Abstract
Xist is an X-linked ribonucleic acid (RNA) gene responsible for the cis induction of X chromosome inactivation (XCI). In cloned mammalian embryos, Xist is ectopically activated at the morula to blastocyst stage on the X chromosome that is supposed to be active, thus resulting in abnormal XCI. Suppression of erroneous Xist expression by injecting small interfering RNA (siRNA) remarkably increased the developmental efficiency of cloned male mouse embryos by approximately 10-fold. However, injection of anti-Xist siRNA resulted in only a slight increase in the developmental ability of injected cloned male pig embryos because the blocking effect of the injected siRNA was not maintained beyond the morula stage, which is 5 days post-activation. To develop a more effective approach for suppressing the ectopic expression of Xist in cloned pig embryos, we compared the silencing effect of short hairpin RNA (shRNA) and siRNA on Xist expression and the effects of these two Xist knockdown methods on the developmental competence of cloned male pig embryos. Results indicated that an shRNA-based RNA interference (RNAi) has a longer blocking effect on Xist expression than an siRNA-mediated RNAi. Injection of anti-Xist shRNA plasmid into two-cell-stage cloned male pig embryos effectively suppressed Xist expression, rescued XCI at the blastocyst stage, and improved the in vitro developmental ability of injected cloned embryos. These positive effects, however, were not observed in cloned male pig embryos injected with anti-Xist siRNA. This study demonstrates that vector-based rather than siRNA-mediated RNAi of Xist expression can be employed to improve pig cloning efficiency.
Keywords: Cloned, Pig, Ribonucleic acid interference (RNAi), Somatic cell nuclear transfer (SCNT), Xist
Xist is an X-linked noncoding RNA gene responsible for the cis induction of mammalian X chromosome inactivation (XCI) [1, 2]. To equalize X-linked gene dosage between male and female mammals, Xist-triggered imprinted and random XCI only occur in females but not in males during normal development [3,4,5]. Therefore, normal Xist expression is vital for mammalian development.
In cloned mammalian embryos or animals generated by somatic cell nuclear transfer (SCNT), the Xist gene exhibits an aberrant expression pattern [6,7,8,9,10,11]. The ectopic expression of the Xist gene on the putative active X chromosome was observed in both male and female mouse SCNT embryos, which resulted in a large-scale downregulation of X-linked genes resembling XCI [11]. Suppression of aberrant Xist expression by deletion of the Xist allele on the putative active X chromosome not only abolished the dysregulation of X-linked genes, but also resulted in an eight- to nine-fold increase in full-term developmental efficiency of mouse SCNT embryos [11]. Inhibition of erroneous Xist expression in early cloned male mouse embryos via injection of Xist-specific small interfering RNA (siRNA) resulted in a 10-fold improvement of cloning efficiency [12].
Previous studies suggested that maternal Xist alleles are aberrantly activated in cloned pig embryos or fetuses because they have a significantly higher Xist mRNA level than in vivo fertilization-derived counterparts [13, 14, 27]. Suppression of aberrant Xist expression by knockout of Xist significantly enhanced the developmental competence of cloned male pig embryos [27]. However, the injection of anti-Xist siRNA into one-cell-stage male pig SCNT embryos resulted in only a slight increase in the developmental ability of injected SCNT embryos [14]. This is because the blocking effect of injected siRNA on Xist expression could not be maintained beyond the morula stage (at 5 days post-activation), at which Xist starts to be ectopically activated in cloned pig embryos [14].
Short hairpin RNA (shRNA) expression plasmid-based RNA interference (RNAi) can provide more persistent and stable gene silencing than siRNA-mediated RNAi [15,16,17,18]. To investigate a more effective method to repress ectopic expression of Xist and to improve pig cloning efficiency, in this study, we (i) compared the silencing effect of shRNA and siRNA on Xist expression in cloned male pig embryos, and (ii) investigated the effects of these two Xist knockdown methods on the developmental competence of male pig SCNT embryos.
Materials and Methods
Ethics statements
This study was performed in strict accordance with the regulations of the Instructive Notions with Respect to Caring for Laboratory Animals issued by the Ministry of Science and Technology of China. The animal experiment protocol was approved by the Institutional Animal Care and Use Committee of South China Agricultural University. All efforts were made to minimize the suffering of the tested animals.
Preparation of siRNAs and chemically modified siRNA
Three siRNA duplexes were designed according to the cDNA sequence of porcine Xist gene and synthesized by the GenePharma Company (Suzhou, China). Their sequences are shown in Table 1. Chemically modified siRNA1 (CM-siRNA1) and negative control siRNA (NC-siRNA) were synthesized by GenePharma Company as well. The anti-Xist siRNA was modified by two types of chemical modifications, including 5’-Chol modification at the 5’ end of the sense strand and 2’-OMe modification at position 2 of the antisense strand.
Table 1. Sequences of three designed siRNA duplexes targeting porcine Xist gene.
| Sense strand | Antisense strand | |
|---|---|---|
| siRNA1 | 5′- GCAUCUGACUGUUAUGUUUTT -3′ | 5′- AAACAUAACAGUCAGAUGCTT -3′ |
| siRNA2 | 5′- GCAUGUGCCCUCGUGAUAATT -3′ | 5′- UUAUCACGAGGGCACAUGCTT -3′ |
| siRNA3 | 5′- CCACAAGACUGUUAAGUUUTT -3′ | 5′- AAACUUAACAGUCUUGUGGTT -3′ |
Construction of shRNA expression plasmid
An anti-porcine Xist shRNA fragment was synthesized in accordance with the sequences of anti-Xist siRNA1 and inserted into multiple cloning sites between BbsI and BamHI of the supersilencing™ shRNA expression plasmid (GenePharma Company) to generate the pU6-shRNA plasmid (Fig. 3).
Fig. 3.
Construction of anti-Xist shRNA expression plasmid. A: Structural illustration of anti-Xist shRNA expression plasmid. B: Partial sequencing results of anti-Xist shRNA expression plasmid.
Transfection
Female porcine kidney (PK-21) cells were grown at 37°C in Dulbecco’s modified Eagle medium (Gibco, Grand Island, NY) containing 10% fetal bovine serum (Gibco). The PK-21 cells were seeded into 24-well plates at a density of 0.5–2.0 × 105 cells/well with fresh medium (500 µl/well) without antibiotics, 24 h prior to transfection.
The PK-21 cells were transfected with siRNA/CM-siRNA (40 pmol) or pU6-shRNA plasmid (40 pmol) using Lipofectamine RNAi MAX Reagent (Invitrogen, Carlsbad, CA) in accordance with the manufacturer’s instructions.
Real-time quantitative PCR (qPCR)
The total RNA was isolated from the transfected cells or microinjected embryos using an RNeasyPlus Micro Kit (Qiagen, Gaithersburg, MD). The cDNA was synthesized using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Tokyo, Japan). SYBR Premix Ex Taq (TaKaRa) and Eco™ Real-time PCR system (Illumine, San Diego, CA) were used for qPCR. All PCR runs were performed at an annealing temperature of 60°C for 50 cycles. The sequences of the primers used in this study are shown in Table 3.
Table 3. Sequences of the primers used for real-time qPCR.
| Gene name | Forward primers | Reverse primers |
|---|---|---|
| Xist | 5′-CTTGCCGCAATCGAAAACAT-3′ | 5′-ACCAATTCCACCACCCTTTC-3′ |
| Clic2 | 5′-GGGCTGTAACCTCTTTGCCA-3′ | 5′-AACCGTGAGTTCCTCAGCAC-3′ |
| Tbliy | 5′-TGCAGCACGGACATGTGTAT-3′ | 5′-GTTCCAGCACACCTCGAAGA-3′ |
| Tlr7 | 5′-CTGGAGGCATTCCCACCAAT-3′ | 5′-GCTGGAGTGATGCTCGCTAT-3′ |
| Las1l | 5′-AAGGTGCAGAGCTGGATGTC-3′ | 5′-AGCAGTTCTCGGGCTCTTTC-3′ |
| Tsr2 | 5′-CTGCAGAGGAGTCTCTAACGC-3′ | 5′-GTGGCGGAACATGGTCTGTA-3′ |
| Gapdh | 5′-TGACCCCTTCATTGACCTCC-3′ | 5′-CTCCGCCTTGACTGTGCC-3′ |
| β-actin | 5′-CCACGAGACCACCTTCAACTC-3′ | 5′-TGATCTCCTTCTGCATCCTGT-3′ |
Somatic cell nuclear transfer
The male SCNT embryos were produced as previously described [19]. Briefly, porcine ovaries were purchased from the Guangzhou Tianhe slaughterhouse located at Tianhe District, Guangzhou City, Guangdong Province, China. Cumulus–oocyte complexes (COCs) were aspirated from the ovaries and matured in vitro for 42–44 h. Matured COCs were freed from cumulus cells by repeated pipetting in 0.10% hyaluronidase. In-vitro-matured oocytes with the first polar body were selected for enucleation. The mature oocyte was aspirated firmly into a holding pipette (outer diameter = 100–120 µm, inner diameter = 20–30 µm) to ensure immobility. The enucleation pipette (inner diameter = 15 µm) was inserted through the zona pellucida. The first polar body and adjacent cytoplasm, which were presumed to contain the entire chromosome, were aspirated into the enucleation pipette. After the fibroblast cells were digested with trypsin, a single fibroblast cell was separated by pipetting and then microinjected into the perivitelline space of the oocytes. The oocyte–donor cell complexes were cultured in PZM3 at 39°C, 5% CO2, 5% O2, 90% N2, and 100% humidity for 1.5 h. The cell complexes were activated to fuse in a medium containing 250 mM of mannitol, 0.1 mM of CaCl2·2H2O, 0.1 mM of MgCl2·6H2O, 0.5 of mM HEPES, and 0.01% polyvinyl alcohol through two successive abbr (DC) pulses each at 1.2 kV/cm for 30 µsec using an electrofusion instrument (model: CF-150/B, Biological Laboratory Equipment Maintenance and Service, Budapest, Hungary). The activated cloned embryos were then cultured in PZM3 containing cytochalasin B (5 µg/ml) for 4 h. After the post-activation treatment, the reconstructed embryos were cultured in PZM3 at 39°C, 5% CO2, 7% O2, 88% N2, and 100% humidity.
Microinjection
Microinjections of shRNA plasmid and siRNA1 were performed using a micropipette driven by the Piezo (PiezoXpert, Eppendorf, Germany). Ten picoliters of 5 or 50 µM siRNA and 5 ng/µl or 10 ng/µl anti-Xist shRNA plasmid was injected into the cytoplasm of each blastomere of two-cell-stage SCNT embryos. Control SCNT embryos were injected with 10 pl of water (Sigma, St. Louis, MO, USA). The injected embryos were cultured in vitro in PZM3 at 39°C, 5% CO2, 7% O2, 88% N2, and 100% humidity.
Immunofluorescence
Injected male pig embryos were collected at the blastocyst stage, washed 3–5 times in PBS, and fixed in Immunol Staining Fix Solution (Beyotime Biotechnology Company, Shanghai, China) for 15 min by incubation at 25°C. After permeabilization using Immunostaining Permeabilization Buffer with Triton X-100 (Beyotime Biotechnology Company) for 30 min, the embryos were blocked in the Immunol Staining Blocking Buffer (Beyotime Biotechnology Company) for 1 h at room temperature and washed 3–5 times followed by an overnight incubation at 4°C with primary antibodies against H3K27me3 (1:200; Thermo Fisher Scientific, Waltham, MA, USA A15024). After 3–5 washes with PBS, the embryos were incubated at room temperature and shielded from light for 1 h with Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody (1:500; Thermo Fisher Scientific, A11008). After washing 3–5 times in PBS, DNA staining was performed using DAPI (Thermo Fisher Scientific) for 5 min at room temperature and shielded from light. After washing, the embryos were mounted on glass slides and observed using an inverted fluorescence microscope (Nikon, Tokyo, Japan).
Statistical analysis
Data were analyzed using the SPSS software version 20 (SPSS, Chicago, IL, USA). The relative transcription levels determined by qPCR were analyzed by the Student’s t-test for comparing group means. One-way ANOVA followed by Tukey’s post hoc test was performed to determine the difference between the developmental rates of five groups of microinjected SCNT embryos. The average number of cells per blastocyst and the blastocyst rates were reported as the mean ± standard error of the mean. A P value less than 0.05 was considered statistically significant.
Results
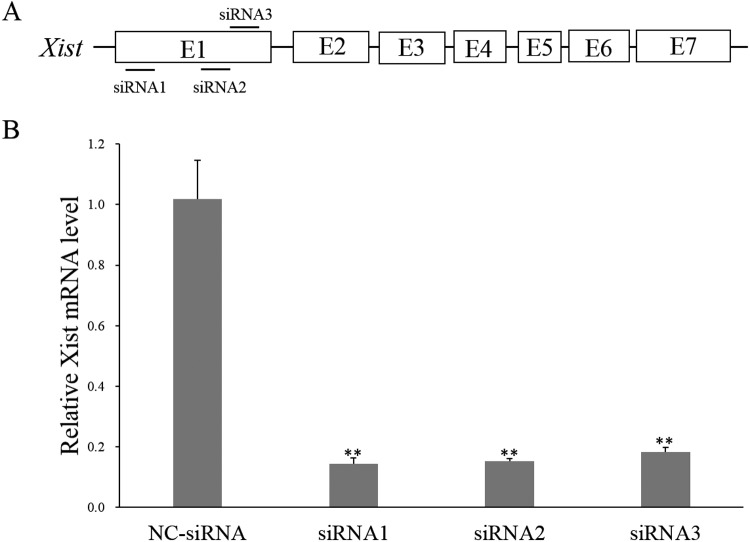
Three siRNAs targeting porcine Xist, namely siRNA1, siRNA2, and siRNA3, were designed and synthesized. Their sequences and target sites are shown in Table 1 and Fig. 1A, respectively. The qPCR results demonstrated that all three Xist-targeted siRNAs effectively suppressed pig Xist expression at 48 h after transfection into female PK21 cells, and that siRNA1 was the most effective siRNA for blocking Xist expression level by over 85% (Fig. 1B). Therefore, siRNA1 and CM-siRNA1 were used for the subsequent experiments.
Fig. 1.
Screening of effective anti-Xist siRNA. A: Target sites of three designed anti-Xist siRNAs on porcine Xist gene. B: Inhibition of Xist expression in female pig kidney cells transfected with three different siRNAs at 48 h post-transfection. ** represents that the mean value calculated from three replicates is significantly different from that of the NC-siRNA group at P < 0.01.
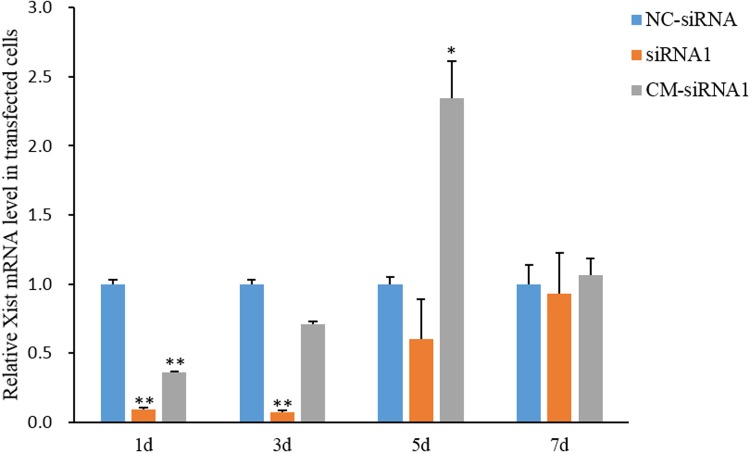
The qPCR results demonstrated that in transfected female PK-21 cells, siRNA1 and CM-siRNA1 maintained their inhibitory effects on Xist expression for at least 3 and 1 days, respectively but lost their silencing effects on Xist expression before 5 and 3 days post-transfection, respectively (Fig. 2). This finding suggests that the injection of siRNA1 or CM-siRNA1 into one-cell-stage cloned male pig embryos could not suppress the ectopic activation of Xist from the morula to blastocyst stage (at 5 to 7 days post-activation).
Fig. 2.
Inhibition of Xist expression in female pig kidney cells at different time points after transfection with anti-Xist siRNA1 and CM-siRNA1. * and ** represent that the mean values calculated from three replicates are significantly different from that of the NC-siRNA group at P < 0.05 and P < 0.01, respectively.
To develop a more effective Xist knockdown method, a plasmid called pU6-shRNA, which expresses anti-Xist siRNA1-derived shRNA, was constructed (Fig. 3). This plasmid effectively blocked Xist transcription for over 5 days after transfection into female PK-21 cells (Fig. 4). This result indicates that shRNA vector-mediated RNAi has a longer silencing action on Xist expression than siRNA-based RNAi in pig cells.
Fig. 4.
Inhibition of Xist expression in female pig kidney cells at different time points after transfection with anti-Xist shRNA expression plasmid. * and ** represent that the mean values calculated from three replicates are significantly different from that of the NC-shRNA group at P < 0.05 and P < 0.01, respectively.
To compare the effects of injecting anti-Xist siRNA1 and shRNA on Xist expression in pig SCNT embryos, 5 ng/µl of pU6-shRNA plasmid, 10 ng/µl of pU6-shRNA plasmid, 5 µM of siRNA1, and 50 µM of siRNA1 were injected into two-cell-stage cloned male pig embryos. Two groups of SCNT embryos injected with anti-Xist siRNA1 did not show a decrease in Xist mRNA level but an increase of 4 X-linked gene mRNA levels at the blastocyst stage compared with the control SCNT embryos injected with water (Fig. 5A). This finding suggests that siRNA1 failed to block Xist expression and upregulate X-linked gene expression at the blastocyst stage of the injected cloned male pig embryos. However, the injections of 5 and 10 ng/µl of pU6-shRNA plasmid into cloned male pig embryos resulted in an almost significant (P = 0.07) and significant decrease in Xist transcription level at the blastocyst stage, respectively (Fig. 5B). Furthermore, the expression levels of 4 X-linked genes in the two groups of SCNT embryos injected with pU6-shRNA plasmid were significantly elevated at the blastocyst stage. These results imply that the injection of pU6-shRNA plasmid into cloned male pig embryos can rescue abnormal XCI by suppressing Xist expression at the blastocyst stage.
Fig. 5.
Effects of injection of anti-Xist siRNA1 (A) and anti-Xist shRNA (B) on the expression of Xist and four randomly selected X-linked genes at the blastocyst stage of injected cloned male pig embryos. The mRNA levels of each injection group were measured from a mixture of 10 to 20 embryos collected at the blastocyst stage (168 h post-activation). The mRNA levels of the 5 or 50 μM siRNA1-injected and 5 or 10 ng/μl shRNA-injected groups were normalized to that of the control group, which was defined as 1. * and ** represent that the mean values calculated from three replicates are significantly different from that of the control group at P < 0.05 and P < 0.01, respectively.
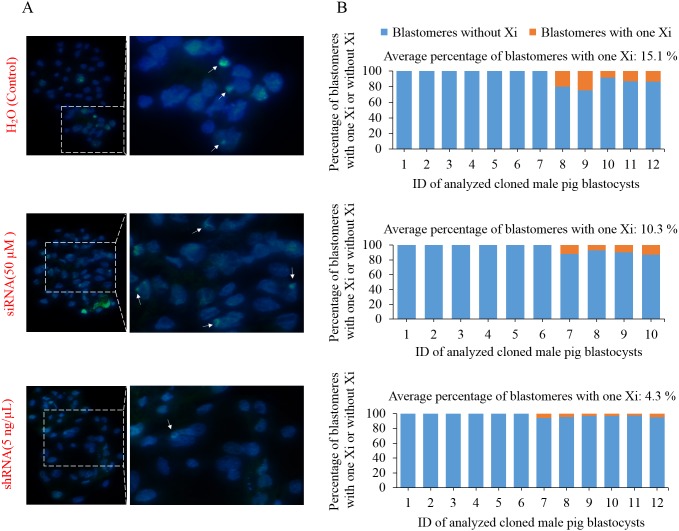
To further compare the effects of injecting anti-Xist siRNA1 and shRNA on XCI in pig SCNT embryos, we performed an immunofluorescence analysis of H3K27me3, which is a marker of XCI, on control, 50 µM anti-Xist siRNA-injected, and 5 ng/µl anti-Xist shRNA plasmid-injected cloned male pig blastocysts. The results showed that in all three groups, approximately half of the cloned male pig blastocysts displayed one inactive X chromosome (Xi) signal in their nuclei (Fig. 6). The average percentages of blastomeres with one Xi signal in control, 50 µM anti-Xist siRNA-injected, and 5 ng/µl anti-Xist shRNA plasmid-injected male pig blastocysts were 15.1%, 10.3%, and 4.3%, respectively (Fig. 6). This suggests that the injection of anti-Xist shRNA expression plasmid can inhibit ectopic X chromosome inactivation in cloned male pig embryos. Moreover, our results indicated that in 12 analyzed cloned male pig blastocysts from the control group, only five blastocysts exhibited one Xi signal; furthermore, in these five Xi signal-positive blastocysts, only 15.1% of blastomeres showed one Xi signal (Fig. 6). The low occurrence of XCI in cloned male pig blastocysts could be attributed to the initiation but incomplete XCI at the blastocyst stage of pig embryos, which has been reported in some studies [28,29,30].
Fig. 6.
Effects of injection of anti-Xist siRNA and anti-Xist shRNA expression plasmid on XCI in cloned male pig blastocysts. A: Immunostaining of an XCI marker H3K27me3 (green) in DAPI (blue)-stained nuclei of injected cloned male pig embryos at the blastocyst stage. B: Percentage of blastomeres with one Xi or without Xi at the blastocyst stage of analyzed injected male pig embryos.
Cloned male pig embryos injected with 5 and 10 ng/µl of pU6-shRNA plasmid showed a significantly higher cell number per blastocyst than the control cloned embryo injected with water (Table 2). The blastocyst rate exhibited an upward trend (P = 0.079) in 5 ng/µl of pU6-shRNA plasmid-injected cloned male embryos compared with that in the control embryos (Table 2). Nevertheless, anti-Xist siRNA1-injected cloned embryos exhibited a similar cell number per blastocyst and blastocyst rate with control embryos (Table 2). These results indicate that the in vitro developmental competence of cloned male pig embryos can be improved by the injection of anti-Xist shRNA expression vector but not by that of anti-Xist siRNA1 at the two-cell stage.
Table 2. Effects of injection of anti-Xist siRNA1 and anti-Xist shRNA on the in vitro developmental efficiency of cloned male pig embryos.
| Total no. of activated cloned embryos |
Total no. of cleaved cloned embryos (two-cell stage) |
Injection groups | Repetition no. |
No. of injected two-cell-stage embryos |
No. of blastocyst/ blastocyst rate (%) |
No. of cells per blastocyst |
|---|---|---|---|---|---|---|
| 1809 | 721 | Control (water) | 4 | 148 | 25/15.53 ± 5.01 | 34.57 ± 1.24 (n = 10) |
| siRNA1 (5 µM) | 3 | 96 | 15/15.94 ± 5.75 | 35.86 ± 3.91 (n = 10) | ||
| siRNA1 (50 µM) | 3 | 101 | 16/16.31 ± 7.71 | 36.33 ± 1.71 (n = 10) | ||
| ShRNA (5 ng/µl) | 4 | 137 | 40/28.78 ± 4.59 § | 40.59 ± 1.68 ** (n = 10) | ||
| ShRNA (10 ng/µl) | 3 | 137 | 34/24.10 ± 3.52 | 39.56 ± 1.91 * (n = 10) |
* and ** represent significant difference at P < 0.05 and P < 0.01, respectively, compared with the control group. § represents statistical difference at P = 0.079 (close to 0.05) compared with the control group.
Discussion
In this study, we demonstrated that the injection of anti-Xist siRNA1 and CM-siRNA1 could not increase the in vitro developmental ability of cloned male pig embryos because the duration of action of injected anti-Xist siRNA1 and CM-siRNA1 was shorter than 5 days. Similar results have been reported in a previous study [14]. In addition, other studies indicated that the inhibitory effects of siRNAs on target gene expression lasted for only 2 to 5 days [20,21,22]. Although CM siRNAs generally have a prolonged silencing effect on target gene expression compared with non-CM siRNA, they were less effective than non-CM siRNAs [23, 24] in some studies, as discovered in this study. Chemical modifications can improve the resistance to enzymatic digestion and thermal stability for siRNA; nonetheless, they could negatively affect the functions of siRNA by changing the conformation of siRNA.
We demonstrated that the blocking action of plasmid-expressed shRNA on Xist expression lasted for over 5 days. This is consistent with the findings of other studies where shRNA expressed from vectors can provide a longer duration of gene silencing than siRNAs [15, 16]. The prolonged gene silencing effect of plasmid-expressed shRNA may be due to the higher stability of plasmid and/or shRNA than siRNA in living cells [25, 26].
The prolonged silencing effect of injected anti-Xist shRNA expression plasmid might be the primary reason for the significant suppression of Xist transcription at the blastocyst stage and for the significant improvement of developmental indexes of injected cloned male pig embryos. However, the injection of shRNA expression plasmid into cloned embryos at the two-cell stage, instead of at the one-cell stage, might have also contributed to effective blockage of Xist expression at the blastocyst stage and the improvement in developmental efficiency of the injected cloned embryos.
Suppression of ectopic Xist activation and enhancement of developmental efficiency of cloned pig embryos can also be achieved by the knockout of the maternal Xist allele in nuclear donor cells [27], similar to that in cloned mouse embryos [11]. Nevertheless, mutation of the Xist gene in cloned embryos results in inheritable genetic modifications, which are undesired in most cases, especially when wild-type cloned embryos or animals are required for subsequent use. If the RNAi-based knockdown of Xist and the knockout of Xist have a similar effect on increasing cloning efficiency, the former method is more favorable because it does not mutate the Xist gene. However, the RNAi-based method is disadvantageous because it can only enhance the developmental competence of male but not female mouse SCNT embryos [28], whereas the knockout of maternal Xist allele can improve the developmental efficiency of both male and female mouse SCNT embryos [11]. Interestingly, a recent study reported that a new method via the epigenetic modification of the Xist gene can increase the developmental ability of mouse embryos [31].
In summary, Xist expression at the blastocyst stage is inhibited and developmental ability is enhanced in cloned male pig embryos by injecting anti-Xist shRNA expression plasmid but not by injecting anti-Xist siRNA at the two-cell stage. This finding indicates that vector-based RNAi rather than siRNA-mediated RNAi of Xist can be used to improve pig cloning efficiency.
Acknowledgments
This study was supported by a grant from the National Natural Science Foundation of China (grant number 31772554) and three grants from the Department of Science and Technology of Guangdong Province, China (grant numbers 2018B030314004, 2018ML1101, and 2018B020203002).
References
- 1.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet 2011; 12: 429–442. [DOI] [PubMed] [Google Scholar]
- 2.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961; 190: 372–373. [DOI] [PubMed] [Google Scholar]
- 3.Ng K, Pullirsch D, Leeb M, Wutz A. Xist and the order of silencing. EMBO Rep 2007; 8: 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahakyan A, Yang Y, Plath K. The role of Xist in X-chromosome dosage compensation. Trends Cell Biol 2018; 28: 999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gayen S, Maclary E, Hinten M, Kalantry S. Sex-specific silencing of X-linked genes by Xist RNA. Proc Natl Acad Sci USA 2016; 113: E309–E318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao S, Miyoshi N, Okamoto I, Jenuwein T, Heard E, Azim Surani M. Initiation of epigenetic reprogramming of the X chromosome in somatic nuclei transplanted to a mouse oocyte. EMBO Rep 2005; 6: 748–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, Li Y, Du W, Zhang L, Yu S, Dai Y, Zhao C, Li N. Aberrant gene expression in organs of bovine clones that die within two days after birth. Biol Reprod 2005; 72: 258–265. [DOI] [PubMed] [Google Scholar]
- 8.Nolen LD, Gao S, Han Z, Mann MR, Gie Chung Y, Otte AP, Bartolomei MS, Latham KE. X chromosome reactivation and regulation in cloned embryos. Dev Biol 2005; 279: 525–540. [DOI] [PubMed] [Google Scholar]
- 9.Shao GB, Ding HM, Gong AH, Xiao DS. Inheritance of histone H3 methylation in reprogramming of somatic nuclei following nuclear transfer. J Reprod Dev 2008; 54: 233–238. [DOI] [PubMed] [Google Scholar]
- 10.Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet 2002; 31: 216–220. [DOI] [PubMed] [Google Scholar]
- 11.Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, Sawai K, Otte AP, Tian XC, Yang X, Ishino F, Abe K, Ogura A. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 2010; 330: 496–499. [DOI] [PubMed] [Google Scholar]
- 12.Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, Ogura A. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci USA 2011; 108: 20621–20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Wu D, Liu D, Shi J, Zhou R, He X, Quan J, Cai G, Zheng E, Wu Z, Li Z. Mutation of the XIST gene upregulates expression of X-linked genes but decreases the developmental rates of cloned male porcine embryos. Mol Reprod Dev 2017; 84: 525–534. [DOI] [PubMed] [Google Scholar]
- 14.Zeng F, Huang Z, Yuan Y, Shi J, Cai G, Liu D, Wu Z, Li Z. Effects of RNAi-mediated knockdown of Xist on the developmental efficiency of cloned male porcine embryos. J Reprod Dev 2016; 62: 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh S, Narang AS, Mahato RI. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm Res 2011; 28: 2996–3015. [DOI] [PubMed] [Google Scholar]
- 16.Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ, Kay MA. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc Natl Acad Sci USA 2011; 108: 9208–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 2004; 427: 645–649. [DOI] [PubMed] [Google Scholar]
- 18.Takabatake Y, Isaka Y, Mizui M, Kawachi H, Takahara S, Imai E. Chemically modified siRNA prolonged RNA interference in renal disease. Biochem Biophys Res Commun 2007; 363: 432–437. [DOI] [PubMed] [Google Scholar]
- 19.Shi J, Zhou R, Luo L, Mai R, Zeng H, He X, Liu D, Zeng F, Cai G, Ji H, Tang F, Wang Q, Wu Z, Li Z. Influence of embryo handling and transfer method on pig cloning efficiency. Anim Reprod Sci 2015; 154: 121–127. [DOI] [PubMed] [Google Scholar]
- 20.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res 2003; 31: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res 2002; 30: 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuschl T. Expanding small RNA interference. Nat Biotechnol 2002; 20: 446–448. [DOI] [PubMed] [Google Scholar]
- 23.Czauderna F, Fechtner M, Dames S, Aygün H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res 2003; 31: 2705–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenski DM, Willingham AT, Haringsma HJ, Li JJ, Flanagan WM. In vivo activity and duration of short interfering RNAs containing a synthetic 5′-phosphate. Nucleic Acid Ther 2012; 22: 90–95. [DOI] [PubMed] [Google Scholar]
- 25.Amarzguioui M, Rossi JJ, Kim D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett 2005; 579: 5974–5981. [DOI] [PubMed] [Google Scholar]
- 26.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 2002; 16: 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruan D, Peng J, Wang X, Ouyang Z, Zou Q, Yang Y, Chen F, Ge W, Wu H, Liu Z, Zhao Y, Zhao B, Zhang Q, Lai C, Fan N, Zhou Z, Liu Q, Li N, Jin Q, Shi H, Xie J, Song H, Yang X, Chen J, Wang K, Li X, Lai L. XIST derepression in active X chromosome hinders pig somatic cell nuclear transfer. Stem Cell Reports 2018; 10: 494–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oikawa M, Matoba S, Inoue K, Kamimura S, Hirose M, Ogonuki N, Shiura H, Sugimoto M, Abe K, Ishino F, Ogura A. RNAi-mediated knockdown of Xist does not rescue the impaired development of female cloned mouse embryos. J Reprod Dev 2013; 59: 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang JY, Oh JN, Park CH, Lee DK, Lee CK. Dosage compensation of X-chromosome inactivation center-linked genes in porcine preimplantation embryos: Non-chromosome-wide initiation of X-chromosome inactivation in blastocysts. Mech Dev 2015; 138: 246–255. [DOI] [PubMed] [Google Scholar]
- 30.Zou H, Yu D, Du X, Wang J, Chen L, Wang Y, Xu H, Zhao Y, Zhao S, Pang Y, Liu Y, Hao H, Zhao X, Du W, Dai Y, Li N, Wu S, Zhu H. No imprinted XIST expression in pigs: biallelic XIST expression in early embryos and random X inactivation in placentas. Cell Mol Life Sci 2019; 76: 4525–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Gao X, Yang J, Fan X, Wang W, Liang Y, Fan L, Han H, Xu X, Tang F, Bao S, Liu P, Li X. Xist intron 1 repression by transcriptional-activator-like effectors designer transcriptional factor improves somatic cell reprogramming in mice. Stem Cells 2019; 37: 599–608. [DOI] [PubMed] [Google Scholar]