Abstract
Bisulfite sequencing is a powerful technique to detect 5-methylcytosine in DNA that has immensely contributed to our understanding of epigenetic regulation in plants and animals. Meanwhile, research on other base modifications, including 6-methyladenine and 4-methylcytosine that are frequent in prokaryotes, has been impeded by the lack of a comparable technique. Bisulfite sequencing also suffers from a number of drawbacks that are difficult to surmount, among which DNA degradation, lack of specificity, or short reads with low sequence diversity. In this review, we explore the recent refinements to bisulfite sequencing protocols that enable targeting genomic regions of interest, detecting derivatives of 5-methylcytosine, and mapping single-cell methylomes. We then present the unique advantage of long-read sequencing in detecting base modifications in native DNA and highlight the respective strengths and weaknesses of PacBio and Nanopore sequencing for this application. Although analysing epigenetic data from long-read platforms remains challenging, the ability to detect various modified bases from a universal sample preparation, in addition to the mapping and phasing advantages of the longer read lengths, provide long-read sequencing with a decisive edge over short-read bisulfite sequencing for an expanding number of applications across kingdoms.
Keywords: base modifications, bisulfite, epigenetics, Nanopore, PacBio
Introduction
Genomic DNA is composed of the canonical DNA bases A, T, C, and G. Modified DNA bases do not change the underlying sequence, but instead carry an extra layer of information that often dictates how that DNA sequence is utilised: for example identifying sequences as endogenous or modulating transcription [1–3]. The DNAmod database catalogs 43 DNA modifications encountered in natural DNA [4], some with regulatory roles but the majority resulting from DNA damage [5].
N6-methyladenine (6mA), 4-methylcytosine (4mC), and 5-methylcytosine (5mC) are frequent in bacteria and have roles not only in cellular defence but also in the regulation of gene expression, with effects on virulence and physiology [1]. 5mC is also the most frequent, most studied and best understood modification in plants and animals [6–8]. This is in part due to the accurate bisulfite-based short-read sequencing techniques available to measure 5mC [9,10]. However, bisulfite sequencing suffers from a number of limitations and is not easily applicable to the detection of other base modifications.
On the other hand, emerging long-read sequencing techniques offer exciting possibilities to study a wide range of modifications, with the advantages inherent to long reads and single-molecule sequencing. In this review, we will discuss the current gold-standard in detection of 5mC and its oxidised derivatives and compare it to the current and future possibilities offered by long-read sequencing from Pacific Biosciences and Oxford Nanopore technologies. We note, however, that there are many alternative methods available to measure 5mC that we will not discuss here, each of which may be useful in specific applications and warrant consideration (reviewed in [11–13]).
Sequencing of bisulfite-converted DNA
DNA methylation marks remain intact upon DNA extraction. Treatment of genomic DNA with sodium bisulfite results in deamination of unmethylated cytosines to uracil, leaving methylated cytosines intact [14] (Figure 1). Treated DNA is subsequently PCR-amplified with a uracil-tolerant polymerase to provide sufficient template for analysis, causing uracil to convert to thymine. DNA methylation can therefore be read directly by traditional Sanger or Illumina short-read sequencing through comparison to a reference or untreated sequence, providing a readout that is highly quantitative and with base-pair resolution.
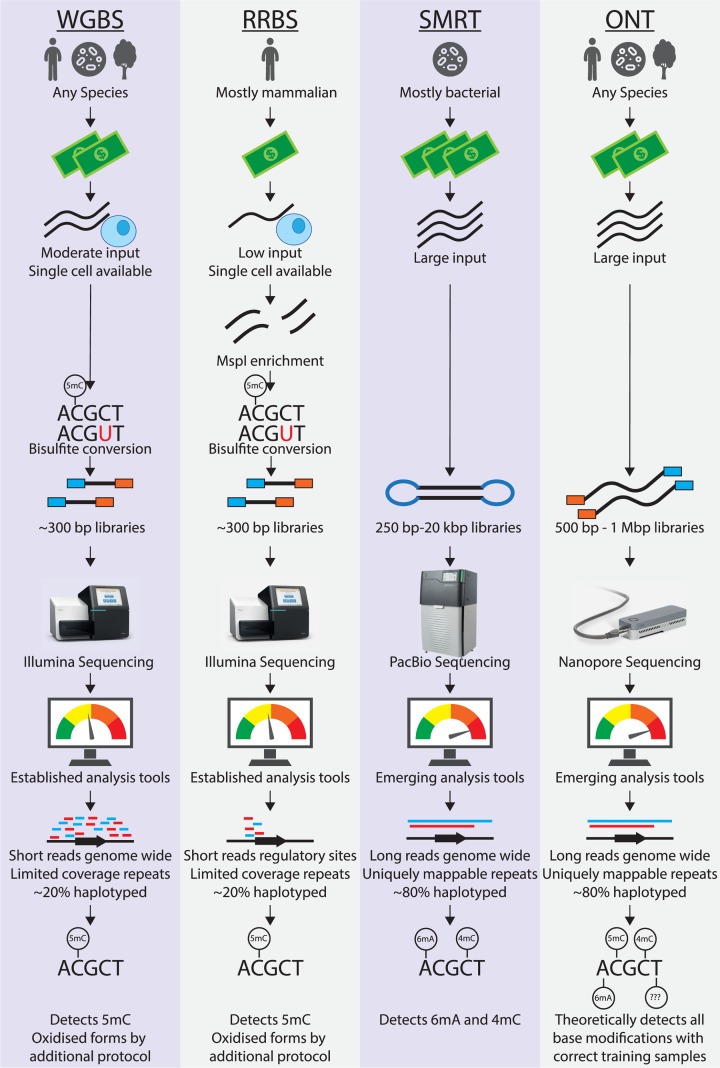
Figure 1. Detection of base modifications by bisulfite and long-read sequencing.
Whole-genome bisulfite sequencing (WGBS) provides accurate binary calls of cytosine methylation status at nucleotide resolution, but cannot distinguish between 5mC and 5hmC or detect other oxidised forms without additional techniques. The short reads have limitations in mapping to repeated sequences and haplotyping. Reduced representation bisulfite (RRBS) restricts the sequencing space to CpG islands. Both bisulfite protocols are compatible with single-cell genomics. PacBio single molecule real-time (SMRT) sequencing generates 250 b-20 kb reads with relatively low accuracy probabilistic calls for 4mC and 6mA modifications. Oxford Nanopore Technologies (ONT) nanopore sequencing produces 500 b-1 Mb reads with higher accuracy probabilistic calls for a range of modifications, including 5mC, 4mC, and 6mA. Long-read technologies offer a simpler library preparation workflow that avoids amplification biases, but they require more input material and more advanced data analysis. The longer read lengths improve mapping and haplotyping (estimates based on our experience with C57BL/6 x Cast/Eij F1 mice on Nanopore [62]).
In animals, 5mC is most often enriched in the CpG dinucleotide context [7,15,16]; however, 5mC also appears in the CHG and CHH contexts (where H is either A, C or T) where studies also suggest possible functional roles [17–20]. In plants, methylation is abundant in all CG, CHG and CHH contexts although with extensive variation across species [21]. The fact that bisulfite sequencing informs on all cytosines regardless of context can therefore be highly beneficial. These properties have made sequencing of bisulfite-converted DNA the gold standard of 5mC detection techniques; however, there are some downfalls for consideration. Firstly, the bisulfite treatment is very harsh, leading to degradation and DNA that is harder to PCR amplify, therefore large amounts of input DNA are often required. Recently, New England Biolabs reported a technique called Enzymatic Methyl-seq (EM-seq) that uses enzymatic deamination of cytosine by APOBEC, thereby producing sequence identical to bisulfite treatment, but avoiding the need for the harsh chemical treatment. Additionally, bisulfite sequence data require more sophisticated bioinformatic analysis techniques than are required for unconverted DNA, as sequence must be compared to a bisulfite-converted reference genome before methylation calls can be inferred. There are a number of excellent tools designed specifically to process bisulfite sequence data (reviewed in [13]). Illumina sequencing of bisulfite DNA also suffers from the same problems as all short-read data, particularly mapping issues to repeating or low complexity regions, including regions of pertinence to 5mC such as heavily GC rich regulatory regions and repetitive DNA (Figure 1). These issues are further compounded by the loss of sequence diversity due to the bisulfite conversion [22–24]. Additionally, short reads are difficult to haplotype as the short nature of the read reduces the likelihood of it containing an informative single-nucleotide polymorphism (SNP) [25] (Figure 1). These limitations of short-read sequencing are abrogated with long-read sequencing, discussed in detail below.
Illumina sequencing of total genomic DNA is known as whole-genome bisulfite sequencing (WGBS) and provides the most comprehensive and unbiased survey of 5mC currently possible [9,10]; however, obtaining such comprehensive data requires a high number of reads, with a coverage of 5–15× recommended (∼160–480M 100-bp reads for a haploid human genome, pooling both DNA strands) [26].
Enrichment techniques for bisulfite sequencing
In order to mitigate the high costs associated with obtaining the necessary reads for quality WGBS data in large genomes, techniques that enrich for regions of interest have been developed. For a small number of genomic loci (≤ 20), amplicon sequencing is straightforward and cost-effective [27].
DNA is first bisulfite-treated before amplification by specific primers and barcoding, then sequenced as a multiplex [28]. For larger numbers of regions, capture-sequencing avoids the labour-intensive design of primer pairs; however, they require the synthesis of a probe panel. Capture by hybridisation to specific probes can be performed either before (Agilent Sure-Select Methyl-Seq, TruSeq Methyl Capture, [29]) or after bisulfite conversion (Roche SeqCap Epi, [30–32]). In the latter case, there is a risk that preferential binding of probes to certain methylation states of the target fragments could introduce biases in the quantitation of methylation. Custom panels can be expensive for one-off applications, but there are commercially available panels that perform well for the human genome [33].
In mammals, reduced representation bisulfite sequencing (RRBS, Figure 1) offers a cost-efficient solution [34,35], enabling enrichment of regions where regulation by mC is more likely such as CpG Islands: regions of high CpG density known to frequently correspond to differentially methylated gene regulatory regions such as enhancers and promoters [36]. By utilising MspI, a 5mC agnostic restriction enzyme that cuts at CCGG motifs, RRBS has been found to be informative for 85% of CpG islands, representing < 3% of the genome and therefore greatly reducing sequencing costs [37,38].
The obvious drawback to RRBS is that it is by design limited to loci containing MspI cut sites. Regions of moderate CpG density that flank CpG islands, known as CpG island shores, are also found to be frequently differentially methylated [39,40] and these can be captured by sequencing the longer restriction fragments, in a technique known as enhanced RRBS [41]. Another consideration with RRBS is that the MspI cut creates a lack of diversity at the start of sequencing reads, which can interfere with calibration and cluster detection on the latest Illumina sequencers. However, this can be overcome by masking the first bases of each read from the sequencer (known as dark sequencing) [42], using adapters that contain diversity bases, or spiking in libraries of high diversity.
Identification of oxidised forms of methylation by bisulfite sequencing
In mammals, active DNA demethylation is achieved through the oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by the TET family of dioxygenases, with demethylation being completed by the excision of 5fC or 5caC by the DNA glycosylase TDG (reviewed in [43,44]). Like 5mC, 5hmC is protected from bisulfite-induced deamination and therefore bisulfite sequencing is unable to differentiate the two forms, while 5fC and 5caC are vulnerable to deamination and thus read as unmethylated cytosine [45]. This is a major caveat to be considered for any bisulfite sequencing experiment; however, adapted bisulfite sequencing methods have been developed to distinguish these oxidised forms. Oxidative bisulfite sequencing (OxBS-seq) utilises specific chemical oxidation of 5hmC to 5fC, such that only 5mC remains protected from bisulfite-induced deamination. Therefore OxBS-seq informs specifically on 5mC without confounding 5hmC. Positions of 5hmC can be determined through a subtraction process of OxBS-seq from unoxidized bisulfite sequencing [46]. TET-assisted bisulfite sequencing (TAB-seq) uses the TET1 enzyme to oxidise 5mC to 5caC while protecting 5hmC from oxidation through the addition of a glucose, thus rendering positions of 5mC, but not 5hmC, susceptible to bisulfite induced deamination [47]. Therefore, TAB-seq provides a direct measurement of 5hmC, with positions of 5mC able to be determined by subtracting TAB-seq signal from standard bisulfite sequencing. Identification of 5fC positions can be determined from bisulfite sequencing either by protecting 5fC from bisulfite-induced deamination chemically (fCAB-seq [48]) or by the selective reduction to 5hmC (redBS-Seq [49]). Carboxylcytosine can be protected from deamination by chemical modifications rendering it also detectable by bisulfite sequencing (CAB-seq [50]).
Single-cell bisulfite sequencing
Until recently, large input requirements have meant genomic assays, including bisulfite sequencing, could only provide average measurements across bulk cell populations. Now single-cell genomics has begun to offer unprecedented insight into cell-to-cell variation. Here, bisulfite sequencing has major advantages over other methods of 5mC detection. Firstly, changing the order of adapter ligation to after bisulfite treatment (known as post bisulfite adapter tagging or PBAT), vastly reduced the input requirement to that of a single nucleus [51,52], although adapter ligation is not free from biases and chimeric reads tend to be formed [53]. Secondly, whereas techniques that utilise read count-based statistics as measurements suffer from the low coverage typical of single-cell sequencing, bisulfite sequence data contains the measurement within the read, meaning every mappable read is informative. Techniques are available for both single-cell WGBS [52] and single-cell RRBS [54] (Figure 1). Excitingly, RNA-seq from the cytoplasmic fraction is being combined with bisulfite sequencing of the nucleus from within the same single cell, giving unprecedented comparison of the functional link between the epigenome and the transcriptome. Known as single-cell multi-omics, methods currently exist to measure combinations of 5mC, RNA, copy number and nucleosome positioning [55–57]. Detailed discussion of multi-omic methods are available in [58–61].
Detecting DNA methylation through single molecule long-read sequencing
Long-read sequencing offers a solution to the mappability problem of short reads. Two long-read sequencing technologies are currently available: nanopore sequencing from Oxford Nanopore Technologies (ONT) and single molecule real-time (SMRT) sequencing from Pacific Biosciences (PacBio) (Figure 1). Both strategies can be applied to bisulfite-treated and amplified DNA to provide a readout similar to short-read bisulfite sequencing, but their main advantage lies in their ability to sequence native DNA and infer base modifications from their impact on the raw sequencing signal. DNA degradation due to bisulfite conversion is then avoided, as are amplification biases. More generally there is no longer a need for enzymatic or chemical treatments specific to each base modification of interest, opening up the range of assayable modifications and reducing experimental complexity. The downside of this feature is that the DNA cannot be amplified; therefore, input amounts can be limiting (200 fmol for Nanopore, or about 1 μg of 8 kb fragments; 5 μg for a typical PacBio library, although 100 ng can be sufficient [63]). In cases where only small DNA amounts are available because of experimental design (e.g. single small organism, microdissected tissue, single cells) or because of the preciousness of the original tissue (e.g. biopsies or paleogenomic samples), Nanopore and PacBio native DNA sequencing will not be feasible. These approaches are therefore best suited to bulk samples, constraining the granularity of the analyses and making cell-to-cell variation difficult to evaluate.
When only parts of the genome are of interest, PCR-free, CRISPR-based enrichment techniques are available for both PacBio and ONT. Hundred- or thousand-fold enrichments are achievable [64–66], providing cost- and sequencing-effective targeted genetic and epigenetic assays.
As opposed to short-read bisulfite sequencing, calling base modifications at a single-base, single-read level from long-read sequencing is not accurate. Accurate estimates are thus derived by aggregating statistics over multiple passes (PacBio only), or summarising at genomic positions (requiring sufficient sequencing depth), regions or motifs when applicable. PacBio and ONT have distinct strengths and limitations that influence their respective use cases (Figure 1).
SMRT sequencing
PacBio’s SMRT sequencing relies on sequencing-by-synthesis, where the sequence of a circular DNA template is determined from the succession of fluorescence pulses, each resulting from the addition of one labelled nucleotide by a polymerase fixed to the bottom of a well. Base modifications thus do not affect the basecalled sequence, but they affect the kinetics of the polymerase. By considering the inter-pulse duration, base modifications can be inferred from the comparison of a modified template to an in silico model or an unmodified template [67] (Figure 1). For example, presence of a 6mA in the template strand tends to delay the incorporation of the complementary T by the polymerase. The patterns of kinetic perturbations can be more complex and context-dependent, while the magnitude of the perturbations also depend on the base modification [68].
Because the signal-to-noise ratio is low, the detection of base modifications in single-molecule is inaccurate and often requires summarising at the genomic position-level. As 6mA and 4mC produce strong kinetic signatures, a coverage of 25× per strand is recommended [68]. However the subtle effects of 5mC and 5hmC increase the requirements to 250×, unless they are enriched or modified to produce a larger kinetic effect by glycosylation or TET-conversion to 5-carboxylcytosine [68,69]. PacBio sequencing therefore only achieves single-molecule resolution for certain marks and on relatively short fragments (≤ 2 kb) that can be read a large number of times by the polymerase [70]. With longer fragments, cell-to-cell variability cannot be investigated in detail. PacBio’s price per Gb, sensitivity to particular modifications, resolution and high coverage requirements make this technology particularly suited to bacterial genomes, where 6mA and 4mC are frequent and often concentrated on specific motifs (Figure 1). The use of SMRT sequencing since 2012 has greatly expanded the number of known methyltransferases [71,72]. SMRT sequencing has also been applied to the detection of base J (β-d-glucosyl-hydroxymethyluracil) in Leishmania [73], and has the potential to discover unknown modifications [74,75]. In 2019, the introduction of PacBio’s Sequel II sequencer and v2 chemistry greatly improved the throughput and affordability of SMRT sequencing, generating up to 160 Gb per SMRT cell.
Nanopore sequencing
ONT’s nanopore sequencing measures the variation in ionic current through a biological nanopore as a single-stranded nucleic acid is ratcheted through. Neural networks translate the current trace into nucleotides in a process named basecalling. Base modifications on the DNA introduce deviations in the raw signal, making them detectable (Figure 1).
Detection of base modifications in ONT data commonly involves three steps: (1) basecalling with canonical bases (e.g. with ONT’s Guppy basecaller [76]), (2) anchoring the raw signal to a genomic reference, and (3) weighing the evidence that a base is modified. Nanopolish [77] is a popular software to detect 5mCG with a pre-trained algorithm, showing good correlation with bisulfite data on human and mouse genomes [62,77,78]. Because Nanopolish incorporates a model for 5mCG, there is no need to sequence a PCR-amplified, unmodified control in addition to the sample of interest. Nanopolish outputs the probability that a base is modified at a single-read, almost single-nucleotide (actually single k-mer) resolution. Other available tools differ in the underlying algorithms and the modifications they are trained to detect: signalAlign demonstrated detection of 6mA, 5mC and 5hmC [79], mCaller, DeepSignal, DeepMod and Megalodon detect 6mA and 5mCG [80–83], and D-Nascent and RepNano are tailored towards BrdU detection [84,85]. ONT-developed Tombo provides a model for 5mC and 6mA [86]. Detection of 6mA is generally less accurate than for 5mCG, although gains may still be obtained from improved algorithms and training data [79,80]. Similarly to the principles of base modification detection in PacBio, when no pre-trained algorithms are available for the base modification of interest, it can be inferred by comparison to an in silico reference signal or, more effectively, to a PCR-amplified control devoid of modifications (Tombo, NanoMod [87]).
Only very recently has it become possible to directly basecall modifications from the raw signal, without genomic anchoring (from Guppy v3.2.1 [76]). This technique is very promising, limiting the need for computationally intensive downstream analysis; however, it is for the moment not benchmarked and restricted to 5mC in the CG and CC(A/T)GG contexts and 6mA in the GATC context.
The performances of methods that use prior knowledge about the expected deviations in signal depend notably on the training data used, which is typically composed of a fully unmodified sample (PCR-amplified or synthesised) and a fully modified sample (synthesised or modified in vitro by enzymes). Motifs that are not represented in the training set or that contain mixtures of modified and unmodified bases lead to suboptimal performance [80]. For example, on a motif such as CGCGT, Nanopolish only reports a likelihood that the whole group is methylated, rather than probabilities for individual cytosines [77].
The price of whole-genome nanopore sequencing is comparable to that of whole-genome bisulfite sequencing. Detection of base modifications by nanopore sequencing is still an area of active development. We do not yet know the full extent of modifications that can be distinguished, what the limits to sensitivity are and we lack generalised algorithms able to call many modifications at the same time. Independent benchmarks of established and emerging tools are needed to understand the stability of performance across species and sequencing batches. Every time ONT upgrades the pore chemistry, the raw signal changes and the algorithms have to be trained again. Fortunately, many tools offer the possibility for users to train the algorithms on their own data, both at the basecalling stage (e.g. Taiyaki [88], Chiron [89]) or post-alignment (Nanopolish, DeepSignal, mCaller, Tombo). Recent advances in basecalling hold the promise that genetic and epigenetic information will soon come directly out of the sequencer without the need for extra processing.
Discussion
Bisulfite sequencing provides a quantitative and sensitive assay for DNA methylation at base-resolution (Figure 1). The high-cost of deep coverage can be avoided by targeted sequencing techniques. However, standard bisulfite conversion cannot distinguish between 5mC and 5hmC and specialised protocols are required to specifically detect each of these marks. This is also true for 5caC and 5fmC and library preparation and sequencing have to be performed separately for each mark of interest.
By contrast, SMRT and nanopore long-read sequencing have the capacity to detect a range of base modifications simultaneously, without additional sample preparation. SMRT most sensitively detects 4mC and 6mA, particularly relevant to bacterial epigenomics. Nanopore sequencing so far performs best at detecting 5mCG, although accuracy on m6A is comparable with PacBio [80], and the lower cost per Gb compared to SMRT make it suitable for larger genomes. Both technologies are compatible with hypothesis-free testing for new base modifications. The accuracy of the long-read technologies in detecting 5mC remains lower than that of bisulfite sequencing, a trend likely to hold true for other marks. This becomes particularly problematic for rare modifications, outside of motifs, where a high false positive rate may hide the signal in background noise. Orthogonal validation is therefore recommended [72]. Focusing on specific motifs where the mark of interest is abundant is a successful strategy to increase the signal-to-noise ratio. Improving the accuracy and range of detectable modifications depends on the generation of appropriate training data, where DNA containing known base modifications are present at known positions, in all biologically relevant motifs. Unfortunately, our ability to synthesise DNA trails our capacity to sequence it. However, increases in long-read sequencing throughput and progress in applying machine learning suggest that there are still accuracy gains to be made, both for PacBio and ONT sequencing. Nanopore basecallers that include modified bases are starting to emerge [76,85], foreshadowing a near future where base modifications are a standard component of DNA and RNA sequencing.
Optimisation of bisulfite sequencing for low input requirements have made it suitable for single-cell sequencing (Figure 1). While SMRT and nanopore sequencing are single-molecule techniques, they are not single-cell techniques and currently require in excess of 100 ng of DNA. The loss of base modifications during PCR is a major hurdle to adapting long-read sequencing to single-cell epigenomics.
In addition to the detection of base modifications not amenable to bisulfite sequencing, a major advantage of long-read sequencing is the ability to phase epigenetic and genetic information, providing allele-specific 5mC patterns that allow insight into the effect of mutations, structural variants, or parental origin on gene regulation [62,65,66,78]. Long-read sequencing also provides genetic and epigenetic information over repeat-rich regions that are refractory to short-read sequencing. There are a number of human diseases linked to repeat expansions and failed epigenetic regulation, which have been difficult to study with short reads [90]. Long-read sequencing is expected to greatly contribute to the diagnostic and molecular understanding of these conditions.
Rapid iterations over ONT nanopore chemistry, protocols and software are both exciting and challenging. Contrary to bisulfite sequencing, there are few established analytical pipelines for long-read epigenetics. Benchmarking efforts are crucial to evaluate the performances of the available tools. While we are still a long way from reading out the 43 base modifications listed in DNAmod [4], long-read sequencing brings us closer to obtaining full-length, complete, and phased epigenomes.
Summary
Variations of short-read bisulfite sequencing allow the mapping of 5mC, 5hmC, 5fC, and 5caC at base resolution.
Single-cell methylomes can be obtained with bisulfite sequencing and combined with other omics to study epigenetic regulation at single-cell resolution.
PacBio most sensitively detects 6mA and 4mC and is most adapted to bacterial epigenomics.
Nanopore sequencing can resolve 6mA, 5mC, 5hmC, and BrdU in single molecules and is under active development.
Long reads improve phasing, genomic coverage and completeness of epigenomes without specialised chemistries, compared to bisulfite sequencing.
Abbreviations
- ONT
Oxford Nanopore Technologies
- PacBio
Pacific Biosciences
- RRBS
reduced representation bisulfite sequencing
- SMRT sequencing
single molecule real time
- SNP
single-nucleotide polymorphism
- WGBS
whole-genome bisulfite sequencing
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
Q.G. and A.K. contributed equally to the design and writing of the review.
Funding
A.K. receives funding from the National Health and Medical Research Council [grant number 1140976].
References
- 1.Sánchez-Romero M.A., Cota I. and Casadesús J. (2015) DNA methylation in bacteria: from the methyl group to the methylome. Curr. Opin. Microbiol. 25, 9–16 10.1016/j.mib.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 2.Zhang H., Lang Z. and Zhu J.-K. (2018) Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489 10.1038/s41580-018-0016-z [DOI] [PubMed] [Google Scholar]
- 3.Li E. and Zhang Y. (2014) DNA methylation in mammals. Cold Spring Harbor Perspect. Biol. 6, a019133 10.1101/cshperspect.a019133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sood A.J., Viner C. and Hoffman M.M. (2019) DNAmod: the DNA modification database. J. Cheminformat. 11, 30 10.1186/s13321-019-0349-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korlach J. and Turner S.W. (2012) Going beyond five bases in DNA sequencing. Curr. Opin. Struct. Biol. 22, 251–261 10.1016/j.sbi.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 6.Feng S., Jacobsen S.E. and Reik W. (2010) Epigenetic reprogramming in plant and animal development. Science 330, 622–627 10.1126/science.1190614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones P.A. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 8.Smith Z.D. and Meissner A. (2013) DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- 9.Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D. et al. (2008) Shotgun bisulphite sequencing of the arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 10.1038/nature06745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lister R., O’Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H. et al. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 10.1016/j.cell.2008.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurdyukov S. and Bullock M. (2016) DNA methylation analysis: choosing the right method. Biology (Basel) 5, 3 10.3390/biology5010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olkhov-Mitsel E. and Bapat B. (2012) Strategies for discovery and validation of methylated and hydroxymethylated DNA biomarkers. Cancer Med. 1, 237–260 10.1002/cam4.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yong W.S., Hsu F.M. and Chen P.Y. (2016) Profiling genome-wide DNA methylation. Epigenetics Chromatin 9, 26 10.1186/s13072-016-0075-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W. et al. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 89, 1827–1831 10.1073/pnas.89.5.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bewick A.J., Vogel K.J., Moore A.J. and Schmitz R.J. (2016) Evolution of DNA methylation across Insects. Mol. Biol. Evol. 34, 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao F., Liu X., Wu X.-P., Wang X.-L., Gong D., Lu H. et al. (2012) Differential DNA methylation in discrete developmental stages of the parasitic nematode Trichinella spiralis. Genome Biol. 13, R100 10.1186/gb-2012-13-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J.U., Su Y., Shin J.H., Shin J., Li H., Xie B. et al. (2014) Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 17, 215–222 10.1038/nn.3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G. et al. (2011) Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 10.1038/nature09798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma H., Morey R., O’Neil R.C., He Y., Daughtry B., Schultz M.D. et al. (2014) Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511, 177–183 10.1038/nature13551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziller M.J., Muller F., Liao J., Zhang Y., Gu H., Bock C. et al. (2011) Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet 7, e1002389 10.1371/journal.pgen.1002389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niederhuth C.E., Bewick A.J., Ji L., Alabady M.S., Kim K.D., Li Q. et al. (2016) Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17, 194 10.1186/s13059-016-1059-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkan C., Coe B.P. and Eichler E.E. (2011) Genome structural variation discovery and genotyping. Nat. Rev. Genet. 12, 363–376 10.1038/nrg2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts R.J., Carneiro M.O. and Schatz M.C. (2013) The advantages of SMRT sequencing. Genome Biol. 14, 405 10.1186/gb-2013-14-6-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treangen T.J. and Salzberg S.L. (2011) Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13, 36–46 10.1038/nrg3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaneau O., Howie B., Cox A.J., Zagury J.F. and Marchini J. (2013) Haplotype estimation using sequencing reads. Am. J. Hum. Genet. 93, 687–696 10.1016/j.ajhg.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziller M.J., Hansen K.D., Meissner A. and Aryee M.J. (2015) Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat. Methods 12, 230–232, 1 p following 232 10.1038/nmeth.3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masser D.R., Berg A.S. and Freeman W.M. (2013) Focused, high accuracy 5-methylcytosine quantitation with base resolution by benchtop next-generation sequencing. Epigenetics Chromatin 6, 33 10.1186/1756-8935-6-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masser D.R., Stanford D.R. and Freeman W.M. (2015) Targeted DNA methylation analysis by next-generation sequencing. JoVE (J. Visual. Exp.) 96, e52488 10.3791/52488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee E.-J., Pei L., Srivastava G., Joshi T., Kushwaha G., Choi J.-H. et al. (2011) Targeted bisulfite sequencing by solution hybrid selection and massively parallel sequencing. Nucleic Acids Res. 39, e127–e127 10.1093/nar/gkr598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q., Suzuki M., Wendt J., Patterson N., Eichten S.R., Hermanson P.J. et al. (2015) Post-conversion targeted capture of modified cytosines in mammalian and plant genomes. Nucleic Acids Res. 43, e81–e81 10.1093/nar/gkv244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masser D.R., Stanford D.R., Hadad N., Giles C.B., Wren J.D., Sonntag W.E. et al. (2016) Bisulfite oligonucleotide-capture sequencing for targeted base-and strand-specific absolute 5-methylcytosine quantitation. Age 38, 49 10.1007/s11357-016-9914-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wendt J., Rosenbaum H., Richmond T.A., Jeddeloh J.A. and Burgess D.L. (2018) Targeted bisulfite sequencing using the SeqCap Epi enrichment system. DNA Methylation Protocols, pp. 383–405, Springer; [DOI] [PubMed] [Google Scholar]
- 33.Kacmarczyk T.J., Fall M.P., Zhang X., Xin Y., Li Y., Alonso A. et al. (2018) “Same difference”: comprehensive evaluation of four DNA methylation measurement platforms. Epigenetics Chromatin 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meissner A., Gnirke A., Bell G.W., Ramsahoye B., Lander E.S. and Jaenisch R. (2005) Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 10.1093/nar/gki901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A. et al. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 10.1038/nature07107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deaton A.M. and Bird A. (2011) CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022 10.1101/gad.2037511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu H., Smith Z.D., Bock C., Boyle P., Gnirke A. and Meissner A. (2011) Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 6, 468–481 10.1038/nprot.2010.190 [DOI] [PubMed] [Google Scholar]
- 38.Smith Z.D., Gu H., Bock C., Gnirke A. and Meissner A. (2009) High-throughput bisulfite sequencing in mammalian genomes. Methods 48, 226–232 10.1016/j.ymeth.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doi A., Park I.H., Wen B., Murakami P., Aryee M.J., Irizarry R. et al. (2009) Differential methylation of tissue- and cancer-specific cpg island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41, 1350–1353 10.1038/ng.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P. et al. (2009) The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissuespecific CpG island shores. Nat. Genet. 41, 178–186 10.1038/ng.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akalin A., Garrett-Bakelman F.E., Kormaksson M., Busuttil J., Zhang L., Khrebtukova I. et al. (2012) Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 8, e1002781 10.1371/journal.pgen.1002781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyle P., Clement K., Gu H., Smith Z.D., Ziller M., Fostel J.L. et al. (2012) Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 13, R92 10.1186/gb-2012-13-10-r92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Branco M.R., Ficz G. and Reik W. (2011) Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13 10.1038/nrg3080 [DOI] [PubMed] [Google Scholar]
- 44.Wu X. and Zhang Y. (2017) TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 10.1038/nrg.2017.33 [DOI] [PubMed] [Google Scholar]
- 45.Huang Y., Pastor W.A., Shen Y., Tahiliani M., Liu D.R. and Rao A. (2010) The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5, e8888 10.1371/journal.pone.0008888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Booth M.J., Branco M.R., Ficz G., Oxley D., Krueger F., Reik W. et al. (2012) Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 10.1126/science.1220671 [DOI] [PubMed] [Google Scholar]
- 47.Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A. et al. (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 10.1016/j.cell.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song C.X., Szulwach K.E., Dai Q., Fu Y., Mao S.Q., Lin L. et al. (2013) Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691 10.1016/j.cell.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Booth M.J., Marsico G., Bachman M., Beraldi D. and Balasubramanian S. (2014) Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat. Chem. 6, 435–440 10.1038/nchem.1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu X., Song C.X., Szulwach K., Wang Z., Weidenbacher P., Jin P. et al. (2013) Chemical modification-assisted bisulfite sequencing (CAB-seq) for 5-carboxylcytosine detection in DNA. J. Am. Chem. Soc. 135, 9315–9317 10.1021/ja4044856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miura F., Enomoto Y., Dairiki R. and Ito T. (2012) Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 10.1093/nar/gks454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smallwood S.A., Lee H.J., Angermueller C., Krueger F., Saadeh H., Peat J. et al. (2014) Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 10.1038/nmeth.3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu P., Gao Y., Guo W. and Zhu P. (2019) Using local alignment to enhance single cell bisulfite sequencing data efficiency. Bioinformatics 35, 3273–3278 10.1093/bioinformatics/btz125 [DOI] [PubMed] [Google Scholar]
- 54.Guo H., Zhu P., Guo F., Li X., Wu X., Fan X. et al. (2015) Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat. Protoc. 10, 645–659 10.1038/nprot.2015.039 [DOI] [PubMed] [Google Scholar]
- 55.Angermueller C., Clark S.J., Lee H.J., Macaulay I.C., Teng M.J., Hu T.X. et al. (2016) Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 10.1038/nmeth.3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clark S.J., Argelaguet R., Kapourani C., Stubbs T.M., Lee H.J., Alda-Catalinas C. et al. (2018) scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 10.1038/s41467-018-03149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo F., Li L., Li J., Wu X., Hu B., Zhu P. et al. (2017) Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 27, 967–988 10.1038/cr.2017.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bock C., Farlik M. and Sheffield N.C. (2016) Multi-omics of single cells: Strategies and applications. Trends Biotechnol. 34, 605–608 10.1016/j.tibtech.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clark S.J., Lee H.J., Smallwood S.A., Kelsey G. and Reik W. (2016) Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 17, 72 10.1186/s13059-016-0944-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelsey G., Stegle O. and Reik W. (2017) Single-cell epigenomics: Recording the past and predicting the future. Science 358, 69–75 10.1126/science.aan6826 [DOI] [PubMed] [Google Scholar]
- 61.Macaulay I.C., Ponting C.P. and Voet T. (2017) Single-cell multiomics: Multiple measurements from single cells. Trends Genet. 33, 155–168 10.1016/j.tig.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gigante S., Gouil Q., Lucattini A., Keniry A., Beck T., Tinning M. et al. (2019) Using long-read sequencing to detect imprinted DNA methylation. Nucleic Acids Res. 47, e46 10.1093/nar/gkz107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kingan S.B., Heaton H., Cudini J., Lambert C.C., Baybayan P., Galvin B.D. et al. (2019) A high-quality de novo genome assembly from a single mosquito using PacBio sequencing. Genes 10, 62 10.3390/genes10010062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai Y.-C., Greenberg D., Powell J., Höijer I., Ameur A., Strahl M. et al. (2017) Amplification-free, CRISPR-Cas9 targeted enrichment and SMRT sequencing of repeat-expansion disease causative genomic regions. bioRxiv 10.1101/203919 [DOI] [Google Scholar]
- 65.Gilpatrick T., Lee I., Graham J.E., Raimondeau E., Bowen R., Heron A. et al. (2019) Targeted nanopore sequencing with Cas9 for studies of methylation, structural variants, and mutations. bioRxiv 10.1101/604173 [DOI] [Google Scholar]
- 66.Gießelmann P., Brändl B., Raimondeau E., Bowen R., Rohrandt C., Tandon R. et al. (2018) Repeat expansion and methylation state analysis with nanopore sequencing. bioRxiv 10.1101/480285 [DOI] [PubMed] [Google Scholar]
- 67.Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C., Clark T.A., Korlach J. et al. (2010) Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461 10.1038/nmeth.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pacific Biosciences (2015) Detecting DNA base modifications using single molecule, real-time sequencing. Tech. Rep. https://www.pacb.com/wp-content/uploads/2015/09/WP_Detecting_DNA_Base_Modifications_Using_SMRT_Sequencing.pdf [Accessed 13/11/2019] [Google Scholar]
- 69.Clark T.A., Lu X., Luong K., Dai Q., Boitano M., Turner S.W. et al. (2013) Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 11, 4 10.1186/1741-7007-11-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beaulaurier J., Zhang X.-S., Zhu S., Sebra R., Rosenbluh C., Deikus G. et al. (2015) Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat. Commun. 6, 7438 10.1038/ncomms8438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clark T.A., Murray I.A., Morgan R.D., Kislyuk A.O., Spittle K.E., Boitano M. et al. (2011) Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 40, e29–e29 10.1093/nar/gkr1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beaulaurier J., Schadt E.E. and Fang G. (2018) Deciphering bacterial epigenomes using modern sequencing technologies. Nat. Rev. Genet. 20, 157–172 10.1038/s41576-018-0081-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genest P.-A., Baugh L., Taipale A., Zhao W., Jan S., van Luenen H.G.A.M. et al. (2015) Defining the sequence requirements for the positioning of base J in DNA using SMRT sequencing. Nucleic Acids Res. 43, 2102–2115 10.1093/nar/gkv095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schadt E.E., Banerjee O., Fang G., Feng Z., Wong W.H., Zhang X. et al. (2013) Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 23, 129–141 10.1101/gr.136739.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pacfic Biosciences (2019) Methylome analysis technical note. https://github.com/PacificBiosciences/Bioinformatics-Training/wiki/Methylome-Analysis-Technical-Note [Online; accessed 22 October 2019] [Google Scholar]
- 76.Oxford Nanopore Technologies. (2019) Guppy, https://community.nanoporetech.com/downloads/guppy/release_notes [Accessed 13/11/2019] [Google Scholar]
- 77.Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J. and Timp W. (2017) Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 10.1038/nmeth.4184 [DOI] [PubMed] [Google Scholar]
- 78.Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A. et al. (2018) Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 10.1038/nbt.4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rand A.C., Jain M., Eizenga J.M., Musselman-Brown A., Olsen H.E., Akeson M. et al. (2017) Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 14, 411–413 10.1038/nmeth.4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McIntyre A.B.R., Alexander N., Grigorev K., Bezdan D., Sichtig H., Chiu C.Y. et al. (2019) Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat. Commun. 10, 579 10.1038/s41467-019-08289-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ni P., Huang N., Zhang Z., Wang D.-P., Liang F., Miao Y. et al. (2019) DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics 10.1093/bioinformatics/btz276 [DOI] [PubMed] [Google Scholar]
- 82.Liu Q., Fang L., Yu G., Wang D., Xiao C.-l. and Wang K. (2019) Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 10, 2449 10.1038/s41467-019-10168-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oxford Nanopore Technologies (2019) Megalodon. https://github.com/nanoporetech/megalodon [Online; accessed 22 October 2019] [Google Scholar]
- 84.Müller C.A., Boemo M.A., Spingardi P., Kessler B.M., Kriaucionis S., Simpson J.T. et al. (2019) Capturing the dynamics of genome replication on individual ultra-long nanopore sequence reads. Nat. Methods 16, 429–436 10.1038/s41592-019-0394-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hennion M., Arbona J.-M., Cruaud C., Proux F., Tallec B.L., Novikova E. et al. (2018) Mapping DNA replication with nanopore sequencing. bioRxiv 10.1101/426858 [DOI] [Google Scholar]
- 86.Stoiber M.H., Quick J., Egan R., Lee J.E., Celniker S.E., Neely R. et al. (2017) De novo Identification of DNA Modifications Enabled by Genome-Guided Nanopore Signal Processing. bioRxiv 094672 10.1101/094672 [DOI] [Google Scholar]
- 87.Liu Q., Georgieva D.C., Egli D. and Wang K. (2019) NanoMod: a computational tool to detect DNA modifications using Nanopore long-read sequencing data. BMC Genomics 20, 78 10.1186/s12864-018-5372-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oxford Nanopore Technologies (2019) Taiyaki. https://github.com/nanoporetech/taiyaki [Online; accessed 22 October 2019] [Google Scholar]
- 89.Teng H., Cao M.D., Hall M.B., Duarte T., Wang S. and Coin L.J.M. (2018) Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. GigaScience 7, 10.1093/gigascience/giy037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paulson H. (2018) Repeat expansion diseases. Handbook of Clinical Neurology, vol. 147, pp. 105–123, Elsevier; [DOI] [PMC free article] [PubMed] [Google Scholar]