Abstract
Dysbiosis of the gut microbiota in inflammatory bowel disease (IBD) patients is of great interest. It has been reported that Crohn's disease (CD) is associated with a general decrease in microbial diversity [1]. Altered microbial composition and function in CD results in imbalance in host-bacteria interaction and increased immune stimulation [2]. It is shown that microbiota in CD is characterized by increased proportion of E. coli in human gut in contrast to healthy individuals [3]. However, the overall qualitative and quantitative diversity of E. coli strains in CD is not fully understood. Here, we present a dataset of whole-genome sequences of E. coli's.
Keywords: Escherichia coli, Crohn's disease, Whole-genome sequencing, Human gut microbiota
Specifications Table
| Subject | Immunology and microbiology |
| Specific subject area | Microbiology |
| Type of data | Whole-genome sequencing data, table, figure |
| How data were acquired | Whole-genome sequencing on Illumina MiSeq platform. Bioinformatics approaches: genome assembler SPAdes v.3.11.1, rapid prokaryotic genome annotation Prokka v.1.12, pan genome Roary pipeline v.3.12.0, FastTree v.2.1.11 tool, SerotypeFinder-2.0 tool. |
| Data format | Raw, analyzed, deposited data |
| Parameters for data collection | Whole genomes of E. coli isolates from patients with diagnosed Crohn's disease and healthy individuals were sequenced, assembled and annotated |
| Description of data collection | Dataset covers 64 samples (E. coli isolates from stool samples of 18 healthy individuals and 14 Crohn's disease patients) |
| Data source location | Kazan Federal University, Kazan, Russian Federation |
| Data accessibility | The whole genome sequencing data have been deposited to NCBI BioProject with the dataset identifier PRJNA560176 https://www.ncbi.nlm.nih.gov/bioproject/560176 |
| Related research article | Miquel S., Peyretaillade E., Claret L., De Vallée A., Dossat C., Vacherie B., Zineb E., Segurens B., Barbe V., Sauvanet P., Neut C., Colombel, J., Medigue C., Mojica F., Peyret P., Bonnet R., Darfeuille-Michaud A. Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82, PloS one, 5(9) (2010), p. e12714, https://doi.org/10.1371/journal.pone.0012714 |
Value of the Data
|
1. Data
Previous studies showed that CD patient's immune system has aberrant response to gut microbiota resulting in decreased bacterial diversity accompanied by enrichment of Enterobacteriaceae family [1], [2], [3].
In the present article, we report whole genome data of cultivated E. coli strains isolated from stool samples of 14 CD patients and 18 controls (listed in Supplementary Table 1). Out of 97 sequenced genomes, 33 duplicates were revealed using the comparative genome analysis, i.e. isolates sequenced more than once due to varying colony phenotypes. Thus, 64 unique E. coli genomes were obtained: 27 from CD patients (6 from patients with diagnosed ileitis, 14 – colitis, 7 – ileocolitis), and 37 from the control group (Supplementary Table 2). E. coli draft genome assemblies were submitted to NCBI (BioProject ID PRJNA560176).
Phylogenetic group analysis, performed according to Clermont [4], revealed that E. coli strains of E and F groups were observed only in healthy donors.
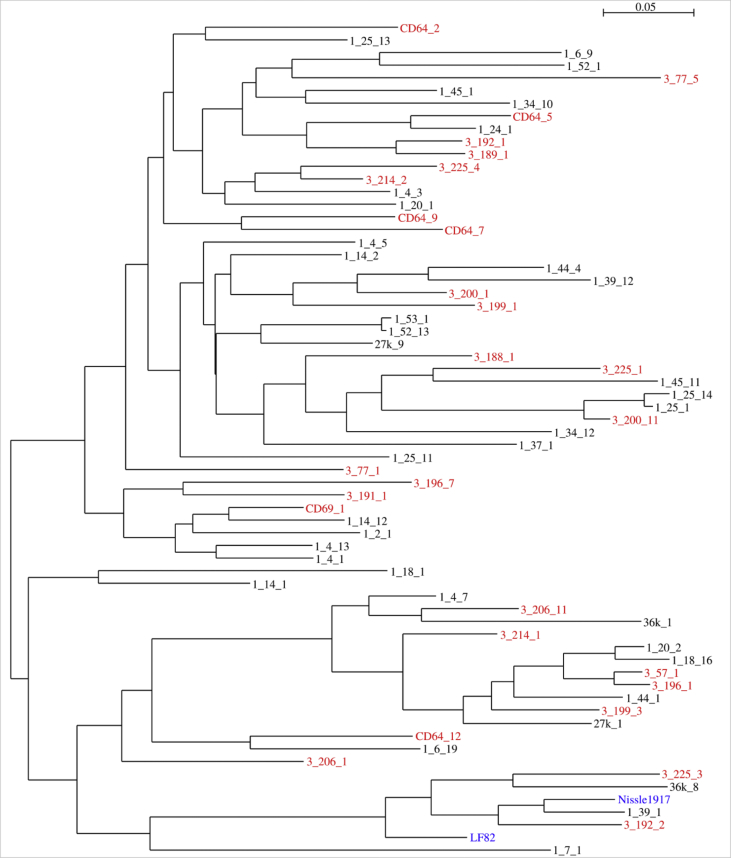
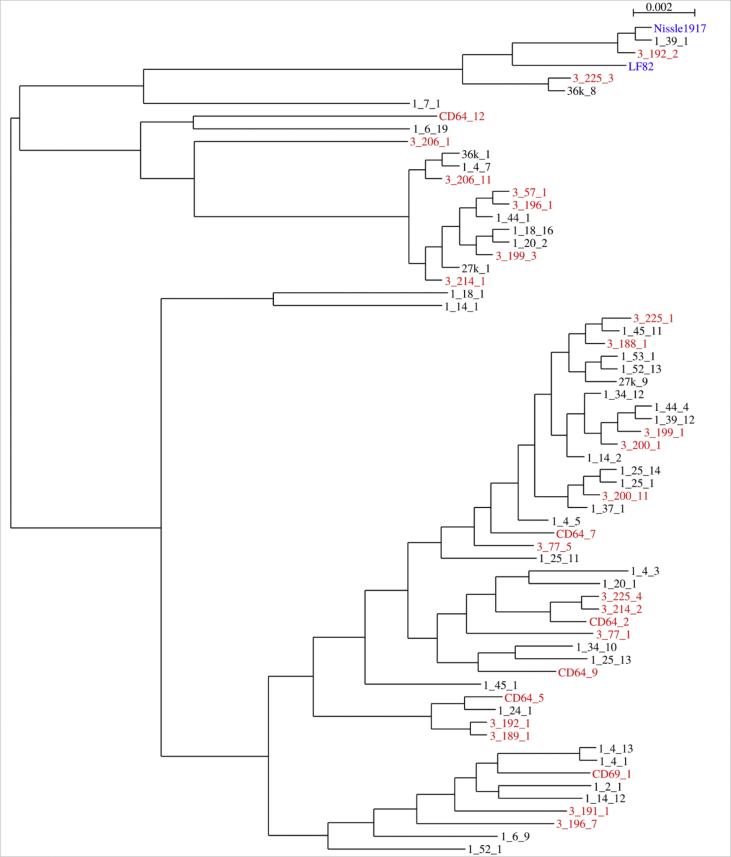
Phylogenetic trees analysis based on core and accessory genes did not reveal any specific E. coli group associated with the disease. For comparison LF82 strain associated with ileal CD [5] and widely studied probiotic strain Nissle 1917 [6] were included as references (Fig. 1, Fig. 2).
Fig. 1.
Phylogenetic tree of E. coli strains from CD patients (red), healthy individuals (black) and reference genomes (blue) based on accessory gene content in genome assemblies.
Fig. 2.
Phylogenetic tree of E. coli strains from CD patients (red), healthy individuals (black) and reference genomes (blue) based on core genes in genome assemblies.
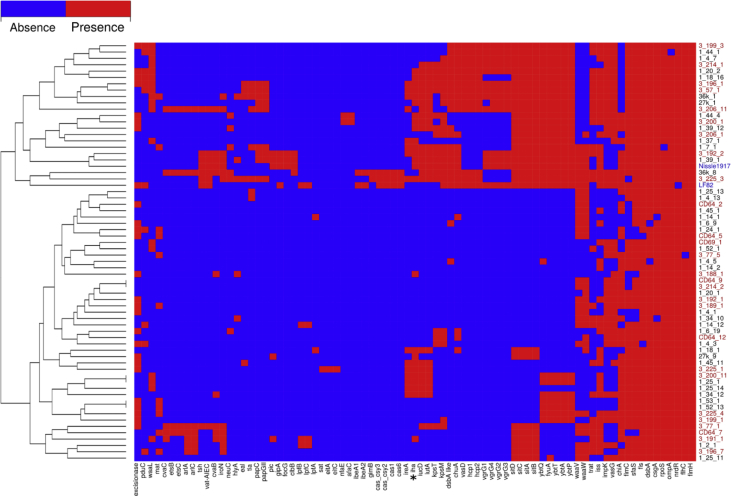
Analysis of 98 previously reported genes associated with pathogenicity and virulence in E. coli [7,8] revealed that the frequency of occurrence of iha gene coding bifunctional enterobactin receptor/adhesin protein among strains from patients with ileitis was higher than with colitis and ileocolitis (exact Fisher test, P = 0.044 with, P value with Benjamini-Hochberg adjustment) (Fig. 3).
Fig. 3.
Distribution of virulence and pathogenicity genes of E. coli from CD patients (red), healthy individuals (black) and reference strains LF82 and Nissle 1917 (blue). Genes present or absent in all analyzed strains are not displayed. Gene with differential distribution in strains from patients with ileitis vs patients with colitis and ileocolitis is marked with asterisk (*).
In silico serotyping showed a vast diversity of E. coli serotypes in both studied cohorts. However, no serotype associated with the disease was found. Strains of 5 serological types were represented both in CD group and control one - O17/O44:H18, O144:H45, O6:H1, O25:H18, O1:H7.
2. Experimental design, materials, and methods
2.1. Sample collection
A total of 32 stool samples, 14 from patients with Crohn's disease diagnosed by colonoscopic examination and confirmed histologically, and 18 from healthy individuals were taken for the analysis. The samples were collected at the Kazan Federal University Hospital (Kazan, Russia) and stored at −80 °C until needed.
2.2. Isolation and identification of E. coli strains
Serial ×10 fold dilutions in PBS solution were made from 0.1 g of stool sample. 0.1 ml of suspension (×102–103 fold) was poured onto Endo agar medium and incubated at 37 °C for 19–20 hours. The total number of colonies was counted and colony morphology (color, shape, size, metallic luster) was registered. Up to 10 representative from each sample lactose-positive colonies (dark red color) were randomly picked up for cultivation in LB medium at 37 °C for 19–20 hours. The identification of the E. coli-like colonies was confirmed using MALDI Biotyper System (Bruker, Germany). Lactose-negative colonies after testing against polyvalent anti-Shigella sera were added to the collection for further sequencing (Agnolla, Russia). In addition, the ability to hemolyze red blood cells was assessed by the presence of clear zones around colonies on blood agar medium after 24 hours of incubation at 37 °C. Relative and absolute abundances of isolated strains are represented in Supplementary Table 2. The mean CFU/g of feces from healthy individuals and CD patients were 3.4*105 and 3.8*105, respectively (one strain with extremely high abundance was excluded).
In total 521 isolates were collected and stored in tryptic soy broth containing 50% glycerol at −80 °C until further phylotype screening.
2.3. DNA extraction and E. coli phylotyping
Genomic DNA was extracted from colonies with PureLink Genomic DNA Mini Kit (Invitrogen) following the manufacturer instructions and quantified using Qubit 2.0 Fluorometer (Invitrogen). The E. coli phylogroup (A, B1, B2, C, D, E, F) of each colony was determined by the quadruplex PCR [4].
2.4. Genome sequencing and analysis
Selected 97 isolates assigned to different phylogenetic groups and/or morphology were subjected to the whole-genome sequencing. DNA libraries were prepared using NEBNext Ultra II Kit (New England BioLabs, USA) according to the manufacturer's recommendations. DNA-library size was evaluated on the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). The sequencing was performed on Illumina MiSeq platform (300 bp paired-end mode).
After adapters removal and filtering by length and quality using cutadapt [9] paired-end reads were de novo assembled using SPAdes v.3.11.1 (http://cab.spbu.ru/software/spades/) [10]. Genome annotation was performed using Prokka v.1.12 [11] and pangenome analysis was performed with Roary pipeline v.3.12.0 [12]. Phylogenetic trees based on core and accessory genes was constructed using FastTree v.2.1.11 [13]. Serotypes were assigned using SerotypeFinder-2.0 tool [14].
Acknowledgments
This work was funded by Russian Foundation for Basic Research according to the research project №17-00-00433.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.104948.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., Roca J., Dore J. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santor R.B. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat. Rev. Gastroenterol. 2006;3(7):390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 3.Kotlowski R., Bernstein C.N., Sepehri S., Krause D.O. High prevalence of Escherichia coli belonging to the B2+ D phylogenetic group in inflammatory bowel disease. Gut. 2007;56(5):669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clermont O., Christenson J.K., Denamur E., Gordon D.M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013;5(1):58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 5.Miquel S., Peyretaillade Eric, Claret L., de Vallée A., Dossat C., Vacherie B., Zineb E.H., Segurens B., Barbe V., Sauvanet P., Neut C., Colombel J.-F., Medigue C., Mojica F.J.M., Peyret P., Bonnet R., Darfeuille-Michaud A. Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82. PLoS One. 2010;5(9):e12714. doi: 10.1371/journal.pone.0012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reister M., Hoffmeier K., Krezdorn N., Rotter B., Liang C., Rund S., Dandekar T., Sonnenborn U., Oelschlaeger T.A. Complete genome sequence of the gram-negative probiotic Escherichia coli strain Nissle 1917. J. Biotechnol. 2014;187:106–107. doi: 10.1016/j.jbiotec.2014.07.442. [DOI] [PubMed] [Google Scholar]
- 7.Fang X., Monk J.M., Nurk S., Akseshina M., Zhu Q., Gemmell C., Gianetto-Hill C., Leung N., Szubin R., Sanders J., Beck P.L., Li W., Sandborn W.J., Gray-Owen S.D., Knight R., Allen-Vercoe E., Palsson B.O., Smarr L. Metagenomics-based, strain-level analysis of Escherichia coli from a time-series of microbiome samples from a Crohn's disease patient. Front. Microbiol. 2018;9:2559. doi: 10.3389/fmicb.2018.02559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camprubí-Font C., Ewers C., Lopez-Siles M., Martinez-Medina M. Genetic and phenotypic features to screen for putative Adherent-Invasive Escherichia coli. Front. Microbiol. 2019;10:108. doi: 10.3389/fmicb.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17(1):10–12. [Google Scholar]
- 10.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. [Google Scholar]
- 12.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price M.N., Dehal P.S., Arkin A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joensen K.G., Tetzschner A.M., Iguchi A., Aarestrup F.M., Scheutz F. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J. Clin. Microbiol. 2015;53(8):2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.