Supplemental Digital Content is available in the text
Keywords: ABCC family, gastric cancer, Kaplan–Meier plotter, molecular mechanism, prognostic significance
Abstract
Gastric cancer (GC) is one of the major leading causes of tumor-related deaths worldwide. Adenosine triphosphate-binding cassette subfamily C (ABCC) consists of 13 members, ABCC1 to 13, which were examined for their associations with GC.
The online Kaplan–Meier Plotter database was used to determine the prognostic significance of ABCC subfamily members in GC. Stratified analyses were performed using gender, disease stage, degree of tumor differentiation, expression of human epidermal growth factor receptor 2 (HER2), and Lauren classification. Molecular mechanisms were examined using the database for annotation, visualization, and integrated discovery database.
ABCC1, ABCC3, ABCC7, ABCC8, ABCC9, and ABCC10 expression showed prognostic significance in the whole population and in male and female subpopulations (all P ≤ .05). Furthermore, high expression of most ABCC family members always suggested a poor prognosis, except for ABCC7 (P > .05). Stratified analyses revealed that ABCC1, ABCC3, ABCC7, ABCC8, ABCC9, and ABCC10 expression showed prognostic significance for the whole population, as well as male and female populations. ABCC2 and ABCC9 were significantly correlated with all disease stages, while ABCC2 and ABCC6 were significantly correlated with all Lauren classifications. Expression of ABCC1, ABCC3, ABCC5, ABCC7, ABCC8, ABCC9, and ABCC10 was significantly correlated with either negative or positive of HER2 status (all P ≤ .05). Enrichment analysis indicated that these genes were involved in ATPase activity, transmembrane transport, or were ABC transporters (all P ≤ .05).
ABCC1, ABCC3, ABCC7, ABCC8, ABCC9, and ABCC10 may be potential prognosis biomarkers for GC, acting as ABC transporters and via ATPase activity.
1. Introduction
Accumulating evidence has indicated that gastric cancer (GC) is one of the major leading causes of malignancy-related deaths worldwide.[1] It was estimated that there were approximately 1 million new GC patient cases in 2018 and about half of new cases occurred in China.[2] The relative 5-year overall survival (OS) rate of GC is less than 25% due to disease invasion and metastasis, compared to an OS of more than 90% for patients diagnosed at an early stage of GC.[3–5] Although early diagnosis and treatment of GC, including surgery, targeted therapy, adjuvant therapy, and radio-chemotherapy have greatly improved outcomes, the prognosis of GC patients with advanced stage GC remains poor and unsatisfactory.[1,6] Currently, research has identified several oncogenic and tumor suppressor genes, microRNAs and proteins as potential biomarkers for GC.[7] These biomarkers have been associated with tumorigenesis, tumor progression, and aggressiveness. Nonetheless, these novel biomarkers need further validation in other cohorts. Therefore, identification of new biomarkers for early diagnosis and prognosis prediction for GC is very important.
The adenosine triphosphate (ATP)-binding cassette (ABC) transporters are a superfamily of membrane proteins which play significant roles in transporting various exogenous and endogenous substances across membranes against concentration gradients through ATP hydrolysis.[8] The ABC gene family has been divided into 7 subfamilies, the ABCA, ABCB, ABCC, ABCD, ABCE, ABCF, and ABCG subfamilies.[8]
The ABCC subfamily consists of 13 member genes: ABCC1, ABCC2, ABCC3, ABCC4, ABCC5, ABCC6, ABCC7 (also known as CFTR), ABCC8, ABCC9, ABCC10, ABCC11, ABCC12, and ABCC13 (https://www.genenames.org/data/genegroup/#!/group/807).[9] Nine of these transporters are well known as multidrug resistance proteins (MRPs).[10]ABCC1, also known as MRP1, is responsible for drug and xenobiotic disposition in organisms and for protecting human organs and tissues from cytotoxic assault.[11]ABCC2, also known as MRP2, plays a pivotal role in biliary elimination of several endogenous substances such as leukotriene C4 and conjugated bilirubins.[12,13]ABCC3, also known as MRP3, compensates for MRP2 deficiency in liver tissue.[14]ABCC4, also known as MRP4, plays an important role in cellular efflux of cyclic adenosine monophosphate (cAMP), cyclic guanosine monophosphate (cGMP) and some secondary messengers.[15] Similar to MRP4, MRP5 (ABCC5), is not only a nucleotide organic anion transporter, but also regulates the efflux of substances such as cAMP, cGMP, and several purine analogs.[16]MRP7 (ABCC10) is highly expressed in colon tissue, skin, and testes.[17]MRP8 (ABCC11) was first reported to be highly expressed in breast cancer via a database mining and prediction program.[18]MRP9 (ABCC12) may also play important roles during meiotic prophase and spermatid development in males.[19]
Currently, little is known about the association between GC and ABCC subfamily members. Therefore, we conducted this study to investigate the prognostic significance of 13 ABCC family members in the OS of GC patients.
2. Material and methods
2.1. Data source
We used the online database, Kaplan–Meier plotter (http://kmplot.com/analysis/), to analyze the prognostic significance of mRNA expression of ABCC family members in GC.[20] This database contains transcriptomic data from 1065 patients based on datasets from 3 major medical centers in Berlin, Bethesda, and Melbourne.[20] In the present study, OS was evaluated to determine prognostic significance in a total of 882 GC patients. Differences in OS were analyzed after stratification by gender and status of human epidermal growth factor receptor 2 (HER2) expression (HER2-positive or negative). Subtype analysis was performed based on cancer stage (stage 1, 2, 3, or 4), Lauren classification (intestinal, diffuse, or mixed), and tumor differentiation (poor, moderate, or well-differentiated). Probe numbers used in the study of ABCC1 to 13 were 202804-at, 206155-at, 214979-at, 203196-at, 226363-at, 214033-at, 205043-at, 210245-at, 208561-at, 213485-s-at, 224146-s-at, 1552590-a-at, and 1552582-at, respectively. In addition, low and high expression groups were divided by median mRNA expression.
2.2. Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
2.3. Gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of ABCC family members involved in GC
To explore potential GO terms and KEGG pathways, we used the online database for annotation, visualization, and integrated discovery, version 6.8 (DAVID; https://david.ncifcrf.gov/).[21,22] DAVID provides users with a comprehensive set of functional annotations of biological processes (BPs) for lists of genes.
2.4. Gene-gene interaction (GGI), protein-protein interaction (PPI), and GO networks
The GGI network was constructed using the geneMANIA plugin in Cytoscape software.[23,24]ABCC13 was not recognized in the GGI network and ABCC7 was recognized as cystic fibrosis transmembrane conductance regulator (CFTR). The PPI network of ABCC members was constructed using the STRING online database (https://string-db.org/).[25] ABCC11 was not recognized in the PPI network. Visualization of GO networks including BP, cellular component (CC), and molecular function (MF) networks were constructed using the BinGO plugin in Cytoscape software.[26]
2.5. Statistical analysis
SPSS version 24.0 (IBM, Armonk, NY) was used for statistical analysis. A P-value ≤ .05 was considered statistically significant. The 95% confidence interval (CI) and hazard ratio (HR) were used for risk assessment in Cox proportional hazards regression analysis.
3. Results
3.1. The association of ABCC family members with OS
The prognostic value of ABCC family members in GC was determined by their association with OS. ABCC1 and ABCC2 showed prognostic differences between the low and high expression groups (both P < .01, Fig. 1A and D). In addition, high expression of these genes always indicated a poor prognosis. Prognostic differences were also observed between high and low expression of ABCC3, but not for ABCC4 (ABCC3: P = 3.2E-14, ABCC4: P = 0.12, Fig. 2A and D). Moreover, high expression of ABCC3 was associated with poor prognosis. ABCC5 showed a prognostic difference between low and high expression groups, whereas ABCC6 did not (ABCC5: P = 9.1E-5, ABCC6: P = 0.22, Fig. 3A and D). High expression of ABCC5 was associated with poor prognosis. ABCC7 and ABCC8 also showed prognostic differences (both P < 0.01, Fig. 4A and D), but in this case low expression of ABCC7 and high expression of ABCC8 indicated poor prognosis. ABCC9 and ABCC10 as well as ABCC11 and ABCC12 showed prognostic differences, where high expression was consistently associated with poor prognosis (all P < .01, Figs. 5 and 6AFigs. 5A and D and 6A and D). Finally, a prognostic difference was observed for ABCC13 in which high expression was associated with decreased OS (P = .031, Fig. 7A).
Figure 1.
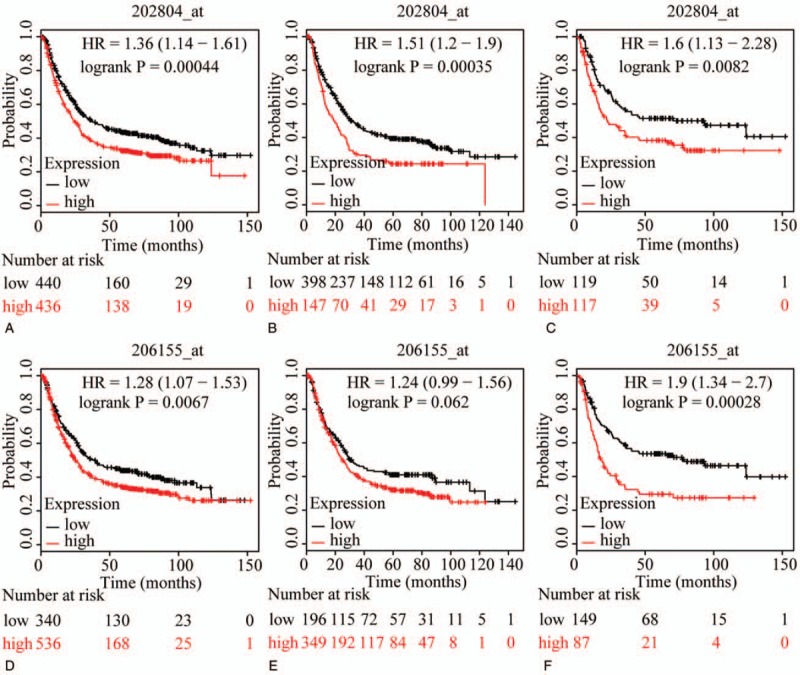
Survival plots of GC patients expressing ABCC1 and ABCC2. (A–C) Survival plots for ABCC1 (202804_at) in whole, male and female populations, respectively. (D–F) Survival plots for ABCC2 (206155_at) in whole, male and female populations, respectively. ABCC = adenosine triphosphate-binding cassette subfamily C, GC = gastric cancer.
Figure 2.
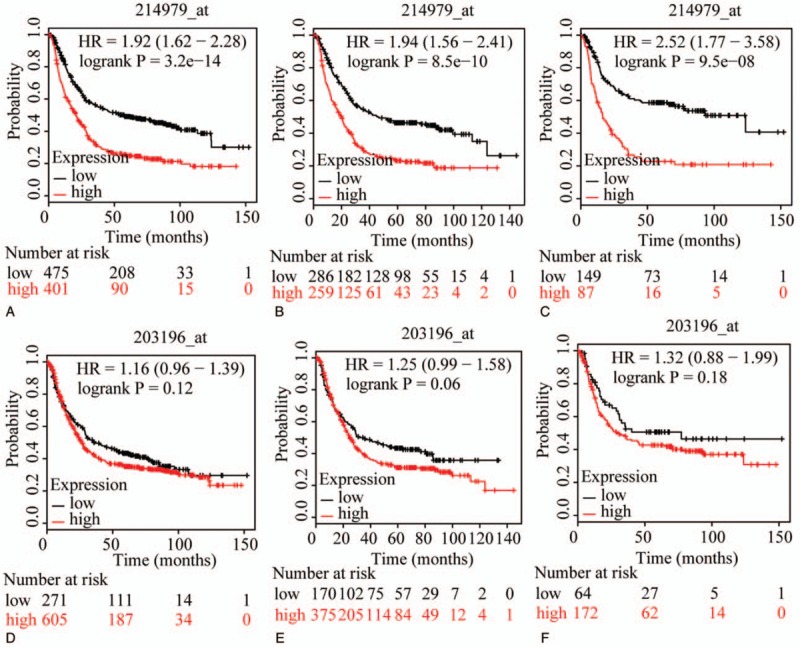
Survival plots of GC patients expressing ABCC3 and ABCC4. (A–C) Survival plots for ABCC3 (214979_at) in whole, male and female populations, respectively. (D–F) Survival plots for ABCC4 (203196_at) in whole, male and female populations, respectively. ABCC = adenosine triphosphate-binding cassette subfamily C, GC = gastric cancer.
Figure 3.
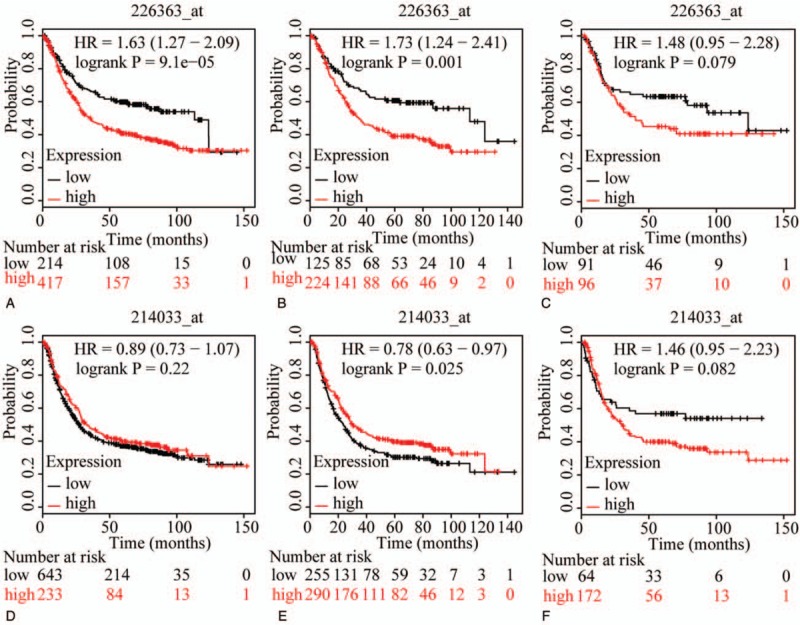
Survival plots of GC patients expressing ABCC5 and ABCC6. (A–C) Survival plots for ABCC5 (226363_at) in whole, male and female populations, respectively. (D–F) Survival plots for ABCC6 (214033_at) in whole, male and female populations, respectively. ABCC = adenosine triphosphate-binding cassette subfamily C, GC = gastric cancer.
Figure 4.
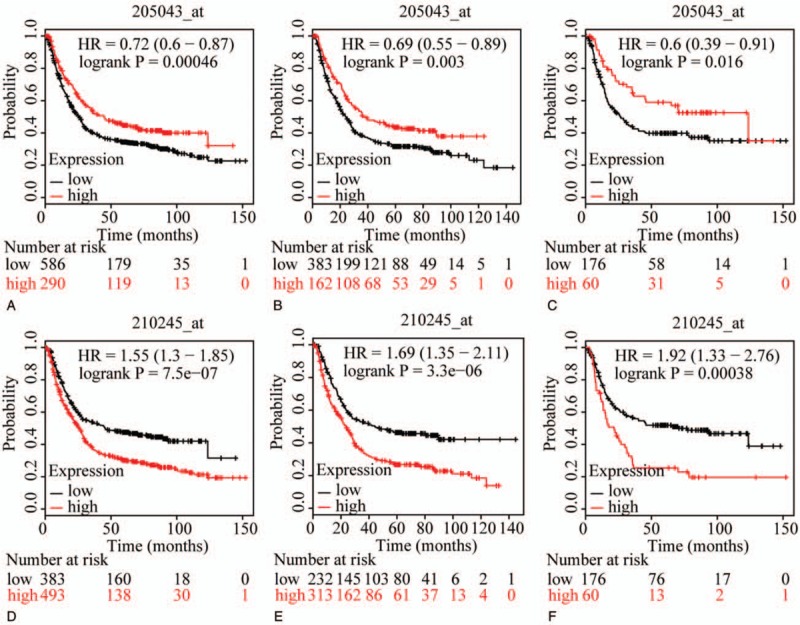
Survival plots of GC patients expressing ABCC7 and ABCC8. (A–C) Survival plots for ABCC7 (205043_at) in whole, male and female populations, respectively. (D–F) Survival plots for ABCC8 (210245_at) in whole, male and female populations, respectively. ABCC = adenosine triphosphate-binding cassette subfamily C, GC = gastric cancer.
Figure 5.
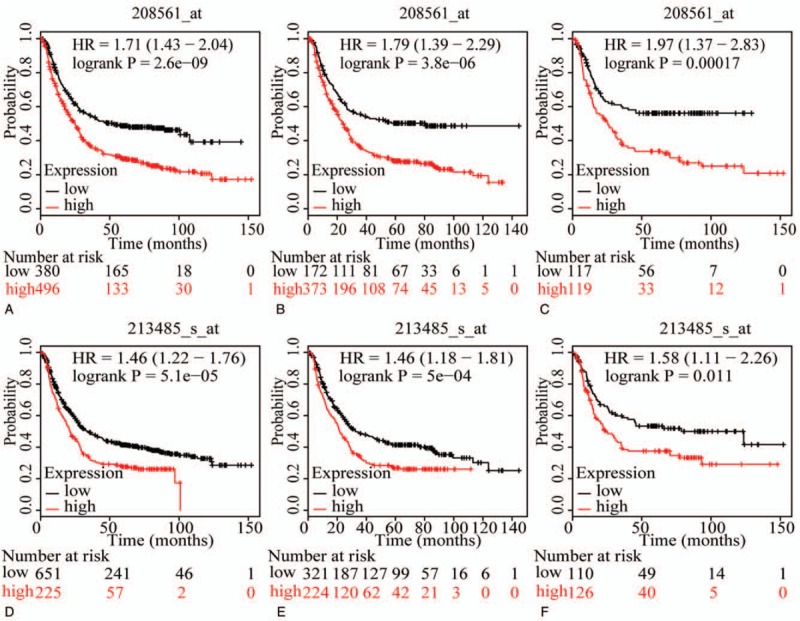
Survival plots of GC patients expressing ABCC9 and ABCC10. (A–C) Survival plots for ABCC9 (208561_at) in whole, male and female populations, respectively. (D–F) Survival plots for ABCC10 (203196_at) in whole, male and female populations, respectively. ABCC = adenosine triphosphate-binding cassette subfamily C, GC = gastric cancer.
Figure 6.
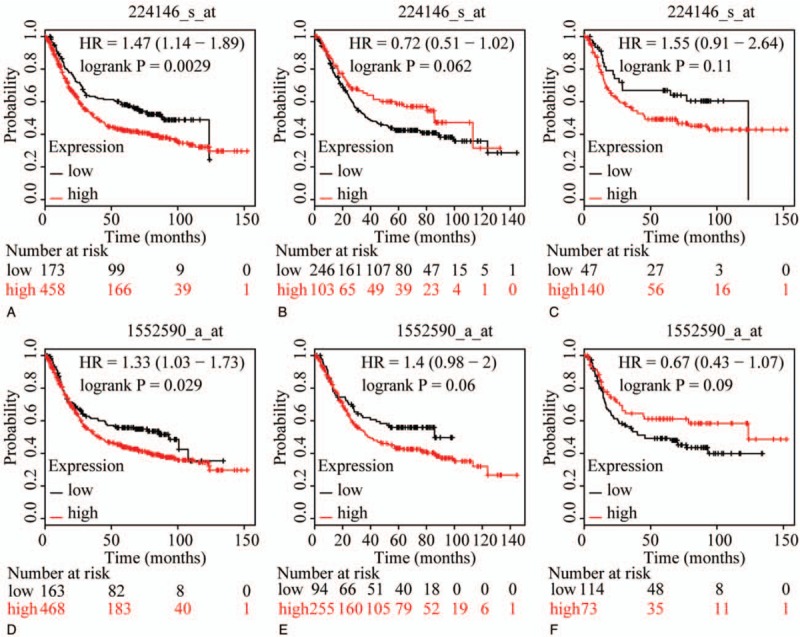
Survival plots of GC patients expressing ABCC11 and ABCC12. (A–C) Survival plots for ABCC11 (214979_at) in whole, male and female populations, respectively. (D–F) Survival plots for ABCC12 (213485_at) in whole, male and female populations, respectively. ABCC = adenosine triphosphate-binding cassette subfamily C, GC = gastric cancer.
Figure 7.
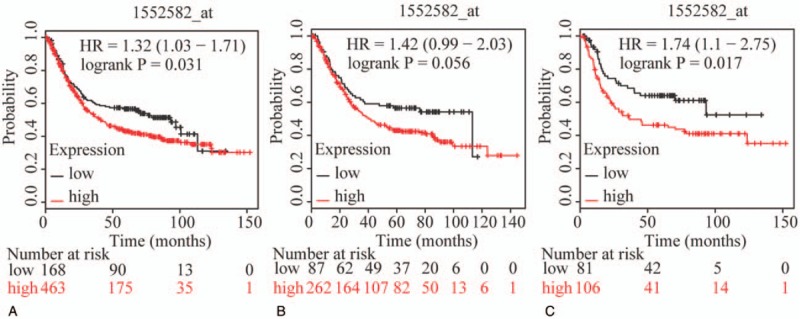
Survival plots of GC patients expressing ABCC13, and maps of gene-gene interaction and protein-protein interaction networks. (A–C) Survival plots for ABCC13 (214979_at) in whole, male and female populations, respectively. ABCC = adenosine triphosphate-binding cassette subfamily C, GC = gastric cancer.
3.2. 3.2 The association of ABCC family members with OS, stratified by gender
To further explore sex-related differences in the OS of patients expressing ABCC genes, OS plots for ABCC family members were depicted by gender. ABCC1, ABCC3, ABCC7, ABCC8, ABCC9, and ABCC10 showed prognostic significance in both male and female populations (all P ≤ .05, Figs. 1B and C, 2B and C, 4B, C, E, and F, 5B, C, E, and F), whereas the association of ABCC4, ABCC11, and ABCC12 with OS was not significant (all P>0.05, Figs. 2E, F and 6C, E, F). ABCC5 and ABCC6 showed prognostic significance in males alone (both P ≤ .05, Fig. 3B and E), while ABCC2 and ABCC13 showed prognostic significance in females only (both P ≤ .05, Figs. 1F and 7C).
In summary, ABCC1, ABCC3, ABCC7, ABCC8, ABCC9, and ABCC10 showed prognostic significance in both the whole population and in segregated male and female populations, whereas the association of ABCC4 with OS was not significant in any population. However, ABCC2, ABCC5, ABCC6, ABCC11, ABCC12, and ABCC13 showed prognostic significance in only some of these populations.
3.3. 3.3 Analysis of GGI, PPI, BP, CC, and MF networks
GGI network analysis indicated that most of the ABCC family members were associated with other ABCA, ABCB, and ABCD family members in shared protein domains. ABCC3 and ABCC5 were co-expressed, as were ABCC8 and ABCC9. ABCC2 was co-expressed with ABCC1, ABCA12, and ABCB6 (Supplementary Fig. 1A). In the GGI network, CFTR showed known interactions with ABCC4, ABCC10 and ABCC1 and ABCC8 showed a known interaction with ABCC9. ABCC5 was co-expressed with ABCC1, ABCC2, ABCC3, ABCC6, ABCC8, and ABCC9. ABCC1 was co-expressed with ABCC4, ABCC5, ABCC10, and ABCC12 (Supplementary Fig. 1B).
In the BP network, transmembrane transport, establishment of localization, ion transport, transport, potassium ion transport, and response to drug, among others, were enriched in the network (Supplementary Fig. 2). In the CC network, integral to plasma membrane, intrinsic to membrane, apical part of cell, membrane fraction, cell fraction, cytoplasmic vesicle part, and other functions were enriched in the network (Supplementary Fig. 3). In the MF network, purine nucleotide binding, ATP binding, adenyl ribonucleotide binding, substrate-specific transporter activity, and primary active transmembrane transporter activity were enriched (Supplementary Fig. 4).
3.4. Correlation analysis between ABCC family members and disease stage, Lauren classification, tumor differentiation, and HER2 status
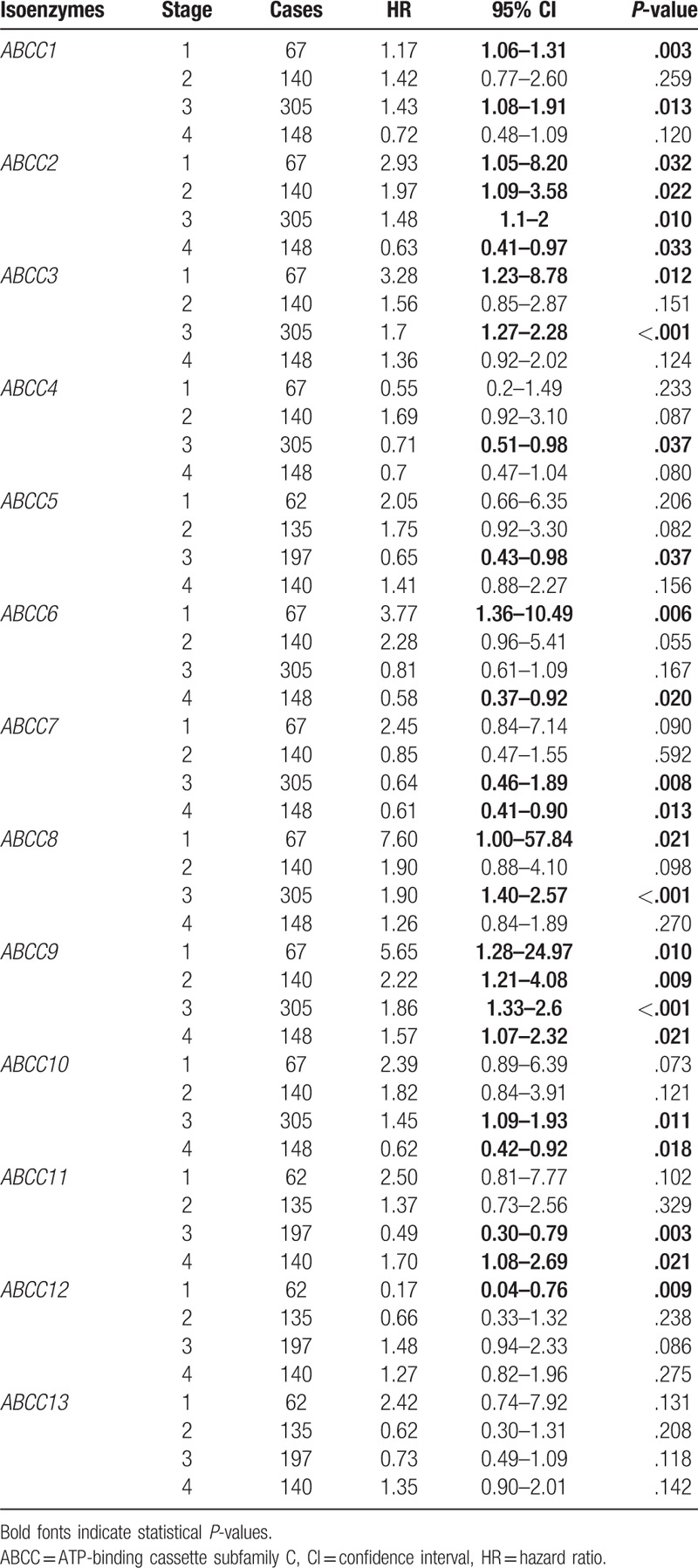
Correlation analysis between ABCC family members and cancer stage indicated that all stages were significantly correlated with the expression of ABCC members, ABCC2 and ABCC9 (all P ≤ .05), but none were significantly correlated with any stage. Most of the other family members were significantly correlated with some of the stages (Table 1). Remarkably, a higher stage always indicated a protective role for ABCC2 and ABCC9 compared to lower stages, which suggested that low stage may be a poor prognosis predictor. These results were consistently observed for other members as well.
Table 1.
Correlation analysis between ABCC family and stage.
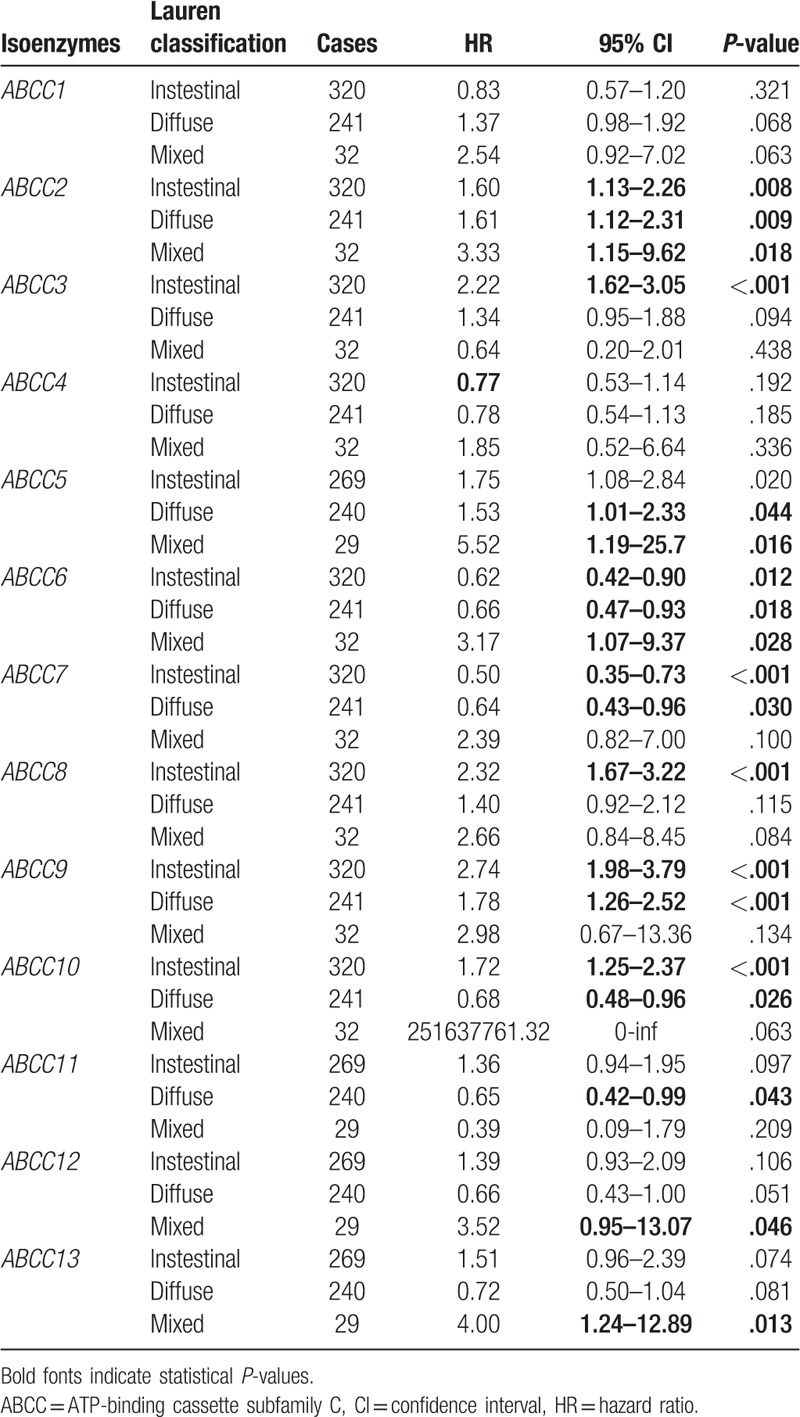
Correlation analysis between ABCC family members and Lauren classification indicated that all of the Lauren classifications were significantly correlated with expression of ABCC members ABCC2 and ABCC6 (all P ≤ .05), whereas none of the Lauren classifications were significantly correlated with ABCC1 and ABCC4 (all P > .05). Other ABCC family members showed a significant correlation with only some of the Lauren classifications (Table 2). Strangely, HRs were not consistent for ABCC2 and ABCC6: all the HRs were >1 for ABCC2, but HRs were not consistent for the 3 Lauren classifications of ABCC6.
Table 2.
Correlation analysis between ABCC family and Lauren classification.
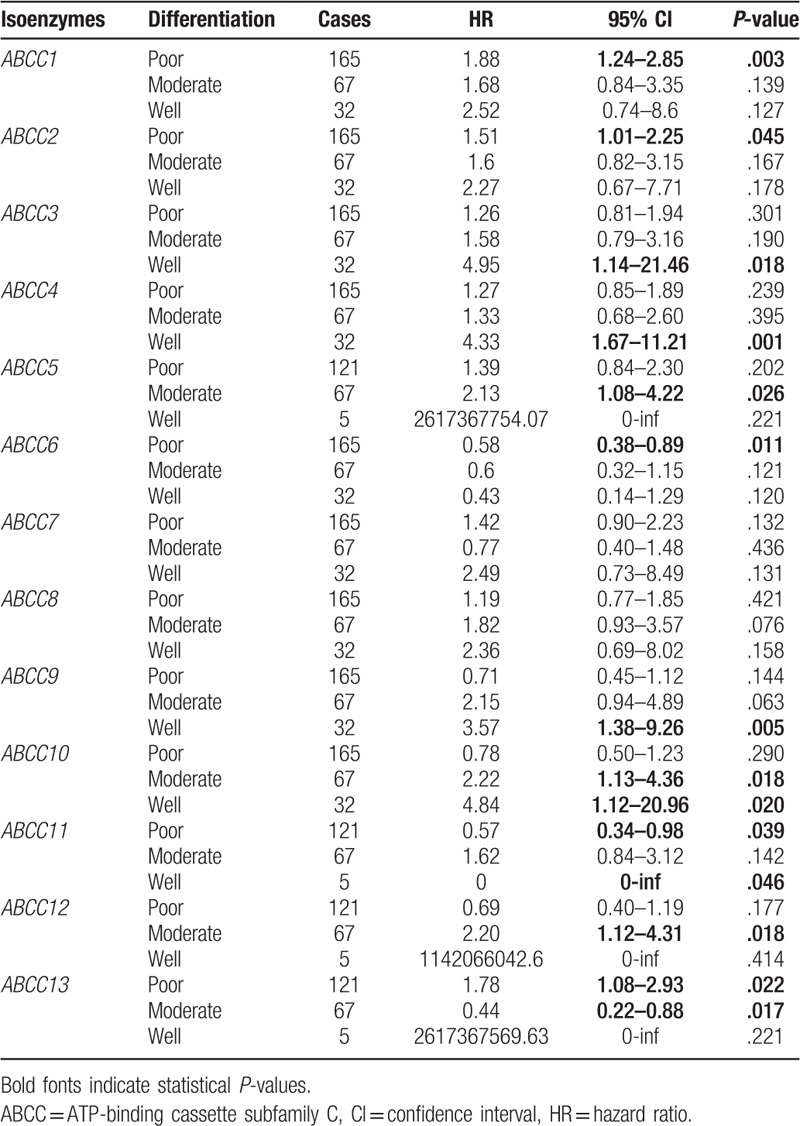
Correlation analysis between ABCC family members and tumor differentiation status indicated that none of the members showed significant correlations with any of the differentiation subtypes. Except for ABCC7 and ABCC8, which did not show any correlation with any of the differentiation subtypes, most ABCC members were correlated with some of the differentiation subtypes (Table 3).
Table 3.
Correlation analysis between ABCC family and differentiation.
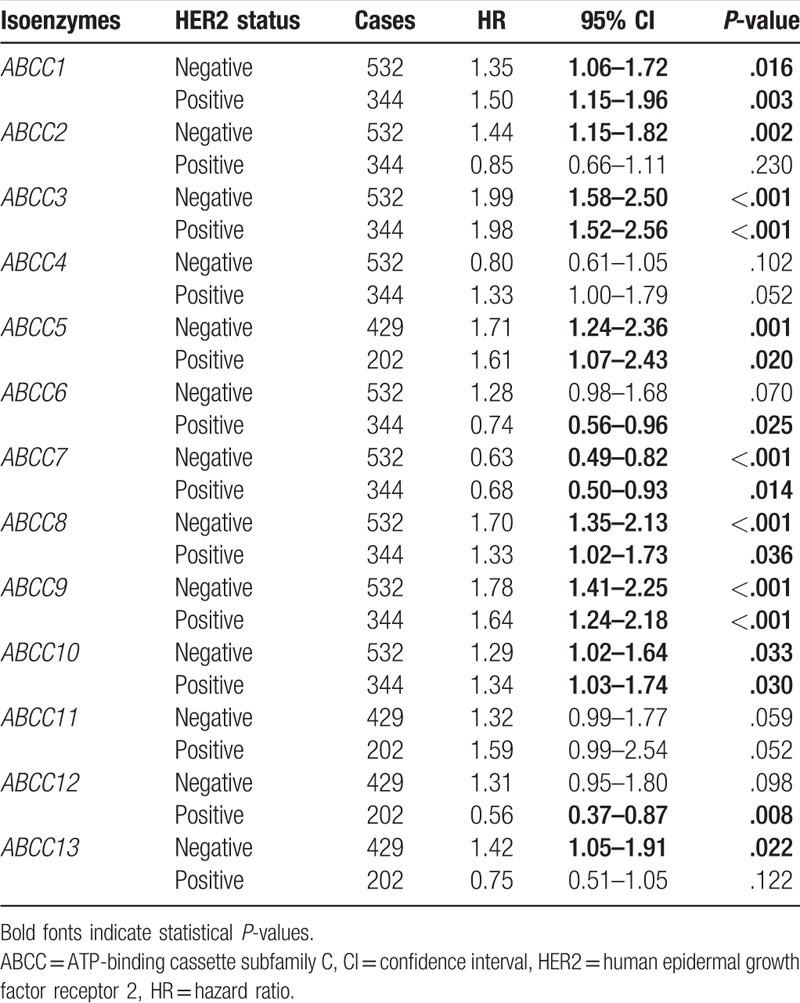
Correlation analysis between ABCC family members and HER2 status indicated that either negative or positive HER2 status was significantly correlated with ABCC members ABCC1, ABCC3, ABCC5, ABCC7, ABCC8, ABCC9, and ABCC10 (all P ≤ .05). Moreover, all the HRs were consistent for these members. With the exception of ABCC4 and ABCC11, which were not correlated with any status, other ABCC members were correlated with either positive or negative HER2 status (Table 4).
Table 4.
Correlation analysis between ABCC family and HER2 status.
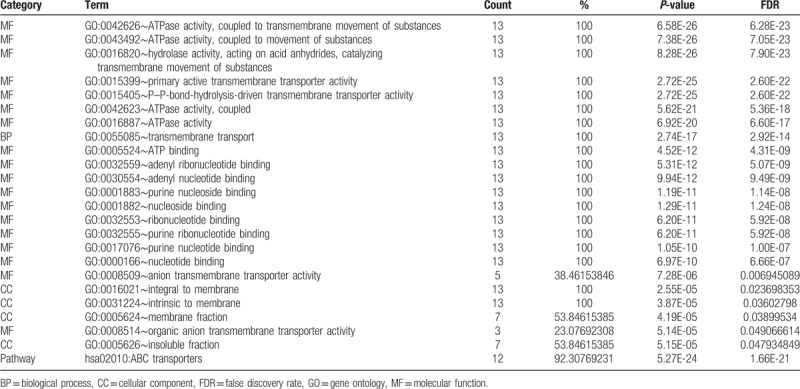
3.5. Enrichment results of GO terms and KEGG pathways
Enrichment analysis was performed using DAVID. Significant GO terms were enriched for BP, CC and MF, and included ATPase activity, coupled to transmembrane movement of substances, hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances, transmembrane transport, integral to membrane, intrinsic to membrane, purine ribonucleotide binding, and others (Table 5). These results were consistent with BinGO results (data not shown). The significant KEGG pathway was enriched in ABC transporters (Table 5).
Table 5.
Enrichment analysis of gene ontologies and KEGG pathways of ABCC family.
4. Discussion
In the present study, we investigated the association of ABCC family members ABCC1 to 13 with OS of GC patients. We found that ABCC1, ABCC2, ABCC3, ABCC5, ABCC6, ABCC7, ABCC8, ABCC9, ABCC10, ABCC11, ABCC12, and ABCC13 exerted a significant effect on OS. Furthermore, high expression of any of the above genes always suggested a poor prognosis, except for ABCC7. Stratified analysis by gender revealed that ABCC1, ABCC3, ABCC7, ABCC8, ABCC9, and ABCC10 showed prognostic significance for the whole population as well as male and female subpopulations; in contrast ABCC4 expression was not significantly different among the above 3 populations. Moreover, ABCC2, ABCC5, ABCC6, ABCC11, ABCC12, and ABCC13 showed prognostic significance in some of these populations. Subtype analysis revealed that ABCC2 and ABCC9 were significantly correlated with all tumor stages; ABCC2 and ABCC6 were significantly correlated with all the Lauren classifications; and ABCC1, ABCC3, ABCC5, ABCC7, ABCC8, ABCC9, and ABCC10 were significantly correlated with either negative or positive HER2 status. Enrichment analysis indicated that these genes were involved with the GO terms ATPase activity, coupled to transmembrane movement of substances, catalyzing transmembrane movement of substances, transmembrane transport, purine ribonucleotide binding, and others, and were enriched in the KEGG pathway of ABC transporters.
ABC transporters make up a large superfamily of membrane proteins and have been found in many living species from bacteria to human beings.[27] Most of these membrane proteins play a pivotal role in transporting various ATP-dependent substances across lipid membranes, such as sugars, lipids, vitamins, sterols, amino acids, xenobiotics as well as some chemotherapeutic drugs.[28,29] ABC transporters possess highly conserved domains as do all ABC superfamily members, consisting of a highly conserved cytosolic nucleotide-binding domain, while the majority of the superfamily members share less conserved domains.[30] RNA-seq analysis indicated that the AGAPOO8236 gene, belonging to ABCC subfamily, was greatly upregulated in a deltamenthrin-resistant strain of Anopheles gambiae.[31]
ABCC subfamily members are well-known as MRPs and have been reported to be involved in the transportation of many drugs, ions, toxins, and endogenous substances.[28,32,33]ABCC1 and ABCC2 protein expression have been detected in mouse and human tissues with chronic pancreatitis.[34]ABCC1 expression has been associated with chemoresistance of small cell lung cancer by microRNA-7 regulation.[35] A single nucleotide polymorphism of variant alleles of rs12762549 in ABCC2 has been associated with the risk of anemia.[36] Perego et al demonstrated that ABCC1 expression was not only associated with tumor grade, but maybe a potential biomarker for aggressiveness of epithelial ovarian cancer, while ABCC4 may be a poor prognosis predictor in ovarian cancer outcome.[37] Our present study showed results consistent with previous reports that ABCC1 and ABCC2 expression were associated with tumor prognosis. In addition, we found that high expression of ABCC1 and ABCC2 indicated a poor prognosis for GC.
ABCC3 protein expression has been reported in normal pancreatic tissues and pancreatic adenocarcinoma.[38] Keppler et al suggested that mRNA expression of ABCC3 was upregulated in pancreatic carcinoma tissues and was associated with tumor stage and grading.[39]ABCC5 was upregulated in pancreatic carcinoma tissues as well, while ABCC1, ABCC4, and ABCG1 were not.[39] These authors further concluded that ABCC3 and ABCC5 participated in drug resistance of pancreatic carcinoma and their expression could be used to predict patient response to chemotherapy.[39]ABCC6 has been found mainly in the basolateral plasma membrane of liver and kidney cells and its mutation has been associated with pseudoxanthoma elasticum, an autosomal recessive disease characterized by progressive ectopic calcification of elastic fibers in vascular, ocular and dermal tissues.[40] Our results indicated that ABCC3 showed prognostic value in the low and high expression groups, whereas ABCC4 did not. Moreover, high expression of ABCC3 was associated with poor prognosis of GC. ABCC5 also showed prognostic value in the low and high expression groups, but ABCC6 did not. In addition, high expression of ABCC5 was associated with poor prognosis of GC.
ABCC7, also named CFTR, is located on 7q31.2 and has been reported to entail more than 1900 different, heterogeneous mutations in various populations (http://www.genet.sickkids.on.ca/cftr/). ABCC7 is the main gene contributing to the development of cystic fibrosis.[41,42]ABCC8 is located on chromosome 11 and its mutation has been related to type 2 diabetes, gestational diabetes and maturity-onset diabetes of the young, which is associated with gene mutations giving rise to abnormalities in pancreatic β cells.[43] Some mutations of ABCC8 lead to hyperinsulinemia in newborns.[44,45]
Hirota et al reported that ABCA13, ABCB6, ABCC1, and ABCC3 expression were regulated by cigarette smoke exposure and that ABCA13, ABCC1, ABCC2, and ABCC9 were differentially expressed in chronic obstructive pulmonary diseases and asthma.[46] Human ABCC11, named MRP8 and located on chromosome 16q12.1, was reported in an investigation of the association between human wet and dry earwax types in geriatric Japanese populations.[47] Dissimilar to other MRP family members, ABCC11 has some orthologous genes in mammals except for primates.[48,49] Protein expression of ABCC11 has been identified in axons in both central and peripheral nerve system neurons and it may play significant roles in the efflux of neuromodulatory steroids.[50] High expression of ABCC11 was correlated with poor OS in acute myeloid leukemia patients, which indicates that ABCC11 may be a predictive biomarker for treatment outcome.[51] Endo et al suggested that high expression of ABCC11 in breast cancer tissues was significantly associated with poor disease-free survival and aggressive subtypes.[52] Our findings are consistent with these 2 reports indicating that high expression of ABCC11 is associated poor tumor prognosis.
ABCC12, also known as MRP9, has been localized next to ABCC11 on chromosome 16q12.1, 20 and is oriented in a tail-to-head position, which indicates that ABCC12 may have originated in a gene duplication event.[48] One of the longest mRNA transcripts of ABCC12 encodes a protein of 1359 amino acids.[48] Tandemly duplicated on chromosome 16q12 in a region harboring genes for paroxysmal kinesigenic choreoathetosis, ABCC11 and ABCC12 are positional candidate biomarkers of this disease.[49] Identified by 2 major transcripts of 4.5 kb and 1.3 kb, ABCC12 is highly expressed in breast cancer tissue and is not expressed at detectable levels in normal tissue; thus, it may represent a potential target for immunotherapy of breast cancer.[53]ABCC13, found on chromosome 21q11.2 and consisting of 14 exons, spans roughly 70 kb and is highly expressed in human fetal liver.[54] In addition to its similarity to ABCC2, the ABCC13 transcript is made up of 6 exons with a total length of 1.1 kb.[55] The open reading frame of this transcript encodes a polypeptide of 274 amino acids, as compared to other ABC-related transporters with more than 1500 amino acids.[55] Furthermore, the truncated ABCC13 transcript is specifically expressed in fetal liver, bone marrow, and colon.[55] However, little is known about the association between expression of ABCC12, ABCC13, and tumors. We are the first to our knowledge to report its prognostic significance in GC patients.
Although we believe we are the first to establish the prognostic significance of ABCC family members in GC, there are still some shortcomings in present study. First, our findings need further validation in other cohorts. Second, multivariate cox regression model should be used for further analysis. Third, experiments in vitro and in vivo should be performed to validate the functions of these prognosis-related genes. In addition, diagnostic significance, including sensitivity and specificity, of ABCC members in GC need to be explored in the future studies. This study also has potential bias, including Affymetrix ID choose, cutoff split and subtypes analysis, need to be recognized.
5. Conclusion
Our study has explored the possible relationship of 13 ABCC family members, ABCC1 to 13, with OS of GC patients. Our study found that expression of ABCC1, ABCC2, ABCC3, ABCC5, ABCC6, ABCC7, ABCC8, ABCC9, ABCC10, ABCC11, ABCC12, and ABCC13 could predict OS in GC patients. High expression of any of these genes always suggested a poor prognosis except for ABCC7. Stratified analysis by gender revealed that ABCC1, ABCC3, ABCC7, ABCC8, ABCC9, and ABCC10 showed prognostic significance in the whole population as well as in male and female subpopulations. Subtype analysis revealed that ABCC2 and ABCC9 were significantly correlated with all disease stages; ABCC2 and ABCC6 were significantly correlated with all of the Lauren classifications; and ABCC1, ABCC3, ABCC5, ABCC7, ABCC8, ABCC9, and ABCC10 were significantly correlated with both negative and positive HER2 status. Enrichment analysis indicated that these genes were involved in GO terms of ATPase activity, coupled to transmembrane movement of substances, transmembrane transport, purine ribonucleotide binding, and the KEGG pathway of ABC transporters. Nonetheless, although we are the first to report the prognostic significance of ABCC family members in GC, our findings need further validation from other cohorts, and experiments in vitro and in vivo should be used to validate the functions of these prognosis-related genes.
Acknowledgment
The authors thank the contributors of Kaplan–Meier Plotter database for their contribution to share data on open access. The authors would like to acknowledge invaluable comments from peer reviewers.
Author contributions
Conceptualization: Guangzhi Zhu, Xianshuang Mao, Zhenhua He, Yongchu Huang.
Data curation: Xianshuang Mao, Zhenhua He, Fengsheng Zhou.
Formal analysis: Xianshuang Mao, Zhenhua He, Fengsheng Zhou.
Investigation: Guangzhi Zhu, Xianshuang Mao.
Methodology: Guangzhi Zhu, Xianshuang Mao, Zhenhua He, Fengsheng Zhou.
Project administration: Guangzhi Zhu.
Resources: Xianshuang Mao, Yongchu Huang.
Software: Xianshuang Mao, Zhenhua He, Fengsheng Zhou, Yongchu Huang.
Supervision: Guangzhi Zhu.
Validation: Guangzhi Zhu, Xianshuang Mao, Zhenhua He, Yongchu Huang.
Visualization: Zhenhua He, Fengsheng Zhou, Yongchu Huang.
Writing – original draft: Xianshuang Mao.
Writing – review and editing: Guangzhi Zhu.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: ABC = ATP-binding cassette, ABCC adenosine triphosphate-binding cassette subfamily C, ATP = adenosine triphosphate, BP = biological process, CC = cellular component, CI = confidence interval, DAVID = database for annotation, visualization and integrated discovery, GC = gastric cancer, GGI = gene–gene interaction, GO = gene ontology, HER2 = human epidermal growth factor receptor 2, HR = hazard ratio, KEGG = Kyoto Encyclopedia of Genes and Genomes, MF = molecular function, MRPs = multidrug resistance proteins, OS = overall survival.
How to cite this article: Mao X, He Z, Zhou F, Huang Y, Zhu G. Prognostic significance and molecular mechanisms of adenosine triphosphate-binding cassette subfamily C members in gastric cancer. Medicine. 2019;98:50(e18347).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Thapa S, Chetry M, Huang K, et al. Significance of aquaporins’ expression in the prognosis of gastric cancer. Biosci Rep 2018;38:BSR20171687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [3].Hartgrink HH, Jansen EP, van Grieken NC, et al. Gastric cancer. Lancet (London, England) 2009;374:477–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin 2008;58:71–96. [DOI] [PubMed] [Google Scholar]
- [5].Shen L, Huang Y, Sun M, et al. Clinicopathological features associated with lymph node metastasis in early gastric cancer: analysis of a single-institution experience in China. Can J Gastroenterol 2009;23:353–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lee JH, Kim KM, Cheong JH, et al. Current management and future strategies of gastric cancer. Yonsei Med J 2012;53:248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jiang B, Li Z, Zhang W, et al. miR-874 Inhibits cell proliferation, migration and invasion through targeting aquaporin-3 in gastric cancer. J Gastroenterol 2014;49:1011–25. [DOI] [PubMed] [Google Scholar]
- [8].Chen ZS, Tiwari AK. Multidrug resistance proteins (MRPs/ABCCs) in cancer chemotherapy and genetic diseases. FEBS J 2011;278:3226–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Braschi B, Denny P, Gray K, et al. Genenames.org: the HGNC and VGNC resources in 2019. Nucleic Acids Res 2019;47:D786–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dean M, Allikmets R. Complete characterization of the human ABC gene family. J Bioenerg Biomembr 2001;33:475–9. [DOI] [PubMed] [Google Scholar]
- [11].Leslie EM, Deeley RG, Cole SP. Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol Appl Pharmacol 2005;204:216–37. [DOI] [PubMed] [Google Scholar]
- [12].Nies AT, Keppler D. The apical conjugate efflux pump ABCC2 (MRP2). Pflugers Arch 2007;453:643–59. [DOI] [PubMed] [Google Scholar]
- [13].Kawabe T, Chen ZS, Wada M, et al. Enhanced transport of anticancer agents and leukotriene C4 by the human canalicular multispecific organic anion transporter (cMOAT/MRP2). FEBS Lett 1999;456:327–31. [DOI] [PubMed] [Google Scholar]
- [14].Konig J, Rost D, Cui Y, et al. Characterization of the human multidrug resistance protein isoform MRP3 localized to the basolateral hepatocyte membrane. Hepatology 1999;29:1156–63. [DOI] [PubMed] [Google Scholar]
- [15].van Aubel RA, Smeets PH, Peters JG, et al. The MRP4/ABCC4 gene encodes a novel apical organic anion transporter in human kidney proximal tubules: putative efflux pump for urinary cAMP and cGMP. J Am Soc Nephrol 2002;13:595–603. [DOI] [PubMed] [Google Scholar]
- [16].Jedlitschky G, Burchell B, Keppler D. The multidrug resistance protein 5 functions as an ATP-dependent export pump for cyclic nucleotides. J Biol Chem 2000;275:30069–74. [DOI] [PubMed] [Google Scholar]
- [17].Hopper E, Belinsky MG, Zeng H, et al. Analysis of the structure and expression pattern of MRP7 (ABCC10), a new member of the MRP subfamily. Cancer Lett 2001;162:181–91. [DOI] [PubMed] [Google Scholar]
- [18].Bera TK, Lee S, Salvatore G, et al. MRP8, a new member of ABC transporter superfamily, identified by EST database mining and gene prediction program, is highly expressed in breast cancer. Mol Med 2001;7:509–16. [PMC free article] [PubMed] [Google Scholar]
- [19].Ono N, Van der Heijden I, Scheffer GL, et al. Multidrug resistance-associated protein 9 (ABCC12) is present in mouse and boar sperm. Biochem J 2007;406:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Szasz AM, Lanczky A, Nagy A, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016;7:49322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [23].Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Montojo J, Zuberi K, Rodriguez H, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 2010;26:2927–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005;21:3448–9. [DOI] [PubMed] [Google Scholar]
- [27].He Q, Yan Z, Si F, et al. ATP-binding cassette (ABC) transporter genes involved in pyrethroid resistance in the malaria vector anopheles sinensis: genome-wide identification, characteristics, phylogenetics, and expression profile. Int J Mol Sci 2019;20:E1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 2001;11:1156–66. [DOI] [PubMed] [Google Scholar]
- [29].Higgins CF. ABC transporters: from microorganisms to man. Ann Rev Cell Biol 1992;8:67–113. [DOI] [PubMed] [Google Scholar]
- [30].Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev Mol Cell Biol 2009;10:218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bonizzoni M, Afrane Y, Dunn WA, et al. Comparative transcriptome analyses of deltamethrin-resistant and -susceptible Anopheles gambiae mosquitoes from Kenya by RNA-Seq. PLoS One 2012;7:e44607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sheppard DN, Welsh MJ. Structure and function of the CFTR chloride channel. Physiol Rev 1999;79: 1 Suppl: S23–45. [DOI] [PubMed] [Google Scholar]
- [33].Moreau C, Prost AL, Derand R, et al. SUR, ABC proteins targeted by KATP channel openers. J Mol Cell Cardiol 2005;38:951–63. [DOI] [PubMed] [Google Scholar]
- [34].Schaarschmidt T, Merkord J, Adam U, et al. Expression of multidrug resistance proteins in rat and human chronic pancreatitis. Pancreas 2004;28:45–52. [DOI] [PubMed] [Google Scholar]
- [35].Liu H, Wu X, Huang J, et al. miR-7 modulates chemoresistance of small cell lung cancer by repressing MRP1/ABCC1. Int J Exp Pathol 2015;96:240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lambrechts S, Lambrechts D, Despierre E, et al. Genetic variability in drug transport, metabolism or DNA repair affecting toxicity of chemotherapy in ovarian cancer. BMC Pharmacol Toxicol 2015;16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bagnoli M, Beretta GL, Gatti L, et al. Clinicopathological impact of ABCC1/MRP1 and ABCC4/MRP4 in epithelial ovarian carcinoma. Biomed Res Int 2013;2013:143202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Scheffer GL, Kool M, de Haas M, et al. Tissue distribution and induction of human multidrug resistant protein 3. Lab Invest 2002;82:193–201. [DOI] [PubMed] [Google Scholar]
- [39].Konig J, Hartel M, Nies AT, et al. Expression and localization of human multidrug resistance protein (ABCC) family members in pancreatic carcinoma. Int J Cancer 2005;115:359–67. [DOI] [PubMed] [Google Scholar]
- [40].Martinelli F, Cuviello F, Pace MC, et al. Extracellular ATP regulates CD73 and ABCC6 expression in HepG2 Cells. Front Mol Biosci 2018;5:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Riordan JR, Rommens JM, Kerem B, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 1989;245:1066–73. [DOI] [PubMed] [Google Scholar]
- [42].Veit G, Avramescu RG, Chiang AN, et al. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol Biol Cell 2016;27:424–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ovsyannikova AK, Rymar OD, Shakhtshneider EV, et al. ABCC8-related maturity-onset diabetes of the young (MODY12): clinical features and treatment perspective. Diabetes Ther 2016;7:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Haghverdizadeh P, Sadat Haerian M, Haghverdizadeh P, et al. ABCC8 genetic variants and risk of diabetes mellitus. Gene 2014;545:198–204. [DOI] [PubMed] [Google Scholar]
- [45].Baier LJ, Muller YL, Remedi MS, et al. ABCC8 R1420H loss-of-function variant in a southwest american indian community: association with increased birth weight and doubled risk of type 2 diabetes. Diabetes 2015;64:4322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Aguiar JA, Tamminga A, Lobb B, et al. The impact of cigarette smoke exposure, COPD, or asthma status on ABC transporter gene expression in human airway epithelial cells. Sci Rep 2019;9:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sato T, Amano T, Ono H, et al. Allele frequencies of the ABCC11 gene for earwax phenotypes among ancient populations of Hokkaido, Japan. J Hum Genet 2009;54:409–13. [DOI] [PubMed] [Google Scholar]
- [48].Yabuuchi H, Shimizu H, Takayanagi S, et al. Multiple splicing variants of two new human ATP-binding cassette transporters, ABCC11 and ABCC12. Biochem Biophys Res Commun 2001;288:933–9. [DOI] [PubMed] [Google Scholar]
- [49].Tammur J, Prades C, Arnould I, et al. Two new genes from the human ATP-binding cassette transporter superfamily, ABCC11 and ABCC12, tandemly duplicated on chromosome 16q12. Gene 2001;273:89–96. [DOI] [PubMed] [Google Scholar]
- [50].Bortfeld M, Rius M, Konig J, et al. Human multidrug resistance protein 8 (MRP8/ABCC11), an apical efflux pump for steroid sulfates, is an axonal protein of the CNS and peripheral nervous system. Neuroscience 2006;137:1247–57. [DOI] [PubMed] [Google Scholar]
- [51].Guo Y, Kock K, Ritter CA, et al. Expression of ABCC-type nucleotide exporters in blasts of adult acute myeloid leukemia: relation to long-term survival. Clin Cancer Res 2009;15:1762–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yamada A, Ishikawa T, Ota I, et al. High expression of ATP-binding cassette transporter ABCC11 in breast tumors is associated with aggressive subtypes and low disease-free survival. Breast Cancer Res Treat 2013;137:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bera TK, Iavarone C, Kumar V, et al. MRP9, an unusual truncated member of the ABC transporter superfamily, is highly expressed in breast cancer. Proc Natl Acad Sci USA 2002;99:6997–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yabuuchi H, Takayanagi S, Yoshinaga K, et al. ABCC13, an unusual truncated ABC transporter, is highly expressed in fetal human liver. Biochem Biophys Res Commun 2002;299:410–7. [DOI] [PubMed] [Google Scholar]
- [55].Annilo T, Dean M. Degeneration of an ATP-binding cassette transporter gene, ABCC13, in different mammalian lineages. Genomics 2004;84:34–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


