, the combination of 1 biomarker with a standard screening test marker may be an important diagnostic means in clinic. Albeit above-listed shortcomings, our findings may provide certain basis for future researches on clinical value of miR-135a in early screening of NSCLC.
Author contributions
Conceptualization: Yuanwu Zou.
Data curation: Yuanwu Zou.
Formal analysis: Yuanwu Zou.
Funding acquisition: Ting Wang.
Investigation: Ting Wang.
Methodology: Ting Wang.
Project administration: Chengbao Jing, Ting Wang.
Resources: Chengbao Jing, Ting Wang.
Software: Chengbao Jing, Li Liu, Ting Wang.
Supervision: Chengbao Jing, Li Liu, Ting Wang.
Validation: Chengbao Jing, Li Liu, Ting Wang.
Visualization: Li Liu, Ting Wang.
Writing – original draft: Yuanwu Zou, Li Liu.
Writing – review and editing: Yuanwu Zou, Li Liu.
Footnotes
Abbreviations: AUC = area under the curve, CEA = carcinoembryonic antigen, ELISA = enzyme-linked immunosorbent assay, MicroRNA = miRNA, NSCLC = non-small cell lung cancer, QRT-PCR = quantitative real-time polymerase chain reaction, ROC = receiver operating characteristic, TNM = tumor node metastasis.
How to cite this article: Zou Y, Jing C, Liu L, Wang T. Serum microRNA-135a as a diagnostic biomarker in non-small cell lung cancer. Medicine. 2019;98:50(e17814).
The authors have no funding and conflicts of interest to disclose.
References
- [1].Lui NS, Yang YW, van Zante A, et al. SULF2 expression is a potential diagnostic and prognostic marker in lung cancer. PloS One 2016;11:e0148911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang D, Hao T, Pan Y, et al. Increased expression of SOX4 is a biomarker for malignant status and poor prognosis in patients with non-small cell lung cancer. Mol Cell Biochem 2015;402:75–82. [DOI] [PubMed] [Google Scholar]
- [3].Qiu ZX, Zhao S, Mo XM, et al. Overexpression of PROM1 (CD133) confers poor prognosis in non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:6589–95. [PMC free article] [PubMed] [Google Scholar]
- [4].Wang RJ, Zheng YH, Wang P, et al. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:765–71. [PMC free article] [PubMed] [Google Scholar]
- [5].Fan W, Yang H, Xue H, et al. ELMO3 is a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:5503–8. [PMC free article] [PubMed] [Google Scholar]
- [6].You Q, Guo H, Xu D. Distinct prognostic values and potential drug targets of ALDH1 isoenzymes in non-small-cell lung cancer. Drug Des Devel Ther 2015;9:5087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ren Y, Hou J, Xu A, et al. Diagnostic utility of PAX2 and PAX5 in distinguishing non-small cell lung cancer from small cell lung cancer. Int J Clin Exp Pathol 2015;8:14709–16. [PMC free article] [PubMed] [Google Scholar]
- [8].Cui D, Yu CH, Liu M, et al. Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumour Biol 2016;37:4127–34. [DOI] [PubMed] [Google Scholar]
- [9].Zhou HX, Yang MX, Wang Y, et al. Plasma LUNX mRNA, a non-invasive specific biomarker for diagnosis and prognostic prediction of non-small cell lung cancer. Am J Cancer Res 2016;6:452–8. [PMC free article] [PubMed] [Google Scholar]
- [10].Zhao W, Zhao JJ, Zhang L, et al. Serum miR-21 level: a potential diagnostic and prognostic biomarker for non-small cell lung cancer. Int J Clin Exp Med 2015;8:14759–63. [PMC free article] [PubMed] [Google Scholar]
- [11].Cho IH JY. Lung cancer biomarkers. Adv Clin Chem 2015;72:107–70. [DOI] [PubMed] [Google Scholar]
- [12].Grunnet M, Sorensen JB. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer 2012;76:138–43. [DOI] [PubMed] [Google Scholar]
- [13].Cho S, Mutlu L, Grechukhina O, et al. Circulating microRNAs as potential biomarkers for endometriosis. Fertil Steril 2015;103:1252–60.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hemmesi K, Squadrito ML, Mestdagh P, et al. miR-135a inhibits cancer stem cell-driven medulloblastoma development by directly repressing Arhgef6 expression. Stem Cells 2015;33:1377–89. [DOI] [PubMed] [Google Scholar]
- [15].Deng YQ, Yang YQ, Wang SB, et al. Intranasal administration of lentiviral miR-135a regulates mast cell and allergen-induced inflammation by targeting GATA-3. PloS One 2015;10:e0139322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tribollet V, Barenton B, Kroiss A, et al. miR-135a inhibits the invasion of cancer cells via suppression of ERRalpha. PloS One 2016;11:e0156445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhou H, Guo W, Zhao Y, et al. MicroRNA-135a acts as a putative tumor suppressor by directly targeting very low density lipoprotein receptor in human gallbladder cancer. Cancer Sci 2014;105:956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zeng YB, Liang XH, Zhang GX, et al. miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell Int 2016;16:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Harrandah AM, Mora RA, Chan EKL. Emerging microRNAs in cancer diagnosis, progression, and immune surveillance. Cancer Lett 2018;438:126–32. [DOI] [PubMed] [Google Scholar]
- [20].Yuan HL, Wang T, Zhang KH. MicroRNAs as potential biomarkers for diagnosis, therapy and prognosis of gastric cancer. Oncotargets Ther 2018;11:3891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shi H, Ji Y, Zhang D, et al. MiR-135a inhibits migration and invasion and regulates EMT-related marker genes by targeting KLF8 in lung cancer cells. Biochem Biophys Res Commun 2015;465:125–30. [DOI] [PubMed] [Google Scholar]
- [22].Zhang YK, Sun B, Sui G. Serum microRNA-135a downregulation as a prognostic marker of non-small cell lung cancer. Genet Mol Res 2016;15: [DOI] [PubMed] [Google Scholar]
- [23].Shaw PA, Pepe MS, Alonzo TA, et al. Methods for assessing improvement in specificity when a biomarker is combined with a standard screening test. Stat Biopharm Res 2009;1:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhu W, Zhou K, Zha Y, et al. Diagnostic value of serum mir-182, mir-183, mir-210, and mir-126 levels in patients with early-stage non-small cell lung cancer. PloS One 2016;11:e0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 2013;105:849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sun J, Song K, Feng X, et al. MicroRNA-367 is a potential diagnostic biomarker for patients with esophageal squamous cell carcinoma. Biochem Biophys Res Commun 2016;473:363–9. [DOI] [PubMed] [Google Scholar]
- [27].Shi L, Li X, Wu Z, et al. DNA methylation-mediated repression of miR-181a/135a/302c expression promotes the microsatellite-unstable colorectal cancer development and 5-FU resistance via targeting PLAG1. J Genet Genomics 2018;45:205–14. [DOI] [PubMed] [Google Scholar]
- [28].Mao XP, Zhang LS, Huang B, et al. Mir-135a enhances cellular proliferation through post-transcriptionally regulating PHLPP2 and FOXO1 in human bladder cancer. J Transl Med 2015;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].He F, Peng F, Xia X, et al. MiR-135a promotes renal fibrosis in diabetic nephropathy by regulating TRPC1. Diabetologia 2014;57:1726–36. [DOI] [PubMed] [Google Scholar]
- [30].Wu H, Huang M, Cao P, et al. MiR-135a targets JAK2 and inhibits gastric cancer cell proliferation. Cancer Biol Ther 2012;13:281–8. [DOI] [PubMed] [Google Scholar]


