Summary
Bacteriocins are regarded as important factors mediating microbial interactions, but their exact role in community ecology largely remains to be elucidated. Here, we report the characterization of a mutant strain, derived from Pseudomonas syringae pv. tomato DC3000 (Pst), that was incapable of growing in plant extracts and causing disease. Results showed that deficiency in a previously unannotated gene saxE led to the sensitivity of the mutant to Ca2+ in leaf extracts. Transposon insertions in the bacteriocin gene syrM, adjacent to saxE, fully rescued the bacterial virulence and growth of the ΔsaxE mutant in plant extracts, indicating that syrM‐saxE encode a pair of bacteriocin immunity proteins in Pst. To investigate whether the syrM‐saxE system conferred any advantage to Pst in competition with other SyrM‐sensitive pathovars, we compared the growth of a SyrM‐sensitive strain co‐inoculated with Pst strains with or without the syrM gene and observed a significant syrM‐dependent growth reduction of the sensitive bacteria on plate and in lesion tissues upon desiccation–rehydration treatment. These findings reveal an important biological role of SyrM‐like bacteriocins and help to understand the complex strategies used by P. syringae in adaptation to the phyllosphere niche in the context of plant disease.
SyrM confers advantage to the producer in competition with other sensitive bacteria upon desiccation. Our findinds reveal an important biological role that bacteriocin played during adaptation to the phyllosphere niche in the context of plant disease.

Introduction
Plant leaves make up the majority of phyllosphere and are inhabited by highly complex bacterial communities consisting of both commensal and adversary species (Vorholt, 2012). The relative abundance of phyllosphere bacterial populations is often conditioned collectively by multiple factors including host plants, environmental cues and interactions between diverse microbial species (Hirano and Upper, 2000; Vorholt, 2012; Wagner et al., 2016). When an appropriate relative abundance is achieved, opportunistic pathogens, such as Pseudomonas syringae species, may launch a range of molecular weapons including phytotoxins and proteinaceous effectors to subvert host immunity and manipulate plant physiology in favour of disease (Nomura et al., 2006; Zheng et al., 2012; Xin et al., 2016) or frost injury (Lindow et al., 1982). Current understanding on the molecular basis of how P. syringae bacteria predominate the phyllosphere niche prior to the attacks is still limited. Early observations reveal that host‐adapted P. syringae survives better than non‐pathogenic bacteria on leaf surface under UV irradiation or desiccation (Wilson et al., 1999), whereas Lee et al. (2012) found that type three effectors of P. syringae pv. syringae B728a influence the survival and size of the foliar epiphytic bacterial populations (Lee et al., 2012). Microbial competitions involving rapid exploitation of the limiting resource(s) or antibiosis are also likely employed in the complex interactions among bacterial populations coexisting in the phyllosphere. The exploitation competition has been widely exhibited in studies of biological control of detrimental P. syringae strains (Lindow, 1987; Völksch and May, 2001; Wensing et al., 2010; Xie et al., 2017); in contrast, the significance of antibiosis is not well established for bacterial competition with P. syringae on leaf surfaces (Lindow and Brandl, 2003). For instance, the production of three antimicrobial compounds in the biocontrol strain Pss22d is dispensable for its antagonistic effect in planta against P. syringae pv. glycinea that causes bacterial blight of soybean (Braun et al., 2010).
Bacteriocins are bacteria‐secreted proteins that are able to kill the close relatives of the producing strain and are regarded as potential competition factors in microbial ecosystems (Riley and Wertz, 2002). Colicin M‐like bacteriocins are a class of bacteriocins that share high homology with the Colicin M (ColM), a bacteriocin originally identified from Escherichia coli strains and can kill the sensitive bacteria by interfering cell wall synthesis. The mode of action of ColM has been well characterized in E. coli. Extracellular ColM needs to be taken up by an outer membrane receptor and traverses the outer membrane via the TonB‐dependent pathway to reach the periplasm, in which it degrades the substrate lipid II, an important precursor used for the biosynthesis of peptidoglycan, thereby inhibits the cell wall synthesis and triggers bacterial cell death (Barreteau et al., 2012). The expression of ColM is tightly regulated by SOS promoters. Thus, under normal conditions, the ColM encoding gene cma is tightly repressed by the LexA protein, and under SOS conditions, the RecA protease is activated and cleaves the LexA repressor protein, which activates the expression of a group of SOS genes including cma (Cascales et al., 2007). ColM producers can protect themselves by expression of the cognate immunity protein Cmi (Ölschläger et al., 1991; Gérard et al., 2011). ColM‐like bacteriocins have been identified from a range of bacterial species including P. syringae (Barreteau et al., 2009; Grinter et al., 2012; Ghequire and De Mot, 2015), and their biological role in bacterial competition has not been elucidated, although many of them have been shown to exhibit lipid II‐cleaving activity and inhibit the growth of close relatives in culture (Barreteau et al., 2009; Grinter et al., 2012; Ghequire and De Mot, 2015).
During our previous work searching for bacterial sax (survival in A rabidopsis extracts) genes that protect P. syringae against plant‐released antimicrobials, transposon Ω‐Km was used to mutagenize the bacteria, and a number of mutant strains sensitive to plant extracts were isolated. Molecular characterization of the mutants reveals that multiple sax genes (saxCAB/F/D/G) encoding the metabolic enzyme and multidrug efflux pumps are required for P. syringae to overwhelm isothiocyanate‐based defence during the necrotrophic stage of infection of Arabidopsis plants (Fan et al., 2011). Here, we report the molecular characterization of another resulting sax mutant that failed to grow on plant extracts and was unable to cause disease in planta. The investigation revealed that a previously unannotated gene, saxE, acted as an immunity gene of the ColM‐like bacteriocin syringacin M (SyrM) in P. syringae pv. tomato DC3000 (Pst) strain and prevented bacteria from being killed by SyrM in the presence of calcium ion. Furthermore, we demonstrated that the syrM‐saxE system was required for Pst to suppress the population of SyrM‐sensitive P. syringae during desiccation–rehydration cycles. Our findings reveal an important biological role of SyrM‐like bacteriocins and highlight the capacity of P. syringae to integrate environmental stresses into the complex bacterial interactions for maintaining a prevailing population in the context of plant disease.
Results
Isolation of a Pst mutant strain that was unable to grow in leaf extracts and to cause disease
Our previous studies have revealed that the Pst ▵saxAB/F strain, in which an isothiocyanate‐metabolizing enzyme and a multidrug efflux system were disrupted, was partially compromised for bacterial growth in Arabidopsis leaf extracts. To seek additional sax genes, transposon‐mutagenized clones derived from the Pst ▵saxAB/F strain were screened for enhanced sensitivity to the plant extracts (Fan et al., 2011). Out of ~4000 clones, we obtained a mutant strain (25D12) that failed to grow in Arabidopsis extracts (Fig. 1A) but grew well in rich media (Fig. S1). Subsequent disease assay showed that there was no increase in the population of 25D12 when infiltrated into Arabidopsis leaves (Fig. 1B), and no disease symptom could be observed on spray‐inoculated plants compared to the wild‐type Pst or the parental Pst ▵saxAB/F strain (Fig. 1E). When tomato plants, the natural host of Pst, were inoculated with the 25D12 mutant, similar levels of defects in bacterial growth and virulence could be observed (Fig. 1C, D and F). These findings indicate that: (i) in 25D12 mutant strain, a genetic component was compromised, which functions either alone or with other sax genes to support the survival and virulence of Pst during infection; and (ii) the factor(s) in leaf extracts that inhibited the growth of 25D12 mutant may exist in species of diverse plant families.
Figure 1.
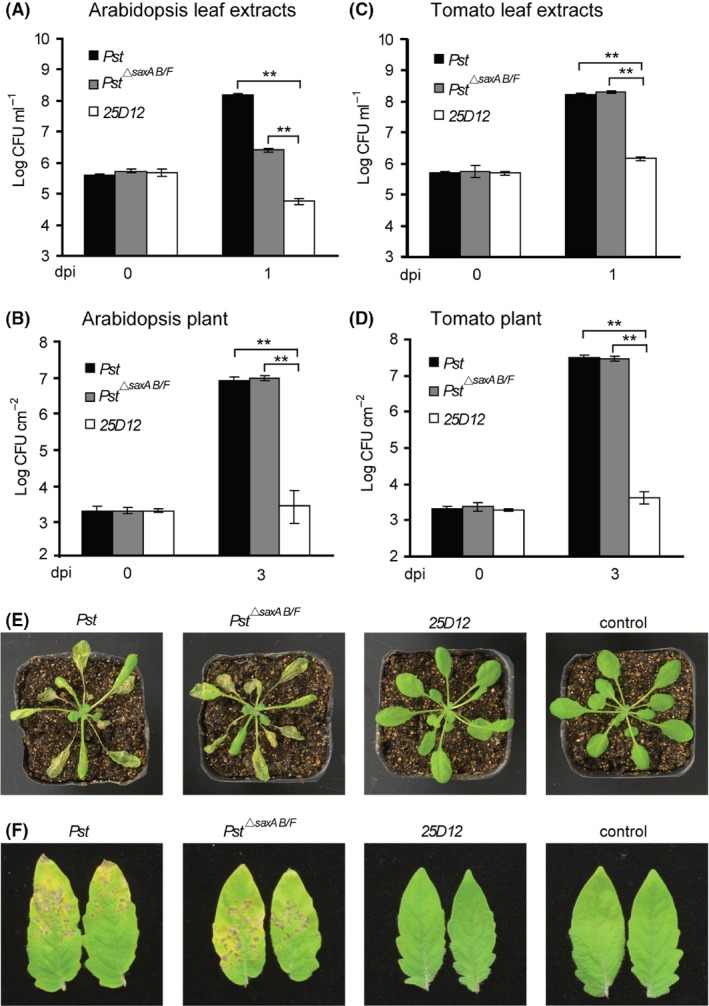
Pst mutant 25D12 was sensitive to plant extracts and unable to cause disease.
For the growth in leaf extracts assay, wild‐type Pst, Pst ▵sax AB /F and 25D12 strains were inoculated into the 100% Arabidopsis (A) and tomato (C) leaf extracts at OD 600 = 0.001. Samples were collected at 0 and 1 day postinoculation (dpi) for colony counts. For the growth in planta assay, bacterial strains were infiltrated into Arabidopsis (B) and tomato (D) leaves at OD 600 = 0.001. Samples were taken at 0 and 3 dpi for colony counts. Arabidopsis (E) and tomato (F) plants were sprayed with the bacteria at OD 600 = 0.2, and disease symptoms were photographed 4 days after inoculation. Non‐sprayed plants were used as control. All experiments were repeated at least three times, and similar results were observed. Data shown are means ± SD. ** indicates statistical significance (t‐test, P < 0.01).
Identification of a previously unannotated gene necessary for Pst to grow in extracts and in planta
Results of TAIL‐PCR showed that the transposon in 25D12 mutant was inserted in a cluster of annotated genes between PSPTO_0572 and PSPTO_0573 (Fig. 2A). DNA fragments downstream the transposon insertion site were thus used to complement the 25D12 mutant, and a minimal region of 562 base pairs of DNA (Cpl‐SacI) containing no annotated gene was unexpectedly found capable of restoring bacterial growth in leaf extracts and in planta (Fig. 2A and B). Further sequence analysis detected several open reading frames (orfs) in the 562 base‐pair region, and the longest one (designated as saxE hereafter) encodes a deduced protein of 135 amino acids (Fig. 4A) similar to a group of hypothetical proteins from several species of Pseudomonas bacteria (Table S1).
Figure 2.
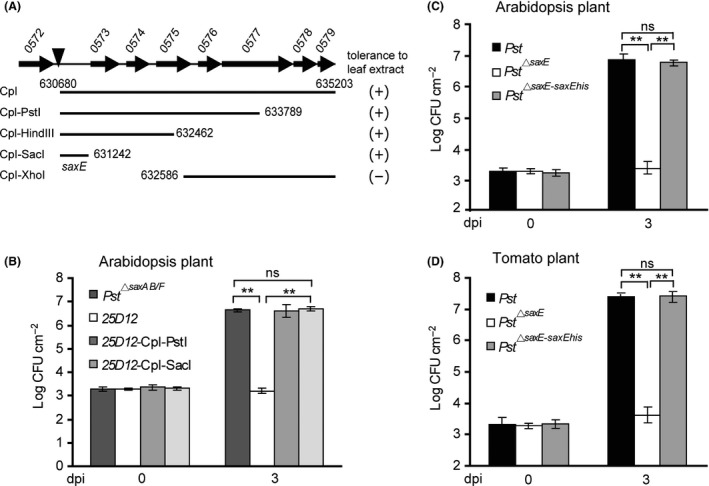
The saxE gene was required for Pst growth in leaf extracts and in planta.
A. A schematic diagram of the complementary tests pinpointing the minimal region required for rescuing the growth of 25D12 mutant in plant leaf extracts. Filled arrows indicate genes annotated by the sequencing project marked with locus tags on the top. The filled triangle shows the insertion site of Ω‐Km transposon in mutant 25D12. Genomic DNA fragments downstream the transposon insertion site were tested for the ability to confer tolerance to leaf extracts.
B. The minimal region that protected 25D12 in leaf extracts also rescued the growth of the mutant on Arabidopsis plant. Bacterial inocula were infiltrated into Arabidopsis leaves at OD 600 = 0.001. Samples were taken at 0 and 3 dpi for colony counts. The saxE orf was required for bacterial virulence on Arabidopsis (C) and tomato (D) plants. Wild‐type Pst, Pst ▵saxE and the complemented strain Pst ▵saxE‐saxEhis were infiltrated into Arabidopsis (C) and tomato (D) leaves at OD 600 = 0.001. Samples were taken at 0 and 3 dpi for colony counts. All experiments were repeated at least three times, and similar results were observed. Data shown are means ± SD. ** indicates significant difference (t‐test, P < 0.01). ns indicates no significant difference (t‐test, P > 0.05).
To investigate whether the saxE orf truly encodes a functional protein, we deleted the entire coding region of saxE from the wild‐type Pst by homologous recombination. The resulting Pst ▵saxE mutant grew well in rich media (Fig. 3A) but failed to grow in plant extracts and during infection (Fig. 2C, D and S2A), indicating that the loss of saxE alone was responsible for the growth defects in 25D12 strain. We modified the Cpl‐SacI DNA fragment by fusing a his‐tag to the C‐terminus of the predicted SaxE protein (saxEhis) and also introduced one nucleotide into the coding region of saxEhis to generate the saxEhis(ins) construct, leading to a frameshift only in the saxE but not other smaller orfs on the DNA fragment. Plasmids harbouring these modified inserts were delivered into the Pst ▵saxE mutant, and resultant strains were subjected to growth assay. Results showed that constructs bearing saxEhis but not saxEhis(ins) were able to restore the growth of the Pst ▵saxE mutant in 100% plant extracts and in planta (Fig. 2C, D and S2A), suggesting that the saxE gene played a key role in protecting bacteria in plant extracts and in planta. The protein gel blot assay detected a protein of the same size as SaxEhis in the strain Pst ▵saxE complemented with saxEhis (Fig. 4B), which corroborated that saxE encodes a protein essential for the growth of Pst in plant extracts and in planta.
Figure 3.
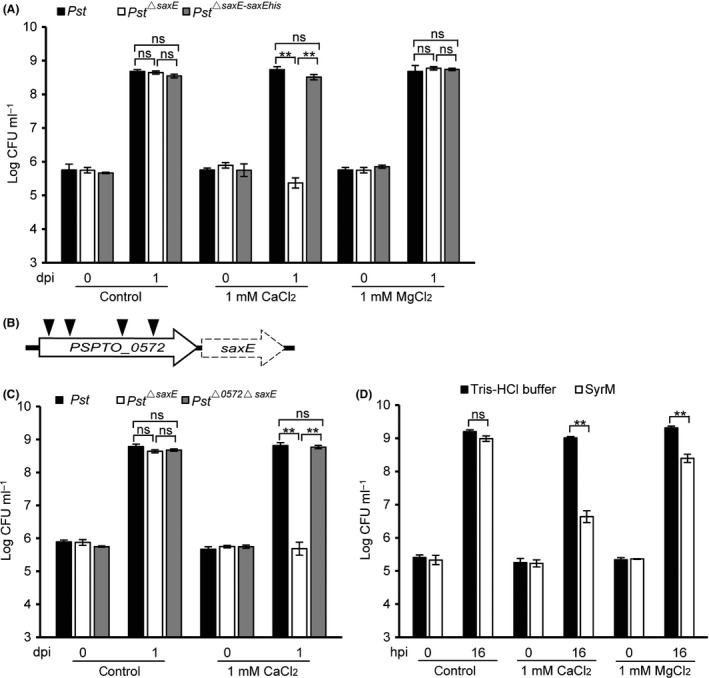
Inhibitory effect of Ca2+ on the growth of strains deficient in the saxE gene.
A. Ca2+ is the inhibitory factor that suppressed the growth of Pst ▵saxE. Bacteria were inoculated into the King's B (KB) liquid medium supplemented with 1 mM CaCl2 or MgCl2 at OD 600 = 0.001. Samples were collected at 0 and 1 dpi for colony counts.
B. A schematic diagram of Ω‐Km transposon insertion events that suppressed the Pst ▵saxE phenotype. Filled triangles indicate the insertion sites that were able to restore the growth of Pst ▵saxE in the presence of Ca2+. The empty and the dashed arrows denote the gene PSPTO_0572 (syrM) and the deleted saxE gene respectively.
C. Deletion of syrM abolished the growth inhibitory effect of Ca2+ on Pst ▵saxE. Bacteria were inoculated into the KB liquid medium supplemented with 1 mM CaCl2 at OD 600 = 0.001, and samples were collected at 0 and 1 dpi for colony counts.
D. Ca2+ and Mg2+ enhanced inhibitory activity of bacteriocin SyrM on the sensitive P. syringae pv. lachrymans‐8 (Psl). Purified SyrM (1 μM) was mixed with 100 μl KB liquid cultures supplemented with 1 mM CaCl2 or MgCl2 inoculated by Psl at OD 600 = 0.0002. Samples were collected at 0 and 16 h postinoculation (hpi) for colony counts. Plain KB medium was used as control. All experiments were repeated at least three times, and similar results were observed. Data shown are means ± SD. ** indicates significant difference (t‐test, P < 0.01). ns indicates no significant difference (t‐test, P > 0.05).
Figure 4.
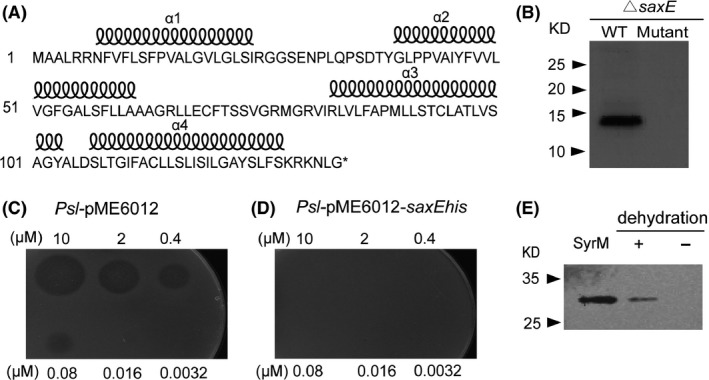
The saxE gene encodes an immunity protein of the bacteriocin SyrM.
A. Amino acid sequence and the predicted secondary structure of SaxE protein. The coils above the amino acids denote the helices predicted by TMHMM (http://www.cbs.dtu.dk/services/ TMHMM/).
B. Gel blot assay of the SaxE protein. Proteins extracted from Pst ▵saxE bacteria harbouring the pME6012 expressing wild‐type SaxEhis (WT) or mutated SaxEhis(ins) (mutant) protein were used for blot assay detected with the his‐tag antibody.
C, D. Purified SyrM protein inhibited the growth of Psl. Five microlitres of serial‐diluted SyrM protein solutions was applied on a plate overlaid with soft agar inoculated with Psl‐pME6012 (C) or Psl‐pME6012‐saxEhis (D). The plates were incubated at 28°C and photographed 1 day after inoculation.
E. SyrM is released upon desiccation treatment. Pst‐pME6012‐syrMhis (OD 600 = 0.01) was applied onto filter paper discs placed on KB plates and cultured at 28°C for 16 h. The discs were desiccated for 4 h (+), and non‐treated samples were used as control (−). Bacteria were washed off from the discs and spun, and the resulting supernatant samples, together with the purified SyrM protein, were used for gel blot assay with the his‐tag antibody.
Ca2+ was the inhibitory component in plant extracts that suppressed the growth of Pst ▵saxE strain
Pseudomonas sax genes have been shown crucial for Pst to overcome the aliphatic isothiocyanate‐based defence in Arabidopsis (Fan et al., 2011). However, tomato plants have not been reported as a source of these metabolites, and we speculated that additional factor in plant extracts conferred the inhibitory effect on Pst ▵saxE strain. Thus, by using ion exchange chromatography we isolated the inhibitory components from leaf extracts and crystallized the active eluates with methanol (see details in Experimental procedures). Nevertheless, the nuclear magnetic resonance (NMR) analysis of these crystals only detected a methyl proton peak (at 1.8 ppm) and two carbon peaks (a strong methyl peak at 23 ppm and a weaker quaternary peak at 181 ppm), reminiscent of the spectra of the acetate ion. Since 10% acetic acid was used for elution during chromatography, we speculated that a metal cation might be the active agent in the crystal. We further tested a panel of metal acetates and found that only calcium acetate preferentially inhibited the bacterial growth of Pst ▵saxE strain (Fig. S2B). We thus dissolved the crystal in water and measured the Ca2+ levels of the solution, and the calcium content of the crystal was thereby determined to be 20.1% (w/w), close to that of Ca(C2H3O2)2·H2O, which is 22.7% (w/w), indicating that the crystal mainly consists of calcium acetate. Subsequent analysis confirmed that 1 mM of Ca2+ but not Mg2+, another divalent cation abundant in plants, strongly inhibited the growth of Pst ▵saxE in rich media, and this inhibition was abolished on the mutant complemented with the wild‐type saxE gene (Fig. 3A). These observations demonstrated that wild‐type Pst strain requires saxE for protection against the Ca2+.
The inhibitory effect of Ca2+ on Pst ▵saxE strain was dependent on the bacterial gene PSPTO_0572
To further dissect the mechanism underlying the inhibitory effect of Ca2+ on the Pst ▵saxE strain, transposon‐mutagenized Pst ▵saxE bacteria were thus subjected to screen for revertant colonies grown on plates supplemented with 10 mM of CaCl2. Thirteen clones were isolated from the screen and TAIL‐PCR revealed that only the locus PSPTO_0572 was disrupted independently in four clones (Fig. 3B), indicating that this locus is required for the Ca2+‐mediated growth suppression of Pst ▵saxE strain. Indeed, the bacterial growth in the presence of 1 mM of CaCl2 was completely restored when PSPTO_0572 was deleted from the Pst ▵saxE background (Fig. 3C). Moreover, by quantitative RT‐PCR analysis, we observed a modest increase in transcript levels of PSPTO_0572 in wild‐type Pst strain upon Ca2+ treatment, whereas the increase was more prominent in the Pst ▵saxE strain following the same treatment (Fig. S3).
The locus PSPTO_0572 encodes a ColM‐like bacteriocin, SyrM (Grinter et al., 2012). Our observations indicated that Ca2+ may be a potent enhancer of this bacteriocin and saxE may function as its immunity gene. We thus expressed the his‐tagged SyrM in E. coli and used the purified protein to test the bacteriocin activity on 16 P. syringae strains. Results showed that the growth of pathovars lachrymans (8 and NCPPB540) and glycinea (race 4) was suppressed by SyrM treatment (Table S2, Fig. 4C), and the inhibitory effect increased significantly by the addition of 1 mM of CaCl2 or MgCl2 (Fig. 3D). We further transformed P. syringae pv. lachrymans‐8 (Psl) with the plasmid carrying the saxE gene from Pst, and the resulting Psl‐saxE strain was able to resist the highest tested concentration (10 μM) of SyrM (Fig. 4D), demonstrating that the Psl‐saxE strain has gained full immunity to the bacteriocin.
The SyrM‐mediated bacterial competition upon dehydration–rehydration treatment
The role of ColM or ColM‐like bacteriocins in bacterial competition has not been described experimentally, although the bactericidal activity of purified proteins on sensitive strains has been demonstrated (Braun et al., 1974; Barreteau et al., 2009). To investigate the potential impact of syrM locus on sensitive bacteria, we co‐cultured Psl with wild‐type Pst and the mutant Pst ▵syrM, respectively, on filter paper discs. Since the expression of syrM appears to be regulated by an SOS promoter, we initially cultured bacteria under SOS‐inducing conditions including UV‐B or mitomycin C (MMC) treatments, but did not find any difference in Psl population sizes between co‐cultures with Pst and Pst ▵syrM strains (not shown). However, we found that a 4‐h dehydration treatment of the overnight culture was able to reduce Pst levels by nearly two orders of magnitude (Fig. 5A) and upregulate the expression of syrM accordingly (Fig. 5B). More importantly, when the desiccated paper discs were further cultured on a fresh plate for another day, population sizes of Pst and Pst ▵syrM rebounded to similar levels, whereas the growth of Psl was significantly reduced when co‐cultured with Pst but not with Pst ▵syrM (compare Fig. 5C and D), indicating the existence of SyrM‐mediated suppression of the sensitive bacteria. Since no difference could be observed in bacterial growth without desiccation treatment (Fig. 5C and D), we further examined whether the desiccation treatment facilitated the release of the bacteriocin from a Pst strain expressing the his‐tagged SyrM. Protein gel blot assays of the supernatants of bacterial wash‐offs detected a band of SyrM from desiccated paper discs but not from the untreated control (Fig. 4E), clearly suggesting a desiccation‐associated release of SyrM into the extracellular space.
Figure 5.
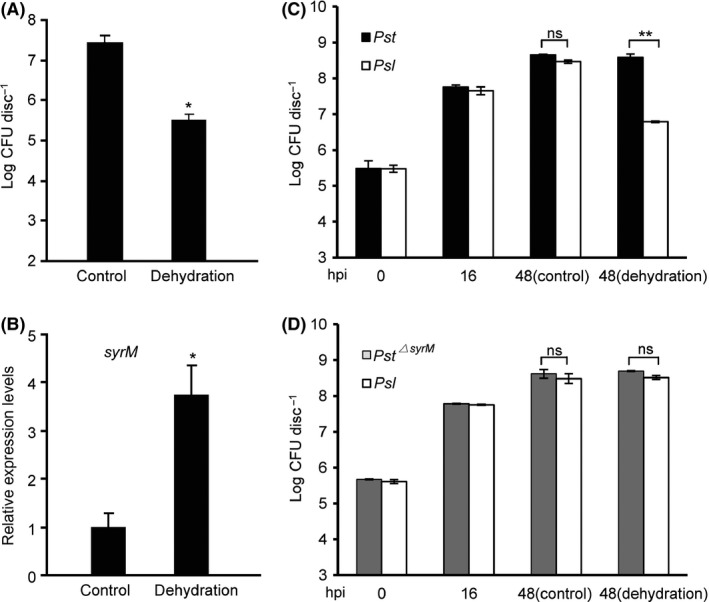
SyrM‐mediated competition between Pst and Psl on plate culture.
A. Desiccation treatment reduced the number of viable bacteria on filter paper discs. Wild‐type Pst (OD 600 = 0.01) was applied onto filter paper discs placed on plates and cultured for 16 h. The inoculated filter paper discs were desiccated for 4 h. Desiccated and untreated paper discs were collected for bacterial counts.
B. Total RNA samples were prepared from bacteria on paper discs collected in (A) and subjected to the quantitative real‐time PCR assay of relative transcript levels of syrM. The recA was used as the reference gene.
C, D. SyrM‐mediated competition depended on dehydration and rehydration treatments. The SyrM‐sensitive Psl were mixed at 1:1 ratio with wild‐type Pst (C) and Pst ▵syrM (D) strains, respectively, and co‐cultured at the same conditions as in (A). After desiccation treatment, the paper discs were rehydrated and cultured to 48 h. Untreated samples were used as controls. All experiments were repeated at least three times, and similar results were observed. Data shown are means ± SD. ** indicates significant difference (t‐test, P < 0.01). * indicates significant difference (t‐test, P < 0.05). ns indicates no significant difference.
SyrM played a key role in suppression of the sensitive bacterial strain in lesion tissue
To further explore whether the observed syrM‐mediated competition may also occur in planta, we first measured the Ca2+ levels of the intercellular fluid during the Pst infection of tomato plants. Results showed that Ca2+ levels reached > 8 mM at 2 dpi when the water‐soaking lesions appeared, about fourfold of the basal levels at 0 dpi, indicating that Ca2+ was released into the extracellular space where bacteria colonize (Fig. 6A). We then inoculated tomato healthy plants with a 1:1 mixture of Pst and Psl, and compared the sizes of bacterial populations with separate inoculation of individual strains. Results showed that at 2 dpi Psl population in the leaves was about 10‐fold higher when co‐inoculated with Pst in comparison with inoculated by Psl alone (Fig. 6B). Hence, no competition between the two populations could be observed during the biotrophic stage of infection. However, when the lesion tissues collected at 2 dpi were repeatedly subjected to first dry conditions (32.5% RH) for 2 days and then wet conditions (100% RH) for 2 days, a fluctuation of nearly 2 orders of magnitude in Pst population could be observed (Fig. 6C). We therefore measured the ratio of Pst/Psl under this condition in comparison with that of Pst ▵syrM/Psl. As shown in Fig. 6D, at 2 dpi, the ratio of Pst/Psl (9:1) was similar to that of Pst ▵syrM/Psl (11:1), whereas at 6 dpi, after one cycle of dry–wet treatment, the ratio of Pst/Psl increased to about 100:1 and that of Pst ▵syrM/Psl still stayed around 10:1, similar to that of 2 dpi. These observations demonstrated that Pst was capable of suppressing the SyrM‐sensitive bacteria during the necrotrophic stage of infection.
Figure 6.
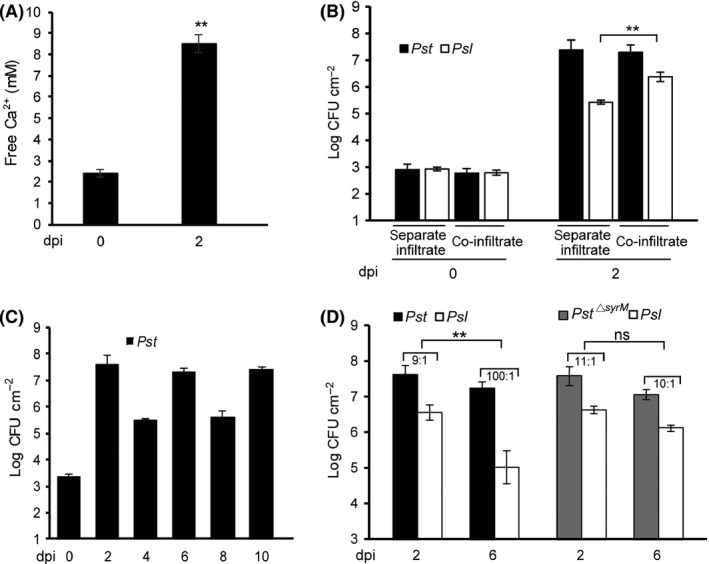
SyrM suppressed the rebound of Psl population during the rehydration of desiccated leaf lesions.
A. Ca2+ released at the late stage of plant–bacteria interaction. Pst was infiltrated into tomato leaves at OD 600 = 0.001. Leaves were collected at 0 and 2 dpi for detection of the concentration of free Ca2+ in the intercellular fluids.
B. Pst infection promoted the growth of Psl on tomato plants. Pst, Psl and a 1:1 mixture of Pst and Psl were infiltrated into tomato leaves at OD 600 = 0.0002 respectively.
C. Bacterial population in leaf lesions fluctuated widely during a dehydration–rehydration regimen. Pst was infiltrated into tomato leaves at OD 600 = 0.001. At 2 dpi, the inoculated leaf lesions were collected and desiccated for 2 day. After dehydration treatment, the desiccated leaf lesions were rehydrated for 2 day. The rehydrated leaf lesions were then treated again as the same desiccation–rehydration treatment.
D. syrM is required for Pst to limit the growth of Psl in rehydrated lesion tissues. The 1:1 mixtures of Pst/Psl and Pst ▵syrM/Psl were infiltrated into tomato leaves respectively. At 2 dpi, the leaf lesions were collected and treated the same as in (C). Lesion samples were taken at 2 and 6 dpi for bacterial counts. The ratio above the columns is means of the ratio between two pathovars in lesion samples. All experiments were repeated at least three times, and similar results were observed. Data shown are means ± SD. ** indicates significant difference (t‐test, P < 0.01). ns indicates no significant difference.
Discussion
Bacteriocins have been widely inferred as important factors mediating interactions between bacterial populations; however, the exact biological role of most bacteriocins remains to be elucidated (Riley and Wertz, 2002). A few studies have demonstrated that colicins participate in the bacterial competition. For instance, ColIb, a pore‐forming colicin, has been shown to confer a competitive advantage in inflammation‐induced overgrowth on a human pathogenic strain of Salmonella enterica against the commensal E. coli (Nedialkova et al., 2014). Nevertheless, the role of ColM or ColM‐like bacteriocins in bacterial community interactions has yet to be unveiled. In this study, we have identified the immunity gene of a ColM‐like bacteriocin SyrM in P. syringae and demonstrated that this bacteriocin is required for the suppression of sensitive bacteria in leaf lesions under a wet–dry–wet regimen (Fig. 6D). Phyllosphere bacteria have to endure the frequent environmental stress caused by desiccation, and it has been well documented in early studies that the size of the epiphytic bacterial population is dependent on the ambient moisture (And and Upper, 1983; Beattie, 2011). Indeed, we observed in the detached leaf lesions a sharp reduction of P. syringae populations under low ambient humidity, and that after rehydration, bacterial numbers rebound to levels similar to those before desiccation treatment (Fig. 6C and S4). The ability to restore the population during desiccation–rehydration cycles is essential for bacteria to colonize the epiphytic habitat and to poise a secondary infection (Leben, 1981; Lindow et al., 1993; Beattie and Lindow, 1994; Wilson et al., 1999; Quesada et al., 2010). Bacterial aggregate formation on the leaf surface is known to help to cope with desiccation (Beattie and Lindow, 1999; Vorholt, 2012). Previous reports demonstrated that the battle of those cells that were in direct contact with other strains was much more likely intense than that with the same strain in the aggregate (Monier and Lindow, 2005,b), in which antibiosis might mediate the bacterial interaction. It is still not clear how P. syringae strains managed the nutrient supply required for the regrowth in lesion tissues (Fig. 6C and S4), but suppression of closely related bacteria with bacteriocin such as SyrM (Fig. 6D) may help to compete for the limited nutrients. ColM‐like bacteriocin has been found in a wide range of bacterial species including P. syringae (Ghequire et al., 2018). Interestingly, the sequences of SyrM‐like proteins from P. syringae species are highly conserved (> 94% of identity), whereas they share much lower similarity with ColM‐like proteins from other Pseudomonas or non‐Pseudomonas species (Table S3), indicating that the SyrM‐mediated competition may be important for P. syringae strains in defining their particular ecological niche. In this study, we have investigated the impact of Pst‐produced SyrM on the population of a single sensitive strain and, more importantly, identified the key role of desiccation–rehydration cycles in exerting the impact of SyrM bacteriocin. As the interactions among phyllosphere bacterial communities are highly complex in nature (Vorholt, 2012), it is therefore intriguing to investigate further the potential influence of this class of bacteriocins on the structure of phyllosphere microbial communities.
The expression of most colicins is regulated by the SOS promoter (Cascales et al., 2007), and we have observed that MMC strongly enhanced the transcript levels of syrM in Pst (data not shown). However, when co‐cultured with Pst, the growth of Psl was not suppressed in either liquid or solid medium containing MMC at levels activating the expression of syrM (data not shown). This lack of suppression could be due to the failure of the release of SyrM protein. It has been shown that the release of some colicins is regulated by colicin release protein (CRP)‐mediated cell lysis (Wal et al., 1995; Cascales et al., 2007) or during the course of phage lysis (Nedialkova et al., 2016). As no SyrM‐associated lysis gene was identified in Pst, we thus speculate that this bacteriocin is released through an alternative mechanism which is still obscure at present. In this study, we observed a mild upregulation of syrM expression and a subsequent release of the protein into the extracellular space upon desiccation treatment (Figs 4E and 5B), and that at the same time, the levels of culturable Pst dropped about two orders of magnitude (Fig. 5A), implying that the release of SyrM was most likely associated with bacterial cell death. Apart from its impact on the gene activation and the release of bacteriocin protein, the dehydration stress might further concentrate the SyrM and enhance the inhibitory effect of the bacteriocin (Chao and Levin, 1981); whether the uptake of SyrM by the sensitive Psl was also enhanced during the dehydration and rehydration cycle remains to be elucidated.
It was unexpected to find that Ca2+ was the active agent inhibiting the growth of Pst ▵saxE strain. Indeed, we observed a mobilization of Ca2+ into the extracellular fluid (8 mM) at a late stage of bacterial infection (Fig. 6A), and the actual Ca2+ levels in the lesion tissue could be much higher during desiccation. In addition, we also found that Ca2+, which has not been regarded as an SOS inducer, activated and enhanced the expression of syrM gene, especially in Pst ▵saxE strain (Fig. S3). Previous reports have demonstrated that divalent cations including Ca2+ and Mg2+ are capable of enhancing the enzymatic activity of ColM or SyrM on degrading lipid II (Schaller et al., 1981a,b; Barreteau et al., 2009; Grinter et al., 2012). Consistent with these findings, our data showed that these cations could enhance the in vitro bactericidal activity of SyrM on Psl (Fig. 3D). However, the growth of Pst ▵saxE strain was inhibited only by Ca2+ but not by Mg2+ (Fig. 3A). Similarly, Ca2+ but not Mg2+ has been found to boost the ColM‐induced cell lysis in liquid culture (Schaller et al., 1981a,b). These observations collectively suggested that factors other than the enzyme activity may account for the Ca2+‐dependent inhibitory effect. A possible explanation is that Ca2+ may enhance the uptake of the ColM‐like bacteriocin as inferred in previous studies (Schaller et al., 1981a,b; Harkness and Braun, 1990). Alternatively, the integrity of bacterial cell envelope is partially compromised by the bacteriocin due to the inhibition of biosynthesis of peptidoglycan polymer and the O‐antigen moiety of lipopolysaccharides (Schaller et al., 1982; Harkness and Braun, 1989a, b; Harkness and Braun, 1989b; Barreteau et al., 2010), which may lead to a detrimental increase in membrane permeability to Ca2+ and result in disruption of bacterial cell physiology. This hypothesis is based on the findings that bacterial cytosolic Ca2+ must be tightly controlled at low levels akin to those found in eukaryotic cells, and the disturbance of calcium ion homeostasis can affect bacterial survival (Dominguez, 2004; Miyamoto et al., 2005). In Gram‐negative bacteria, the periplasmic peptidoglycan polymer and the outer membrane lipopolysaccharides are crucial cell envelope components that are able to bind and prevent the entry of excessive divalent cations such as Mg2+ and Ca2+ into the cytoplasm (Jones et al., 2002; Silhavy et al., 2010; Clifton et al., 2014; Miller and Salama, 2018). Hence, an investigation on the change in levels of bacterial cytosol free Ca2+ in response to SyrM and Ca2+ treatments may help to provide further clues on the validity of this hypothesis.
We did not observe a significant reduction in bacterial population sizes when the 25D12 and Pst ΔsaxE mutant strains were subjected to growth assay in the presence of exudates or 1 mM calcium (Figs 1A–D, and 3A, C, D), suggesting that the SyrM is ineffective to kill the mutant strain under these culture conditions. However, we observed that higher levels of calcium supplemented to the medium (> 25 mM) substantially killed the Pst ΔsaxE mutant; likewise, when the Pst ΔsaxE mutant bacteria were infiltrated into plant leaves at higher levels (108 CFU ml−1), a significant reduction of the bacterial population could be observed (not shown). Hence, the bactericidal activity of the SyrM protein is likely associated with high levels of ambient calcium and the density of bacterial producer, which are two features normally found in desiccated plant necrotic lesions. These observations further strengthen the pivotal role of SyrM in maintaining a predominant population during the necrotrophic stage of infection.
Experimental procedures
Bacterial strains and primers
Bacterial strains and primers used in this study are listed in Table S4 and S5 respectively.
Construction of plasmids
Plasmids used in this study are listed in Table S6. For generation of pDEST6012, the reading frame cassette of pDEST17 (Invitrogen, Carlsbad, CA, USA) containing the attR sites, the chloramphenicol resistance gene (Cmr) and the ccdB gene was amplified by PCR and cloned into the SacI and XhoI sites of the plasmid pME6012 (Fan et al., 2011). To add the his‐tag to SaxE and SyrM, coding regions of saxE and syrM were amplified by PCR from genomic DNA of wild‐type Pst and recombined into pDONR201 (Invitrogen) to generate entry clones via Gateway Technology (Invitrogen). The coding sequence of 6xhis‐tag was added by PCR to the 3′ of the coding region of saxE and syrM genes on the pENTRY vectors respectively. The resulting pENTRY vectors were recombined with pDEST6012 and pDEST14 vector, respectively, to generate pME6012‐saxEhis and pDEST14‐syrMhis. PCR was used to insert an extra nucleotide into the saxE orf on pENTRY‐saxEhis, and the resulting vector was recombined with pDEST6012 to generate pME6012‐saxEhis(ins). For generation of pME6012‐Cpl, the genomic region (630680–635203) of Pst used for complementation test was amplified by PCR and recombined into a pME6012DONR plasmid. The pME6012‐Cpl was further truncated with PstI, HindIII, SacI and XhoI, respectively, and the resulting products were self‐ligated to generate plasmids bearing the truncated regions for complementation test.
Bacterial growth conditions
All Pseudomonas syringae strains were streaked on King's B (KB) plate (Fan et al., 2011) from −80°C stocks and grown overnight in KB medium at 28°C and washed once with sterilized water before use. Bacterial inoculum densities were as indicated in figure legends. For the assay of bacterial growth in the presence of leaf extracts or metal cation, 10 μl of bacterial suspension was mixed with 90 μl KB medium or KB containing the tested components in 96‐well plates and grown at 28°C, 200 rpm for 1 day. Cultures collected at indicated time points were serial‐diluted and spotted on KB plates supplemented with appropriate antibiotics for bacterial counts.
Plant materials and bacterial inoculation
Arabidopsis and tomato plants were grown at 23°C with 9‐h light/15‐h dark cycle and 16‐h light/8‐h dark cycles respectively. Plant extracts were prepared as previously described (Fan et al., 2011). Five‐ to six‐week‐old Arabidopsis and tomato plants were used for bacterial infection experiments. For leaf infiltration assay, bacterial suspension mixed with 10 mM MgCl2 was infiltrated into the Arabidopsis and tomato plants at densities as indicated in figure legends. Unless otherwise noted, plants were covered with a plastic lid to keep high humidity for 1 day. For spray inoculation, plants were sprayed with bacterial suspension (OD600 = 0.2 in 10 mM MgCl2, 0.04% Silwet L‐77) and covered with a plastic lid to keep high humidity for 4 days.
Bacterial growth assay under the desiccation–rehydration treatment
For the assay of bacterial growth on filter paper discs, 5 μl of bacterial suspensions was applied onto discs of 6 mm in diameter, and the inoculated discs were placed on KB plates overlaid with 0.5% soft agar (w/v) containing 1 mM CaCl2. After incubation at 28°C for 16 h, the discs were transferred to an empty petri dish (6 cm) and sealed in a bigger petri dish (15 cm) containing saturated solution of MgCl2 for desiccation treatment (32.5% relative humidity, RH) for 4 h at room temperature (Winston and Bates, 1960). After the treatment, paper discs were returned to a new soft agar plate to resume culture to the 48‐h time point at 28°C. Paper discs not subjected to desiccation treatment were used as controls.
For the assay of bacterial growth in leaf lesions, leaves of tomato plants were infiltrated with bacterial suspensions at indicated concentration and incubated at 25°C, 100% RH for 2 days. Discs of leaf lesion tissue were taken by a cork borer (6 mm in diameter) and transferred to an empty petri dish for desiccation treatment for 2 days as described above. For rehydration, the desiccated leaf lesion discs were misted with deionized water and incubated at 25°C, 100% RH for 2 days.
Purification of the inhibitory component from Arabidopsis leaf extracts
Two hundred grams of Arabidopsis leaves was ground with liquid N2, mixed with 200 ml of water and incubated for 30 min before centrifugation at 10 000 rpm for 10 min. The resultant supernatant was boiled for 10 min and spin again to remove the pellet. After the addition of an equal amount of water into the supernatant, pass the solution through a column packed with 100 g of Dowex 1 resin. The resulting flow‐through was loaded on a weak cation exchanger column packed with 100 g of LEWATIT CNP‐105 resin and washed with 0.1 N NH3H2O. The column was eluted with 10% acetic acid, and the resulting fractions were collected for assays of inhibitory activity. Fractions with high inhibitory activity were freeze‐dried and dissolved with a minimal amount of deionized water. About 9 volume of methanol was added to the resulting solution to crystallize the active component.
Purification of SyrM
An overnight culture of E. coli BL21 (DE3) carrying the plasmid pDEST14‐syrMhis was used to inoculate 2 l of LB broth at the ratio of 1:100. The inoculated broth was cultured at 37°C, 220 rpm till the OD600 reached 0.6 before the addition of 1 mM isopropyl β‐D‐thiogalactoside. The cells were grown for a further 12 h at 24°C and harvested by centrifugation at 4°C for 10 min at 6000 g. The cell pellet was resuspended in cold 20 mM Tris‐HCl buffer, pH 7.5, containing 500 mM NaCl and 5 mM imidazole, and disrupted by sonication using a Branson Digital Sonifier. The resulting suspension was centrifuged at 16 000 g for 30 min at 4°C. The cell‐free lysate was applied to nickel‐nitrilotriacetate (Ni2+‐NTA)‐agarose, and the protein was eluted over a 0–500 mM imidazole gradient. Purified protein was dialysed overnight into 20 mM Tris‐HCl, 100 mM NaCl, pH 8.0, was concentrated to 100 μM and then stored at −80°C after addition of 5% glycerol and filtration sterilization using a 0.22‐μm filter.
Preparation of samples for SDS‐PAGE and protein gel blot analysis
For detection of his‐tagged SaxE in bacterial strains, cells were harvested by centrifugation at 10 000 g for 10 min at 4°C. The supernatant was removed and bacterial pellet was resuspended in protein loading buffer (50 mM Tris‐HCl, pH 7.5, 2% SDS, 0.1% bromphenol blue, 10% glycerol). Samples were then boiled for 10 min at 100°C and centrifuged at 10 000 g for 10 min at 4°C. The resulting supernatant was used for SDS‐PAGE. For detection of his‐tagged SyrM from inoculated filter paper discs, 100 μl of sterilized water was used to vortex and wash off bacteria on paper discs in a 1.5‐ml test tube. The resultant bacterial solutions were subjected to centrifugation, and supernatants were used for protein electrophoresis. For protein gel blot assay, proteins were transferred from the gels onto nitrocellulose membranes at 200 mA for 1.5 h. The membrane was blocked by TBST containing 5% milk powder and probed with mouse anti‐His antibody (1:2000; Sigma, Saint Louis, MO, USA). Goat anti‐Mouse‐HRP (1:10 000; Sigma) was used as secondary antibody. Blots were developed with the enhanced chemiluminescence (ECL) detection system (GE Healthcare, Milwaukee, WI, USA).
Cytotoxicity assay of SyrM
Soft agar overlay method was used for determining the bactericidal activity of SyrM as described (Grinter et al., 2012) with minor modification. Five microlitres of 10 μM purified syringacin M protein was applied on a KB plate overlaid with 3 ml of molten soft agar inoculated with 100 μl of bacteria (OD600 = 0.5) as indicated. Bactericidal activity of SyrM was determined based on the sizes of inhibition zones. Results of all experiments were observed or measured at 1 dpi.
Determination of free Ca2+ levels in the intercellular fluids of tomato leaves
Leaves infiltrated with wild‐type Pst were harvested by cutting petioles with a razor blade, carefully transferred into a 50‐ml centrifuge tube with xylem wound facing upward and centrifuged at 2000 g for 2 min at 4°C. The fluids were collected for measurement of free Ca2+ with a Calcium Colorimetric Assay Kit (NJJC Bioengineering Institute, Nanjing, China) following manufacturer's instructions. Ten microlitres of intercellular fluid was added into 250 μl solutions containing methylthymol blue, and OD610 was measured by Spectra Max i3x (Molecular Devices, Sunnyvale, CA, USA) after incubation for 5 min. Levels of free Ca2+ in intercellular fluids were calculated by an established standard curve.
Quantitative real‐time PCR (qRT‐PCR) assay of syrM gene expression
Total RNA was isolated from bacteria on paper discs with TRIzol reagent (Invitrogen) following the manufacturer's instructions. Recombinant RNase‐free DNase I (Takara, Kusatsu, Japan) and SuperScript III Reverse Transcriptase (Invitrogen) were used to remove genomic DNA and synthesize the first‐strand cDNA respectively. Tenfold diluted solutions of the reaction products were used for the qRT‐PCR assay. The qRT‐PCR was performed in 12 μl volume with Bestar® SYBR Green qPCR MasterMix (DBI® Bioscience, Ludwigshafen, Germany) on ABI QuantStudio 6 Flex (Thermo Fisher, Waltham, MA, USA).
Transposon mutagenesis and thermal asymmetric interlaced PCR (TAIL‐PCR) assay
Pst ▵saxAB/F was mutagenized by electroporation with the suicide plasmid pEJL428 carrying the transposon Omegon‐Km (Ω‐Km; Joseph‐Liauzun et al., 1989; Fan et al., 2011), and the position of Ω‐Km was determined by TAIL‐PCR (Liu and Whittier, 1995).
Conflict of interest
None declared.
Supporting information
Fig. S1. Pst mutant 25D12 grew normally in KB medium.
Fig. S2. Ca2+ is the inhibitory factor that suppressed the growth of Pst strain deficient in the saxE gene.
Fig. S3. Ca2+ enhanced the relative transcript levels of PSPTO_0572 gene.
Fig. S4. Bacterial populations in leaf lesions fluctuated widely during a desiccation‐rehydration regimen.
Table S1. Similarity of SaxE‐like hypothetical proteins in Pseudomonas strains.
Table S2. Sensitivity of the tested P. syringae strains to syringacin M.
Table S3. SyrM‐like sequences in Pseudomonas and other Proteobacteria strains.
Table S4. Bacterial strains used in this study.
Table S5. Primers used in this study.
Table S6. Plasmids used in this study.
Acknowledgements
We thank professors T.C. Zhao and W.C. Yang for providing Psl strains and tomato seeds (OH88119), respectively, and Dr. S. Fairhurst for help with the NMR assay. This work is partially funded by the National Natural Science Foundation of China (31272006) and the National Key R&D Program of China (2017YFD0201106‐01C) to JF.
Microbial Biotechnology (2020) 13(1), 134–147
Funding Information
This work is partially funded by the National Natural Science Foundation of China (31272006) and the National Key R&D Program of China (2017YFD0201106‐01C).
References
- And, S.S.H. , and Upper, C.D. (1983) Ecology and epidemiology of foliar bacterial plant pathogens. Annu Rev Phytopathol 21: 243–270. [Google Scholar]
- Barreteau, H. , Bouhss, A. , Fourgeaud, M. , Mainardi, J.‐L. , Touzé, T. , Gérard, F. , et al (2009) Human‐and plant‐pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M‐like hydrolase activity towards peptidoglycan precursors. J Bacteriol 191: 3657–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreteau, H. , Bouhss, A. , Gérard, F. , Duché, D. , Boussaid, B. , Blanot, D. , et al (2010) Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II‐degrading enzyme. J Biol Chem 285: 12378–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreteau, H. , El Ghachi, M. , Barnéoud‐Arnoulet, A. , Sacco, E. , Touzé, T. , Duché, D. , et al (2012) Characterization of colicin M and its orthologs targeting bacterial cell wall peptidoglycan biosynthesis. Microb Drug Resist 18: 222–229. [DOI] [PubMed] [Google Scholar]
- Beattie, G.A. (2011) Water relations in the interaction of foliar bacterial pathogens with plants. Annu Rev Phytopathol 49: 533–555. [DOI] [PubMed] [Google Scholar]
- Beattie, G.A. , and Lindow, S.E. (1994) Survival, growth, and localization of epiphytic fitness mutants of Pseudomonas syringae on Leaves. Appl Environ Microbiol 60: 3790–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie, G.A. , and Lindow, S.E. (1999) Bacterial colonization of leaves: a spectrum of strategies. Phytopathology 89: 353–359. [DOI] [PubMed] [Google Scholar]
- Braun, V. , Schaller, K. , and Wabl, M.R. (1974) Isolation, characterization, and action of colicin M. Antimicrob Agents Chemother 5: 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, S.D. , Hofmann, J. , Wensing, A. , Weingart, H. , Ullrich, M.S. , Spiteller, D. , and Völksch, B. (2010) In vitro antibiosis by Pseudomonas syringae Pss22d, acting against the bacterial blight pathogen of soybean plants, does not influence in planta biocontrol. J Phytopathol 158: 288–295. [Google Scholar]
- Cascales, E. , Buchanan, S.K. , Duché, D. , Kleanthous, C. , Lloubes, R. , Postle, K. , et al (2007) Colicin biology. Microbiol Mol Biol Rev 71: 158–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, L. , and Levin, B.R. (1981) Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci USA 78: 6324–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton, L.A. , Skoda, M.A. , Brun, A.L. , Ciesielski, F. , Kuzmenko, I. , Holt, S.A. , and Lakey, J.H. (2014) Effect of divalent cation removal on the structure of Gram‐negative bacterial outer membrane models. Langmuir 31: 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, D.C. (2004) Calcium signalling in bacteria. Mol Microbiol 54: 291–297. [DOI] [PubMed] [Google Scholar]
- Fan, J. , Crooks, C. , Creissen, G. , Hill, L. , Fairhurst, S. , Doerner, P. , and Lamb, C. (2011) Pseudomonas sax genes overcome aliphatic isothiocyanate–mediated non‐host resistance in Arabidopsis . Science 331: 1185–1188. [DOI] [PubMed] [Google Scholar]
- Gérard, F. , Brooks, M.A. , Barreteau, H. , Touzé, T. , Graille, M. , Bouhss, A. , et al (2011) X‐ray structure and site‐directed mutagenesis analysis of the Escherichia coli colicin M immunity protein. J Bacteriol 193: 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire, M.G. , and De Mot, R. (2015) Distinct colicin M‐like bacteriocin‐immunity pairs in Burkholderia . Sci Rep 5: 17368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire, M.G. , Buchanan, S.K. , and De Mot, R. (2018) The ColM family, polymorphic toxins breaching the bacterial cell wall. MBio 9: e02267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter, R. , Roszak, A.W. , Cogdell, R.J. , Milner, J.J. , and Walker, D. (2012) The crystal structure of the lipid II‐degrading bacteriocin syringacin M suggests unexpected evolutionary relationships between colicin M‐like bacteriocins. J Biol Chem 287: 38876–38888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness, R.E. , and Braun, V. (1989a) Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J Biol Chem 264: 6177–6182. [PubMed] [Google Scholar]
- Harkness, R.E. , and Braun, V. (1989b) Inhibition of lipopolysaccharide O‐antigen synthesis by colicin M. J Biol Chem 264: 14716–14722. [PubMed] [Google Scholar]
- Harkness, R.E. , and Braun, V. (1990) In vitro peptidoglycan synthesis by envelopes from Escherichia coli tolM mutants is inhibited by colicin M. J Bacteriol 172: 498–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, S.S. , and Upper, C.D. (2000) Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae‐a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64: 624–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, H. , Holland, I. , and Campbell, A. (2002) Direct measurement of free Ca2+ shows different regulation of Ca2+ between the periplasm and the cytosol of Escherichia coli . Cell Calcium 32: 183–192. [DOI] [PubMed] [Google Scholar]
- Joseph‐Liauzun, E. , Fellay, R. , and Chandler, M. (1989) Transposable elements for efficient manipulation of a wide range of Gram‐negative bacteria: promoter probes and vectors for foreign genes. Gene 85: 83–89. [DOI] [PubMed] [Google Scholar]
- Leben, C. (1981) How plant‐pathogenic bacteria survive. Plant Dis 65: 633–637. [Google Scholar]
- Lee, J. , Teitzel, G.M. , Munkvold, K. , Del, P.O. , Martin, G.B. , Michelmore, R.W. , and Greenberg, J.T. (2012) Type III secretion and effectors shape the survival and growth pattern of Pseudomonas syringae on leaf surfaces. Plant Physiol 158: 1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow, S.E. (1987) Competitive exclusion of epiphytic bacteria by ice−Pseudomonas syringae mutants. Appl Environ Microbiol 53: 2520–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow, S.E. , and Brandl, M.T. (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69: 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow, S.E. , Arny, D.C. , and Upper, C.D. (1982) Bacterial ice nucleation: a factor in frost injury to plants. Plant Physiol 70: 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow, S.E. , Andersen, G. , and Beattie, G.A. (1993) Characteristics of insertional mutants of Pseudomonas syringae with reduced epiphytic fitness. Appl Environ Microbiol 59: 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G. , and Whittier, R.F. (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681. [DOI] [PubMed] [Google Scholar]
- Miller, S.I. , and Salama, N.R. (2018) The Gram‐negative bacterial periplasm: size matters. PLoS Biol 16: e2004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, S. , Howes, A.L. , Adams, J.W. , Dorn, G.W. , and Brown, J.H. (2005) Ca2+ dysregulation induces mitochondrial depolarization and apoptosis. J Biol Chem 280: 38505–38512. [DOI] [PubMed] [Google Scholar]
- Monier, J.M. , and Lindow, S.E. (2005. a) Aggregates of resident bacteria facilitate survival of immigrant bacteria on leaf surfaces. Microb Ecol 49: 343–352. [DOI] [PubMed] [Google Scholar]
- Monier, J.M. , and Lindow, S.E. (2005. b) Spatial organization of dual‐species bacterial aggregates on leaf surfaces. Appl Environ Microbiol 71: 5484–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova, L.P. , Denzler, R. , Koeppel, M.B. , Diehl, M. , Ring, D. , Wille, T. , et al (2014) Inflammation fuels colicin Ib‐dependent competition of Salmonella serovar Typhimurium and E. coli in enterobacterial blooms. PLoS Pathog 10, e1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova, L.P. , Sidstedt, M. , Koeppel, M.B. , Spriewald, S. , Ring, D. , Gerlach, R.G. , et al (2016) Temperate phages promote colicin‐dependent fitness of Salmonella enterica serovar Typhimurium. Environ Microbiol 18: 1591–1603. [DOI] [PubMed] [Google Scholar]
- Nomura, K. , Debroy, S. , Yong, H.L. , Pumplin, N. , Jones, J. , and He, S.Y. (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223. [DOI] [PubMed] [Google Scholar]
- Ölschläger, T. , Turba, A. , and Braun, V. (1991) Binding of the immunity protein inactivates colicin M. Mol Microbiol 5: 1105–1111. [DOI] [PubMed] [Google Scholar]
- Quesada, J.M. , Penyalver, R. , Pérezpanadés, J. , Salcedo, C.I. , Carbonell, E.A. , and López, M.M. (2010) Dissemination of Pseudomonas savastanoi pv. savastanoi populations and subsequent appearance of olive knot disease. Plant Pathol 59: 262–269. [Google Scholar]
- Riley, M.A. , and Wertz, J.E. (2002) Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56: 117–137. [DOI] [PubMed] [Google Scholar]
- Schaller, K. , Dreher, R. , and Braun, V. (1981a) Structural and functional properties of colicin M. J Bacteriol 146: 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, K. , Krauel, A. , and Braun, V. (1981b) Temperature‐sensitive, colicin M‐tolerant mutant of Escherichia coli . J Bacteriol 147: 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, K. , Höltje, J. , and Braun, V. (1982) Colicin M is an inhibitor of murein biosynthesis. J Bacteriol 152: 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy, T.J. , Kahne, D. , and Walker, S. (2010) The bacterial cell envelope. Cold Spring Harb Perspect Biol 2: a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völksch, B. , and May, R. (2001) Biological control of Pseudomonas syringae pv. glycinea by epiphytic bacteria under field conditions. Microb Ecol 41: 132–139. [DOI] [PubMed] [Google Scholar]
- Vorholt, J.A. (2012) Microbial life in the phyllosphere. Nat Rev Microbiol 10: 828–840. [DOI] [PubMed] [Google Scholar]
- Wagner, M.R. , Lundberg, D.S. , Del Rio, T.G. , Tringe, S.G. , Dangl, J.L. , and Mitchell‐Olds, T. (2016) Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun 7: 12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wal, F.J.V.D. , Luirink, J. , and Oudega, B. (1995) Bacteriocin release proteins: mode of action, structure, and biotechnological application. FEMS Microbiol Rev 17: 381–385. [DOI] [PubMed] [Google Scholar]
- Wensing, A. , Braun, S. , Buttner, P. , Expert, D. , Volksch, B. , Ullrich, M. , and Weingart, H. (2010) Impact of siderophore production by Pseudomonas syringae pv. syringae 22d/93 on epiphytic fitness and biocontrol activity against Pseudomonas syringae pv. glycinea 1a/96. Appl Environ Microbiol 76: 2704–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, M. , Hirano, S.S. , and Lindow, S.E. (1999) Location and survival of leaf‐associated bacteria in relation to pathogenicity and potential for growth within the leaf. Appl Environ Microbiol 65: 1435–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, P.W. , and Bates, D.H. (1960) Saturated solutions for the control of humidity in biological research. Ecology 41: 232–237. [Google Scholar]
- Xie, J. , Shu, P. , Strobel, G. , Chen, J. , Wei, J. , Xiang, Z. , and Zhou, Z. (2017) Pantoea agglomerans SWg2 colonizes mulberry tissues, promotes disease protection and seedling growth. Biol Control 113: 9–17. [Google Scholar]
- Xin, X.F. , Nomura, K. , Aung, K. , Velásquez, A.C. , Jian, Y. , Boutrot, F. , et al (2016) Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539: 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. , Spivey, N.W. , Zeng, W. , Liu, P.P. , Fu, Z.Q. , Klessig, D.F. , et al (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that Inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Pst mutant 25D12 grew normally in KB medium.
Fig. S2. Ca2+ is the inhibitory factor that suppressed the growth of Pst strain deficient in the saxE gene.
Fig. S3. Ca2+ enhanced the relative transcript levels of PSPTO_0572 gene.
Fig. S4. Bacterial populations in leaf lesions fluctuated widely during a desiccation‐rehydration regimen.
Table S1. Similarity of SaxE‐like hypothetical proteins in Pseudomonas strains.
Table S2. Sensitivity of the tested P. syringae strains to syringacin M.
Table S3. SyrM‐like sequences in Pseudomonas and other Proteobacteria strains.
Table S4. Bacterial strains used in this study.
Table S5. Primers used in this study.
Table S6. Plasmids used in this study.


