Abstract
DNA methylation has been widely studied for associations with exposures and health outcomes. Both 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) are epigenetic marks that may function differently to impact gene expression; however, the most commonly used technology to assess methylation for population studies in blood use are the Illumina 450K and EPIC BeadChips, for which the traditional bisulfite conversion does not differentiate 5mC and 5hmC marks. We used a modified protocol originally developed by Stewart et al. to analyse oxidative bisulfite-converted and conventional bisulfite-converted DNA for the same subject in parallel by the EPIC chip, allowing us to isolate the two measures. We measured 5mC and 5hmC in cord blood of 41 newborn participants of the Center for Health Assessment of Mothers and Children of Salinas (CHAMACOS) birth cohort and investigated differential methylation of 5mC + 5hmC, isolated 5mC and isolated 5hmC with sex at birth as an example of a biological variable previously associated with DNA methylation. Results showed low levels of 5hmC throughout the epigenome in the cord blood samples in comparison to 5mC. The concordance of autosomal hits between 5mC + 5hmC and exclusive 5mC analyses were low (25%); however, overlap was larger with increased effect size difference. There were 43 autosomal cytosine nucleotide followed by a guanine nucleotide (CpG) sites where 5hmC was associated with sex, 21 of which were unique to 5hmC after adjustment for cell composition. 5hmC only accounts for a small portion of overall methylation in cord blood; however, it has the potential to impact interpretation of combined 5hmC + 5mC studies in cord blood, especially given that effect sizes of differential methylation analyses are often small. Several significant CpG sites were unique to 5hmC, suggesting some functions distinct from 5mC. More studies of genome-wide 5hmC in children are warranted.
Introduction
Epigenetic changes such as DNA methylation associated with prenatal exposures may be one mechanism impacting health outcomes later in life (1). The most common genome-wide measure of DNA methylation used in health studies comes from arrays such as the Illumina 450K and EPIC BeadChips. The traditional bisulfite conversion protocol for methylation assessment by these chips provides combined methylation levels for 5-hydroxymethylcytosine (5hmC) and 5-methylcytosine (5mC). 5hmC is the first oxidative product in the active demethylation of 5mC (2). Since these two types of methylation marks may function differently, it is important to tease apart associations of exposures and health outcomes with each of the two epigenetic measures.
The production of 5hmC seems to serve an important function, promoting gene expression during active demethylation during early stages of embryogenesis (3). 5hmC can also be associated with decreased gene expression depending on interactions with different regulatory elements such as TET family proteins and histone deacetylase complexes (6). In the brain and embryonic stem cells, 5hmC is enriched in promoters and gene bodies (5–7). 5hmC is also correlated with gene expression, with studies reporting increased 5hmC in gene bodies being associated with increased gene expression, while increased 5hmC in promoter regions has been associated with decreased gene expression (8,9). In mammals, 5hmC has been shown to regulate pluripotency of embryonic stem cells as well as other developmental processes (9). In humans, 5hmC levels are higher in the brain, liver, kidney and colorectal tissues, while low in the heart, breast and placenta (10). There is suggestive evidence that 5hmC may also play a role in brain development and may be associated with cancers (11,12) and age (13), as well as exposures to lead (14), arsenic (15) and mercury (16) in blood. A prior study in cord blood showed much lower levels of global 5hmC methylation compared to 5mC methylation with a ratio of 24:1 (5mC:5hmC) (16). Levels of site-specific 5hmC in cord blood have not been accessed using array technology.
Until recently, wide-scale research on 5hmC has been hindered by limitations of available methodologies, which have not been able to distinguish between 5mC and 5hmC, and, as a result, there are little data available in children regarding site-specific genome-wide changes in 5hmC in relation to either exposure or health outcomes. The majority of 5hmC studies have focussed on adults, have been mostly limited to cancers and have assessed global rather than site-specific DNA methylation. For example, one study reported lower levels of 5hmC in leukocyte DNA of patients with inflammatory bowel disease and colorectal cancer compared to healthy subjects (12), while another human study reported that 5hmC levels in circulating cell-free DNA could be used as a biomarker for different cancers (12). It also has been shown that blood levels of global 5hmC decreased with age, especially in those over age 70 (13). Blood levels of mercury in cord and early childhood blood were associated with lower 5hmC levels but were not significantly associated with 5mC levels (16). Lead exposure appeared to affect levels of regional 5hmC in the cord blood of newborns assessed with the 450K chip (14), while another study in adults reported an association between arsenic and global 5mC and 5hmC in adult blood (15). Both of these studies found no association between 5hmC and sex of the subjects.
In the present study, we use oxidative bisulfite conversion in combination with the EPIC array to measure 5hmC in DNA from the cord blood of newborns. This method was first described for adult whole blood and male brain tissue with the 450K array (17). By running bisulfite-converted and oxidative bisulfite-converted samples in parallel, it is possible to isolate the 5mC proportion of methylation from the 5hmC. We describe the distribution of 5hmC in cord blood DNA of the participants of the Center for Health Assessment of Mothers and Children of Salinas (CHAMACOS) birth cohort study using EPIC arrays. Our previous studies have established an association between child sex and DNA methylation at individual cytosine nucleotide followed by a guanine nucleotide (CpG) sites and regions (18) and with Alu and LINE-1 (19) repetitive elements in newborns and older children in CHAMACOS cohort. We now investigate the association between DNA methylation and sex after separating 5mC and 5hmC and compare these results to the associations with the combined measure of DNA methylation obtained by conventional use of the EPIC BeadChip.
Materials and methods
Study participants
Subjects were a convenience sample of the CHAMACOS longitudinal birth cohort study. A detailed description of the CHAMACOS cohort has previously been published (20). Briefly, women were eligible for study if they were Spanish or English speaking, at least 18 years of age, less than 20 weeks gestation and were receiving prenatal care at community clinics. From 1999 to 2000, 601 pregnant women were enrolled and 526 women were followed to their delivery of a live-born singleton. Study protocols were approved by the University of California, Berkeley Committee for Protection of Human Subjects and written informed consent was obtained from all mothers. For this study, 41 newborn subjects with sufficient quantity and high quality DNA (260/280 ratios between 1.8 and 2.2) were selected. Their characteristics did not differ from the rest of the cohort.
Blood collection and processing
Cord blood specimens were collected by hospital staff at time of delivery in vacutainers without heparin (red-top), separated into clots and serum and stored at −80°C until analysis. The protocol for cord blood collection for the CHAMACOS study was piloted to assure that it does not result in maternal blood admixture. Specifically, two assays were conducted: Fluorescent in situ hybridisation (FISH) analysis for sex chromosomes and PCR analysis using human leukocyte antigen markers. Cord blood samples of 30 randomly selected boys were analysed, and none of the assays indicated any contamination with maternal blood.
DNA preparation
DNA was isolated from the banked non-heparinised umbilical cord blood clot samples using QIAamp DNA Blood Maxi Kits (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol with minor modifications as previously described (21).
EPIC 5mC AND 5hmC analysis
Since bisulfite conversion cannot differentiate between 5mC and 5hmC, we used the method termed oxidative bisulfite (oxBS)-450K outlined in Stewart et al. with small modifications (17). Each sample was run on EPIC arrays at the same time using both traditional bisulfite-converted DNA as well as oxidative bisulfite-converted DNA to differentiate the 5hmC and 5mC levels. Briefly, 1 µg of DNA was used per subject, with equal quantities used for traditional bisulfite (BS) conversion and the oxBS protocol. DNA was processed using the TrueMethyl Array Kit (CEGX, Cambridge, UK) and applied to Illumina EPIC chips. Samples were denatured and 1 μl of oxidant solution was added to the oxBS samples converting hydroxymethylated cytosines to formylcytosines, while 1 μl of ultra-pure water was added to the BS sample for a mock reaction not affecting hydoxymethylation. Bisulfite reagent and additive were then added to the samples and reactions were incubated using bisulfite thermal cycling conditions as per manufacturer’s instructions. For the oxBS sample, the oxidated-induced 5fCs were deaminated to uracils so that only the original 5mC bases remained cytosines. In the conventional BS samples, both 5hmCs and 5mCs remained cytosines since the conversion does not discriminate between the two modifications of this base. DNA was whole genome amplified, enzymatically fragmented, purified and applied to the Illumina Infinium MethylationEPIC BeadChips (Illumina, San Diego, CA, USA) according to the Illumina methylation protocol. To maximise the amount of template used, 7 μl of recovered TrueMethyl template was used with 1 μl of 0.4-N NaOH, instead of using 4 μl of template and 4 μl of 0.1-N NaOH.
For quality control, available DNA from a single human pre-frontal cortex sample was put through the 5hmC protocol four times. We also included three BS replicates and four oxBS replicates of cord samples. DNA was run in two batches, randomised across plates and chips. oxBS and BS conversion samples for the same subject were always on the same chip.
Methylation data was processed using the R package minfi (22). First, IDAT files were loaded and processed for background subtraction and dye correction using preprocessNoob and then quantile normalised using preprocessQuantile to adjust for the different Infinium chemistry used for Type I and Type II probes. Samples where >95% of probes had a detection P > 0.01 were removed. Probes were filtered for >95% of samples with detection P > 0.01, Mexican ancestry from Los Angeles single nucleotide polymorphisms, cross-reactive and probes identified as poor performing by Illumina were removed, leaving a total of 790 721 probes for analysis.
For each CpG site on the EPIC array, a β value between 0 (completely unmethylated) and 1 (completely methylated), representing percent of methylation of that site, is produced from a ratio of methylated probe intensity to overall probe intensity. One method to distinguish 5mC and 5hmC values is to simply subtract oxBS methylation β values from the BS methylation values; however, when using this method, noise in the β values can lead to negative estimates of 5hmC β values. Instead, we used the R package OxyBS, which performs maximum likelihood estimation to determine levels of 5mC and 5hmC from the signal intensities (23). This method produced positive β values for all 5mC and 5hmC estimates.
Statistical analyses
Differential methylation by sex was assessed using a limma linear model with empirical Bayes variance shrinkage (24), adjusting for seven cord blood cell types, including nucleated red blood cells, estimated in minfi (25). Sensitivity analyses adjusting for six blood cell types using an adult reference data set were also conducted to assess the impact of using a cord blood reference data set. Pathway analyses were performed using the R package missMethyl (26). Fisher’s exact tests were conducted to assess enrichment for 5hmC at CpG sites in 3’UTR, Body, TSS1500, 5’UTR, intergenic, first Exon and TSS200 regions according to Illumina annotation. All statistical analyses were conducted using R version 3.4.1 (27).
Results
Study participants
A total of 41 unique subjects were included for the analysis with N = 24 (59%) girls and N = 17 (41%) boys. Characteristics of this subsample did not differ from other participants of the CHAMACOS cohort (Table 1). The distribution of combined 5hmC and 5mC in this subset of 41 samples was similar (mean r > 0.99) to the methylation profile of 109 CHAMACOS cord samples previously analysed on the EPIC BeadChip (28).
Table 1.
Characteristics of CHAMACOS newborns
| Included subjects | All CHAMACOS newborns | P-value | |
|---|---|---|---|
| N (%) or mean (SD) | N (%) or mean (SD) | ||
| Child Sex | 0.40 | ||
| Boy | 17(41) | 266 (50) | |
| Girl | 24 (59) | 270 (50) | |
| Birthweight (g) | 3414 (530) | 3417 (550) | 0.97 |
EPIC 5mC and 5hmC analysis
Distribution of 5hmC
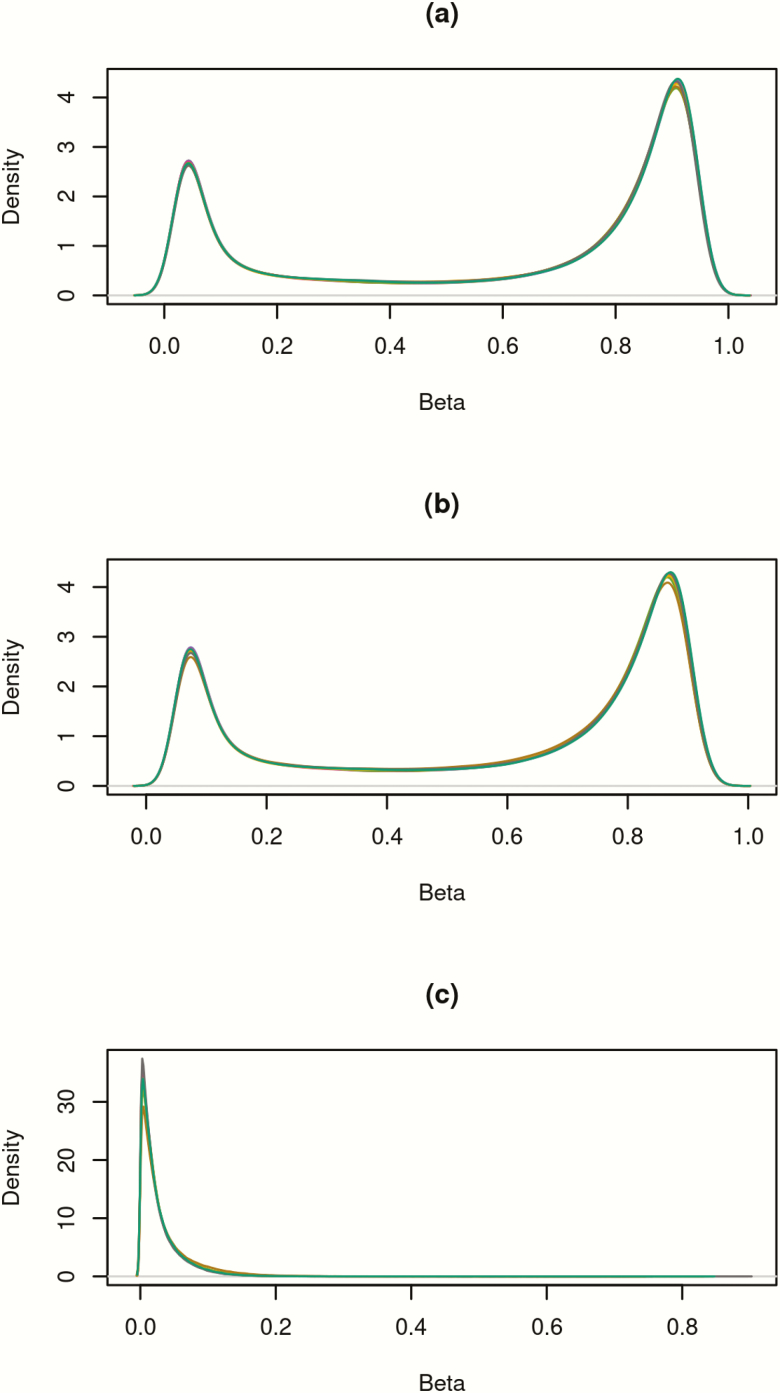
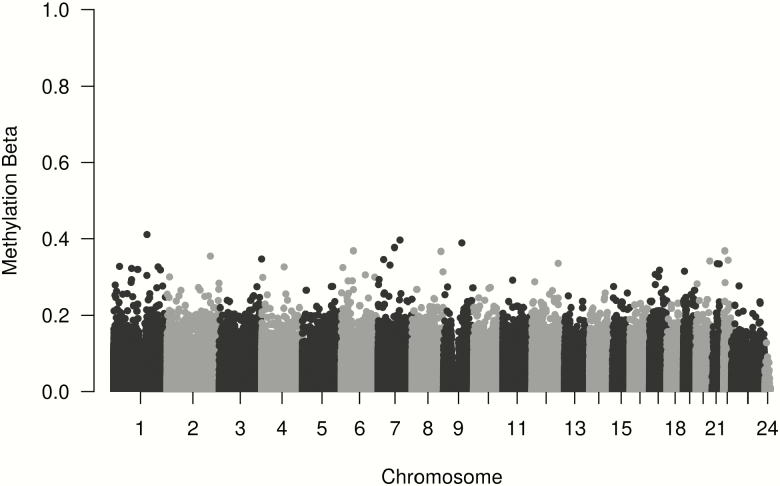
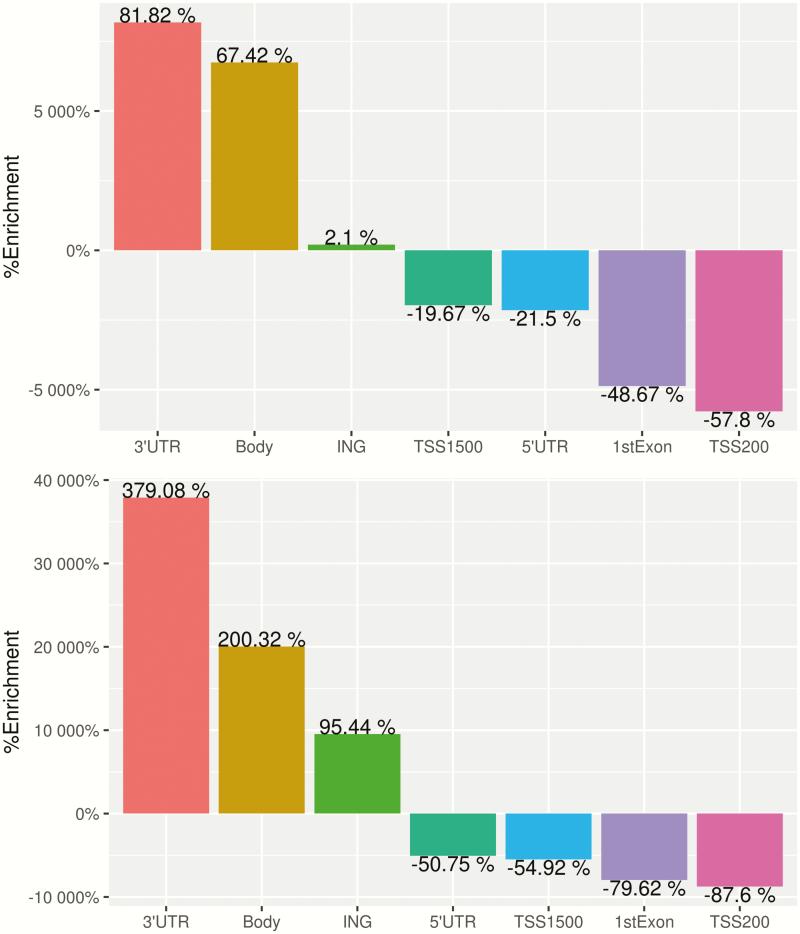
Since levels of 5hmC are known to be higher in brain samples, the assay was initially run with four pre-frontal cortex replicates for quality control purposes. There was strong replication between the four runs of the pre-frontal cortex sample with correlations of 0.96–0.97 for oxBS and 0.99 for all BS conversion samples. There was also high correlation between the four oxBS and three BS cord samples, all with correlations >0.99 and root mean square errors <0.037 for all replicate pairs. Overall, 5hmC levels were much lower than 5mC values (Figure 1). A total of 11 644 probes had 5hmC >0.1; 91 probes had >0.25 and only eight probes >0.35 (Table 2). On average, 5hmC made up only 3.7% of the total methylation at each site. Figure 2 shows the mean methylation β values of probes with average 5hmC across all individuals >0, with 5hmC evenly distributed throughout the genome. Average 5hmC and 5mC β values were weakly correlated with r = 0.22. Figure 3 shows percent enrichment of (i) 5hmC and (ii) 5mC in different areas of genes. Notably, we observe significant enrichment (81.82%) of 5hmC in the 3’UTR regions and significantly decreased enrichment (−57.8%) in TSS200 regions. This also highlights the differences in levels of enrichment between 5hmC and 5mC throughout the epigenome.
Figure 1.
(a) β values of combined 5mC and 5hmC. (b) β values of 5mC (after subtracting away 5hmC). (c) β values of 5hmC. Each line represents the density of β values for each individual (N = 41).
Table 2.
Distribution of methylation β-values*
| Min | 25% | 50% | 75% | Max | |
|---|---|---|---|---|---|
| β 5mC + 5hmC | 0.012 | 0.185 | 0.795 | 0.896 | 0.976 |
| β 5mC | 0.039 | 0.223 | 0.746 | 0.850 | 0.946 |
| β 5mC in probes with mean 5hmC >0.1 (11 644 probes) |
0.092 | 0.534 | 0.622 | 0.689 | 0.808 |
| β 5mC in probes with mean 5hmC >0.25 (91 probes) | 0.157 | 0.290 | 0.397 | 0.475 | 0.548 |
| β 5mC in probes with mean 5hmC >0.35 (eight probes) | 0.254 | 0.313 | 0.402 | 0.456 | 0.487 |
| β 5hmC (all probes) | 0.000 | 0.002 | 0.008 | 0.022 | 0.411 |
*Methylation β-values represent the percent of methylation at CpG sites between 0 (completely unmethylated) and 1 (completely methylated).
Figure 2.
Average 5hmC β-value across all individuals. Probes with 5hmC are distributed evenly throughout the epigenome.
Figure 3.
Bar plot of percent enrichment of (a) 5hmC and (b) 5mC according to genomic region of the site.
Differential methylation analysis by sex
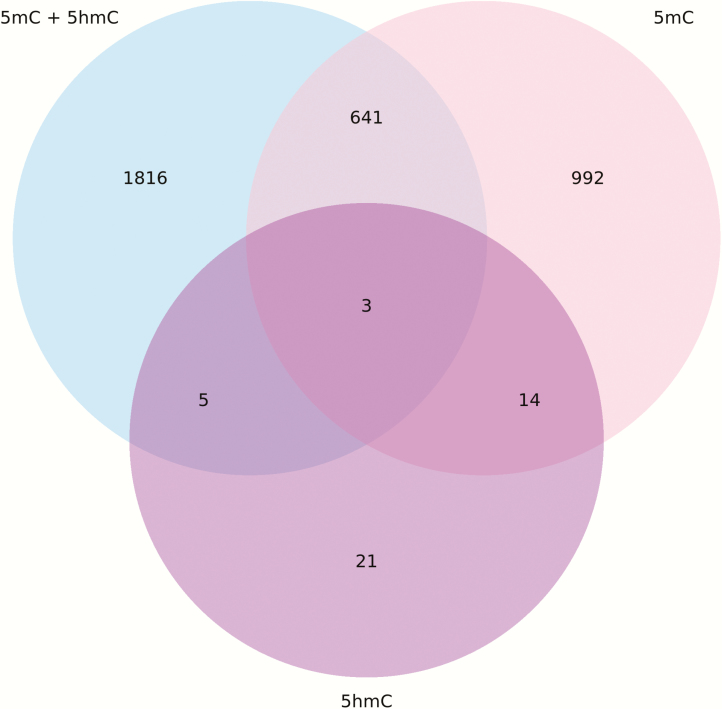
Previously we reported significant differences in DNA methylation by 450K in CHAMACOS boys and girls at birth (18,19). Now we expanded to include 5hmC as an additional epigenetic mark in children. Figure 4 summarises the results of the analysis of differential methylation associated with sex at birth for autosomal CpG sites only. The differentially methylated position analysis showed 2465 significant autosomal CpG sites (14 641 in sex chromosomes) with combined 5mC and 5hmC values. After subtracting away 5hmC values to isolate 5mC, there were 1650 significant autosomal sites (13 962 in sex chromosomes), with 644 autosomal sites (13 323 in sex chromosomes) in common with those identified by the combined methylation measure. 5hmC assessed alone was associated with fewer sites different between boys and girls with 43 significant in autosomes (855 in sex chromosomes), of which 21 in autosomes (28 in sex chromosomes) were not identified by the combined or exclusive 5mC models. A list of all 5hmC sites differentially methylated by sex can be found in Supplementary Table 1.
Figure 4.
Venn diagram showing overlap of hits differentially methylated by sex for 5mC + 5hmC versus 5mC versus 5hmC on autosomes.
There were large differences when examining the impact of 5hmC in CpG sites on all chromosomes (including sex chromosomes) compared to only autosomes. For all chromosomes, 82% of false discovery rate (FDR) significant hits overlapping between 5hmC + 5mC compared to 5mC; however, when looking only at autosomal sites, there is much lower concordance with only 26% overlap of FDR significant hits. If we look specifically at CpG sites which had differential methylation >10%, there is much higher concordance amongst autosomes, with 75% of significant hits overlapping between 5mC and 5hmC + 5mC measures.
In total, 788 of the 898 (88%) of significant 5hmC sites were common between use of an adult blood reference data set for cell-type adjustment compared to the cord blood reference data set; however, there were only 25 autosomal hits when using the adult reference compared to the 43 hits using the cord blood reference, with almost complete overlap of the adult adjusted hits within in the cord adjusted hits (24/25, 96%).
Further analysis of the 49 unique significant 5hmC CpG sites showed 21 autosomal CpG sites that were not previously identified in the CHAMACOS 450K analyses (18), and all 21 CpG sites were specific to the EPIC chip that has approximately double the coverage of the DNA methylome than the 450K chip. These CpG sites were not in genes enriched for any particular biologic pathways.
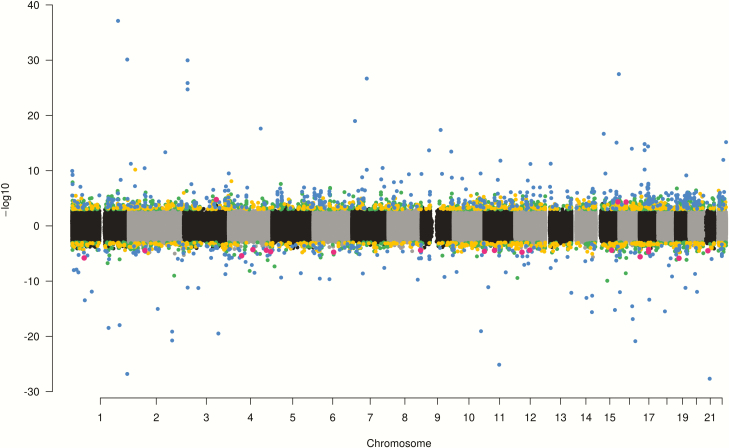
Significant hypermethylated and hypomethylated CpG sites are shown in the Manhattan plot (Figure 5). The majority of combined 5hmC + 5mC (67%) and 5mC (64%) autosomal hits were hypomethylated in boys with a mean percent methylation difference of 2.6% amongst 5hmC + 5mC hits and 2.3% amongst 5mC hits. The same trend was seen for hits on sex chromosomes with 67% hypomethylated in boys (in comparison to girls) for both 5hmC + 5mC and 5mC models, with a mean percent methylation difference of 6.7% for 5hmC + 5mC and 7.7% for 5mC. In contrast, the opposite trend was seen in 5hmC hits, where the majority of CpG sites were hypermethylated in boys in both autosomes (88%) and sex chromosomes (67%) with a mean methylation difference of 2.3% in autosomes and 3.1% in sex chromosomes.
Figure 5.
Manhattan plot of results of analyses for differential methylation by sex at birth. CpG sites that were hypermethylated in boys compared to girls are plotted above the y-axis, while hypomethylated sites are below the y-axis. 5mC + 5mC sites are highlighted in green, 5mC sites are yellow, 5hmC sites are red and sites associated with two or more methylation measures are blue.
Discussion
It is important to differentiate 5hmC and 5mC in association studies because these epigenetic marks can be functionally and ontogenetically distinct. In this paper, we used oxidative bisulphite conversion in combination with the Illumina HumanMethylation EPIC BeadChip to characterise both 5mC and 5hmC values in newborn cord blood. Our results showed that the 5hmC makes up a very small portion of DNA methylation in cord blood with only 91 probes having 5hmC β values greater than 0.25. Locations with enrichment of CpG sites showing 5hmC methylation are consistent with a prior study by Stewart et al. in adults, which similarly showed increased enrichment within the gene body and decreased enrichment in the TSS200 region. Our results showed that the highest enrichment was seen in the 3’UTR region, whereas the prior study showed more enrichment in the gene body and intergenic regions; however, the sample analysed for enrichment by Stewart et al. was from brain tissue, so we expect differences from our cord blood samples.
Differential analysis using either combined 5mC + 5hmC or isolated 5mC as the methylation measure outcome showed that 5hmC does have a significant impact on calls for differentially methylated CpG sites in interrogating differential methylation by sex. This means there is potential for 5hmC methylation to have similarly large impacts on the various relationships with exposures and health outcomes that have mostly relied on combined 5hmC + 5mC values (by 450K or EPIC arrays). This is an unexpected result given the overall small percentage (3.7%) of total methylation that was attributed to 5hmC in our data. When examining significant sites that had >10% methylation differences by sex in both 5hmC + 5mC and 5mC analyses, the concordance of significant hits was much higher at 75% compared to the overall 25%. With stronger methylation differences, 5hmC has a smaller impact on the results. This suggests that even though 5hmC levels are low, they are high enough to impact results of differential methylation with lower effect sizes, and this may be especially problematic since many differential methylation studies report statistically significant results with low effect sizes.
As expected when interrogating differential methylation by sex, most of the significant sites were observed on sex chromosomes; however, there is a significant number of 5hmC sites differentially methylated by sex on autosomes as was previously reported based on DNA methylation array data for combined 5hmC + 5mC. The majority of significant differentially methylated 5mC and combined 5mC + 5hmC CpG sites were hypomethylated in boys compared to girls. This trend was seen previously in 450K analyses in the CHAMACOS cohort (18). Interestingly, sites where 5hmC was significantly associated with sex showed the opposite direction of effect, with boys typically showing higher 5hmC than girls. A recent study in mice also found 5hmC and 5mC to be associated with age in opposite direction (29).
Although there were relatively low levels of 5hmC in the cord blood samples in our study, we found that 5hmC is significantly associated with sex at birth at 898 CpG sites. However, we also identified 49 sites that were exclusively associated with 5hmC. This may be a possible reflection of early embryonic stages of methylation profiles enriched for 5hmC and could in part be due to the larger proportion of progenitor cells in cord blood compared to adult blood (30). Given the relatively small number of significantly differentially methylated 5hmC CpG sites by sex compared to 5mC + 5hmC, this highlights the importance of using appropriate cell-type reference data sets when conducting differential 5hmC analyses. Using the cord blood reference data set, there were 72% increased number of differentially methylated 5hmC sites compared to the number of hits using the less appropriate adult blood reference data set.
The number of human studies of 5hmC is limited. A recent report on investigating associations between 5hmC and lead exposure in cord blood observed sex-specific differences in response to lead (14). However, this study focussed only on certain 5hmC-immunoprecipitation-enriched regions but not on all CpG sites assessed by the 450K chip. In contrast, our study used the EPIC BeadChip, which nearly doubles the coverage of the 450K chips and, thus, was able to interrogate sites not previously examined for 5hmC sex differences. While the lead study had a similar sample size (N = 48) to our study (N = 41); cell-type heterogeneity adjustment was done using the adult Houseman et al. reference data set, which may not be completely appropriate for studies of cord blood (18). Our study benefitted from a recently developed reference data set (25), which uses sorted cells from newborns and includes estimates for nucleated red blood cells that are commonly found in cord blood samples but not in adult blood (1,31,32). Furthermore, since 5hmC levels may be higher in different tissue types such as the brain, heart and skeletal muscle, whole blood may not provide comprehensive characterisation of 5hmC in newborns or the association with sex. Other studies in both skin cells (33) and adult blood (15) did not uncover an association between 5hmC and sex but they only focussed on global levels of 5hmC and did not characterise site-specific associations nor adjusted for cell-type heterogeneity, which can confound relationships between methylation and health variables or exposures.
Further studies are needed to validate 5hmC EPIC results in newborn blood with larger sample sizes and to investigate the impact 5hmC has on other health outcomes commonly interrogated for differential methylation. Since blood is a common tissue for interrogation of environmental exposures and DNA methylation in cohort studies of newborns and children, it is important to emphasise that, while 5hmC represents only a small fraction of overall DNA methylome, it may significantly impact interpretation and findings of previous studies with DNA methylation arrays that have not taken into account 5hmC compared to 5hmC + 5mC methylation, especially when dealing with small effect sizes.
Funding
This work was supported by grants from the National Institute of Environmental Health Science (P01ES009605, R01ES012503, R01ES021369, R24ES028529, F31ES027751); the Environmental Protection Agency (EPA) (RD83273401, RD83171001); and the National Institutes of Health (NIH) (UG3OD023356). Generous support from the JPB Foundation of New York is appreciated. The content is solely the responsibility of the authors and does not necessarily represent the official views of the EPA, or the NIH.
Supplementary Material
Acknowledgements
We are grateful to CHAMACOS participants and field staff. Contributions of the personnel and students in the Holland, Eskenazi, Kobor, and Meaney research groups are acknowledged.
References
- 1. Breton C. V., Marsit C. J., and Faustman E (2017) Small-magnitude effect sizes in epigenetic end points are important in children’s environmental health studies: the children’s environmental health and disease prevention research center’s epigenetics working group. Environ. Health Perspect., 125, 511–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tahiliani M., Koh K. P., Shen Y., et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khare T., Pai S., Koncevicius K., et al. (2012) 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol., 19, 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams K., Christensen J., Pedersen M. T., Johansen J. V., Cloos P. A., Rappsilber J. and Helin K (2011) TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature, 473, 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pastor W. A., Pape U. J., and Huang Y (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature, 2011;473, 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu Y., Wu F., Tan L., et al. (2011) Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell, 42, 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo J. U., Su Y., Zhong C., Ming G. L. and Song H (2011) Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle, 10, 2662–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nestor C. E., Ottaviano R., Reddington J., et al. (2012) Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res., 22, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi D. Q., Ali I., Tang J. and Yang W. C (2017) New insights into 5hmC DNA modification: generation, distribution and function. Front. Genet., 8, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li W, Liu M (2011) Distribution of 5-hydroxymethylcytosine in different human tissues. J. Nucleic Acids, 2011, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Starczak M., Zarakowska E., Modrzejewska M., et al. (2018) In vivo evidence of ascorbate involvement in the generation of epigenetic DNA modifications in leukocytes from patients with colorectal carcinoma, benign adenoma and inflammatory bowel disease. J. Transl. Med., 16, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li W., Zhang X., Lu X., et al. (2017) 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res., 27, 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buscarlet M., Tessier A., Provost S., Mollica L. and Busque L (2016) Human blood cell levels of 5-hydroxymethylcytosine (5hmC) decline with age, partly related to acquired mutations in TET2. Exp. Hematol., 44, 1072–1084. [DOI] [PubMed] [Google Scholar]
- 14. Sen A., Cingolani P., Senut M. C., Land S., Mercado-Garcia A., Tellez-Rojo M. M., Baccarelli A. A., Wright R. O. and Ruden D. M (2015) Lead exposure induces changes in 5-hydroxymethylcytosine clusters in CpG islands in human embryonic stem cells and umbilical cord blood. Epigenetics, 10, 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Niedzwiecki M. M., Liu X., Hall M. N., et al. (2015) Sex-specific associations of arsenic exposure with global DNA methylation and hydroxymethylation in leukocytes: results from two studies in Bangladesh. Cancer Epidemiol. Biomarkers Prev., 24, 1748–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andres C., Rifas-Shiman S. L. and Lode G (2003) Prenatal exposure to mercury: associations with global DNA methylation and hydroxymethylation in cord blood and in childhood. Environ. Health Perspect., 125, 087022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stewart S. K., Morris T. J., Guilhamon P., Bulstrode H., Bachman M., Balasubramanian S. and Beck S (2015) oxBS-450K: a method for analysing hydroxymethylation using 450K BeadChips. Methods, 72, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yousefi P., Huen K., Davé V., Barcellos L., Eskenazi B. and Holland N (2015) Sex differences in DNA methylation assessed by 450 K BeadChip in newborns. BMC Genomics, 16, 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huen K., Harley K., Kogut K., Rauch S., Eskenazi B. and Holland N (2016) DNA methylation of LINE-1 and Alu repetitive elements in relation to sex hormones and pubertal timing in Mexican-American children. Pediatr. Res., 79, 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eskenazi B., Bradman A., Gladstone E. A., Jaramillo S., Birch K. and Holland N (2003) CHAMACOS, A longitudinal birth cohort study: lessons from the fields. J. Children Health, 1, 3–27. [Google Scholar]
- 21. Holland N., Furlong C., Bastaki M., Richter R., Bradman A., Huen K., Beckman K. and Eskenazi B (2006) Paraoxonase polymorphisms, haplotypes, and enzyme activity in Latino mothers and newborns. Environ. Health Perspect., 114, 985–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., Hansen K. D. and Irizarry R. A (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Houseman E. A., Johnson K. C. and Christensen B. C (2016) OxyBS: estimation of 5-methylcytosine and 5-hydroxymethylcytosine from tandem-treated oxidative bisulfite and bisulfite DNA. Bioinformatics, 32, 2505–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol., 3, Article3. [DOI] [PubMed] [Google Scholar]
- 25. Bakulski K. M., Feinberg J. I., Andrews S. V., et al. (2016) DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics, 11, 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phipson B., Maksimovic J. and Oshlack A (2016) missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics, 32, 286–288. [DOI] [PubMed] [Google Scholar]
- 27. R Core Team. (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 28. Solomon O., MacIsaac J., Quach H., et al. (2018) Comparison of DNA methylation measured by Illumina 450K and EPIC BeadChips in blood of newborns and 14-year-old children. Epigenetics, 13, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kochmanski J., Marchlewicz E. H., Cavalcante R. G., Sartor M. A. and Dolinoy D. C (2018) Age-related epigenome-wide DNA methylation and hydroxymethylation in longitudinal mouse blood. Epigenetics, 13, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng S., Papalexi E., Butler A., Stephenson W. and Satija R (2018) Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol. Syst. Biol., 14, e8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yousefi P., Huen K., Quach H., Motwani G., Hubbard A., Eskenazi B. and Holland N (2015) Estimation of blood cellular heterogeneity in newborns and children for epigenome-wide association studies. Environ. Mol. Mutagen., 56, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Goede O. M., Razzaghian H. R., Price E. M., Jones M. J., Kobor M. S., Robinson W. P. and Lavoie P. M (2015) Nucleated red blood cells impact DNA methylation and expression analyses of cord blood hematopoietic cells. Clin. Epigenetics, 7, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Silva M. B. D., Melo A. R. D. S., Costa L. d. A., Barroso H. and Oliveira N. F. P. d (2017) Global and gene-specific DNA methylation and hydroxymethylation in human skin exposed and not exposed to sun radiation. Anais Brasileiros De Dermatologia, 92, 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.