Abstract
Porous graphitized carbon (PGC) based chromatography achieves high-resolution separation of glycan structures released from glycoproteins. This approach is especially valuable when resolving structurally similar isomers and for discovery of novel and/or sample-specific glycan structures. However, the implementation of PGC-based separations in glycomics studies has been limited because system-independent retention values have not been established to normalize technical variation. To address this limitation, this study combined the use of hydrolyzed dextran as an internal standard and Skyline software for post-acquisition normalization to reduce retention time and peak area technical variation in PGC-based glycan analyses. This approach allowed assignment of system-independent retention values that are applicable to typical PGC-based glycan separations and supported the construction of a library containing >300 PGC-separated glycan structures with normalized glucose unit (GU) retention values. To enable the automation of this normalization method, a spectral MS/MS library was developed of the dextran ladder, achieving confident discrimination against isomeric glycans. The utility of this approach is demonstrated in two ways. First, to inform the search space for bioinformatically predicted but unobserved glycan structures, predictive models for two structural modifications, core-fucosylation and bisecting GlcNAc, were developed based on the GU library. Second, the applicability of this method for the analysis of complex biological samples is evidenced by the ability to discriminate between cell culture and tissue sample types by the normalized intensity of N-glycan structures alone. Overall, the methods and data described here are expected to support the future development of more automated approaches to glycan identification and quantitation.
Introduction
The ability to detect and quantify individual glycan structures present within a sample defines the level of specificity that can be achieved in glycomic analyses.1 Given the high degree of complexity and similarity among glycan structures found within cells and tissues, the ability to discriminate among isomeric glycan structures is often necessary to fully define their glycome. Capillary electrophoresis and liquid chromatography (LC) are separation techniques capable of resolving isomeric glycan structures.2 Of the LC methods available, hydrophilic interaction chromatography (HILIC) can provide robust separations of glycan structures3 while PGC is capable of resolving these glycan isomers further, including those not typically resolved with HILIC.4 However, as the separation mechanism of PGC has not been fully defined, glycan structure retention times (RTs) cannot currently be predicted using in silico models.5 To promote the applicability of PGC for glycomics, glycan retention libraries and elution order rules have been established.6–9 As a result of these efforts, glycan structural identity can be assigned to peaks detected as a result of chromatographic separation, but the RTs accompanying these glycan libraries are not yet widely applicable as they remain system specific values.6–9 Consequently, system-based variations in glycan RTs observed among laboratories and datasets currently impede precise structural determination using PGC alone.
A potential solution to this challenge is the use of an internal retention time standard. Currently, including a dextran ladder with an increasing degree of polymerization allows system-independent retention constants for glycans to be derived, although these values are stationary phase specific.10 Guile et al.11 used a dextran ladder as a glycan retention index for fluorescent detection of glycans separated by HILIC and this has been further developed into a robust and routine method3,12–14 that has now been commercialized by Waters (RapiFluor-MS GU Scientific Glycan Library).
The dextran ladder is a glucose polymer linked through α−1,6-glycosidic bonds15 which enables system-independent retention constants to be measured in glucose units (GU). One successful example of its implementation is the construction of GlycoBase, a database of >375 glycan structures (2-aminobenzamide-labelled) separated on HILIC and assigned with corresponding GU values.16 GlycoBase has recently been incorporated and published in GlycoStore, a curated database with chromatographic and electrophoretic values for glycan structures.9 This database promotes automated structural assignment to observed peaks by combining fluorescent detection and GU values, but is dependent on reductive amination-based glycan labelling.14
Currently, an equivalent RT standardization approach for building a retention index database using PGC separations has not been established. However, considering the high resolving power of PGC for underivatized glycan structures and its compatability with electrospray ionization tandem mass spectrometry (ESI-MS/MS), implementation of new strategies for standardization are expected to provide additional value to a glycomics experiment beyond that which HILIC-based separation or fluorescence-based detection can provide.
To address these limitations, the current study characterized the elution behaviour of a reduced dextran ladder separated by PGC-LC. Subsequently, parameters for using the dextran ladder as an internal standard for RT and peak area normalization of glycans analyzed by PGC-LC-ESI-MS/MS were determined. These results supported the development of a system-independent glycan retention value library that was subsequently expanded and applied to characterize N-glycans released from a complex protein mixture. To aid automation and enhance robustness of data analysis, a spectral library of the dextran ladder was created to allow high throughput RT normalization of complex glycan mixtures, even those containing glycans isomeric to the ladder. Finally, this normalization strategy was applied to glycans released from biological samples ranging in complexity, including secreted protein mixtures, cell lysates and tissue lysates. Results demonstrate that this strategy can be extended to sample classification based on N-glycans alone, providing an efficient strategy for sample specific glycotyping.
Experimental section
Chemicals and materials
All chemicals were sourced from Sigma Aldrich (Sydney, Australia) unless otherwise specified. Peptide: N-glycosidase F (PNGase F, product#: V4831) was obtained from Promega (Sydney, Australia). All solvents used were LC-MS grade and obtained from Merck Millipore (Sydney, Australia). Bovine ribonuclease B (product#: R7884), porcine gastric mucin (product#: M1778), human IgA (product#: I1010), bovine lactoferrin (product#: L9507), human lactoferrin (product#: L0520), bovine fetuin (product#: F3385) and human IgG (product#: I4506) were sourced from Sigma Aldrich (Sydney, Australia). Human neutrophil elastase (product#: 342–40) was sourced from LeeBio (Maryland Heights, USA). Fungal cellobiohydrolase I was isolated as previously described.17 All other chemicals were analytical grade.
Tissue preparation
Pathogen-free adult male Balb/c mice (300–350 g; University of Adelaide, Australia, approval number: M-2013–227) were deeply anesthetized using an i.p. injection of 60 mg kg−1 Lethabarb and then transcardially perfused using cold 0.9% saline. The lumbar enlargement (L3–L5) of the spinal cord was removed; snap frozen and stored at −80 °C until further assessment. A similar method was used for brain sectioning with the brain quickly removed, snap frozen, periaqueductal grey (PAG) and rostral ventromedulla (RVM) containing regions sliced in 50 μm sections in a cryostat and stored at 80 °C until further assessment.
Cell culture
The U87MG, HEK293 and BV2 cell lines were cultured in T-75 flasks with 10 mL of Dulbecco’s Modified Eagle’s Medium supplemented with 10% (v/v) fetal bovine serum. The PC-12 cell line was cultured in similar conditions with slight modifications, using 10 mL of RPMI-1640 Medium supplemented with 10% (v/v) heat-inactivated horse serum, 5% (v/v) fetal bovine serum and 50 ng mL−1 neural growth factor. After the adherent cells reached 70% confluency, the growth medium was removed and adherent cells scraped and collected. The cells were washed with phosphate buffered saline, collected by centrifugation (500g for 10 min), and supernatant was removed for a total of three washes. Following cell lysis, proteins were precipitated using chloroform : methanol : water extraction (10 : 10 : 3, by volume). The protein pellet was collected re-solubilized in 4 M urea, and quantified by the Bradford protein assay.18
For analysis of secreted proteins, EX-CELL 293 (Sigma Aldrich, Sydney, Australia) was reconstituted as specified by the manufacturer. Once cells reached 70% confluency in the standard culture media containing serum, the culture medium was removed, and cells were washed three times with phosphate buffered saline (PBS) before 10 mL of the reconstituted EX-CELL 293 medium was added to the cells. Cells were acclimatized for 48 h before secreted protein collection. Secreted proteins were precipitated using a final concentration of 90% (v/v) acetone, solubilized and quantified as described above.
Preparation of dextran ladder
10 mg of dextran T2000 (Amersham Pharmacia Biotech AB, Uppsala, Sweden) was dissolved in 1 mL of 1 M trifluoroacetic acid (TFA) and held at 80 °C for 30 min. After this partial acid hydrolysis,19 the sample was dried by centrifugal evaporation at room temperature. Once dried, the sample was reduced, desalted and carbon cleaned as described by Jensen et al.4 with the following scale-up modifications: reduction was performed with 500 μL of 2 M NaBH4 in 50 mM KOH, the sample was desalted with 100 μL packed volume of cation exchange resin and enriched with 100 μL packed volume of graphitized carbon. Liquid volumes for desalting and enrichment were increased four-fold from 50 μL to 200 μL and OMIX C18–100 μL pipette tips (Agilent, Santa Clara, CA) were used to contain the increased solid phase resin volumes. The final amount of dextran prepared following graphitized carbon enrichment was quantified using the phenol sulphuric acid method.20
Glycan release
N- and O-Glycans were released from 30 μg of the protein samples as described by Jensen et al.4 Briefly, protein samples were immobilized onto polyvinylidene difluoride membranes (Millipore, Sydney, Australia) and stained with Direct Blue (Sigma-Aldrich, Sydney, Australia). The membrane spots were excised and washed in separate wells in a 96-well plate (Corning Incorporated, NY). N-glycans were released using 1 unit PNGase-F (Promega, Sydney, Australia) at 37 °C, over-night. Following N-glycanremoval, 500 mM NaBH4 in 50 mM KOH solution was added to immobilized protein spots for 16 h at 50 °C to release reduced O-linked glycans by reductive β-elimination. N-glycans were reduced with 1 M NaBH4 in 50 mM KOH solution which was held at 50 °C for 3 hours. Glycan reduction is required to obtain a single chromatographic peak for each glycan structure.4 Both N-glycans and O-glycans were desalted and enriched offline using AG 50W-X8 (Bio-Rad, Sydney, Australia) strong cation exchange followed by PGC solid phase extraction micro-columns (Thermo Scientific, Sydney, Australia). Prior to injection, 26 ng of the dextran ladder was added to each glycan mixture.
Glycan data acquisition
PGC-LC-ESI-MS/MS experiments were performed on an UltiMate3000 high performance liquid chromatography (HPLC) system (Dionex, Sunnyvale, CA, USA) interfaced with an LTQ Velos Pro ion trap (Thermo Fisher Scientific, San Jose, CA, USA). Separations were performed using a PGC LC column (3 μm, 100 mm × 0.18 mm, Hypercarb, Thermo Fisher Scientific) maintained at 50 °C. Mobile phases used for separation were composed of 10 mM ammonium bicarbonate aqueous solution (solvent A) and 10 mM ammonium bicarbonate aqueous solution with 70% (v/v) acetonitrile (MeCN, solvent B) with a flow rate of 4 μl min−1. Four gradients were used for normalization experiments with the first as follows: 0–4.9 min, 1% B; 4.9–50 min, linear increase up to 45.2% B; 50–55 min, held at 99% B; 55–60 min, equilibrated at 1% B for 5 min before next injection for a total run time of 60 min and the second gradient, simulating reduced glycan retention, was the same except 0–4.9 min was at 8% B (Fig. 2). The gradient used for building the GU library was as follows: 0–4.9 min, 1% B; 5 min, 7.8% B; 5–73 min, linear increase to 64% B; 73–78 min, 99% B, 78–83 min, equilibrated at 1% B for 5 min before next injection for a total run time of 83 min. The final gradient used for applying the GU library to typical biological samples was as follows: 0 min, 5% B; 0–110 min, linear increase to 30% B; 110–115 min, 99% B, equilibrated at 1% B for 5 min before next injection for a total run time of 120 min. MS parameters were used as previously optimized and are described by Ashwood et al.21 to obtain ten data points per peak and MS2 spectra capable of discriminating glycan isomers.
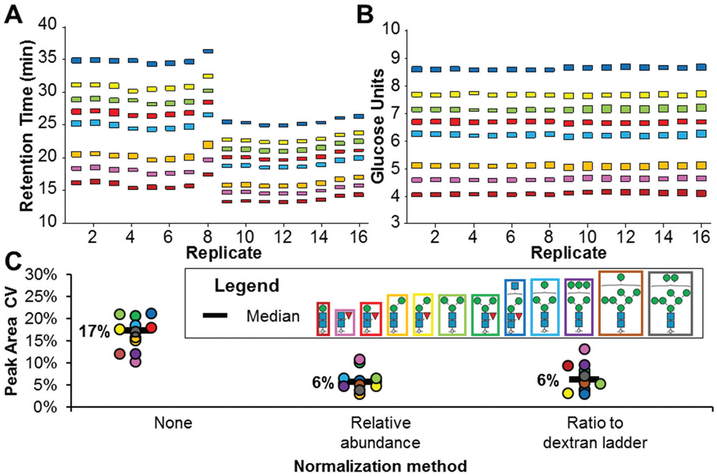
Fig. 2.
Normalizing two aspects of LC-MS variation using a dextran ladder. Glycan structure colour legend corresponds to all panels. (A) Observed RTs of a glycan mixture over 16 technical replicates with a simulated loss of retention for replicates 9–16. (B) Normalized RTs of the 16 technical replicates shown in A (C) Peak area variation comparison of post-acquisition normalization techniques for glycan quantitation. Glycan structures visualized per SNFG guidelines.33
Glycan structural data analysis
Software-assisted glycan identification and annotation of spectra was performed as previously described21 using Xcalibur (v3.0), Proteowizard22 (v3.0.10730), GlycoMod23 and GlycoWorkBench (v2.1).24 O-Glycan structural assignment was specifically performed by searching the UniCarb-DB25 human mucin dataset created by Jin et al.26 Apex RT values used were as identified with Skyline (v4.2.1.19058).27 Peak area quantitation and RT normalization to the dextran ladder was also performed by Skyline (first 3 isotopic peaks integrated from centroided data with EIC extraction width of 200 ppm for theoretical m/z values for each glycan structure). Equation determination and relative signal normalization was performed with Microsoft Excel (v2010). To improve usability and throughput of RT normalization, a script and graphical user interface (GUI) was developed which, when provided run-specific correction values, automatically converts RTs into GU values for LC-MS data files in the vendor-neutral mzML format (https://github.com/chrashwood/mzML-RT-to-iRT-Exponential). The glycan library with GU values represented as RTs was built in Excel and the corresponding Skyline assay was made in “small molecule” mode as previously described by Ashwood et al.21 To compare sample-specific glycan profiles, hierarchical clustering was performed with Morpheus (https://software.broadinstitute.org/morpheus) using Euclidean distance, complete linkages and clustering of structures and samples.
Dextran ladder spectral library
Raw data files were centroided (vendor-provided algorithm) using msconvert and the SSL file was subsequently populated with appropriate fields from the analysis of raw data files using msaccess from Proteowizard.22 Chemical formula and inchikeys were determined using ChemDraw (http://www.perkinelmer.com/category/chemdraw). Unique glycan structure identifiers were assigned to unambiguous glycan structures using GlyTouCan28 (last accessed: 2019-01-14) and the spectral library constructed using Bibliospec.29 MS2 spectral annotation was performed using GlycoWorkBench24 (https://code.google.com/archive/p/glycoworkbench/downloads), manually inspected for verification, and added to the BLIB file using SQLite v3.22 (https://www.sqlite.org/download.html).
Results and discussion
Dextran ladder separated by PGC chromatography
Detailed characterization of dextran ladder separation was completed as a first step in evaluating its utility as a standard in PGC-LC-ESI-MS/MS. Analysis of partially hydrolyzed dextran, a chemically reduced dextran ladder, identified subunit lengths varying between 3–13 GU, with (Glc)6 being the most abundant subunit (Fig. 1A/C). Dextran ladder subunits less than 3 GU were not detected and their loss is attributed to the graphitized carbon clean-up step which has been reported to not retain monosaccharides and some disaccharides.4 As dextran ladder subunits greater than 13 GU were not detected, we hypothesise that an eluent composition greater than 50% (v/v) MeCN/0.1% (v/v) TFA would be required for their elution from the graphitized carbon clean-up. Therefore, this MeCN concentration was purposefully used to maintain the performance of the analytical PGC column by preventing irreversible binding of the longer dextran ladder subunits.
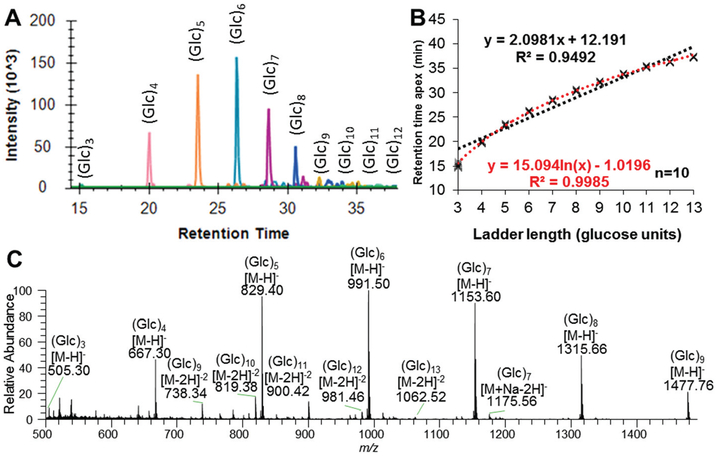
Fig. 1.
Characterization of the dextran ladder using PGC-LC-ESI-MS/MS. (A) Combined extracted ion chromatograms of each observed subunit of the dextran ladder. (B) Best equation fitting of RT for GU assignment (110 data points) with linear equation in black and logarithmic equation in red. (C) Summed MS spectra across the dextran ladder RT range.
With linear gradient elution, the observed retention pattern of the dextran ladder was best described with a logarithmic equation, following the principle of parsimony (Fig. 1B, Table S-1,† R2 > 0.999). The observed RTs of the hydrolyzed dextran (3–13 GU) did not significantly vary over 10 technical replicate injections as evidenced by a median co-efficient of variation (CV) of 0.5% for RT and no observed loss of retention over the 10 analyses/injections (Fig. S-1†). Reproducibility of the analytical platform for MS1-based monoisotopic extracted ion chromatogram (EIC) based quantitation is evidenced by a median peak area CV of 8% (Fig. S-1†). In summary, these data represent the first report of the elution pattern of a dextran ladder separated by PGC-LC and complement a previous report of a logarithmic elution pattern for 2-aminobenzamide labelled dextran ladder by HILIC amide-based chromatography.11 These observed logarithmic trends are in contrast to a linear elution pattern observed for a permethylated dextran ladder separated by C18 reversed phase chromatography. Consequently, these results collectively demonstrate the importance of generating stationary phase-specific normalized retention values.30
Normalizing RT and peak area variation for glycomics
Having established the logarithmic elution pattern of the dextran ladder in PGC-LC, its utility for normalizing RT variation in LC-MS-based glycomics studies was assessed. RT variation was normalized to a GU index using run-specific logarithmic equation corrections based on the dextran ladder internal standard. Evaluating the effectiveness of RT normalization, a batch of 16 samples containing a mixture of paucimannose and high mannose glycans was separated by a linear gradient. An artificial RT shift was introduced by running the last 8 samples with a higher starting organic solvent composition (6% MeCN instead of 1%). As shown in Fig. 2A, this gradient change significantly disrupted glycan RTs. Using GU values instead of retention times for these glycans significantly minimized technical variation (Fig. 2B).
In addition to RT normalization, peak area normalization is also an important step in reducing the technical variation in quantitative glycomic analyses.31 Without normalization, the median co-efficient of variation (CV) in glycan peak areas was 17% (Fig. 2C). Here, the effectiveness of two different post-acquisition normalization methods for reducing peak area technical variation were evaluated using the released glycan mixture with the dextran ladder internal standard. In one strategy, peak area variation of technical replicates was normalized by quantifying glycans based on their EIC peak area as a ratio to the dextran ladder internal standards (Glc4–8). These specific subunits were chosen for normalization as they were the most abundant and the use of multiple point peak area normalization has been demonstrated to be more robust than single point normalization.32
Relative abundance, a normalization method which quantifies individual glycan structures based on their monoisotopic EIC peak area compared to the combined EIC peak areas for all detected glycans, is typically used in LC-MS-based glycomics,4 and more than halved this variation (6% median CV). Quantifying glycans based on their peak area ratios relative to the dextran ladder standards was performed in parallel and found to reduce variation to the same extent (6% median CV).
In contrast to the methods used here, glycan quantitation by relative normalization is a popular strategy for quantifying differences in glycan populations as it does not require internal standards. This form of quantitation partially normalizes technical variation due to each glycan being a percentage of the total glycan signal in a single analytical run.31 However, one inherent limitation of this approach is the inability to quantify changes in the total amount of glycans in a sample, a consideration which may be important for biological studies such as the inhibition of the N-glycosylation pathway.34 Therefore, an advantage of the approach described here is that when peak areas are normalized to internal standards that are added in equal amount to all samples, changes to the total glycan abundance can be monitored.35
Adapting software tools for glycan RT and peak area normalization
Compared to manual approaches, software-based analyses are desirable for automated glycan quantification as it results in improved reproducibility and higher throughput.36 Skyline, an open-source software package for analyzing LC-MS data,27 was used here to normalize both glycan peak areas and RTs. While Skyline is currently limited to linear RT normalization, glycan RT normalization was possible using five point linear calibration (Fig. S-2,† R2 = 0.998). However, until this is formally incorporated within the Skyline platform, glycan GU value assignment can be performed manually for the best possible accuracy. Here, this manual approach was applied using seven point logarithmic calibration and yielded high correlation (Fig. 1B, R2 > 0.999).
In a previous study,11 HILIC-based labelled-glycan separation in conjunction with glycosidase treatment for structure elucidation and RT normalization allowed the observed GU value of a glycan to be a widely applicable parameter for tracking glycan structures within, and among, LC-MS datasets. Here, a similar yet distinct approach was applied. First, glycan structures separated by PGC were elucidated by annotating MS/MS spectra. Subsequently, a glycan structure library with assigned GU values was constructed using Proteowizard22 and Excel (File S-2†). GU value combined with PGC LC separation with subsequent MS/MS acquisition has not previously been described and enables the monitoring of glycan structures regardless of column condition or instrument configuration.
Development of a glycan GU library for PGC-based chromatography
To evaluate the utility of the dextran ladder and normalization approach, and construct a glycan structure library with associated GU values, glycans were released from purified glycoproteins carrying the main classes of N-glycans (high mannose, hybrid, paucimannose, complex) and O-glycans (O-GalNAc multiple core-types and O-mannose). Released glycans were combined with the purified dextran ladder and analyzed by PGC-LC-ESI-MS/MS. From these analyses, 189 N-glycan and 100 O-glycan unique structures were assigned GU values (Table S-2 and File S-2†). Demonstrating a broad range of use, the dextran ladder was suitable for interpolative assignment of GU values for N-glycans from one of the smallest and most hydrophilic paucimannose N-glycans (M3B, 4 GU) up to core-fucosylated tetra-antennary glycans (A4G(4)4F1S4, 13 GU). Notably, several O-glycan structures were outside this range with 5 structures assigned GU values of less than 3 and 4 structures with assigned GU values greater than 13 (Table S-3†). Therefore, the logarithmic elution behaviour of the dextran ladder enables extrapolated GU values for glycans outside 3–13 GU, and serves as a suitable strategy until further improvements that allow detection of an increased dextran ladder length on PGC-LC-ESI-MS/MS are achieved.
To eliminate redundancy in the library, glycans with precursor values within m/z 0.5 and retention values within 0.2 GU of glycan library entries were excluded. In the future, more stringent filtering could be performed with longer elution gradients which would give more accurate RT/GU values and identify glycan structures that are not baseline separated with this current LC elution gradient. In addition to assignment of GU to glycan structures (represented by GlyTouCan accession values28), other important metadata (MIRAGE compliant parameters {LC, MS and sample preparation guidelines37–39}) are included with the glycan library and supplementary files, improving the accessibility of this library to other LC-MS-based platforms for glycomics.37
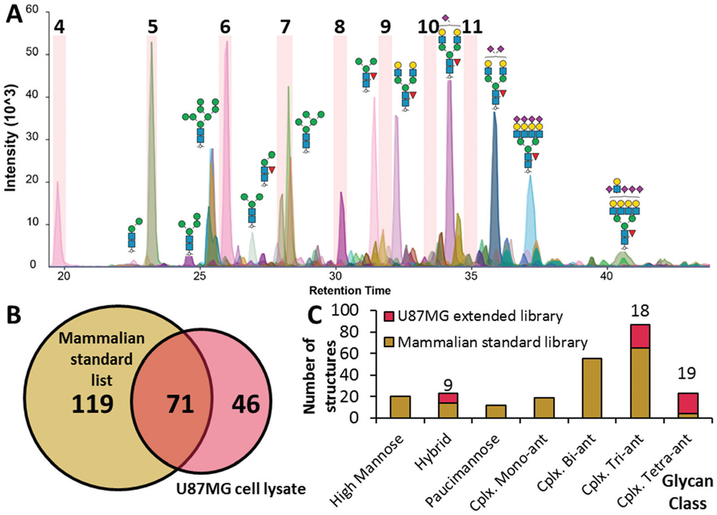
To evaluate utility of this library for analyzing biologically relevant glycan mixtures, N-glycans were released from a total protein extract from U87MG cell line lysate and mixed with the dextran ladder and analyzed by PGC-LC-ESI-MS/MS. After filtering for unique structures and using derived GU values and precursor masses, 71 glycan structures were matched to the standards in the library, utilising 1/3rd of our mammalian GU library (Fig. 3A and B). In parallel to the glycan GU library method, these data were analyzed manually and 46 previously unidentified N-glycanstructures were characterized and identified from the cell lysate sample, which were subsequently added to the existing glycan library. These N-glycanstructures were predominantly complex tri-antennary and tetra-antennary N-glycans which did not exist in our original library due to different combinations of sialic acid linkages and/or the presence of the LacDiNAc epitope which were not detected in the analysis of purified glycoproteins (Fig. 3C).
Fig. 3.
Application of a PGC-based GU library for identification of N-glycans released from a U87MG cell lysate. (A) Extracted ion chromatograms of U87MG glycans mixed with the dextran ladder, only dextran subunits 4–11 highlighted. (B) Coverage of the GU library comprising glycans from purified mammalian glycoprotein standards. (C) Comparison of glycan class coverage following extension of GU library with cell lysate N-glycans.
Altogether, this workflow provides the first PGC-based GU library for automated glycan characterization by LC-MS and, as a result of significantly reducing RT technical variation, the library is applicable for PGC-based LC separations regardless of mass spectrometer and column condition. While GU values obtained using PGC chromatography appear to be suitable for discriminating between glycan isomers (e.g. glycan structures differing in sialic acid linkage), the additional use of MS/MS derived diagnostic ions can further increase confidence in glycan structure assignment.21
Prediction of GU values for experimentally undescribed glycans
Computational methods estimate over one million glycan structures are possible from the N-glycosylation biosynthetic pathway40 but fewer than 4000 N-glycanstructures (GlyConnect,41 accessed 2019-01-26) have been described experimentally. Importantly, a composition-based (precursor m/z only) approach for experimental detection is not sufficient to bridge this gap, as a significant number of these undetected N-glycanstructures are isomers of glycans that have been identified experimentally (an example is shown in Table S-4†).
To facilitate the identification of these undetected glycan structures, expected GU value prediction for these undetected glycan structures can inform the experimental search space and provide additional confidence in the identification of new glycan structures by demonstrating when observed glycans, represented by m/z, GU value and MS/MS spectra, do not match experimentally described glycan structures. Demonstrating a proof of concept for GU glycan structure prediction using our glycan library, the derived GU values for N-glycans with and without substructural features, core-fucose and bisecting GlcNAc, were compared and strong linear correlative trends were observed (Fig. 4, R2 > 0.9). By using the experimentally derived GU values of related biosynthetic structures for GU prediction of undetected structures, our models account for experimental variation which is an expected contributing factor for our observed imperfect correlation values.
Fig. 4.
Identification of equations to predict glycan GU values by correlation of base structure GU value with (top) bisected and (bottom) core-fucosylated glycan structures.
Development of a dextran ladder spectral library
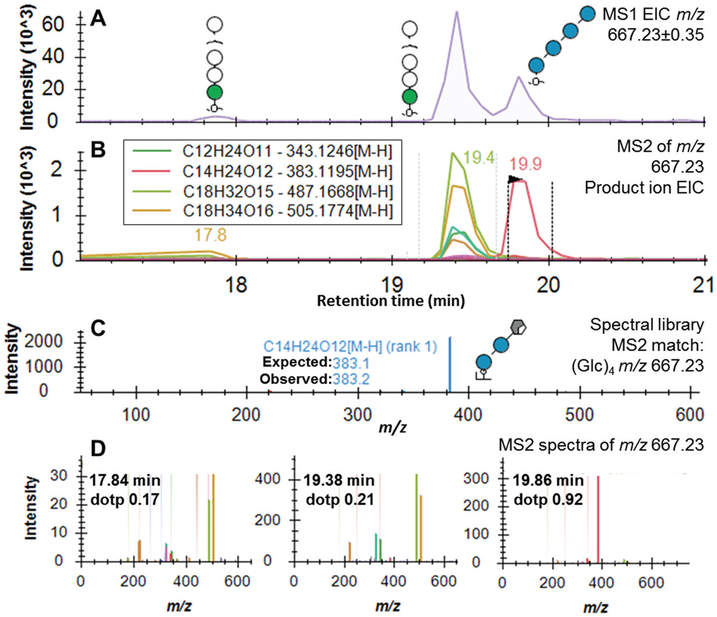
A significant challenge in using the dextran ladder in isomer-containing mixtures is the assignment of the GU ladder to the correct isomeric peaks, given that MS1-level information is not sufficient to confidently assign structural identity. To resolve this, we developed a spectral library of the 3–13 units of the dextran ladder. To assess the spectral library approach, a challenging sample containing the dextran ladder mixed with O-mannose glycans from cellobiohydrolase I was selected due to the mannosidic glycans being isomeric with the dextran ladder.17 As shown in Fig. 5A, three peaks were detected with masses consistent with a composition of a hexose-based tetrasaccharide (m/z 667.23 ± 0.35) and, as multiple MS2 scans were collected for each isomer, extracted ion chromatograms for each theoretically expected product ion were constructed (Fig. 5B). Based on the MS2 spectra acquired when performing characterization of the pure dextran ladder, an annotated spectral library was generated (Fig. 5C). Of the three glycan peaks, the spectral matching feature in Skyline identified the peak at 19.9 min to be the most probable match to the dextran ladder library. The strong match between the peak at 19.9 minutes (dotp 0.92) and our spectral library (Fig. 5C) was supported by a single prominent peak at m/z 383.1 representing a cross-ring fragment of a glucose tetramer which is specific for dextran rather than the isomeric O-mannosylated glycans.42 The same correct peak picking was achieved for all other hexose isomers ((Hex)3, (Hex)4, (Hex)5 and (Hex)6) due to the characteristic cross-ring fragmentation pattern of the dextran glucose (Fig. S-3†).
Fig. 5.
Automated glucose-based dextran ladder peak assignment in a glycan sample containing isomeric mannose-based glycans using a spectral library approach. (A) MS1 extracted ion chromatograms for the glycan composition (Hex)4 m/z 667.2302 [M − H]−, structures identified. (B) MS2 extracted ion chromatograms of abundant product ions for the glycan composition defined in (A). (C) Annotated MS2 spectra of dextran standard, (Glc)4 m/z 667.2302 [M − H]− for use in spectral matching. (D) Isomeric (Hex)4 MS2 spectra with spectral similarity score shown as dotp value.
The assigned dotP score, as defined by Toprak et al.,43 uses the normalized spectral contrast angle comparison between library and unknown spectra and has previously been used successfully for targeted peptide spectral matching. The normalized spectral contrast angle comparison was found to sensitively quantify the similarity of peptide spectra and serves as a useful foundation for future studies in spectral matching methods. In summary, the development of an annotated spectral library, combined with the unique fragmentation pattern of hydrolyzed dextran, allows automated assignment of the dextran ladder subunits to discrete RTs without requiring manual peak selection, even in challenging isomeric mixtures.
Application of the approach for glycomic analysis of complex biological samples
To demonstrate the application of this glycan GU library for glycan structure profiling of complex biological samples, protein extracts from cultured cells, proteins secreted by these cell lines, and central nervous system tissue (brain and spinal cord) (Table S-5†) were subjected to the previously described analytical method, including use of the dextran ladder internal standard. Samples were analyzed in parallel to provide a quantitative comparison among sample types. In these examples, the dextran ladder spectral library was used to automatically derive RT values for 4–9 GU with only 7% of peaks requiring manual intervention.
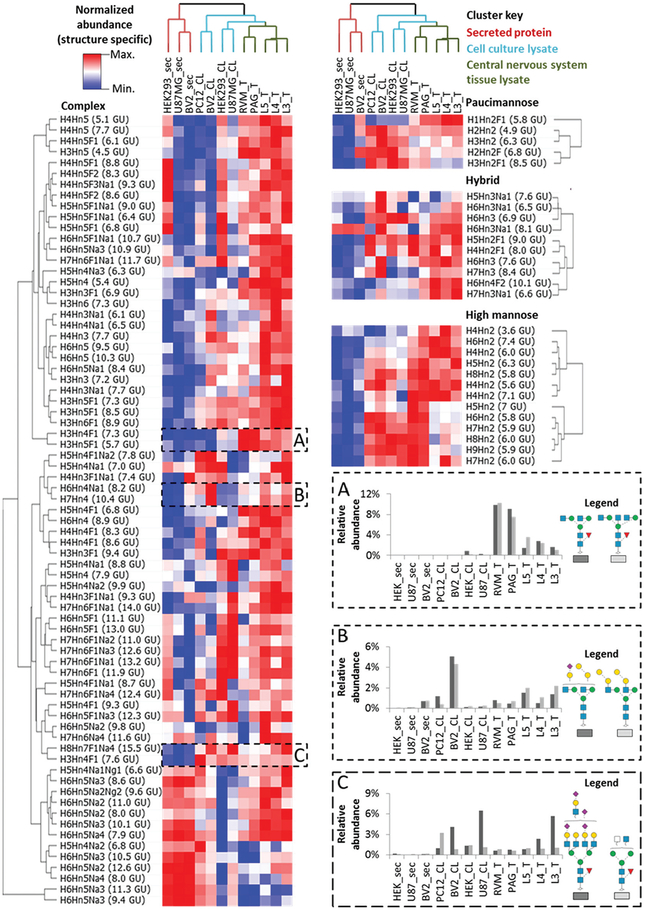
The glycan GU library was used to quantify observed N-glycans (a total of 101 N-glycans quantified across all samples, normalized peak areas for each structure can be found in ESI file S-4†), and the dextran ladder was used to normalize their RTs and peak areas. The normalized glycan profiles of each sample type was sorted by hierarchical clustering and N-glycanstructures alone were found to be sufficient for unsupervised discrimination of sample types (Fig. 6).
Fig. 6.
Hierarchical clustered heatmap of glycan abundances from a wide variety of biological samples. Glycan structures are clustered by class, with the most abundant glycan for each class represented in notation (H = Hex, Hn = HexNAc, F = Fucose, Na = NeuAc and Ng = NeuGc) followed by observed GU value. (A) Complex N-glycans detected in tissue samples. (B) Di-galactosylated N-glycans. (C) Complex N-glycans not detected in secreted glycoprotein samples.
In this evaluation of sample specific N-glycans, glycans derived from secreted proteins demonstrated distinct glycan features such as a relatively low amount of high mannose, hybrid and paucimannose structures, as well as some complex structures (Fig. 6C). Lysate from cultured cells contains relatively high levels of most paucimannosidic N-glycanstructures. The cultured cells also have similar relative abundances for some large (Man6–Man9) high mannose N-glycanstructures compared to brain-derived mouse tissue sections although not compared to spinal cord tissue sections. As two of these cell lines, U87MG and BV2, represent cell types found in the brain, these data suggest that select N-glycanstructures may be shared between immortalized cell lines and the respective tissue regions from where the primary cells originate.
In addition to glycan structures that are shared between cell lines and tissue, many N-glycans were found in greater abundance in CNS tissue compared to their representative cultured cell types (Fig. 6A). One such structure, H3Hn4F1 7.3 GU, has a closely related isomer at 7.6 GU that is not only found in tissue samples. Thus, the ability to resolve these two glycan structures with different sample specificities justifies the value of a structure-specific approach as opposed to composition only. These structures, in addition to the larger high mannose glycan structures mentioned earlier, contribute to the clustering of tissue sections-based on their CNS location (e.g. PAG/RVM or L3/L4/L5). Additionally, these analyses detected glycans possessing an alpha-galactose motif in secreted, cell line and tissue samples, which have not been found in human and old world primate samples (Fig. 6B).44 This structure contributed to the mouse (BV2) and rat cell lysate samples (PC12) being clustered together. Importantly, the inability to detect significant amounts of these structures in the secreted protein samples also provides confirmation that these glycan profiles are not complicated by the presence of animal products used in cell culture, such as fetal bovine serum, which has been reported to contain glycans with the alpha-galactose motif.45
Conclusions
These results demonstrate the first example of system-independent assignment of glycan structures based only on precursor mass and GU value using PGC-based LC-MS, with orthogonal confirmation by MS/MS. Initial characterization of the dextran ladder internal standard uncovered a logarithmic elution pattern and this was exploited to construct a glycan library from glycan structures released from mammalian glycoprotein standards. RT and peak area normalization to the dextran ladder internal standard achieved a significant minimalization in technical variation and enabled the development of algorithms for predicting the retention properties of unobserved glycan structures based on the retention properties of structurally similar structures.
To improve the robustness of this normalization strategy, we developed an annotated spectral library which exploits the unique fragmentation pattern of the dextran ladder to automate peak assignment and subsequent RT normalization, even in challenging isomeric mixtures. Finally, we applied this glycan GU library for structural profiling of complex biological samples which was found to effectively discriminate between samples types (cultured cells, secreted proteins and CNS tissue). As a result of the library’s open-access structure, it can be further extended into a comprehensive glycan structure library for automated glycan identification and quantitation. Although in some cases assigned glycan structures are partially ambiguous, continuing efforts in glycan characterization by alternative MS/MS modes46 and ion-mobility/spectroscopy47 in combination with our described normalization techniques will allow these partially ambiguous glycan structures to be tracked across laboratories and methods.
Supplementary Material
Acknowledgements
This work was supported by the Australian Research Council Centre of Excellence for Nanoscale Biophotonics (CE140100003). Portions of this research were supported by grants from the National Institute of General Medical Sciences (R01 GM103551) and National Heart Lung and Blood Institute (R01 HL134010). We thank M. R. Hutchinson and V. Staikopoulos for providing mouse tissue sections.
Footnotes
Ethical statement
1. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of the University of Adelaide and approved by the Animal Ethics Committee of the University of Adelaide.
2. All cell lines were originally from the ATCC. All of these cell lines were then cultured at Macquarie University, Australia.
Conflicts of interest
There are no conflicts to declare.
Electronic supplementary information (ESI) available: Complete LC-MS method parameters are provided in file S-1 in addition to all supplementary figures and tables. The glycan GU libraries are available in file S-2. The raw data files, Skyline assays, glycan GU libraries and the annotated dextran ladder spectral library can be found on Panorama Public (https://panoramaweb.org/GlycoGU.url). Glycan MS2 spectra annotated by GlycoWorkBench are found in file S-3. In accordance with MIRAGE guidelines for liquid chromatography, mass spectrometry, sample preparation reporting37–39 are provided in file S-5.
References
- 1.Rudd PM, Rudan I and Wright AF, High-Throughput Glycome Analysis Is Set to Join High-Throughput Genomics, J. Proteome Res, 2009, 8(3), 1105, DOI: 10.1021/pr900040s. [DOI] [PubMed] [Google Scholar]
- 2.Ashwood C, Shathili AMM and Packer NH, Human Disease Glycomics: Technology Advances Enabling Protein Glycosylation Analysis–part 2, Expert Rev. Proteomics, 2018, 15(4), 341–352, DOI: 10.1080/14789450.2018.1421946. [DOI] [PubMed] [Google Scholar]
- 3.Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, Kim YG, Henry GW, Shadick NA, Weinblatt ME, et al. , HPLC-Based Analysis of Serum N-glycans on a 96-Well Plate Platform with Dedicated Database Software, Anal. Biochem, 2008, 376(1), 1–12, DOI: 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Jensen PH, Karlsson NG, Kolarich D and Packer NH, Structural Analysis of N- and O-Glycans Released from Glycoproteins, Nat. Protoc, 2012, 7, 1299–1310, DOI: 10.1038/nprot.2012.063. [DOI] [PubMed] [Google Scholar]
- 5.Melmer M, Stangler T, Premstaller A and Lindner W, Comparison of Hydrophilic-Interaction, Reversed-Phase and Porous Graphitic Carbon Chromatography for Glycan Analysis, J. Chromatogr. A, 2011, 1218(1), 118–123, DOI: 10.1016/j.chroma.2010.10.122. [DOI] [PubMed] [Google Scholar]
- 6.Pabst M, Bondili JS, Stadlmann J, Mach L and Altmann F, Mass+Retention Time=Structure: A Strategy for the Analysis of N-glycans by Carbon LC-ESI-MS and Its Application to Fibrin N-glycans, Anal. Chem, 2007, 79, 5051–5057, DOI: 10.1021/ac070363i. [DOI] [PubMed] [Google Scholar]
- 7.Pabst M and Altmann F, Influence of Electrosorption, Solvent, Temperature, and Ion Polarity on the Performance of LC-ESI-MS Using Graphitic Carbon for Acidic Oligosaccharides, Anal. Chem, 2008, 80, 7534–7542, DOI: 10.1021/ac801024r. [DOI] [PubMed] [Google Scholar]
- 8.Aldredge D, An HJ, Tang N, Waddell K and Lebrilla CB, Annotation of a Serum N-glycan Library for Rapid Identification of Structures, J. Proteome Res, 2012, 11(3), 1958–1968, DOI: 10.1021/pr2011439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abrahams JL, Campbell MP and Packer NH, Building a PGC-LC-MS N-glycan Retention Library and Elution Mapping Resource, Glycoconjugate J, 2018, 35(1), 15–29, DOI: 10.1007/s10719-017-9793-4. [DOI] [PubMed] [Google Scholar]
- 10.Zhao S, Walsh I, Abrahams JL, Royle L, Nguyen-Khuong T, Spencer D, Fernandes DL, Packer NH, Rudd PM and Campbell MP, GlycoStore: A Database of Retention Properties for Glycan Analysis, Bioinformatics, 2018, 2008(April), 1–2, DOI: 10.1093/bioinformatics/bty319. [DOI] [PubMed] [Google Scholar]
- 11.Guile GR, Rudd PM, Wing DR, Prime SB and Dwek RA, A Rapid High-Resolution High-Performance Liquid Chromatographic Method for Separating Glycan Mixtures and Analyzing Oligosaccharide Profiles, Anal. Biochem, 1996, 240(2), 210–226, DOI: 10.1006/abio.1996.0351. [DOI] [PubMed] [Google Scholar]
- 12.Luo Q, Rejtar T, Wu S-L and Karger BL, Hydrophilic Interaction 10 Mm I.D. Porous Layer Open Tubular Columns for Ultratrace Glycan Analysis by Liquid Chromatography-Mass Spectrometry, J. Chromatogr. A, 2009, 1216(8), 1223–1231, DOI: 10.1016/j.chroma.2008.09.105Hydrophilic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn J, Bones J, Yu YQ, Rudd PM and Gilar M, Separation of 2-Aminobenzamide Labeled Glycans Using Hydrophilic Interaction Chromatography Columns Packed with 1.7 Mm Sorbent, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci, 2010, 878(3–4), 403–408, DOI: 10.1016/j.jchromb.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Stockmann H, Adamczyk B, Hayes J and Rudd PM, Automated, High-Throughput IgG-Antibody Glycoprofiling Platform, Anal. Chem, 2013, 85(18), 8841–8849, DOI: 10.1021/ac402068r. [DOI] [PubMed] [Google Scholar]
- 15.Bundle DR, Jennings HJ and Kenny CP, Studies on the Group-Specific Polysaccharide of Neisseria Meningitidis Serogroup X and an Improved Procedure for Its Isolation, J. Biol. Chem, 1974, 249(15), 4797–4801. [PubMed] [Google Scholar]
- 16.Campbell MP, Royle L, Radcliffe CM, Dwek RA and Rudd PM, GlycoBase and AutoGU: Tools for HPLC-Based Glycan Analysis, Bioinformatics, 2008, 24(9), 1214–1216, DOI: 10.1093/bioinformatics/btn090. [DOI] [PubMed] [Google Scholar]
- 17.Ashwood C, Abrahams JL, Nevalainen H and Packer NH, Enhancing Structural Characterisation of Glucuronidated O -Linked Glycans Using Negative Mode Ion Trap-HCD Mass Spectrometry, Rapid Commun. Mass Spectrom, 2017, 31, 851–858, DOI: 10.1002/rcm.7851. [DOI] [PubMed] [Google Scholar]
- 18.Nanodrop P, Bradford Protein Assay, Methods, 1976, 2003, 1–4, DOI: 10.1101/pdb.prodprot15. [DOI] [Google Scholar]
- 19.Schneider P and Ferguson MAJ, Microscale Analysis of Glycosylphosphatidylinositol Structures, Methods Enzymol, 1995, 250(C), 614–630, DOI: 10.1016/0076-6879(95)50100-2. [DOI] [PubMed] [Google Scholar]
- 20.Masuko T, Minami A, Iwasaki N, Majima T, Nishimura SI and Lee YC, Carbohydrate Analysis by a Phenol-Sulfuric Acid Method in Microplate Format, Anal. Biochem, 2005, 339(1), 69–72, DOI: 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Ashwood C, Lin C-H, Thaysen-Andersen M and Packer NH, Discrimination of Isomers of Released N- and O-Glycans Using Diagnostic Product Ions in Negative Ion PGC-LC-ESI-MS/MS, J. Am. Soc. Mass Spectrom, 2018, 29(6), 1194–1209, DOI: 10.1007/s13361-018-1932-z. [DOI] [PubMed] [Google Scholar]
- 22.Kessner D, Chambers M, Burke R, Agus D and Mallick P, ProteoWizard: Open Source Software for Rapid Proteomics Tools Development, Bioinformatics, 2008, 24(21), 2534–2536, DOI: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper CA, Gasteiger E and Packer NH, GlycoMod–a Software Tool for Determining Glycosylation Compositions from Mass Spectrometric Data, Proteomics, 2001, 1, 340–349, DOI: . [DOI] [PubMed] [Google Scholar]
- 24.Ceroni A, Maass K, Geyer H, Geyer R, Dell A and Haslam SM, GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans, J. Proteome Res, 2008, 7, 1650–1659, DOI: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 25.Hayes CA, Karlsson NG, Struwe WB, Lisacek F, Rudd PM, Packer NH and Campbell MP, UniCarb-DB: A Database Resource for Glycomic Discovery, Bioinformatics, 2011, 27(9), 1343–1344, DOI: 10.1093/bioinformatics/btr137. [DOI] [PubMed] [Google Scholar]
- 26.Jin C, Kenny DT, Skoog EC, Padra M, Adamczyk B, Vitizeva V, Thorell A, Venkatakrishnan V, Lindén SK and Karlsson NG, Structural Diversity of Human Gastric Mucin Glycans, Mol. Cell. Proteomics, 2017, 16(5), 743–758, DOI: 10.1074/mcp.M117.067983. [DOI] [PubMed] [Google Scholar]
- 27.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC and MacCoss MJ, Skyline: An Open Source Document Editor for Creating and Analyzing Targeted Proteomics Experiments, Bioinformatics, 2010, 26(7), 966–968, DOI: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiemeyer M, Aoki K, Paulson J, Cummings RD, York WS, Karlsson NG, Lisacek F, Packer NH, Campbell MP, Aoki NP, et al. , GlyTouCan: An Accessible Glycan Structure Repository, Glycobiology, 2017, 27(10), 915–919, DOI: 10.1093/glycob/cwx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frewen B and MacCoss MJ, Using BiblioSpec for Creating and Searching Tandem MS Peptide Libraries, Curr. Protoc. Bioinf, 2007, 1–12, DOI: 10.1002/0471250953.bi1307s20. [DOI] [PubMed] [Google Scholar]
- 30.Zhou S, Hu Y and Mechref Y, High-Temperature LC-MS/MS of Permethylated Glycans Derived from Glycoproteins, Electrophoresis, 2016, 37(11), 1506–1513, DOI: 10.1002/elps.201500568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moh ESX, Thaysen-Andersen M and Packer NH, Relative versus Absolute Quantitation in Disease Glycomics, Proteomics: Clin. Appl, 2015, 9(3–4), 368–382, DOI: 10.1002/prca.201400184. [DOI] [PubMed] [Google Scholar]
- 32.Mizuno H, Ueda K, Kobayashi Y, Tsuyama N, Todoroki K, Min JZ and Toyo’oka T, The Great Importance of Normalization of LC–MS Data for Highly-Accurate Non-Targeted Metabolomics, Biomed. Chromatogr, 2017, 31(1), 1–7, DOI: 10.1002/bmc.3864. [DOI] [PubMed] [Google Scholar]
- 33.Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, et al. , Symbol Nomenclature for Graphical Representations of Glycans, Glycobiology, 2015, 25(12), 1323–1324, DOI: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elbein A, Glycosylation Inhibitors for N-Linked Glycoproteins, in Methods in Enzymology, 1987, vol. 138, pp. 661–709. [DOI] [PubMed] [Google Scholar]
- 35.Zaia J, Mass Spectrometry and the Emerging Field of Glycomics, Chem. Biol, 2008, 881–892, DOI: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shubhakar A, Reiding KR, Gardner RA, Spencer DIR, Fernandes DL and Wuhrer M, High-Throughput Analysis and Automation for Glycomics Studies, Chromatographia, 2014, 78(5–6), 321–333, DOI: 10.1007/s10337-014-2803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Struwe WB, Agravat S, Aoki-Kinoshita KF, Campbell MP, Costello CE, Dell A, Feizi T, Haslam SM, Karlsson NG, Khoo KH, et al. , The Minimum Information Required for a Glycomics Experiment (MIRAGE) Project: Sample Preparation Guidelines for Reliable Reporting of Glycomics Datasets, Glycobiology, 2016, 26(9), 907–910, DOI: 10.1093/glycob/cww082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J, McBride R, Agravat S, Campbell MP, Kato M, Ranzinger R, et al. , The Minimum Information Required for a Glycomics Experiment (MIRAGE) Project: Improving the Standards for Reporting Mass-Spectrometry-Based Glycoanalytic Data, Mol. Cell. Proteomics, 2013, 12, 991–995, DOI: 10.1074/mcp.O112.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell MP, Abrahams JL, Rapp E, Struwe WB, Costello CE, Novotny M, Ranzinger R, York WS, Kolarich D, Rudd PM, et al. , The Minimum Information Required for a Glycomics Experiment (MIRAGE) Project: LC Guidelines, Glycobiology, 2019, 29(5), 349–354, DOI: 10.3762/mirage.2. [DOI] [PubMed] [Google Scholar]
- 40.Akune Y, Lin CH, Abrahams JL, Zhang J, Packer NH, Aoki-Kinoshita KF and Campbell MP, Comprehensive Analysis of the N-glycan Biosynthetic Pathway Using Bioinformatics to Generate UniCorn: A Theoretical N-glycanstructure Database, Carbohydr. Res, 2016, 431, 56–63, DOI: 10.1016/j.carres.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Alocci D, Mariethoz J, Gastaldello A, Gasteiger E, Karlsson NG, Kolarich D, Packer NH and Lisacek F, GlyConnect : Glycoproteomics Goes Visual, Interactive and Analytical, J. Proteome Res, 2019, 18(2), 664–667, DOI: 10.1021/acs.jproteome.8b00766. [DOI] [PubMed] [Google Scholar]
- 42.Čmelík R, Štikarovská M and Chmelík J, Different Behavior of Dextrans in Positive-Ion and Negative-Ion Mass Spectrometry, J. Mass Spectrom, 2004, 39(12), 1467–1473, DOI: 10.1002/jms.763. [DOI] [PubMed] [Google Scholar]
- 43.Toprak UH, Gillet LC, Maiolica A, Navarro P, Leitner A and Aebersold R, Conserved Peptide Fragmentation as a Benchmarking Tool for Mass Spectrometers and a Discriminating Feature for Targeted Proteomics, Mol. Cell. Proteomics, 2014, 13(8), 2056–2071, DOI: 10.1074/mcp.O113.036475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinke JW, Platts-mills TAE and Commins SP, The Alpha Gal Story: Lessons Learned from Connecting the Dots, J. Allergy Clin. Immunol, 2015, 135(3), 589–597, DOI: 10.1016/j.jaci.2014.12.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langeveld JPM, Noelken ME, Hard K, Todd P, Vliegenthart JFG, Rouse J and Hudson BG, Bovine Glomerular Basement Membrane: Location and Structure of the Asparagine-Linked Oligosaccharide Units, J. Biol. Chem, 1991, 266(4), 2622–2631. [PubMed] [Google Scholar]
- 46.Tang Y, Wei J, Costello CE and Lin C, Characterization of Isomeric Glycans by Reversed Phase Liquid Chromatography-Electronic Excitation Dissociation Tandem Mass Spectrometry, J. Am. Soc. Mass Spectrom, 2018, 1–13, DOI: 10.1007/s13361-018-1943-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manz C and Pagel K, Glycan Analysis by Ion Mobility-Mass Spectrometry and Gas-Phase Spectroscopy, Curr. Opin. Chem. Biol, 2018, 42, 16–24, DOI: 10.1016/j.cbpa.2017.10.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.