Summary
Halogenated aromatics are used widely in various industrial, agricultural and household applications. However, due to their stability, most of these compounds persist for a long time, leading to accumulation in the environment. Biological degradation of halogenated aromatics provides sustainable, low‐cost and environmentally friendly technologies for removing these toxicants from the environment. This minireview discusses the molecular mechanisms of the enzymatic reactions for degrading halogenated aromatics which naturally occur in various microorganisms. In general, the biodegradation process (especially for aerobic degradation) can be divided into three main steps: upper, middle and lower metabolic pathways which successively convert the toxic halogenated aromatics to common metabolites in cells. The most difficult step in the degradation of halogenated aromatics is the dehalogenation step in the middle pathway. Although a variety of enzymes are involved in the degradation of halogenated aromatics, these various pathways all share the common feature of eventually generating metabolites for utilizing in the energy‐producing metabolic pathways in cells. An in‐depth understanding of how microbes employ various enzymes in biodegradation can lead to the development of new biotechnologies via enzyme/cell/metabolic engineering or synthetic biology for sustainable biodegradation processes.
This minireview discusses the molecular mechanisms of the enzymatic reactions for degrading halogenated aromatics which naturally occur in various microorganisms. An in‐depth understanding of how microbes employ various enzymes in biodegradation can lead to the development of new biotechnologies via enzyme/cell/metabolic engineering or synthetic biology for sustainable biodegradation processes.
Introduction
Halogenated aromatic compounds are widely used in various applications including agricultural, dye, chemical and pharmaceutical industries (Arora et al., 2012, 2018; Arora and Bae, 2014). Widespread usage and accumulation of these compounds in the environment is a source of great concern, as they can cause harmful and toxic effects on human and animal health. Exposure to these compounds can cause many severe health problems such as DNA damage, organ damage, cancer or even acute death (Olaniran and Igbinosa, 2011; Georgiadis et al., 2018). Thus, technologies for limiting their spread into the environment, such as more effective methods for degrading these compounds are important for human health. In the past, several physical and biological processes have been explored for detoxification or degradation of chlorinated phenols (CPs) (Pera‐Titus et al., 2004; Field and Sierra‐Alvarez, 2008; Arora and Bae, 2014; Cycon et al., 2017; Huang et al., 2017). In nature, various microbes have evolved metabolic pathways to combat the existence of halogenated aromatics in their habitat. Proper exploitation of microbial biodegradation processes can provide a green and sustainable method for detoxifying these toxic compounds and reduce their harmful levels in freshwater and soil. The purpose of this review is to highlight the current state of knowledge regarding microbial pathways related to halogenated aromatics degradation.
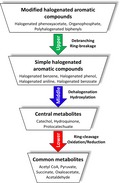
In general, halogenated aromatics commonly used in household or industries can be grouped into three types based on their structural components (Fig. 1). The first type is a class of halogenated aromatics which contain only an aromatic skeleton with substituents such as halogenated benzene (HB), halogenated phenol (HP) [such as 4‐chlorophenol (4‐CP), 2,4‐dichlorophenol (2,4‐DCP), 2,4,5‐trichlorophenol (2,4,5‐TCP), 2,4,6‐trichlorophenol (2,4,6‐TCP), pentachlorophenol (PCP)], halogenated aniline and its derivatives (such as 4‐chloroaniline, 2‐chloro‐4‐aminophenol, 4‐chloro‐2‐aminophenol), 4‐chlorobenzoic acid and tetrachlorohydroquinone (TCHQ). The second type is a class of compounds in which the aromatic rings are attached to extra moieties which often result in increased pesticide potency, but at the same time are more persistent due to their resistance to degradation (Casida and Quistad, 2004; Huang et al., 2017; Mostafalou and Abdollahi, 2017; Supreeth and Raju, 2017). These include 2,4‐dichlorophenoxyacetic acid (2,4‐D), 2,4,5‐trichlorophenoxyacetic acid (2,4,5‐T) and its analogues, including 2‐(4‐chloro‐2‐methylphenoxy) propionic acid (mecoprop), 2‐(2,4‐dichlorophenoxy) propionic acid (dichlorprop), 2‐methyl‐4‐chlorophenoxy acetic acid (MCPA), 4‐(4‐chloro‐2‐methylphenoxy)butanoic acid (MCPB) and organophosphate herbicides such as chlorpyrifos and profenofos. The third type is a class of compounds containing more than one aromatic ring such as polychlorinated biphenyl (PCBs) and polybrominated biphenyl (PBBs). Most of the compounds shown in Fig. 1 have been listed in the priority pollutants list and/or hazardous substances by the United States Environmental Protection Agency (USEPA) (United States Environmental Protection Agency, 2014) and European List of Notified Chemical Substances (ELINCS) published by the European Commission (EC) (Joaquin et al., 2009). The acceptable levels of these compounds in the environment and food‐related samples are tightly regulated.
Figure 1.

Structures of toxic halogenated aromatics generally found as pollutants.
Although most halogenated aromatic compounds are introduced to environment via anthropogenic activities with limited biodegradability, some microbes have evolved enzymes and metabolic pathways to detoxify and utilize these compounds as their sole carbon sources (Reineke and Knackmuss, 1988; Commandeur and Parsons, 1990; Häggblom, 1992; Fetzner and Lingens, 1994; Copley, 1997; Arora and Bae, 2014). Overall microbial degradation pathways can be simplified and arranged into three stages as shown in Fig. 2. The upper pathway deals mainly with the debranching process to remove additional moieties attached to the aromatic ring such as removing an acetyl group from 2,4‐D or dephosphorylation of organophosphate halogenated aromatics. The compounds derived from this stage are mostly derivatives of mono‐aromatic ring compounds such as halogenated benzene, halogenated phenol, halogenated aniline and halogenated benzoate derivatives which serve as substrates for enzymes in the next stage (middle pathway). The middle pathway deals mainly with halide removal and activation of aromatic rings by incorporation of oxygen atom (in the case of aerobes). Halogen atom removal is the key step for detoxification of halogenated aromatics because the recalcitrance of substance for degradation depends on the type and number of halogen substituents (Fetzner and Lingens, 1994). The halogen atom removal can occur via reductive and oxidative modes. For the oxidative mode, it generally involves with oxygen incorporation via various pathways yielding hydroxylated aromatic products such as derivatives of catechol, hydroquinone or protocatechuate which also have low cellular toxicity and higher solubility than the original toxic substrates. The final stage (lower pathway) deals with ring‐cleavage reactions to convert aromatic into aliphatic molecules which can be further cleaved to form common metabolites of central metabolic pathways such as succinate, pyruvate, acetyl CoA, oxaloacetate and acetaldehyde (Commandeur and Parsons, 1990; Harwood and Parales, 1996; Fuchs et al., 2011; Arora and Bae, 2014; Tinikul et al., 2018; Chenprakhon et al., 2019).
Figure 2.

General microbial pathways of halogenated aromatics degradations. Three main catabolic pathways (upper, middle and lower pathways) are involved in overall biodegradation.
Complete degradation pathways of 2,4‐D from Cupriavidus necator JMP134 (Plumeier et al., 2002; Matus et al., 2003; Kumar et al., 2016), chlorpyrifos from Cupriavidus nantongensis X1T (Fang et al., 2019) and polychlorinated biphenyls (PCBs) from Pseudomonas sp. (Furukawa and Miyazaki, 1986; Benning et al., 1996; Copley, 1997; Nováková et al., 2002; Pieper, 2005) are shown in Fig. 3 as common examples of microbial degradation of halogenated aromatics that employ the three‐stage reactions mentioned above. It should be noted that most of the enzymes in the lower pathway do not have much reaction variation because they deal with a certain common set of metabolites from the central pathway. In contrast, enzymes in the upper and middle pathways show high reaction variation because they have to deal with various types of compounds. As most of exclusively reviews for molecular mechanisms and enzymatic reactions of halogenated aromatic biodegradation were published around two decades ago during 1980–1990s (Reineke and Knackmuss, 1988; Fetzner and Lingens, 1994), this review aims to highlight more updated information of the field since then and also discusses the enzymatic reactions used by various microbes to degrade specific halogenated aromatics. We also discuss recent technology for detection of these toxic substances which was developed based on basic knowledge of enzymes degrading halogenated aromatics. Enzymatic reactions are arranged based on the three stages of the degradation process and are organized under the following topics:
Figure 3.
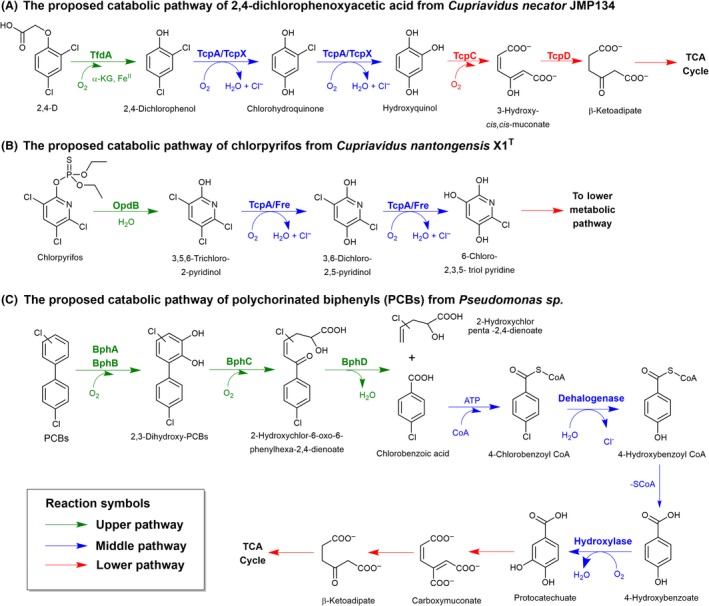
Upper catabolic pathway of halogenated aromatic pesticides.
A. Biodegradation pathway of 2,4‐dichlorophenoxyacetic acid (2,4‐D) from Cupriavidus necator JMP134. The enzymes involved are TfdA, α‐ketoglutarate‐dependent dioxygenase; TcpA, flavin‐dependent chlorophenol monooxygenase; TcpX, flavin reductase; TcpC, hydroxyquinol‐1,2‐dioxygenase; and TcpD, maleylacetate reductase (Plumeier et al., 2002; Matus et al., 2003; Kumar et al., 2016).
B. Biodegradation pathway of chlorpyrifos from Cupriavidus nantongensis XT 1. The enzymes involved are OpdB, organophosphorus hydrolase; TcpA, flavin‐dependent monooxygenase; and Fre, flavin reductase (Fang et al., 2019).
C. Biodegradation pathway of polychlorinated biphenyls (PCBs) from Pseudomonas sp. The enzymes involved are BphA, biphenyl‐2,3‐dioxygenase; BphB, biphenyl‐dihydrodiol‐dehydrogenase; BphC, 2,3‐dihydroxy‐biphenyl‐1,2‐dioxygenase; BphD, 2‐hydroxychlor‐6‐oxo‐6‐phenylhexa‐2,4‐dienoathydrolase; Dehalogenase, 4‐chlorobenzoyl coenzyme A dehalogenase; and Hydroxylase, p‐hydroxybenzoate hydroxylase (Furukawa and Miyazaki, 1986; Benning et al., 1996; Copley, 1997; Nováková et al., 2002; Pieper, 2005). Green, blue and red reactions indicated the upper, middle and lower pathway respectively.
-
The Upper Metabolic Pathways for Degradation of Halogenated aromatic
Debranching of 2,4‐dichlorophenoxyacetic acid (2,4‐D) and its derivatives
Dephosphorylation of organophosphate pesticides
Cleavage of the phenyl moiety from polyhalogenated biphenyl derivatives
-
The Middle Metabolic Pathways for Dehalogenation of Halogenated aromatics
Reductive dehalogenation of halogenated aromatics
Oxidative dehalogenation of halogenated aromatics by monooxygenases
Oxidative dehalogenation of halogenated aromatics by dioxygenases
Hydrolytic dehalogenation of halogenated aromatics
Ring Breakage and Generation of Common Metabolites by the Lower Metabolic Pathways
Technologies of Halogenated aromatic‐degrading Microbes and its Future Perspectives
The upper metabolic pathways for degradation of halogenated aromatics
The upper metabolic pathway generally involves removal of extra moieties to generate simple aromatic compounds that can be easily metabolized by downstream enzymes in the middle and lower pathways. Examples of enzymatic reactions involved in group removals or debranching of 2,4‐D and its derivatives, organophosphate pesticides (chlorpyrifos, profenofos, etc.) and polyhalogenated biphenyls are discussed below. These reactions are critically required prior to the downstream biodegradation process (Fig. 3). However, this upper pathway is not necessary for biodegradation of small halogenated aromatic compounds which can be directly processed by enzymes in the middle pathway.
Debranching of 2,4‐dichlorophenoxyacetic acid (2,4‐D) and its derivatives
Biodegradation of 2,4‐D is among the best understood pathways of halogenated aromatic degradation in bacteria. Several phylogenetically diverse bacterial species were reported to be capable of using 2,4‐D as a sole carbon source (Tonso et al., 1995; Kamagata et al., 1997; Itoh et al., 2002; Kumar et al., 2016). The 2,4‐D biodegradation pathway of C. necator JMP 134 was extensively studied (Don et al., 1985; Laemmli et al., 2000, 2004; Plumeier et al., 2002). The debranching step uses the enzyme α‐ketoglutarate (α‐KG)‐dependent dioxygenase (TfdA) encoded by the gene tfdA in the tfd operon (tfdA‐tfdF). The tfd operon contains the genes involved in 2,4‐D degradation to convert 2,4‐D to 2,4‐dichlorophenol (2,4‐DCP) and other smaller molecules. TfdA catalyses the cleavage of the ether bond that links the acetyl and phenoxyl moieties (green reaction in Fig. 3A). TfdA requires Fe(II) as a cofactor and uses α‐KG, 2,4‐D and oxygen as substrates to produce carbon dioxide, succinate, 2,4‐DCP and glyoxylate as products. Ferrous ion is required for its activity and cannot be replaced by other metals (Hogan et al., 1997). The product from the TfdA reaction, 2,4‐dichlorophenol is a substrate for the next enzyme in the operon, 2,4‐dichlorophenol hydroxylase which further hydroxylates 2,4‐DCP to generate the hydroxylated product (blue reaction in Fig. 3A) (Fukumori and Hausinger, 1993a,b; Saari and Hausinger, 1998; Hegg et al., 1999; Dunning Hotopp and Hausinger, 2002). Additionally, the tfd genes for the degradation of 2,4‐D were also found in diverse bacteria that contain 2,4‐D degrading abilities such as Burkholderia sp. (Baelum et al., 2010), Aeronomas hydrophila IBRB‐36 4CPA (Markusheva et al., 2004) and some aquatic bacteria of the genus Alcaligenes (Amy et al., 1985). The bacterial strain RD5‐C2 which is a member of the Bradyrhizobium‐Agromonas‐Nitrobacter‐Afipia cluster in the α‐proteobacteria was reported to carry tfdA‐like genes that encode proteins belonging to the α‐KG‐dependent dioxygenase family (Itoh et al., 2002).
In addition to the simple 2,4‐D compound described above, some halogenated aromatic herbicides such as mecoprop and dichlorprop are chiral molecules which require chiral‐specific α‐KG dioxygenases to catalyse etherolytic cleavage at the initial degradation steps (Lu et al., 2019). Reactions of enantiomer specific α‐KG dioxygenases such as RdpA and SdpA have been investigated (Nickel et al., 1997; Muller et al., 2004, 2006). The results have shown that RdpA dioxygenase is highly specific to the R enantiomer of (R)‐2‐(4‐chloro‐2‐methylphenoxy) propionic acid) (R‐mecoprop) and has low activity toward the S enantiomer. In contrast, SdpA is enantioselective to the S enantiomer but can also utilize other phenoxyacetate derivatives such as 2,4‐D and MCPA (Westendorf et al., 2003, 2006; Muller et al., 2006).
Dephosphorylation of organophosphate pesticides
For degradation of organophosphate pesticides such as chlorpyrifos and profenofos, the upper metabolic pathway involves cleavage of phosphoester bonds by hydrolase reactions to yield halogenated aromatic compounds in the form of phenol derivatives. Various bacteria including C. nantognensis X1T (Fang et al., 2019), Xanthomonas sp. 4R3‐M1, Pseudomonas sp. 4H1‐M3 (Rayu et al., 2017), Bacillus subtilis (Salunkhe et al., 2013), Stenotrophomonas sp. G1 (Deng et al., 2015), several Pseudomonas sp. (Siripattanakul‐Ratpukdi et al., 2015) and others have been isolated and reported to utilize organophosphate pesticides as their sole carbon sources. Recently, the catabolic pathway of chlorpyrifos degradation in C. nantognensis X1T (Fang et al., 2019) was reported. The initial step of chlorpyrifos biodegradation is hydrolysis at the phosphorus–oxygen bond by organophosphorus hydrolase (OpdB) encoded by the opdB gene to result in 3,5,6‐trichloro‐2‐pyridinol and diethylphosphate as products (green reaction in Fig. 3B). OpdB hydrolase also has a broad spectrum of substrate utilization, allowing it to hydrolyse various organophosphate pesticides including triazophos, parathion, isocarbophos, profenofos, malathion, cadusafos, methidathion and dimethoate. Some of the products, such as 3,5,6‐trichloro‐2‐pyridinol can be further degraded by enzymes in the middle and lower metabolic pathways found in the same bacterial strain.
Cleavage of the phenyl moiety from polyhalogenated biphenyl derivatives
Biodegradation of polyhalogenated biphenyl compounds that contain two aromatic rings require a cleavage between the two phenyl rings to generate single aromatic derivatives which can be further metabolized through enzymes in the middle and lower pathways. Various microbes are known for carrying genes encoding for the catabolic pathway of PCBs (Nováková et al., 2002; Pieper, 2005). Pseudomonas pseudoalcaligenes KF707 (Furukawa and Miyazaki, 1986) and Pseudomonas sp. LB400 (Mondello, 1989) have the gene cluster, bphABCD, which encodes enzymes for biphenyl ring breakage. The main enzymes in the upper catabolic pathway that can catalyse biphenyl ring cleavage of PCBs are two dioxygenases, biphenyl‐2,3‐dioxygenase (BphA) and 2,3‐dihydroxy‐biphenyl‐1,2‐dioxygenase (BphC). BphA is the first enzyme to incorporate two hydroxyl groups into a biphenyl ring, while BphC uses its dioxygenation activity to catalyse a ring‐cleavage reaction to generate 2‐hydroxychlor‐6‐oxo‐6‐phenylhexa‐2,4‐dienoate. Another enzyme, 2‐hydroxychlor‐6‐oxo‐6‐phenylhexa‐2,4‐dienoathydrolase (BphD), then hydrolyses this compound to yield chlorobenzoic acid and 2‐hydroxychloro‐penta‐2,4‐dienoate (green reaction in Fig. 3C) which will then be further metabolized by enzymes from the middle pathway as explained below.
It can be seen that enzymatic reactions in the upper metabolic pathway are diversified, depending of the types of halogenated aromatics and bacterial species. We have shown above three major pathways including debranching by etherolytic of halophenoxyacetate derivatives, phosphoester bond hydrolysis of organophosphate and ring breakage of PCBs. Although these enzymatic reactions are different, the resulting products from these different reactions narrow down to a few types of small halogenated aromatic compounds such as halogenated benzene, halogenated phenol, halogenated benzoate or halogenated aniline. These common compounds can be further degraded by enzymes in the middle pathway.
The middle metabolic pathways for dehalogenation of halogenated aromatics
At the middle metabolic biodegradation pathway, small halogenated aromatics are converted into less toxic and more soluble compounds such as catechol, hydroquinone or protocatechuate by halide removal (dehalogenation) in conjunction with or without group addition reactions (Fig. 2). In oxidative dehalogenation, the concomitant group addition can be catalysed by oxygenases to incorporate molecular oxygen or by hydrolases to incorporate water into the substrates. Because halogen substituents mainly affect the recalcitrance of halogenated aromatic degradation (Fetzner and Lingens, 1994) and they also have electronic and steric effects which reduce electron delocalization efficiency and hinder dioxygenase ring‐cleavage activities in the lower pathway (Copley, 1997), dehalogenation in the form of group removal generally occurs prior to the process catalysed by further enzymes. Therefore, the group addition (especially hydroxylation) in conjunction with dehalogenation is required to prepare the compounds to be ready for ring‐cleavage dioxygenases to convert the aromatic compounds to aliphatic compounds. However, dehalogenation without involvement of oxygen (reductive dehalogenation) was also reported (Fuchs et al., 2011; Boll et al., 2014). The following discussion will highlight the nature of the reaction and the mechanistic features of the dehalogenases that catalyse group removal via reductive, oxidative and hydrolytic reactions.
Reductive dehalogenation of halogenated aromatic compounds
Reductive dehalogenation is the replacement of halogen with hydrogen atom that can be performed in anaerobes and aerobes. Well‐studied anaerobes that are capable of performing reductive dehalogenation are organohalide‐respiring bacteria that are also capable of degrading a broad spectrum of halogenated aromatics including halogenated benzenes, halogenated phenols, halogenated dioxin and also polyhalogenated biphenyls (Adrian et al., 2000; Bunge et al., 2003; Wang et al., 2018). In the strict anaerobes, halogenated aromatics serve as a terminal electron acceptor in the electron transport chain of organohalide‐respiring bacteria with hydrogen gas (H2) as a sole electron donor (Jugder et al., 2016; Schubert et al., 2018; Wang et al., 2018). The key enzymes in this electron transport chain are membrane‐bound reductive dehalogenases (RDs) that receive electrons and pass them through Fe‐S clusters and B12 cofactors to halogenated aromatics as their final electron acceptors via a two‐step electron transfer (Fig. 4A) (Jugder et al., 2016). Because halogenated aromatics generally have high two‐electron reduction potentials, they serve as efficient electron acceptors in the H2‐dependent electron transport chain (Dolfing and Novak, 2015).
Figure 4.
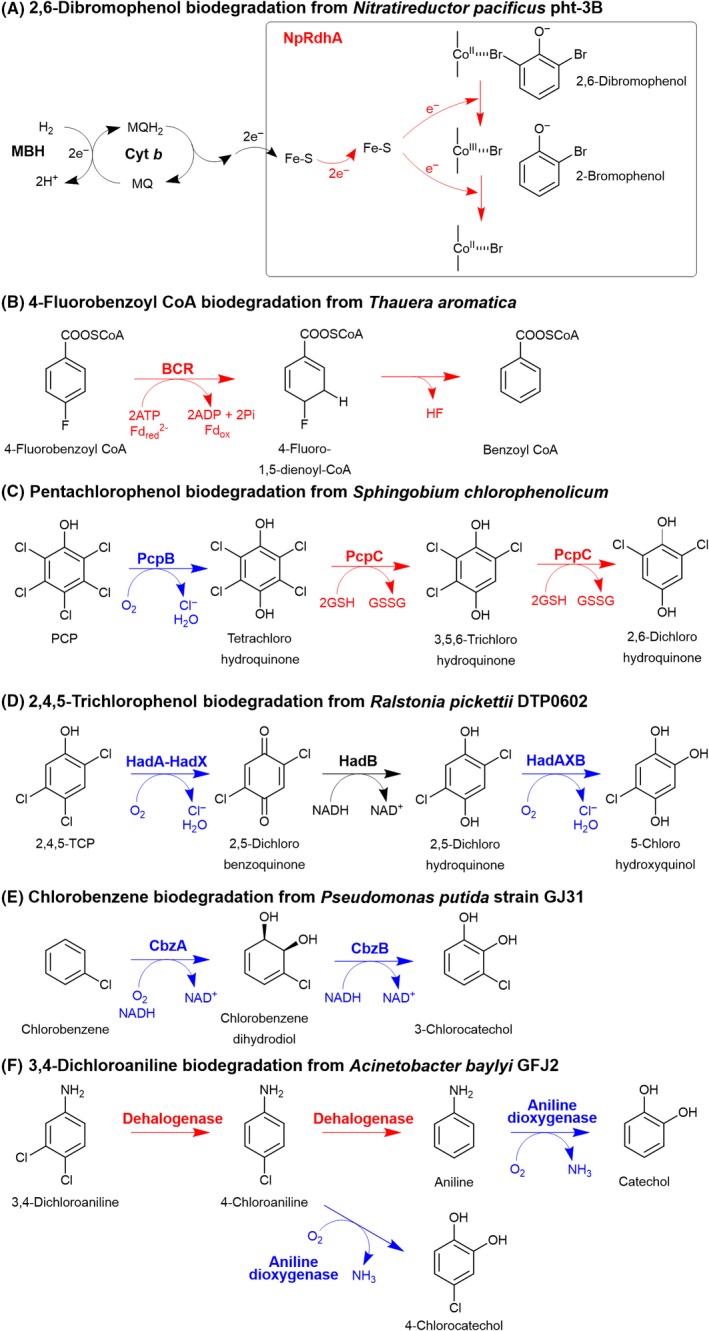
Various reactions in the middle catabolic pathway.
A. Reductive dehalogenation of 2,6‐dibromophenol from Nitratireductor pacificus pht‐3B involves the cobalamin (B12)‐dependent reductive dehalogenase, NpRdhA. Two electrons (2e−) are transferred from H2 through the membrane‐bound hydrogenase (MPH), cytochrome b (Cyt b) and the unidentified electron transfer component prior to their split through two, one electron (e−) reduction steps in the reductive dehalogenation of NpRdhA (Payne et al., 2015; Jugder et al., 2016; Collins et al., 2018).
B. 4‐fluorobenzoyl CoA biodegradation from Thauera aromatica involves the reductive dehalogenation by ATP‐dependent class I benzoyl CoA reductase (BCR) to catalyse dearomatization of 4‐fluorobenzoyl CoA to 4‐fluoro‐1,5‐dienoyl‐CoA prior to elimination of HF to generate benzoyl CoA as a central intermediate (Tiedt et al., 2016).
C. Pentachlorophenol biodegradation pathway from Sphingobium chlorophenolicum is initiated by oxidative dehalogenation involving a single‐component flavin‐dependent pentachlorophenol hydroxylase, PcpB, prior to reductive dehalogenation of tetrachlorohydroquinone by glutathione‐dependent tetrachlorohydroquinone dehalogenase, PcpC (Warner and Copley, 2007a,b; Hlouchova et al., 2012).
D. Oxidative catabolism of 2,4,5‐trichlorophenol from Ralstonia pickettii DTP0602 involves Had enzymes. HadA and HadX are the two‐component flavin‐dependent monooxygenase and reductase, respectively, while HadB is a quinone reductase (Pimviriyakul et al., 2017; Pimviriyakul and Chaiyen, 2018).
E. Chlorobenzene biodegradation pathway from Pseudomonas putida strain GJ31 occurs via a dioxygenation reaction by chlorobenzene dioxygenase (CbzA) and chlorobenzene dihydrodiol dehydrogenase (CbzB) to generate 3‐chlorocatechol as a product.
F. Branching catabolism of 3,4‐dichloroaniline involves both unidentified reductive dehalogenase and aniline dioxygenase respectively (Hongsawat and Vangnai, 2011). The red reaction indicates the dehalogenation via a reductive type reaction, while the blue reaction indicates the dehalogenation via an oxidative type reaction.
Organohalide‐respiring bacteria such as Dehalobacter sp. strain TeCB1 (Alfan‐Guzman et al., 2017), Dehalococcoides mccartyi strain DCMB5 (Poritz et al., 2015), Dehalococcoides mccartyi strain CBDB1 (Adrian et al., 2007; Wagner et al., 2012) were reported to be capable of utilizing halobenzenes. The strain TeCB1 can reductively remove a chlorine atom from 1,2,4,5‐tetrachlorobenzene to form 1,2,4‐trichlorobenzene and 1,3‐ or 1,4‐dichlorobenzene respectively (Alfan‐Guzman et al., 2017). The strain DCMB5 is capable of dehalogenating various chlorobenzenes including pentachlorobenzene, all types of tetrachlorobenzenes, 1,2,3‐trichlorophenol, as well as 1,2,3,4‐tetra‐ and 1,2,4‐trichlorodibenzo‐p‐dioxin, pentachlorophenol, tetrachlorophenol and all types of trichlorophenols (Poritz et al., 2015). Some anaerobes contain RDs that are involved in halogen removal from halogenated phenols. RDs from Dehalococcoides mccartyi strain JNA specifically remove the ortho‐chlorine atom from multi‐substituted chlorophenols (Fricker et al., 2014). A putative chlorophenol RD, cprA, from Dehalobacter sp. strain TCP1 can remove the ortho‐chlorine atom from 2,4,6‐TCP (Wang et al., 2014). Desulfitobacterium hafniense strain PCP‐1 can dehalogenate and convert PCP to 3‐CP (Bisaillon et al., 2010).
RdhA3 from Desulfitobacterium hafniense strain Y51 (Mac Nelly et al., 2014) and NpRdhA from Nitratireductor pacificus pht‐3B (Payne et al., 2015) are RDs which use cobalamin as a cofactor to catalyse halide elimination. The recombinant RdhA3 contains a cobalamin cofactor with two iron–sulfur clusters that can reductively dechlorinate 3,5‐DCP, 2,3‐DCP and 2,4‐DCP to generate 3‐CP, 2‐CP and 4‐CP as dechlorinated products respectively (Mac Nelly et al., 2014). NpRdhA is also a cobalamin (B12)‐dependent enzyme that can catalyse reduction of 2,6‐dibromophenol to 2‐bromophenol via a halide‐cobalt(II) formation. Investigation of the reaction mechanism based on structure determination combined with electron paramagnetic resonance spectroscopy and simulations using 2,6‐dibromophenol as a substrate revealed that a bromide–cobalt(II) intermediate forms before the cleavage of the carbon–bromide bond occurs to eliminate a bromide from the organic substrate (red reaction in Fig. 4A) (Payne et al., 2015). As ferredoxin (Fd) is required as an electron donor, the reaction of NpRdhA requires an additional reaction of ferredoxin reductase to generate reduced Fd. It was demonstrated that E. coli ferredoxin reductase (EcFldr) and spinach ferredoxin (SpFd) can provide electrons to the cobalt centre at active site of NpRdhA. Therefore, the overall reductive halogenation with all involved enzymes is NADPH‐dependent (Collins et al., 2018).
Another type of reductive dehalogenation is involved in the degradation pathway of halogenated benzoyl CoA which is ATP‐dependent (Fuchs et al., 2011; Boll et al., 2014). There are several microbes which contain this ATP‐dependent pathway including Thauera aromatica (Boll and Fuchs, 1995; Tiedt et al., 2016), Thauera chlorobenzoica sp. (Kuntze et al., 2011; Song et al., 2001), Pseudomonas stutzeri (Vargas et al., 2000) and Rhodopseudomonas palustris (Egland et al., 2001). The first step usually involves with the conversion of halogenated benzoate to halogenated benzoyl CoA by an ATP‐dependent benzoate‐CoA ligase (Wischgoll et al., 2005). In the following step, an ATP‐dependent reductive dehalogenase catalyses dearomatization of aromatic ring in concomitant with halide elimination (Boll and Fuchs, 1995; Fuchs et al., 2011; Boll et al., 2014). Recently, the enzyme in class I benzoyl CoA reductase (BCR) from T. aromatica was reported to catalyse reductive defluorination of 4‐fluorobenzoyl CoA to benzoyl CoA and hydrofluoric (HF) (Tiedt et al., 2016). This enzyme is composed of two main modules which are responsible for electron transfer via ferredoxin and [4Fe‐4S] clusters and aromatic ring binding (Fuchs et al., 2011; Boll et al., 2014). In its enzymatic mechanism (red reaction in Fig. 4B), the proposed mechanism was initiated by dearomatization of 4‐fluorobenzoyl CoA to 4‐fluoro‐1,5‐dienoyl‐CoA by receiving two electrons from ferredoxin in concomitant with hydrolysis of ATP to ADP. After that, HF was eliminated and rearomatization occurs to yield benzoyl CoA as a product. The resulting benzoyl CoA central intermediate is metabolized by downstream enzymes in the lower pathway via β‐oxidation‐like reactions to yield acetyl‐CoA and CO2 as final products (Wischgoll et al., 2005; Fuchs et al., 2011; Boll et al., 2014; Tiedt et al., 2016).
Reductive dehalogenation also occurs in aerobes such as Delftia sp. EOB‐17 (Chen et al., 2015) which can reductively dehalogenate 3,5‐dibromo‐4‐hydroxybenzoate (DBHB) to form 4‐hydroxybenzoate as a product. The enzymatic reactions of RDs from aerobes are different from those of the anaerobe RDs explained in the previous section. The well‐known tetrachlorohydroquinone dehalogenase or PcpC found in the pentachlorophenol biodegradation pathway of Sphingobium chlorophenolicum (originally called Flavobacterium sp.), is classified as a member of the glutathione S‐transferase family. PcpC catalyses the dechlorination of tetrachlorohydroquinone to trichlorohydroquinone and then trichlorohydroquinone to dichlorohydroquinone (red reaction in Fig. 4C) (Xun et al., 1992a; McCarthy et al., 1996). The reaction mechanism of PcpC has been extensively investigated (Warner and Copley, 2007a,b). Experiments using transient kinetic approaches (stopped‐flow and rapid‐quench techniques) with spectroscopic detection of PcpC using trichlorohydroquinone (TriCHQ) as a substrate were carried out. The reaction starts with rapid deprotonation of TriCHQ after binding to PcpC followed by delocalization of the electrons of a hydroxyl group to form 3,5,6‐trichloro‐4‐hydroxycyclohexa‐2,4‐dienone (TriCHQ*). In the presence of glutathione (GSH), GSH attacks TriCHQ* to form the 3,5‐dichloro‐2‐S‐glutathionylhydroquinone intermediate (GS‐DCHQ*) before chloride elimination occurs. This intermediate decays by reacting with another GSH to result in the final product of 2,6‐dichlorohydroquinone (2,6‐DCHQ) and glutathione disulfide (red reaction in Fig. 4C).
Oxidative dehalogenation of halogenated aromatic compounds by monooxygenases
Oxidative dehalogenation is another means to decrease the toxicity of halogenated aromatic compounds by incorporation of an –OH group to yield more hydrophilic products with lower toxicity. The addition of the extra hydroxyl group prepares the products for ring‐cleavage enzymes in the following stage of biodegradation. The concomitant incorporation of a hydroxyl group with halide elimination can be catalysed by monooxygenases.
Monooxygenation is the incorporation of one hydroxyl group (–OH) derived from molecular oxygen into organic compounds. Monooxygenases that catalyse monooxygenation are metal‐dependent, pterin‐dependent and flavin‐dependent enzymes (van Berkel et al., 2006; Roberts and Fitzpatrick, 2013; Huijbers et al., 2014; Romero et al., 2018; Banerjee et al., 2019; Chenprakhon et al., 2019). For monooxygenation of halogenated aromatics, the majority of the reactions are catalysed by flavin‐dependent monooxygenases (Arora and Bae, 2014). These enzymes catalyse dehalogenation plus incorporation of a hydroxyl group into halogenated phenols which are generated by enzymes in the upper pathway such as phenoxyacetic acid and organophosphate debranching enzymes previously discussed in the upper pathway section. These enzymes catalyse concomitant hydroxylation with halide elimination of halogenated phenols using reduced flavin (flavinred) and molecular oxygen (O2) as co‐substrates.
Flavin‐dependent dehalogenases can be divided into two types: single‐ and two‐component flavin‐dependent monooxygenases. Generally, all steps of the single‐component monooxygenases are catalysed by one polypeptide which binds oxidized flavin (flavinox) as a cofactor (Ballou et al., 2005; van Berkel et al., 2006; Palfey and McDonald, 2010; Huijbers et al., 2014; Chenprakhon et al., 2019). In contrast, reactions of two‐component flavin‐dependent monooxygenases are carried out by a separate reductase and oxygenase. Oxidized flavin is first reduced by NADH or NADPH on the reductase component before being transferred via diffusion to the oxygenase component (Huijbers et al., 2014; Sucharitakul et al., 2014; Chenprakhon et al., 2019). The rest of the hydroxylation plus dehalogenation reactions occur in the oxygenase component similar to the reaction of single‐component monooxygenases (Fig. 5).
Figure 5.
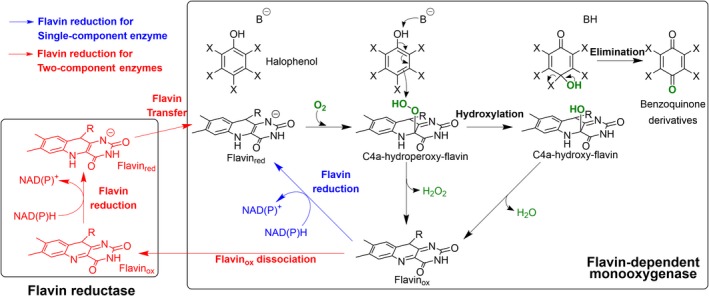
Mechanism of flavin‐dependent monooxygenase for dehalogenation of halogenated phenol. A simple reaction scheme of halogenated phenol oxidative dehalogenation by a flavin‐dependent monooxygenase which occurs via the reaction of the C4a‐hydroperoxy‐flavin intermediate. For a single‐component enzyme like PcpB from S. chlorophenolicum, the substrate is pentachlorophenol and the flavin is reduced via the blue pathway (Hlouchova et al., 2012; Rudolph et al., 2014). For two‐component enzymes, the HadA‐HadX system from R. pickettii, the substrate is 4‐chlorophenol and the flavin is reduced via the red pathway where HadA is a flavin‐dependent monooxygenase and HadX is a flavin reductase (Pimviriyakul et al., 2017; Pimviriyakul and Chaiyen, 2018). B represents a catalytic base for proton abstraction to facilitate electrophilic aromatic substitution.
PcpB from the pentachlorophenol degradation pathway of S. chlorophenolicum represents an extensively investigated single‐component flavin‐dependent monooxygenase (Hlouchova et al., 2012; Rudolph et al., 2014). The enzyme catalyses the conversion of PCP to tetrachlorohydroquinone (blue reaction in Fig. 4C) which can be further reductively dehalogenated by the PcpC discussed in the previous section (red reaction in Fig. 4C) (Hlouchova et al., 2012; Rudolph et al., 2014). In addition, PcpB was also reported to be able to catalyse elimination of other substituents such as bromide, iodide, nitro or cyano from phenolic derivatives (although with very low yield) (Xun et al., 1992b). The reaction mechanism of PcpB has been extensively investigated (Fig. 5). The reaction is initiated by the reduction of a flavin cofactor by NADH or NADPH during a reductive half‐reaction to generate the enzyme‐bound reduced FAD (FADH–). O2 then reacts with FADH– to form the C4a‐hydroperoxy‐flavin intermediate in which its terminal peroxide –OH acts as an electrophile to be incorporated into a phenolic substrate. The hydroxylated product eliminates a halide group in the next step while the resulting C4a‐hydroxyflavin eliminates water to yield the resting state of the cofactor, oxidized FAD, to complete the catalytic cycle (Hlouchova et al., 2012; Rudolph et al., 2014). Similar to PcpB, phenol hydroxylase from Trichosporon cutaneum which normally catalyses hydroxylation of the phenol also has promiscuous activities in hydroxylation and elimination of ortho‐fluoride from pentafluorophenol to yield tetrafluorocatechol (Peelen et al., 1995).
Two‐component monooxygenase‐reductase systems found in various aerobic microbes include the HadA‐HadX system from Ralstonia pickettii DTP006 (blue reaction in Fig. 4D) (Hatta et al., 2012; Pimviriyakul and Chaiyen, 2018), the TcpA‐TcpX system from Ralstonia eutropha JMP134 (blue reaction in Fig. 3A) (Matus et al., 2003; Xun and Webster, 2004; Belchik and Xun, 2008), the TftD‐TftC system from Burkholderia cepacia AC1100 (Gisi and Xun, 2003; Webb et al., 2010), the CphC‐I‐CphB system from Arthrobacter chlorophenolicus A6 (Cho et al., 2017; Kang et al., 2017), the DcmB1‐DcmB2 system from Rhodococcus sp. JT‐3 (Zhang et al., 2018) and the TcpA‐Fre system from C. nantongensis X1T (blue reaction in Fig. 3B) (Fang et al., 2019). These enzymatic systems catalyse the hydroxylation and halide elimination of halogenated phenol derivatives at the para‐ or ortho‐positions to yield either hydroquinone or catechol derivatives as products. It should be mentioned that in addition to dehalogenation, some of these systems can also catalyse elimination reactions of other substituents including denitration of nitrophenol by HadA‐HadX (Pimviriyakul and Chaiyen, 2018; Pimviriyakul et al., 2018) and by CphC‐I‐CphB (Kang et al., 2017).
HadA catalyses biodetoxification of 2,4,5‐TCP by two consecutive dechlorination steps at the para‐ and ortho‐positions to generate 5‐chlorohydroxyquinol as a final product (blue reaction in Fig. 4D). Recently, in‐depth investigation of HadA‐HadX reaction mechanisms using 4‐chlorophenol (4‐CP) as substrate (Pimviriyakul et al., 2017, 2018; Pimviriyakul and Chaiyen, 2018) have been reported (Fig. 5). Based on transient kinetic studies using stopped‐flow and rapid‐quench techniques, the first step of the HadA reaction is the binding of reduced flavin supplied from a reductase partner such as HadX via free diffusion. This step is followed by reaction with O2 to form the C4a‐hydroperoxy‐flavin reactive intermediate. 4‐CP then reacts with the C4a‐hydroperoxy via electrophilic aromatic substitution to result in hydroxylated 4‐CP which then eliminates the chloride group to result in the final products of benzoquinone and the C4a‐hydroxyflavin adduct which decays to oxidized flavin via dehydration (Pimviriyakul et al., 2017). The overall simple reaction scheme of single and two‐component dehalogenases can be summarized as in Fig. 5. Correlation of the rate constants of the individual steps of the HadA reaction with the physicochemical parameters obtained from density functional theory (DFT) analysis indicated that the overall reaction is dictated by the deprotonation ability of the substrates which is likely the step with the highest activation energy (Pimviriyakul et al., 2018). Currently, structures of HadA and its homologous enzymes including TftD and TcpA are only available in the apoform (Webb et al., 2010; Hayes et al., 2012; Pongpamorn et al., 2019). Based on structural modelling of substrate binding, His290 of HadA, H289 of TftD and H290 of TcpA are proposed to be the catalytic base that catalyses proton abstraction from the –OH group of a phenol substrate which is a crucial step for initiating the hydroxylation (B in Fig. 5) (Webb et al., 2010; Hayes et al., 2012; Pimviriyakul et al., 2018; Pongpamorn et al., 2019).
Recent investigations also indicate that two additional reductases are required for completion of HadA activity. HadX, a reductase encoded in the same operon as HadA was shown to be the most efficient and physiologically relevant reductase to provide reduced FAD for the HadA reaction. As the initial hydroxylated product from HadA and other two‐component monooxygenases are benzoquinone derivatives, some microbes possess quinone reductases such as HadB in Ralstonia pickettii DTP006 (Pimviriyakul and Chaiyen, 2018) and TcpB in Ralstonia eutropha JMP134 (Belchik and Xun, 2008) to enzymatically convert benzoquinone to hydroquinone. Hydroquinone is a more stable compound which can be used by downstream enzymes in the lower biodegradation pathway (Fig. 4D). The whole system (HadA monooxygenase, HadX flavin reductase and HadB quinone reductase) can be coupled together as an in vitro enzymatic cascade, resulting in high efficiency of hydroxylation and group elimination (Pimviriyakul and Chaiyen, 2018). The hydroquinone product can be easily metabolized by a downstream HadC 1,2‐hydroquinone dioxygenase (Hatta et al., 1999). The whole cascade of HadA can be used as a basis set for applications in bioremediation processes.
Oxidative dehalogenation of halogenated aromatic compounds by dioxygenases
Dioxygenation is incorporation of two hydroxyl groups (both oxygen atoms are derived from O2) into organic substrates. Reactions are catalysed by dioxygenase enzymes and they are generally found in the biodegradation of halogenated benzenes. Degradation of chlorobenzene (CB) by Ralstonia pickettii strain L2 occurs via two steps of hydroxylation to yield a phenolic intermediate and catecholic product respectively (Zhang et al., 2011). The whole biodegradation pathway for CB in Pseudomonas putida strain GJ31 by the enzymes encoded in the cbz cluster has been proposed (Kunze et al., 2009). CB is first hydroxylated by chlorobenzene dioxygenase (CbzA) to form chlorobenzene dihydrodiol which is then further converted to 3‐chlorocatechol by another enzyme chlorobenzene dihydrodiol dehydrogenase (CbzB) (Fig. 4E). Fluorobenzene (FB) can be metabolized by Burkholderia fungorum FLU100 (DSM 23736) (Strunk and Engesser, 2013) and Labrys portucalensis to result in 3‐fluorocatechol as product. Halogenated catechols can be further cleaved by another enzyme in the pathway, catechol 2,3‐dioxygenase (Kunze et al., 2009; Strunk and Engesser, 2013).
Another type of compounds that can be degraded by dioxygenases is halogenated aniline. Chloroanilines are widely used in the industrial production of dyes, cosmetics, pharmaceutical products and herbicides. Many bacteria were reported to have the ability to degrade chloroaniline. The degradation of monochloroaniline can occur via two proposed pathways (Fig. 4F). In the first pathway, the process initiates via dechlorination of monochloroaniline to generate aniline which is then converted to catechol by aniline dioxygenase. In the other pathway, the initial step of monochloroaniline degradation is a dioxygenation which is then followed by deamination of monochloroaniline to form the corresponding chlorocatechol (Hongsawat and Vangnai, 2011; Arora, 2015). Bacterial degradation of dichloroanilines has also been investigated. Based on product analysis by LC/MS, Bacillus megaterium IMT21 degrades 2,3‐dichloroaniline, 2,4‐dichloroaniline and 2,5‐dichloroaniline via dichloroaminophenol, and 3,4‐ and 3,5‐dichloroaniline via dichloroacetanilide, although information on the enzymes involved in the degradation process is still unclear (Yao et al., 2011). Pseudomonas fluorescens 26‐K degrades 3,4‐dichloroaniline via 4‐amino‐2‐chlorophenol through initial dehalogenation and hydroxylation (Travkin et al., 2003). A strain of Pseudomonas sp. degrades 3,4‐dichloroaniline via 4,5‐dichlorocatechol, 3,4‐dichloromuconate, 3‐chlorobutenolide, 3‐chloromaleylacetate and 3‐chloro‐4‐ketoadipate (You and Bartha, 1982). Variation of the degradation pathway of 3,4‐dichloroaniline was observed in Acinetobacter baylyi strain GFJ2 in which the initial step of degradation involves dehalogenation of 3,4‐dichloroaniline to 4‐chloroaniline. 4‐Chloroaniline can be further degraded via two different routes. In the first route, 4‐chloroaniline undergoes dehalogenation to produce aniline that is further degraded via catechol and the ortho‐cleavage pathway. In the second route, 4‐chloroaniline undergoes dioxygenation to yield 4‐chlorocatechol and ammonia (Fig. 4F). However, specific enzymes involved in each degradation step in A. baylyi strain GFJ2 have not yet been identified (Hongsawat and Vangnai, 2011; Arora, 2015).
Hydrolytic dehalogenation of halogenated aromatic compounds
Hydrolytic dehalogenation is replacement of a halogen by a hydroxyl group derived from H2O. Hydrolytic dehalogenating enzymes catalyse dehalogenation of chlorobenzoate which is part of polychlorinated biphenyls (PCBs) biodegradation pathways (Fig. 3C). Hydrolytic dehalogenation is also found in several organisms such as Pseudomonas sp. CBS3 (Klages et al., 1981), Arthrobacter sp. 4‐CB1 (Crooks and Copley, 1994), Arthrobacter sp. (Zhuang et al., 2003; Zhou et al., 2008), Rhodococcus ruber P25 (Plotnikova et al., 2012), Betaproteobacteria (Chae et al., 2008). 4‐Chlorobenzoate can be used as a substrate for ATP‐dependent 4‐halobenzoate coenzyme A ligase in the cell to generate a reactive metabolite, 4‐chlorobenzoyl coenzyme A (Pieper, 2005). 4‐Chlorobenzoyl coenzyme A dehalogenase can then carry out the next reaction of hydrolytic dehalogenation (blue reaction in Fig. 3C). The reaction mechanism of 4‐chlorobenzoyl CoA dehalogenase was extensively investigated (Copley, 1997; Lau and Bruice, 2001; Luo et al., 2001; Zhang et al., 2001; Xu et al., 2004; Wu et al., 2006) and it was found that the key catalytic residue is Asp145, which performs a nucleophilic attack on the para‐position of 4‐chlorobenzoate CoA where a chlorine atom is present. The chlorine atom was eliminated and replaced by a –OH group derived from H2O (Benning et al., 1996; Copley, 1997). The overall degradation pathway of PCBs, including the reaction of hydrolytic 4‐chlorobenzoyl coenzyme A dehalogenase can be summarized in Fig. 3C. The hydroxylated product 4‐hydroxybenzoyl‐CoA can be hydrolysed to form 4‐hydroxybenzoate (4‐HB) by 4‐hydroxybenzoyl‐CoA thioesterase. 4‐HB can be further metabolized by 4‐hydroxybenzoate hydroxylase and ring‐cleavage dioxygenases (Seibold et al., 1996; Palfey and McDonald, 2010; Tinikul et al., 2018; Lubbers et al., 2019).
Ring breakage and generation of common metabolites by the lower metabolic pathways
To achieve full assimilation of aromatic compounds by bacteria, metabolites from the central pathways such as phenols, hydroquinone, catechol and protocatechuate need to be converted to metabolites commonly found in the central metabolic pathways such as oxo‐acids in the tricarboxylic acid (TCA) cycle. Depending on the type of aromatic derivatives, a specific site of ring cleavage, either intradiol or extradiol cleavage can be catalysed by different sets of dioxygenases which result in diverse products (Fig. 6). The intradiol and extradiol dioxygenases are non‐heme iron‐containing enzymes that have been extensively studied (Fielding et al., 2014; Wang et al., 2017). These enzymes catalyse the incorporation of two oxygen atoms resulting in concomitant C‐C bond fission, in which oxygen activation occurs through a substrate–oxygen–metal coordinated complex (Lipscomb, 2008; Fielding et al., 2014; Pornsuwan et al., 2017).
Figure 6.
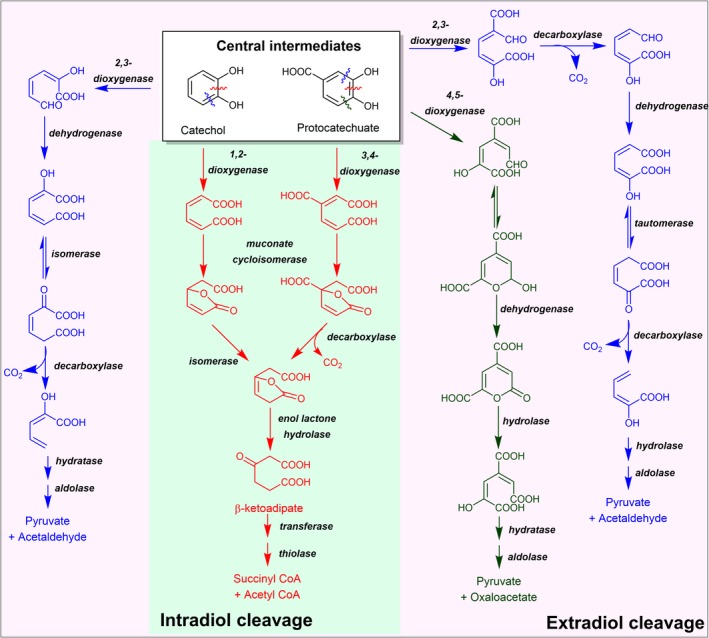
Various reactions in the lower catabolic pathways. Red pathway is an intradiol cleavage pathway initiated by 1,2‐cleavage via 1,2‐dioxygenase for catechol or by 3,4‐cleavage via 3,4‐dioxygenase for protocatechuate. Blue pathway is an extradiol cleavage pathway initiated by 2,3‐cleavage using 2,3‐dioxygenase. Green pathway is extradiol cleavage pathway initiated by 4,5‐cleavage using 4,5‐dioxygenase.
Gene clusters and enzymes from the steps of ring cleavage in the degradation pathways of halogenated aromatics are similar to those found in microbes metabolizing general and non‐halogenated aromatic compounds. Enzymes involved in ring cleavage of aromatic compounds have been extensively studied and many good articles and reviews related to this topic are available (Fuchs et al., 2011; Wells and Ragauskas, 2012; Thotsaporn et al., 2016; Kamimura et al., 2017). In general, the reaction pathways can be divided into the intradiol ring‐cleavage type (green area in Fig. 6) in which bond cleavage occurs between two hydroxyl groups of catecholic compounds or the extradiol ring‐cleavage type in which cleavage occurs outside the dihydroxyl groups (pink area in Fig. 6).
Examples of intradiol dioxygenases (green area in Fig. 6) include catechol‐1,2‐dioxygenase and protocatechuate‐3,4‐dioxygenase (red pathway Fig. 6). Genes coding for enzymes in the intradiol ring‐cleavage pathway have been identified in various bacteria and fungi (Patel et al., 1975; Bull and Ballou, 1981; Hartnett et al., 1990; Murakami et al., 1997; Iwagami et al., 2000; Wang et al., 2006). The cleavage by intradiol dioxygenases resulted in muconate derivatives such as cis,cis‐muconate and β‐carboxy‐cis,cis‐muconate. These compounds can be further lactonized by the enzyme muconate cycloisomerase (Fig. 6). The cleavage reaction pathways of catechol and protocatechuate proceed through a common intermediate at the formation of β‐ketoadipate enol‐lactone step before the lactone ring is hydrolysed by an enzyme enol‐lactone hydrolase to generate β‐ketoadipate (Patel et al., 1975). Further reactions along the pathway include two reactions catalysed by β‐ketoadipate:succinyl‐CoA transferase and β‐ketoadipyl‐CoA thiolase to generate succinyl‐CoA and acetyl‐CoA, which are common intermediates of the TCA cycle (Harwood and Parales, 1996).
For the extradiol ring‐cleavage pathway (pink area in Fig. 6), dioxygenases catalyse C‐C bond breaking outside the two hydroxyl groups either between the C2‐C3 positions of catechol or the C2‐C3 and C4‐C5 positions of protocatechuate. The catechol‐2,3‐dioxygenase cleaves the catecholic ring to yield 2‐hydroxymuconic semialdehyde (blue pathways in Fig. 6). This molecule can be further metabolized by the subsequent reactions catalysed by dehydrogenase, isomerase and decarboxylase encoded in the same gene cluster to generate 2‐hydroxypenta‐2,4‐dienoate. With additional reactions of hydratase and aldolase, 2‐hydroxypenta‐2,4‐dienoate is hydroxylated and cleaved to generate pyruvate and acetaldehyde. For the protocatechuate cleavage pathway, two possible cleavage sites can be metabolized by different enzymes to yield either pyruvate and oxaloacetate or pyruvate and acetyl‐CoA (blue and green pathways in Fig. 6) (Wolgel et al., 1993; Maruyama et al., 2004) which are common metabolites in the central metabolic pathway. After being cleaved by protocatechuate‐4,5‐dioxygenase or protocatechuate‐2,3‐dioxygenase, both paths use similar sets of dehydrogenase, isomerase, hydratase and aldolase to generate the final products. It is also noted that two additional reactions of decarboxylases are required in the protocatechol‐2,3‐cleavage route (Kasai et al., 2009) (blue pathway in Fig. 6). The organism uses a simpler pathway of protocatechol‐4,5‐cleavage rather than the protocatechuate‐2,3‐cleavage, as the gene cluster of the 2,3‐cleavage pathway is rarely found in nature. Recently, Paenibacillus sp. was reported to contain a protocatechuate‐2,3‐position cleavage enzyme (Kasai et al., 2009).
In terms of mechanistic investigation, only ring‐cleavage dioxygenases have been extensively explored (Lipscomb, 2008; Fielding et al., 2014). Knowledge related to the reaction mechanisms of the enzymes succeed these dioxygenases is limited and remains an area needing further exploration. More in‐depth understanding of these enzymes should be useful for future applications in bioremediation and biocatalysis.
Technologies of halogenated aromatic‐degrading microbes and future perspectives
Microbes capable of biodegradation of halogenated aromatics are highly sought after for their ability to perform bioremediation through clean and green processes. Thus, several technologies have been developed to improve their degradation efficiency. The most straight forward approach is to use the microbe directly. However, several parameters related to bioattenuation, biostimulation and bioaugmentation processes need to be adjusted to obtain optimal efficiency (Megharaj et al., 2011). For example, improvement of halogenated aromatic degradation by simply mixing microbial consortia that have synergetic effects on the degradation of halogenated aromatics has been investigated. Reports on pentachlorophenol and chlorobenzene detoxification by mixtures of microbes showed a higher consumption as compared to a single organism (Hechmi et al., 2016; Cheng et al., 2017). On the other hand, bioaugmentation through the induction of native strains was also demonstrated to be useful. The anaerobe Dehalococcoides mccartyi was induced by preculture under the condition containing chloroethane as an alternative electron acceptor for 1 week prior to its use in PCBs degradation. This process increased the dechlorination rate of PCBs by the induced microbes by 30‐times compared to the non‐induced microbes (Chen and He, 2018). In some case, biostimulation or growth of the microbes in favourable environments such as nutrients and its substrate is performed to increase microbe amount before applying the microbes in the treatment process (Megharaj et al., 2011).
More advanced techniques for biodegradation development involve enzyme engineering, metabolic engineering and cell engineering. The use of these techniques for developing powerful halogenated aromatic‐degrading microbes has been drawing great interest (Dvorak et al., 2017; Sharma et al., 2018; Sheldon and Woodley, 2018; Bilal et al., 2019; Liu et al., 2019). Based on basic knowledge of the molecular mechanisms of the degradation pathways, selected genes from various organisms are overexpressed in the same host to improve its degradation ability. Recently, the HadA monooxygenase which can catalyse hydroxylation plus halide and nitro group elimination was engineered to increase its thermostability based on computational prediction. A single‐point mutation of the G513Y variant resulted in the engineered enzyme that has significantly improved stability relative to the wild‐type enzyme and has equivalent activity. G513Y has an activity half‐life 72‐fold (50°C) and 160‐fold (45°C) longer than the wild‐type enzyme (Pongpamorn et al., 2019). The engineered E. coli strain that contains the constructed pathway (mainly enzymes in the middle and lower pathways) has the ability to degrade aromatic derivatives while the native one cannot (Clarkson et al., 2017; Wang et al., 2019). Other engineering processes such as cell/protein immobilization, enzyme cross‐linking and process optimization have also been investigated (Sheldon and Woodley, 2018). As the major purpose of these microbes is for eventual use in situ for the decontamination of pollutants mainly in hazardous contaminated sites, most of the development of halogenated aromatic‐degrading microbes has been done in whole cell engineering because microbes are more robust than individual enzymes. One of the most efficient approaches is cell surface engineering to localize the enzymes of interested on the cell surface to increase the chances of substrate utilization (Ueda, 2016). The immobilization of laccase to the cell surface combined with the cellular enzymes of Pseudomonas putida resulted in an ability to degrade chlorpyrifos without accumulation of its metabolites (Liu et al., 2016).
In addition to biodegradation, biodetection of halogenated aromatics is also important for controlling the level of these compounds in environment. Bio‐based detection of toxic halogenated aromatic pesticides based on direct and indirect measurements have been developed (Rao et al., 2014; Nigam and Shukla, 2015; Xu et al., 2018). Enzyme‐based biosensors that directly detect the amount of organophosphate pesticides have been invented based on the use of organophosphate hydroxylases from the upper pathway to immobilize the enzyme on an electrode (Gothwal et al., 2014). The contamination of halogenated aromatic pollutants can be determined by the coexpression of genes promoters responsive to specific contaminants and biomarker genes such as luciferase gene (luc) or green fluorescent protein gene (gfp). The light signal is generated in the presence of contaminants (Kumar et al., 2018). Recently, a novel one‐pot chemo‐enzymatic cascade based on the reaction of HadA monooxygenase was coupled to a de novo synthesis of firefly D‐luciferin which is a valuable reagent used in biomedical research. When firefly luciferase was added into the reaction cascade, the glow bioluminescence signal allows the detection of halogenated phenols and nitrophenols in ppb level. This technology can also detect trace amount of halogenated phenols and nitrophenols in biological samples such as urine and serum without requiring sample pretreatment, illustrating its promise as a powerful platform for high throughput biomedical screening to identify farmers or workers that are at risk of occupational hazards from dealing with these toxicants (Watthaisong et al., 2019).
Although a substantial amount of basic knowledge regarding the degradation of halogenated aromatics is available, biotechnological innovations based on this knowledge are still limited. This is largely due to the low efficiency and instability of relevant enzymes and microbes. Further development using enzyme engineering approaches to improve efficiency and stability are needed so that these biomolecules and cells can be used more effectively in biodegradation applications.
Conflict of interest
None declared.
Acknowledgements
Research in the authors laboratories are supported by the grants from Thailand Science Research and Innovation [MRG6280185 (to P. P.), MRG6080234 (to T. W.), MRG6180151 (to R. T.) and RTA5980001 (to P. C.)], grants from Mahidol University (to R. T.) and grants from Vidyasirimedhi Institute of Science and Technology (to T. W. and P. C.).
Microbial Biotechnology (2020) 13(1), 67–86
Funding information
Research in the authors laboratories are supported by the grants from Thailand Science Research and Innovation [MRG6280185 (to P. P.), MRG6080234 (to T. W.), MRG6180151 (to R. T.) and RTA5980001 (to P. C.)], grants from Mahidol University (to R. T.) and grants from Vidyasirimedhi Institute of Science and Technology (to T. W. and P. C.).
use and distribution in any medium, provided the original work is properly cited, the use is non‐commercial and no modifications or adaptations are made.
References
- Adrian, L. , Szewzyk, U. , Wecke, J. , and Görisch, H. (2000) Bacterial dehalorespiration with chlorinated benzenes. Nature 408: 580–583. [DOI] [PubMed] [Google Scholar]
- Adrian, L. , Rahnenfuhrer, J. , Gobom, J. , and Holscher, T. (2007) Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl Environ Microbiol 73: 7717–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfan‐Guzman, R. , Ertan, H. , Manefield, M. , and Lee, M. (2017) Isolation and characterization of Dehalobacter sp. strain TeCB1 including identification of TcbA: a novel tetra‐ and trichlorobenzene reductive dehalogenase. Front Microbiol 8: 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amy, P.S. , Schulke, J.W. , Frazier, L.M. , and Seidler, R.M. (1985) Characterization of aquatic bacteria and cloning of genes specifying partial degradation of 2,4‐dichlorophenoxyacetic acid. Appl Environ Microbiol 49: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, P.K. (2015) Bacterial degradation of monocyclic aromatic amines. Front Microbiol 6: 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, P.K. , and Bae, H. (2014) Bacterial degradation of chlorophenols and their derivatives. Microb Cell Fact 13: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, P.K. , Sasikala, C. , and Ramana, C.V. (2012) Degradation of chlorinated nitroaromatic compounds. Appl Microbiol Biotechnol 93: 2265–2277. [DOI] [PubMed] [Google Scholar]
- Arora, P.K. , Srivastava, A. , Garg, S.K. , and Singh, V.P. (2018) Recent advances in degradation of chloronitrophenols. Bioresour Technol 250: 902–909. [DOI] [PubMed] [Google Scholar]
- Baelum, J. , Jacobsen, C.S. , and Holben, W.E. (2010) Comparison of 16S rRNA gene phylogeny and functional tfdA gene distribution in thirty‐one different 2,4‐dichlorophenoxyacetic acid and 4‐chloro‐2‐methylphenoxyacetic acid degraders. Syst Appl Microbiol 33: 67–70. [DOI] [PubMed] [Google Scholar]
- Ballou, D.P. , Entsch, B. , and Cole, L.J. (2005) Dynamics involved in catalysis by single‐component and two‐component flavin‐dependent aromatic hydroxylases. Biochem Biophys Res Commun 338: 590–598. [DOI] [PubMed] [Google Scholar]
- Banerjee, R. , Jones, J.C. , and Lipscomb, J.D. (2019) Soluble methane monooxygenase. Annu Rev Biochem 88: 409–431. [DOI] [PubMed] [Google Scholar]
- Belchik, S.M. , and Xun, L. (2008) Functions of flavin reductase and quinone reductase in 2,4,6‐trichlorophenol degradation by Cupriavidus necator JMP134. J Bacteriol 190: 1615–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning, M.M. , Taylor, K.L. , Liu, R.Q. , Yang, G. , Xiang, H. , Wesenberg, G. , et al (1996) Structure of 4‐chlorobenzoyl coenzyme A dehalogenase determined to 1.8 Å resolution: an enzyme catalyst generated via adaptive mutation. Biochemistry 35: 8103–8109. [DOI] [PubMed] [Google Scholar]
- van Berkel, W.J. , Kamerbeek, N.M. , and Fraaije, M.W. (2006) Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol 124: 670–689. [DOI] [PubMed] [Google Scholar]
- Bilal, M. , Asgher, M. , Cheng, H. , Yan, Y. , and Iqbal, H.M.N. (2019) Multi‐point enzyme immobilization, surface chemistry, and novel platforms: a paradigm shift in biocatalyst design. Crit Rev Biotechnol 39: 202–219. [DOI] [PubMed] [Google Scholar]
- Bisaillon, A. , Beaudet, R. , Lepine, F. , Deziel, E. , and Villemur, R. (2010) Identification and characterization of a novel CprA reductive dehalogenase specific to highly chlorinated phenols from Desulfitobacterium hafniense strain PCP‐1. Appl Environ Microbiol 76: 7536–7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll, M. , and Fuchs, G. (1995) Benzoyl‐coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur J Biochem 234: 921–933. [DOI] [PubMed] [Google Scholar]
- Boll, M. , Loffler, C. , Morris, B.E. , and Kung, J.W. (2014) Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl‐coenzyme A esters: organisms, strategies and key enzymes. Environ Microbiol 16: 612–627. [DOI] [PubMed] [Google Scholar]
- Bull, C. , and Ballou, D.P. (1981) Purification and properties of protocatechuate 3,4‐dioxygenase from Pseudomonas putida. A new iron to subunit stoichiometry. J Biol Chem 256: 12673–12680. [PubMed] [Google Scholar]
- Bunge, M. , Adrian, L. , Kraus, A. , Opel, M. , Lorenz, W.G. , Andreesen, J.R. , et al (2003) Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421: 357. [DOI] [PubMed] [Google Scholar]
- Casida, J.E. , and Quistad, G.B. (2004) Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem Res Toxicol 17: 983–998. [DOI] [PubMed] [Google Scholar]
- Chae, J.C. , Song, B. , and Zylstra, G.J. (2008) Identification of genes coding for hydrolytic dehalogenation in the metagenome derived from a denitrifying 4‐chlorobenzoate degrading consortium. FEMS Microbiol Lett 281: 203–209. [DOI] [PubMed] [Google Scholar]
- Chen, C. , and He, J. (2018) Strategy for the rapid dechlorination of polychlorinated biphenyls (PCBs) by Dehalococcoides mccartyi strains. Environ Sci Technol 52: 13854–13862. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Jian, S. , Huang, L. , Ruan, Z. , Li, S. , and Jiang, J. (2015) Reductive dehalogenation of 3,5‐dibromo‐4‐hydroxybenzoate by an aerobic strain of Delftia sp. EOB‐17. Biotechnol Lett 37: 2395–2401. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.W. , Li, C. , Kennes, C.T. , Ye, J.X. , Chen, D.Z. , Zhang, S.H. , et al (2017) Improved biodegradation potential of chlorobenzene by a mixed fungal‐bacterial consortium. Int Biodeterior Biodegradation 123: 276–285. [Google Scholar]
- Chenprakhon, P. , Wongnate, T. , and Chaiyen, P. (2019) Monooxygenation of aromatic compounds by flavin‐dependent monooxygenases. Protein Sci 28: 8–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S.Y. , Kwean, O.S. , Yang, J.W. , Cho, W. , Kwak, S. , Park, S. , et al (2017) Identification of the upstream 4‐chlorophenol biodegradation pathway using a recombinant monooxygenase from Arthrobacter chlorophenolicus A6. Bioresour Technol 245: 1800–1807. [DOI] [PubMed] [Google Scholar]
- Clarkson, S.M. , Giannone, R.J. , Kridelbaugh, D.M. , Elkins, J.G. , Guss, A.M. , and Michener, J.K. (2017) Construction and optimization of a heterologous pathway for protocatechuate catabolism in Escherichia coli enables bioconversion of model aromatic compounds. Appl Environ Microbiol 83: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, F.A. , Fisher, K. , Payne, K.A.P. , Gaytan Mondragon, S. , Rigby, S.E.J. , and Leys, D. (2018) NADPH‐driven organohalide reduction by a nonrespiratory reductive dehalogenase. Biochemistry 57: 3493–3502. [DOI] [PubMed] [Google Scholar]
- Commandeur, L.C. , and Parsons, J.R. (1990) Degradation of halogenated aromatic compounds. Biodegradation 1: 207–220. [DOI] [PubMed] [Google Scholar]
- Copley, S.D. (1997) Diverse mechanistic approaches to difficult chemical transformations: microbial dehalogenation of chlorinated aromatic compounds. Chem Biol 4: 169–174. [DOI] [PubMed] [Google Scholar]
- Crooks, G.P. , and Copley, S.D. (1994) Purification and characterization of 4‐chlorobenzoyl CoA dehalogenase from Arthrobacter sp. strain 4‐CB1. Biochemistry 33: 11645–11649. [DOI] [PubMed] [Google Scholar]
- Cycon, M. , Mrozik, A. , and Piotrowska‐Seget, Z. (2017) Bioaugmentation as a strategy for the remediation of pesticide‐polluted soil: a review. Chemosphere 172: 52–71. [DOI] [PubMed] [Google Scholar]
- Deng, S. , Chen, Y. , Wang, D. , Shi, T. , Wu, X. , Ma, X. , et al (2015) Rapid biodegradation of organophosphorus pesticides by Stenotrophomonas sp. G1. J Hazard Mater 297: 17–24. [DOI] [PubMed] [Google Scholar]
- Dolfing, J. , and Novak, I. (2015) The Gibbs free energy of formation of halogenated benzenes, benzoates and phenols and their potential role as electron acceptors in anaerobic environments. Biodegradation 26: 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don, R.H. , Weightman, A.J. , Knackmuss, H.J. , and Timmis, K.N. (1985) Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4‐dichlorophenoxyacetic acid and 3‐chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J Bacteriol 161: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp, J.C. , and Hausinger, R.P. (2002) Probing the 2,4‐dichlorophenoxyacetate/alpha‐ketoglutarate dioxygenase substrate‐binding site by site‐directed mutagenesis and mechanism‐based inactivation. Biochemistry 41: 9787–9794. [DOI] [PubMed] [Google Scholar]
- Dvorak, P. , Nikel, P.I. , Damborsky, J. , and de Lorenzo, V. (2017) Bioremediation 3.0: engineering pollutant‐removing bacteria in the times of systemic biology. Biotechnol Adv 35: 845–866. [DOI] [PubMed] [Google Scholar]
- Egland, P.G. , Gibson, J. , and Harwood, C.S. (2001) Reductive, coenzyme A‐mediated pathway for 3‐chlorobenzoate degradation in the phototrophic bacterium Rhodopseudomonas palustris . Appl Environ Microbiol 67: 1396–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, L. , Shi, T. , Chen, Y. , Wu, X. , Zhang, C. , Tang, X. , et al (2019) Kinetics and catabolic pathways of the insecticide chlorpyrifos, annotation of the degradation genes, and characterization of enzymes TcpA and Fre in Cupriavidus nantongensis X1T . J Agric Food Chem 67: 2245–2254. [DOI] [PubMed] [Google Scholar]
- Fetzner, S. , and Lingens, F. (1994) Bacterial dehalogenases: biochemistry, genetics, and biotechnological applications. Microbiol Rev 58: 641–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, J.A. , and Sierra‐Alvarez, R. (2008) Microbial degradation of chlorinated phenols. Rev Environ Sci Bio 7: 211–241. [DOI] [PubMed] [Google Scholar]
- Fielding, A.J. , Lipscomb, J.D. , and Que, L. Jr (2014) A two‐electron‐shell game: intermediates of the extradiol‐cleaving catechol dioxygenases. J Biol Inorg Chem 19: 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker, A.D. , LaRoe, S.L. , Shea, M.E. , and Bedard, D.L. (2014) Dehalococcoides mccartyi strain JNA dechlorinates multiple chlorinated phenols including pentachlorophenol and harbors at least 19 reductive dehalogenase homologous genes. Environ Sci Technol 48: 14300–14308. [DOI] [PubMed] [Google Scholar]
- Fuchs, G. , Boll, M. , and Heider, J. (2011) Microbial degradation of aromatic compounds – from one strategy to four. Nat Rev Microbiol 9: 803–816. [DOI] [PubMed] [Google Scholar]
- Fukumori, F. , and Hausinger, R.P. (1993a) Alcaligenes eutrophus JMP134 2,4‐dichlorophenoxyacetate monooxygenase is an alpha‐ketoglutarate‐dependent dioxygenase. J Bacteriol 175: 2083–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumori, F. , and Hausinger, R.P. (1993b) Purification and characterization of 2,4‐dichlorophenoxyacetate/alpha‐ketoglutarate dioxygenase. J Biol Chem 268: 24311–24317. [PubMed] [Google Scholar]
- Furukawa, K. , and Miyazaki, T. (1986) Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes . J Bacteriol 166: 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis, N. , Tsarouhas, K. , Tsitsimpikou, C. , Vardavas, A. , Rezaee, R. , Germanakis, I. , et al (2018) Pesticides and cardiotoxicity. Where do we stand? Toxicol Appl Pharmacol 353: 1–14. [DOI] [PubMed] [Google Scholar]
- Gisi, M.R. , and Xun, L. (2003) Characterization of chlorophenol 4‐monooxygenase (TftD) and NADH:flavin adenine dinucleotide oxidoreductase (TftC) of Burkholderia cepacia AC1100. J Bacteriol 185: 2786–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothwal, A. , Beniwal, P. , Dhull, V. , and Hooda, V. (2014) Preparation of electrochemical biosensor for detection of organophosphorus pesticides. Int J Anal Chem 2014: 303641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häggblom, M.M. (1992) Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol Rev 9: 29–71. [DOI] [PubMed] [Google Scholar]
- Hartnett, C. , Neidle, E.L. , Ngai, K.L. , and Ornston, L.N. (1990) DNA sequences of genes encoding Acinetobacter calcoaceticus protocatechuate 3,4‐dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol 172: 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood, C.S. , and Parales, R.E. (1996) The beta‐ketoadipate pathway and the biology of self‐identity. Annu Rev Microbiol 50: 553–590. [DOI] [PubMed] [Google Scholar]
- Hatta, T. , Nakano, O. , Imai, N. , Takizawa, N. , and Kiyohara, H. (1999) Cloning and sequence analysis of hydroxyquinol 1,2‐dioxygenase gene in 2,4,6‐trichlorophenol‐degrading Ralstonia pickettii DTP0602 and characterization of its product. J Biosci Bioeng 87: 267–272. [DOI] [PubMed] [Google Scholar]
- Hatta, T. , Fujii, E. , and Takizawa, N. (2012) Analysis of two gene clusters involved in 2,4,6‐trichlorophenol degradation by Ralstonia pickettii DTP0602. Biosci Biotechnol Biochem 76: 892–899. [DOI] [PubMed] [Google Scholar]
- Hayes, R.P. , Webb, B.N. , Subramanian, A.K. , Nissen, M. , Popchock, A. , Xun, L. , and Kang, C. (2012) Structural and catalytic differences between two FADH2‐dependent monooxygenases: 2,4,5‐TCP 4‐monooxygenase (TftD) from Burkholderia cepacia AC1100 and 2,4,6‐TCP 4‐monooxygenase (TcpA) from Cupriavidus necator JMP134. Int J Mol Sci 13: 9769–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechmi, N. , Bosso, L. , El‐Bassi, L. , Scelza, R. , Testa, A. , Jedidi, N. , and Rao, M.A. (2016) Depletion of pentachlorophenol in soil microcosms with Byssochlamys nivea and Scopulariopsis brumptii as detoxification agents. Chemosphere 165: 547–554. [DOI] [PubMed] [Google Scholar]
- Hegg, E.L. , Whiting, A.K. , Saari, R.E. , McCracken, J. , Hausinger, R.P. , and Que, L. Jr (1999) Herbicide‐degrading alpha‐keto acid‐dependent enzyme TfdA: metal coordination environment and mechanistic insights. Biochemistry 38: 16714–16726. [DOI] [PubMed] [Google Scholar]
- Hlouchova, K. , Rudolph, J. , Pietari, J.M. , Behlen, L.S. , and Copley, S.D. (2012) Pentachlorophenol hydroxylase, a poorly functioning enzyme required for degradation of pentachlorophenol by Sphingobium chlorophenolicum . Biochemistry 51: 3848–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, D.A. , Buckley, D.H. , Nakatsu, C.H. , Schmidt, T.M. , and Hausinger, R.P. (1997) Distribution of the tfdA gene in soil bacteria that do not degrade 2,4‐dichlorophenoxyacetic acid (2,4‐d). Microb Ecol 34: 90–96. [DOI] [PubMed] [Google Scholar]
- Hongsawat, P. , and Vangnai, A.S. (2011) Biodegradation pathways of chloroanilines by Acinetobacter baylyi strain GFJ2. J Hazard Mater 186: 1300–1307. [DOI] [PubMed] [Google Scholar]
- Huang, X. , He, J. , Yan, X. , Hong, Q. , Chen, K. , He, Q. , et al (2017) Microbial catabolism of chemical herbicides: microbial resources, metabolic pathways and catabolic genes. Pestic Biochem Physiol 143: 272–297. [DOI] [PubMed] [Google Scholar]
- Huijbers, M.M. , Montersino, S. , Westphal, A.H. , Tischler, D. , and van Berkel, W.J. (2014) Flavin dependent monooxygenases. Arch Biochem Biophys 544: 2–17. [DOI] [PubMed] [Google Scholar]
- Itoh, K. , Kanda, R. , Sumita, Y. , Kim, H. , Kamagata, Y. , Suyama, K. , et al (2002) tfdA‐like genes in 2,4‐dichlorophenoxyacetic acid‐degrading bacteria belonging to the Bradyrhizobium‐Agromonas‐Nitrobacter‐Afipia cluster in alpha‐Proteobacteria . Appl Environ Microbiol 68: 3449–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwagami, S.G. , Yang, K. , and Davies, J. (2000) Characterization of the protocatechuic acid catabolic gene cluster from Streptomyces sp. strain 2065. Appl Environ Microbiol 66: 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joaquin, B.F. , Heidi, O. and Birgit, S.K. (2009) JRC Scientific and Technical report : European list of notified chemical substances (ELINCS). URL http://publications.jrc.ec.europa.eu/repository/bitstream/JRC52455/reqno_jrc52455.pdf.
- Jugder, B.E. , Ertan, H. , Bohl, S. , Lee, M. , Marquis, C.P. , and Manefield, M. (2016) Organohalide respiring bacteria and reductive dehalogenases: key tools in organohalide bioremediation. Front Microbiol 7: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamagata, Y. , Fulthorpe, R.R. , Tamura, K. , Takami, H. , Forney, L.J. , and Tiedje, J.M. (1997) Pristine environments harbor a new group of oligotrophic 2,4‐dichlorophenoxyacetic acid‐degrading bacteria. Appl Environ Microbiol 63: 2266–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura, N. , Takahashi, K. , Mori, K. , Araki, T. , Fujita, M. , Higuchi, Y. , et al (2017) Bacterial catabolism of lignin‐derived aromatics: new findings in a recent decade: update on bacterial lignin catabolism. Environ Microbiol Rep 9: 679–705. [DOI] [PubMed] [Google Scholar]
- Kang, C. , Yang, J.W. , Cho, W. , Kwak, S. , Park, S. , Lim, Y. , et al (2017) Oxidative biodegradation of 4‐chlorophenol by using recombinant monooxygenase cloned and overexpressed from Arthrobacter chlorophenolicus A6. Bioresour Technol 240: 123–129. [DOI] [PubMed] [Google Scholar]
- Kasai, D. , Fujinami, T. , Abe, T. , Mase, K. , Katayama, Y. , Fukuda, M. , et al (2009) Uncovering the protocatechuate 2,3‐cleavage pathway genes. J Bacteriol 191: 6758–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages, U. , Markus, A. , and Lingens, F. (1981) Degradation of 4‐chlorophenylacetic acid by a Pseudomonas species. J Bacteriol 146: 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A. , Trefault, N. , and Olaniran, A.O. (2016) Microbial degradation of 2,4‐dichlorophenoxyacetic acid: Insight into the enzymes and catabolic genes involved, their regulation and biotechnological implications. Crit Rev Microbiol 42: 194–208. [DOI] [PubMed] [Google Scholar]
- Kumar, V. , Shahi, S.K. , and Singh, S. (2018) Bioremediation: An eco‐sustainable approach for restoration of contaminated sites In Microbial Bioprospecting for Sustainable Development. Singh J., Sharma D., Kumar G., and Sharma N.R. (eds). Singapore: Springer Singapore, pp. 115–136. [Google Scholar]
- Kuntze, K. , Kiefer, P. , Baumann, S. , Seifert, J. , von Bergen, M. , Vorholt, J.A. , et al (2011) Enzymes involved in the anaerobic degradation of meta‐substituted halobenzoates. Mol Microbiol 82: 758–769. [DOI] [PubMed] [Google Scholar]
- Kunze, M. , Zerlin, K.F. , Retzlaff, A. , Pohl, J.O. , Schmidt, E. , Janssen, D.B. , et al (2009) Degradation of chloroaromatics by Pseudomonas putida GJ31: assembled route for chlorobenzene degradation encoded by clusters on plasmid pKW1 and the chromosome. Microbiology 155: 4069–4083. [DOI] [PubMed] [Google Scholar]
- Laemmli, C.M. , Leveau, J.H. , Zehnder, A.J. , and van der Meer, J.R. (2000) Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutropha JMP134(pJP4). J Bacteriol 182: 4165–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, C. , Werlen, C. , and van der Meer, J.R. (2004) Mutation analysis of the different tfd genes for degradation of chloroaromatic compounds in Ralstonia eutropha JMP134. Arch Microbiol 181: 112–121. [DOI] [PubMed] [Google Scholar]
- Lau, E.Y. , and Bruice, T.C. (2001) The active site dynamics of 4‐chlorobenzoyl‐CoA dehalogenase. Proc Natl Acad Sci USA 98: 9527–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb, J.D. (2008) Mechanism of extradiol aromatic ring‐cleaving dioxygenases. Curr Opin Struct Biol 18: 644–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Tan, L. , Wang, J. , Wang, Z. , Ni, H. , and Li, L. (2016) Complete biodegradation of chlorpyrifos by engineered Pseudomonas putida cells expressing surface‐immobilized laccases. Chemosphere 157: 200–207. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Bilal, M. , Duan, X. , and Iqbal, H.M.N. (2019) Mitigation of environmental pollution by genetically engineered bacteria ‐ current challenges and future perspectives. Sci Total Environ 667: 444–454. [DOI] [PubMed] [Google Scholar]
- Lu, Q. , Qiu, L. , Yu, L. , Zhang, S. , de Toledo, R.A. , Shim, H. , et al (2019) Microbial transformation of chiral organohalides: distribution, microorganisms and mechanisms. J Hazard Mater 368: 849–861. [DOI] [PubMed] [Google Scholar]
- Lubbers, R.J.M. , Dilokpimol, A. , Visser, J. , Mäkelä, M.R. , Hildén, K.S. and de Vries, R.P. (2019) A comparison between the homocyclic aromatic metabolic pathways from plant‐derived compounds by bacteria and fungi. Biotechnol Adv, in press. [DOI] [PubMed] [Google Scholar]
- Luo, L. , Taylor, K.L. , Xiang, H. , Wei, Y. , Zhang, W. , and Dunaway‐Mariano, D. (2001) Role of active site binding interactions in 4‐chlorobenzoyl‐coenzyme A dehalogenase catalysis. Biochemistry 40: 15684–15692. [DOI] [PubMed] [Google Scholar]
- Mac Nelly, A. , Kai, M. , Svatos, A. , Diekert, G. , and Schubert, T. (2014) Functional heterologous production of reductive dehalogenases from Desulfitobacterium hafniense strains. Appl Environ Microbiol 80: 4313–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markusheva, T.V. , Zhurenko, E. , Galkin, E.G. , Korobov, V.V. , Zharikova, N.V. , and Gafiiatova, L.R. (2004) Identification and characterization of a plasmid in strain Aeronomas hydrophila IBRB‐36 4CPA carrying genes for catabolism of chlorophenoxyacetic acids. Genetika 40: 1469–1474. [PubMed] [Google Scholar]
- Maruyama, K. , Shibayama, T. , Ichikawa, A. , Sakou, Y. , Yamada, S. , and Sugisaki, H. (2004) Cloning and characterization of the genes encoding enzymes for the protocatechuate meta‐degradation pathway of Pseudomonas ochraceae NGJ1. Biosci Biotechnol Biochem 68: 1434–1441. [DOI] [PubMed] [Google Scholar]
- Matus, V. , Sanchez, M.A. , Martinez, M. , and Gonzalez, B. (2003) Efficient degradation of 2,4,6‐trichlorophenol requires a set of catabolic genes related to tcp genes from Ralstonia eutropha JMP134(pJP4). Appl Environ Microbiol 69: 7108–7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, D.L. , Navarrete, S. , Willett, W.S. , Babbitt, P.C. , and Copley, S.D. (1996) Exploration of the relationship between tetrachlorohydroquinone dehalogenase and the glutathione S‐transferase superfamily. Biochemistry 35: 14634–14642. [DOI] [PubMed] [Google Scholar]
- Megharaj, M. , Ramakrishnan, B. , Venkateswarlu, K. , Sethunathan, N. , and Naidu, R. (2011) Bioremediation approaches for organic pollutants: a critical perspective. Environ Int 37: 1362–1375. [DOI] [PubMed] [Google Scholar]
- Mondello, F.J. (1989) Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J Bacteriol 171: 1725–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafalou, S. , and Abdollahi, M. (2017) Pesticides: an update of human exposure and toxicity. Arch Toxicol 91: 549–599. [DOI] [PubMed] [Google Scholar]
- Muller, T.A. , Byrde, S.M. , Werlen, C. , van der Meer, J.R. , and Kohler, H.P. (2004) Genetic analysis of phenoxyalkanoic acid degradation in Sphingomonas herbicidovorans MH. Appl Environ Microbiol 70: 6066–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, T.A. , Fleischmann, T. , van der Meer, J.R. , and Kohler, H.P. (2006) Purification and characterization of two enantioselective alpha‐ketoglutarate‐dependent dioxygenases, RdpA and SdpA, from Sphingomonas herbicidovorans MH. Appl Environ Microbiol 72: 4853–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, S. , Kodama, N. , Shinke, R. , and Aoki, K. (1997) Classification of catechol 1,2‐dioxygenase family: sequence analysis of a gene for the catechol 1,2‐dioxygenase showing high specificity for methylcatechols from Gram+ aniline‐assimilating Rhodococcus erythropolis AN‐13. Gene 185: 49–54. [DOI] [PubMed] [Google Scholar]
- Nickel, K. , Suter, M.J. , and Kohler, H.P. (1997) Involvement of two alpha‐ketoglutarate‐dependent dioxygenases in enantioselective degradation of (R)‐ and (S)‐mecoprop by Sphingomonas herbicidovorans MH. J Bacteriol 179: 6674–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam, V.K. , and Shukla, P. (2015) Enzyme based biosensors for detection of environmental pollutants‐a review. J Microbiol Biotechnol 25: 1773–1781. [DOI] [PubMed] [Google Scholar]
- Nováková, H. , Vošahlíková, M. , Pazlarová, J. , Macková, M. , Burkhard, J. and Demnerová, K. (2002) PCB metabolism by Pseudomonas sp. P2. Int Biodeterior Biodegradation 50: 47–54. [Google Scholar]
- Olaniran, A.O. , and Igbinosa, E.O. (2011) Chlorophenols and other related derivatives of environmental concern: properties, distribution and microbial degradation processes. Chemosphere 83: 1297–1306. [DOI] [PubMed] [Google Scholar]
- Palfey, B.A. , and McDonald, C.A. (2010) Control of catalysis in flavin‐dependent monooxygenases. Arch Biochem Biophys 493: 26–36. [DOI] [PubMed] [Google Scholar]
- Patel, R.N. , Mazumdar, S. , and Ornston, L.N. (1975) Beta‐ketoadipate enol‐lactone hydrolases I and II from Acinetobacter calcoaceticus . J Biol Chem 250: 6567. [PubMed] [Google Scholar]
- Payne, K.A. , Quezada, C.P. , Fisher, K. , Dunstan, M.S. , Collins, F.A. , Sjuts, H. , et al (2015) Reductive dehalogenase structure suggests a mechanism for B12‐dependent dehalogenation. Nature 517: 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peelen, S. , Rietjens, I.M. , Boersma, M.G. , and Vervoort, J. (1995) Conversion of phenol derivatives to hydroxylated products by phenol hydroxylase from Trichosporon cutaneum. A comparison of regioselectivity and rate of conversion with calculated molecular orbital substrate characteristics. Eur J Biochem 227: 284–291. [DOI] [PubMed] [Google Scholar]
- Pera‐Titus, M. , Garcia‐Molina, V. , Banos, M.A. , Gimenez, J. , and Esplugas, S. (2004) Degradation of chlorophenols by means of advanced oxidation processes: a general review. Appl Catal B 47: 219–256. [Google Scholar]
- Pieper, D.H. (2005) Aerobic degradation of polychlorinated biphenyls. Appl Microbiol Biotechnol 67: 170–191. [DOI] [PubMed] [Google Scholar]
- Pimviriyakul, P. , and Chaiyen, P. (2018) A complete bioconversion cascade for dehalogenation and denitration by bacterial flavin‐dependent enzymes. J Biol Chem 293: 18525–18539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimviriyakul, P. , Thotsaporn, K. , Sucharitakul, J. , and Chaiyen, P. (2017) Kinetic mechanism of the dechlorinating flavin‐dependent monooxygenase HadA. J Biol Chem 292: 4818–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimviriyakul, P. , Surawatanawong, P. , and Chaiyen, P. (2018) Oxidative dehalogenation and denitration by a flavin‐dependent monooxygenase is controlled by substrate deprotonation. Chem Sci 9: 7468–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova, E.G. , Solyanikova, I.P. , Egorova, D.O. , Shumkova, E.S. , and Golovleva, L.A. (2012) Degradation of 4‐chlorobiphenyl and 4‐chlorobenzoic acid by the strain Rhodococcus ruber P25. Microbiology 81: 143–153. [PubMed] [Google Scholar]
- Plumeier, I. , Perez‐Pantoja, D. , Heim, S. , Gonzalez, B. , and Pieper, D.H. (2002) Importance of different tfd genes for degradation of chloroaromatics by Ralstonia eutropha JMP134. J Bacteriol 184: 4054–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongpamorn, P. , Watthaisong, P. , Pimviriyakul, P. , Jaruwat, A. , Lawan, N. , Chitnumsub, P. , et al (2019) Identification of a hotspot residue for improving the thermostability of a flavin‐dependent monooxygenase. ChemBioChem, in press. 10.1002/cbic.201900413 [DOI] [PubMed] [Google Scholar]
- Poritz, M. , Schiffmann, C.L. , Hause, G. , Heinemann, U. , Seifert, J. , Jehmlich, N. , et al (2015) Dehalococcoides mccartyi strain DCMB5 respires a broad spectrum of chlorinated aromatic compounds. Appl Environ Microbiol 81: 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornsuwan, S. , Maenpuen, S. , Kamutira, P. , Watthaisong, P. , Thotsaporn, K. , Tongsook, C. , et al (2017) 3,4‐Dihydroxyphenylacetate 2,3‐dioxygenase from Pseudomonas aeruginosa: An Fe(II)‐containing enzyme with fast turnover. PLoS ONE 12: e0171135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, M.A. , Scelza, R. , Acevedo, F. , Diez, M.C. , and Gianfreda, L. (2014) Enzymes as useful tools for environmental purposes. Chemosphere 107: 145–162. [DOI] [PubMed] [Google Scholar]
- Rayu, S. , Nielsen, U.N. , Nazaries, L. , and Singh, B.K. (2017) Isolation and molecular characterization of novel chlorpyrifos and 3,5,6‐trichloro‐2‐pyridinol‐degrading bacteria from sugarcane farm soils. Front Microbiol 8: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke, W. , and Knackmuss, H.J. (1988) Microbial degradation of haloaromatics. Ann Rev Microbiol 42: 263–287. [DOI] [PubMed] [Google Scholar]
- Roberts, K.M. , and Fitzpatrick, P.F. (2013) Mechanisms of tryptophan and tyrosine hydroxylase. IUBMB Life 65: 350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, E. , Gomez Castellanos, J.R. , Gadda, G. , Fraaije, M.W. , and Mattevi, A. (2018) Same substrate, many Reactions: oxygen activation in flavoenzymes. Chem Rev 118: 1742–1769. [DOI] [PubMed] [Google Scholar]
- Rudolph, J. , Erbse, A.H. , Behlen, L.S. , and Copley, S.D. (2014) A radical intermediate in the conversion of pentachlorophenol to tetrachlorohydroquinone by Sphingobium chlorophenolicum . Biochemistry 53: 6539–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari, R.E. , and Hausinger, R.P. (1998) Ascorbic acid‐dependent turnover and reactivation of 2,4‐dichlorophenoxyacetic acid/alpha‐ketoglutarate dioxygenase using thiophenoxyacetic acid. Biochemistry 37: 3035–3042. [DOI] [PubMed] [Google Scholar]
- Salunkhe, V.P. , Sawant, I.S. , Banerjee, K. , Rajguru, Y.R. , Wadkar, P.N. , Oulkar, D.P. , et al (2013) Biodegradation of profenofos by Bacillus subtilis isolated from grapevines (Vitis vinifera). J Agric Food Chem 61: 7195–7202. [DOI] [PubMed] [Google Scholar]
- Schubert, T. , Adrian, L. , Sawers, R.G. , and Diekert, G. (2018) Organohalide respiratory chains: composition, topology and key enzymes. FEMS Microbiol Ecol 94: 1–17. [DOI] [PubMed] [Google Scholar]
- Seibold, B. , Matthes, M. , Eppink, M.H. , Lingens, F. , Van Berkel, W.J. , and Muller, R. (1996) 4‐Hydroxybenzoate hydroxylase from Pseudomonas sp. CBS3: purification, characterization, gene cloning, sequence analysis and assignment of structural features determining the coenzyme specificity. Eur J Biochem 239: 469–478. [DOI] [PubMed] [Google Scholar]
- Sharma, B. , Dangi, A.K. , and Shukla, P. (2018) Contemporary enzyme based technologies for bioremediation: a review. J Environ Manage 210: 10–22. [DOI] [PubMed] [Google Scholar]
- Sheldon, R.A. , and Woodley, J.M. (2018) Role of biocatalysis in sustainable chemistry. Chem Rev 118: 801–838. [DOI] [PubMed] [Google Scholar]
- Siripattanakul‐Ratpukdi, S. , Vangnai, A.S. , Sangthean, P. , and Singkibut, S. (2015) Profenofos insecticide degradation by novel microbial consortium and isolates enriched from contaminated chili farm soil. Environ Sci Pollut Res Int 22: 320–328. [DOI] [PubMed] [Google Scholar]
- Song, B. , Palleroni, N.J. , Kerkhof, L.J. , and Häggblom, M.M. (2001) Characterization of halobenzoate‐degrading, denitrifying Azoarcus and Thauera isolates and description of Thauera chlorobenzoica sp. nov. Int J Syst Evol Microbiol 51: 589–602. [DOI] [PubMed] [Google Scholar]
- Strunk, N. , and Engesser, K.H. (2013) Degradation of fluorobenzene and its central metabolites 3‐fluorocatechol and 2‐fluoromuconate by Burkholderia fungorum FLU100. Appl Microbiol Biotechnol 97: 5605–5614. [DOI] [PubMed] [Google Scholar]
- Sucharitakul, J. , Tinikul, R. , and Chaiyen, P. (2014) Mechanisms of reduced flavin transfer in the two‐component flavin‐dependent monooxygenases. Arch Biochem Biophys 555–556: 33–46. [DOI] [PubMed] [Google Scholar]
- Supreeth, M. , and Raju, N.S. (2017) Biotransformation of chlorpyrifos and endosulfan by bacteria and fungi. Appl Microbiol Biotechnol 101: 5961–5971. [DOI] [PubMed] [Google Scholar]
- Thotsaporn, K. , Tinikul, R. , Maenpuen, S. , Phonbuppha, J. , Watthaisong, P. , Chenprakhon, P. , et al (2016) Enzymes in the p‐hydroxyphenylacetate degradation pathway of Acinetobacter baumannii . J Mol Catal B Enzym 134: 353–366. [Google Scholar]
- Tiedt, O. , Mergelsberg, M. , Boll, K. , Muller, M. , Adrian, L. , Jehmlich, N. , et al (2016) ATP‐dependent C‐F bond cleavage allows the complete degradation of 4‐fluoroaromatics without oxygen. MBio 7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinikul, R. , Chenprakhon, P. , Maenpuen, S. , and Chaiyen, P. (2018) Biotransformation of plant‐derived phenolic acids. Biotechnol J 13: e1700632. [DOI] [PubMed] [Google Scholar]
- Tonso, N.L. , Matheson, V.G. , and Holben, W.E. (1995) Polyphasic characterization of a suite of bacterial isolates capable of degrading 2,4‐D. Microb Ecol 30: 3–24. [DOI] [PubMed] [Google Scholar]
- Travkin, V.M. , Solyanikova, I.P. , Rietjens, I.M. , Vervoort, J. , van Berkel, W.J. , and Golovleva, L.A. (2003) Degradation of 3,4‐dichloro‐ and 3,4‐difluoroaniline by Pseudomonas fluorescens 26‐K. J Environ Sci Health B 38: 121–132. [DOI] [PubMed] [Google Scholar]
- Ueda, M. (2016) Establishment of cell surface engineering and its development. Biosci Biotechnol Biochem 80: 1243–1253. [DOI] [PubMed] [Google Scholar]
- United States Environmental Protection Agency . (2014) Priority pollutant list. URL https://www.epa.gov/eg/toxic-and-priority-pollutants-under-clean-water-act#priority.
- Vargas, C. , Song, B. , Camps, M. , and Häggblom, M.M. (2000) Anaerobic degradation of fluorinated aromatic compounds. Appl Microbiol Biotechnol 53: 342–347. [DOI] [PubMed] [Google Scholar]
- Wagner, A. , Cooper, M. , Ferdi, S. , Seifert, J. , and Adrian, L. (2012) Growth of Dehalococcoides mccartyi strain CBDB1 by reductive dehalogenation of brominated benzenes to benzene. Environ Sci Technol 46: 8960–8968. [DOI] [PubMed] [Google Scholar]
- Wang, C.L. , You, S.L. , and Wang, S.L. (2006) Purification and characterization of a novel catechol 1,2‐dioxygenase from Pseudomonas aeruginosa with benzoic acid as a carbon source. Process Biochem 41: 1594–1601. [Google Scholar]
- Wang, S. , Zhang, W. , Yang, K.L. , and He, J. (2014) Isolation and characterization of a novel Dehalobacter species strain TCP1 that reductively dechlorinates 2,4,6‐trichlorophenol. Biodegradation 25: 313–323. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Li, J. , and Liu, A. (2017) Oxygen activation by mononuclear nonheme iron dioxygenases involved in the degradation of aromatics. J Biol Inorg Chem 22: 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Qiu, L. , Liu, X. , Xu, G. , Siegert, M. , Lu, Q. , et al (2018) Electron transport chains in organohalide‐respiring bacteria and bioremediation implications. Biotechnol Adv 36: 1194–1206. [DOI] [PubMed] [Google Scholar]
- Wang, B. , Xu, J. , Gao, J. , Fu, X. , Han, H. , Li, Z. , et al (2019) Construction of an Escherichia coli strain to degrade phenol completely with two modified metabolic modules. J Hazard Mater 373: 29–38. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. , and Copley, S.D. (2007a) Mechanism of the severe inhibition of tetrachlorohydroquinone dehalogenase by its aromatic substrates. Biochemistry 46: 4438–4447. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. , and Copley, S.D. (2007b) Pre‐steady‐state kinetic studies of the reductive dehalogenation catalyzed by tetrachlorohydroquinone dehalogenase. Biochemistry 46: 13211–13222. [DOI] [PubMed] [Google Scholar]
- Watthaisong, P. , Pongpamorn, P. , Pimviriyakul, P. , Maenpuen, S. , Ohmiya, Y. and Chaiyen, P. (2019) A chemo‐enzymatic cascade for smart detection of nitro and halogenated phenols. Angew Chem Inter Ed Engl, in press. 10.1002/anie.201904923 [DOI] [PubMed] [Google Scholar]
- Webb, B.N. , Ballinger, J.W. , Kim, E. , Belchik, S.M. , Lam, K.S. , Youn, B. , et al (2010) Characterization of chlorophenol 4‐monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100. J Biol Chem 285: 2014–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, T. Jr , and Ragauskas, A.J. (2012) Biotechnological opportunities with the beta‐ketoadipate pathway. Trends Biotechnol 30: 627–637. [DOI] [PubMed] [Google Scholar]
- Westendorf, A. , Müller, R.H. , and Babel, W. (2003) Purification and characterisation of the enantiospecific dioxygenases from Delftia acidovorans MC1 initiating the degradation of phenoxypropionate and phenoxyacetate herbicides. Acta Biotechnol 23: 3–17. [Google Scholar]
- Westendorf, A. , Benndorf, D. , Pribyl, T. , Harms, H. , and Müller, R.H. (2006) Kinetic traits and enzyme form patterns of (R)‐2‐(2,4‐dichlorophenoxy)propionate/α‐ketoglutarate dioxygenase (RdpA) after expression in different bacterial strains. Eng Life Sci 6: 552–559. [Google Scholar]
- Wischgoll, S. , Heintz, D. , Peters, F. , Erxleben, A. , Sarnighausen, E. , Reski, R. , et al (2005) Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens . Mol Microbiol 58: 1238–1252. [DOI] [PubMed] [Google Scholar]
- Wolgel, S.A. , Dege, J.E. , Perkins‐Olson, P.E. , Jaurez‐Garcia, C.H. , Crawford, R.L. , Munck, E. , et al (1993) Purification and characterization of protocatechuate 2,3‐dioxygenase from Bacillus macerans: a new extradiol catecholic dioxygenase. J Bacteriol 175: 4414–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Xu, D. , Lu, X. , Wang, C. , Guo, H. , and Dunaway‐Mariano, D. (2006) Contributions of long‐range electrostatic interactions to 4‐chlorobenzoyl‐CoA dehalogenase catalysis: a combined theoretical and experimental study. Biochemistry 45: 102–112. [DOI] [PubMed] [Google Scholar]
- Xu, D. , Wei, Y. , Wu, J. , Dunaway‐Mariano, D. , Guo, H. , Cui, Q. , et al (2004) QM/MM studies of the enzyme‐catalyzed dechlorination of 4‐chlorobenzoyl‐CoA provide insight into reaction energetics. J Am Chem Soc 126: 13649–13658. [DOI] [PubMed] [Google Scholar]
- Xu, Y.‐L. , Li, F.‐Y. , Ndikuryayo, F. , Yang, W.‐C. , and Wang, H.‐M. (2018) Cholinesterases and engineered mutants for the detection of organophosphorus pesticide residues. Sensors 18: 4281–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun, L. , and Webster, C.M. (2004) A monooxygenase catalyzes sequential dechlorinations of 2,4,6‐trichlorophenol by oxidative and hydrolytic reactions. J Biol Chem 279: 6696–6700. [DOI] [PubMed] [Google Scholar]
- Xun, L. , Topp, E. , and Orser, C.S. (1992a) Diverse substrate range of a Flavobacterium pentachlorophenol hydroxylase and reaction stoichiometries. J Bacteriol 174: 2898–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun, L. , Topp, E. , and Orser, C.S. (1992b) Purification and characterization of a tetrachloro‐p‐hydroquinone reductive dehalogenase from a Flavobacterium sp . J Bacteriol 174: 8003–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X.F. , Khan, F. , Pandey, R. , Pandey, J. , Mourant, R.G. , Jain, R.K. , et al (2011) Degradation of dichloroaniline isomers by a newly isolated strain, Bacillus megaterium IMT21. Microbiology 157: 721–726. [DOI] [PubMed] [Google Scholar]
- You, I.S. , and Bartha, R. (1982) Stimulation of 3,4‐dichloroaniline mineralization by aniline. Appl Environ Microbiol 44: 678–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Wei, Y. , Luo, L. , Taylor, K.L. , Yang, G. , Dunaway‐Mariano, D. , et al (2001) Histidine 90 function in 4‐chlorobenzoyl‐coenzyme a dehalogenase catalysis. Biochemistry 40: 13474–13482. [DOI] [PubMed] [Google Scholar]
- Zhang, L.L. , Leng, A.Q. , Zhu, R.Y. , and Chen, J.M. (2011) Degradation of chlorobenzene by strain Ralstonia pickettii L2 isolated from a biotrickling filter treating a chlorobenzene‐contaminated gas stream. Appl Microbiol Biotechnol 91: 407–415. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Yu, T. , Li, J. , Wang, Y.R. , Wang, G.L. , Li, F. , et al (2018) Two dcm gene clusters essential for the degradation of diclofop‐methyl in a microbial consortium of Rhodococcus sp. JT‐3 and Brevundimonas sp. JT‐9. J Agric Food Chem 66: 12217–12226. [DOI] [PubMed] [Google Scholar]
- Zhou, L. , Poh, R.P. , Marks, T.S. , Chowdhry, B.Z. , and Smith, A.R. (2008) Structure and denaturation of 4‐chlorobenzoyl coenzyme A dehalogenase from Arthrobacter sp. strain TM‐1. Biodegradation 19: 65–75. [DOI] [PubMed] [Google Scholar]
- Zhuang, Z. , Gartemann, K.H. , Eichenlaub, R. , and Dunaway‐Mariano, D. (2003) Characterization of the 4‐hydroxybenzoyl‐coenzyme A thioesterase from Arthrobacter sp. strain SU. Appl Environ Microbiol 69: 2707–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]


