Abstract
Monoallelic gene expression occurs in diploid cells when only one of the two alleles of a gene is active. There are three main classes of genes that display monoallelic expression in mammalian genomes: (1) imprinted genes that are monoallelically expressed in a parent-of-origin dependent manner; (2) X-linked genes that undergo random X-chromosome inactivation in female cells; (3) random monoallelically expressed single and clustered genes located on autosomes. The heritability of monoallelic expression patterns during cell divisions implies that epigenetic mechanisms are involved in the cellular memory of these expression states. Among these, methylation of CpG sites on DNA is one of the best described modification to explain somatic inheritance. Here, we discuss the relevance of DNA methylation for the establishment and maintenance of monoallelic expression patterns among these three groups of genes, and how this is intrinsically linked to development and cellular states.
Keywords: DNA methylation, epigenetics, imprinting, monoallelic expression, X-chromosome inactivation
Introduction
In diploid organisms, somatic cells possess two alleles of each gene that are in most cases expressed at the same time and at similar levels. However, some genes can be expressed either strictly or preferentially from either one of the two alleles. This phenomenon, known as monoallelic expression, is stable and clonally inherited during cell divisions. Monoallelic expression is often associated with DNA sequence polymorphisms, within regulatory regions for example, which may render one of the two alleles of the gene less expressed or completely silent. However, monoallelic expression can also arise from differential epigenetic marks decorating at least one of the two alleles of a gene, without any changes in the underlying DNA sequence. Epigenetically based monoallelic expression is usually associated with a programmed necessity to regulate gene dosage during development, as biallelic expression can be associated with severe phenotypes, or as a means to enhance cellular diversity and specificity [1,2].
Epigenetically based monoallelic expression can be imprinted; in this case, a gene is expressed in a parent-of-origin dependent manner from either the paternal or the maternal allele in all cells [3]. Monoallelic expression can also be random, where a gene can be expressed either from the paternal or from the maternal allele in different cells. The most classic example of random monoallelic expression (RME) concerns X-linked genes that undergo X-chromosome inactivation (XCI) during early embryonic development in female cells [4]. RME also affects large families of autosomal genes located in clusters, such as antigen receptors (AgRs), olfactory receptors (ORs) or protocadherins (Pcdh), which are expressed in a highly cell type-specific manner [2]. Moreover, RME also occurs at the level of individual autosomal genes that belong to a wide variety of gene ontologies, which were identified through genome-wide studies in polymorphic clonal cell populations [5,6].
The stable maintenance and heritability of monoallelic expression during cell divisions imply that epigenetic mechanisms are at play to maintain these expression states. These mechanisms include nuclear organisation, DNA replication timing, histone modifications and also DNA methylation [1,2], the best-described modification for somatic inheritance of transcriptional states [7]. DNA methylation consists of the addition of methyl groups to specific nucleotides on the DNA molecule. In mammals, the 5-methylcytosine (5mC – fifth carbon of the pyrimidine ring) is the major form of DNA modification, which occurs predominantly on both complementary strands of the palindromic CpG dinucleotide [8]. In higher eukaryote genomes, 5mC is mostly targeted to repetitive sequences, but is also present on gene bodies of active genes. Interestingly, CpG dinucleotides are highly methylated; however, 5mC is generally absent from CpG-dense regions, commonly known as CpG islands (CGI), frequently present at promoters [9]. There are a few exceptions to this rule particularly in developmental and disease contexts [10,11]. A notable example discussed in this review is the case of monoallelically expressed genes, where methylation of the promoter is often associated with the silent allele [12].
5mC is established and maintained through cell division by respectively, de novo (DNMT3A, DNMT3B and the sperm-specific DNMT3C) and maintenance DNA methyltransferases (DNMT1) [8,13]. Their actions are assisted by specific accessory proteins such as the DNMT3-like (DNMT3L) and the ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) [14–16]. Removal of 5mC can be achieved by passive loss through DNA replication in the absence of DNMT1 or through active pathways involving the conversion of 5mC into 5-hydroxymethylcytosine by the ten-eleven translocation (TET) enzymes (TET1, TET2 and TET3) or by less-understood DNA repair pathways involving the activation-induced deaminase (AID) and thymine-DNA glycosylase [17,18]. In somatic cells, DNA methylation patterns are remarkably stable and only subtle changes can be induced through environmental causes or ageing [19,20]. In contrast, DNA methylation is highly dynamic during development. For instance, during the mammalian life cycle, two major waves of global DNA demethylation of the genome occur: (1) during the early specification of the germline lineage, which is followed by sex-specific remethylation in the specialised gametes; (2) during pre-implantation stage, which is then followed by stage- and tissue-specific re-establishment of DNA methylation after implantation [8,11,21]. Interestingly, monoallelic expression is established at different stages of the DNA methylation cycle. In this review, we summarise the current understanding about the role of DNA methylation in regulating expression of the three classes of monoallelically expressed genes in mammalian cells.
Genomic imprinting: parental allele-specific gene expression determined by DNA methylation
Genomic imprinting is an epigenetic mechanism affecting over 100 genes that are expressed from only one of the two parental alleles [3]. Imprinted genes exert important roles in growth and development of the foetus and placenta during intra-uterine life, as well as in brain functions and metabolic pathways in adults [3,22]. Dysregulation of the dosage of imprinted genes is associated with several (neuro)-developmental, such as Angelman or Beckwith–Weidemann syndromes [23].
Imprinted genes tend to be located near each other in genomic regions called imprinted clusters. These regions contain both maternally and paternally imprinted genes and sometimes non-imprinted genes [3]. Interestingly, imprinted expression across these clusters is coordinated and depends on shared regulatory DNA regions, of which the most important are imprinting control regions (ICRs). These are CpG-dense regions that have been defined for all the imprinted clusters described in the mammalian genome [3]. Deletion of ICRs affects imprinted expression and results in either biallelic or no expression of several genes within the same cluster [24,25], confirming their crucial role in cis-acting long-range imprinting control. The most distinctive feature of ICRs is their differential DNA methylation state between the maternally and paternally inherited alleles, which is established in the germline (Figure 1A,B). Besides ICRs, there are other differentially methylated regions (DMRs) between the parental alleles within imprinted clusters. Those, known as somatic DMRs, acquire differential DNA methylation after implantation and are hierarchically dependent on the methylation status of the ICRs [26,27].
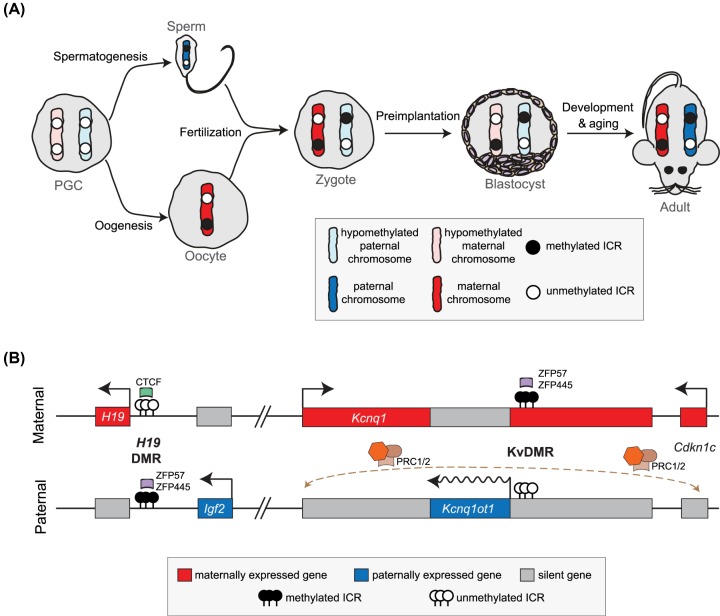
Figure 1. Genomic imprinting regulation by parental allele-specific DNA methylation.
(A) The imprinting cycle: the scheme illustrates the overall changes in DNA methylation at ICRs (both at ICRs methylated on the maternal inherited allele and unmethylated on the paternal inherited allele and vice versa) from PGC (primordial germ cells), mature germ cells (sperm and oocyte), zygote, blastocyst to the adult mouse; overall methylation status is simplified in two states represented with graded colours for both the maternally and paternally inherited chromosomes: maternal/paternal chromosome corresponds to normal methylation levels of somatic cells; hypomethylated maternal/paternal chromosomes correspond to reduced methylation levels upon major waves of DNA demethylation. (B) Representation of Igf2-H19 and Kcnq1/Kcnq1ot1 imprinted clusters on distal end of mouse chromosome 7 or on human 11p15.5 chromosomal region. Igf2-H19 imprinting is regulated by the H19 DMR, an intergenic ICR methylated on the paternal allele and unmethylated on the maternal allele; the unmethylated allele is bound by CTCF, which is involved in 3D chromatin looping and important for imprinting regulation at this cluster; the methylated allele is bound by both ZFP57 and/or ZFP445 that are crucial to protect the methylated state during the major epigenetic remodelling observed in pre-implantation stages. The Kcnq1/Kcnq1ot1 cluster is represented here in a simplified fashion; this imprinted cluster is regulated by the KvDMR, an ICR located at the promoter of the Kcnq1ot1 lncRNA, which is methylated and bound by ZFP57 and/or ZFP445 on the maternal allele. Paternally expressed Kcnq1ot1 lncRNA is believed to recruit Polycomb repressive complexes 1 and 2 (PRC1/2) (dashed arrows) to silence protein-coding genes such as Kcnq1 and Cdkn1c. This scheme is not drawn to scale.
Imprinting cycle
A role for DNA methylation as a central epigenetic mechanism in genomic imprinting was unveiled shortly after the first imprinted genes were discovered. Consistent with the parental allele-specific differences in DNA methylation encountered at imprinted loci [28–31], Dnmt1−/− embryos were shown to be unable to sustain imprinted expression [30]. Moreover, DNA methylation was also shown to be crucial for the establishment of genomic imprinting. Indeed, DNMT3A and its cofactor DNMT3L are necessary for the correct establishment of methylation marks or imprints at ICRs in the germline [15,32]. The majority of mouse ICRs (22 out of 25) acquire DNA methylation in the oocyte and map to CpG-dense promoters, while only 3 ICRs, located in intergenic regions, have sperm-derived imprints (http://www.mousebook.org/mousebook-catalogs/imprinting-resource). Interestingly, DNA methylation does not seem to be specifically imposed on maternal and paternal imprints. This rather occurs as part of the genome-wide DNA methylation programme that targets preferentially transposons and intergenic regions during spermatogenesis [33,34], and gene bodies and intragenic CGI during oogenesis [34–37]. One distinctive feature of ICRs compared with the rest of gametic-specific DMRs is their ability to resist the genome-wide DNA demethylation wave during pre-implantation development [38] (Figure 1A). Several factors including DNMT1, PGC7/STELLA/DPPA3, KAP1/TRIM28 and NAA10P have been implicated [39–43]. But the specificity of ICRs to escape the genome-wide demethylation seems to be associated with the binding of the ZFP57 and ZNF445/ZFP445 zinc-finger proteins to methylated ICRs [41,44]. This mechanism is well established for the ZFP57 protein, which binds the TGCCGC motif present at ICRs in a methylation-dependent manner and recruits epigenetic modifiers through its association with the KAP1 cofactor [45,46]. Both ZFP57 and ZNF445 seem to be required to protect imprinting memory, but their requirement varies for different imprinted clusters [41,44]. Interestingly, there are a few documented cases, where gametic-specific DMRs are lost [34,47]. This occurs, for example at the Gpr1/Zdbf2 locus, where differential parental allele-specific methylation is lost due to gain of methylation of the paternal allele around implantation [48]. This is, nonetheless, sufficient to establish life-long imprinted expression [48,49]. Conversely, unmethylated ICRs also resist global de novo DNA methylation around the time of implantation [38] (Figure 1A). The mechanisms for this protection remain poorly studied. In the case of the Igf2/H19 locus, binding of the zinc finger protein CTCF or the OCT4/SOX2 pluripotency factors to the unmethylated maternal allele have been implicated in this protection [50–52], but it is not known whether this applies to other imprinted loci. It is also during this period that methylation of most somatic DMRs is established [26,27].
Once surviving the dynamic changes occurring during early development, parental allele-specific differences in DNA methylation at ICRs and at most somatic DMRs remain remarkably stable in somatic cells throughout life [53] (Figure 1A). Interestingly, many genes exhibit tissue-specific imprinted expression [54]. This relies on secondary transcriptional and chromatin states, which are nonetheless initially imposed by the methylation status of the ICR. In contrast to somatic cells, aberrations in methylation at ICRs were reported in cancer cells, leading to loss of monoallelic expression of many imprinted genes [55,56]. Interestingly, a recent study suggests that DNA methylation changes at ICRs in cancer cells are mostly caused by locus-specific copy number aberrations rather than epigenetic alterations [57], suggesting a less labile methylation pattern at ICRs than originally thought. Environmental factors are also known to influence epigenetic states [19], but genomic imprinting is not believed to be neither more vulnerable nor protected from environmental perturbations during development [58].
To complete the imprinting cycle, parental allele-specific DNA methylation differences in the ICRs are erased early in the germline lineage to be reset during gametogenesis according to the sex of the individual (Figure 1A). This occurs during the second major wave of DNA demethylation from which imprints do not escape [59–62]. This starts from embryonic day 8 in the mouse and involved multiple passive, such as the down-regulation of UHRF1, and active mechanisms, through the action of AID and TET1/TET2 proteins, which are part of the major epigenetic reprogramming events that lead to massive chromatin changes in primordial germ cells [63].
Regulation of imprinted clusters
ICRs are enigmatic cis-acting DNA regions that dictate imprinted expression across an imprinted cluster, which can contain up to ten genes and span up to 4 Mb in size. How this is mechanistically regulated is not completely understood and might vary from loci to loci. There are two main models to explain the coordinated regulation of imprinted expression within a cluster: the insulator and the lncRNA models [64]. The insulator model was proposed to explain imprinting regulation at the Igf2-H19 cluster (Figure 1B). The intergenic ICR, located between the two genes, acts as a binding site for CTCF only on the unmethylated maternal allele. CTCF, which is a major regulator of the 3D chromatin structure [65], induces chromatin loops believed to prevent interaction of Igf2 with downstream enhancers on the maternal allele [66,67]. Absence of CTCF binding to the methylated paternal ICR allows Igf2 to interact with its enhancers, which results in its paternal-specific expression (Figure 1B). This is illustrative of how parental allele-specific epigenetic differences at ICRs might reshape the 3D conformation of imprinted regions and affect gene expression. The lncRNA model results from the fact that many imprinted clusters contain genes encoding lncRNAs (e.g. Airn, Kcnq1ot1, Nespas), which are themselves subject to imprinted expression. In these clusters, ICRs are often located around the promoter region of a lncRNA gene, which is expressed from the unmethylated allele, while protein-coding genes are preferentially expressed from the opposite allele [64], as illustrated by the Kcnq1-Kcnq1ot1 imprinted cluster (Figure 1B). Deletion experiments for some lncRNAs result in biallelic expression of the protein-coding genes, suggesting that these lncRNAs function as cis-acting silencers [68–70]. Two major mechanisms, not mutually exclusive, have been proposed to explain how imprinted lncRNAs silence neighbouring genes. This could occur through transcriptional interference of sense–antisense pairs of protein-coding/lncRNA transcripts [71]; or formation of a silent compartment reminiscent of the one induced by the Xist lncRNA on the inactive X chromosome (Xi) [72], as proposed for the Kcnq1ot1 and Airn lncRNAs [73,74]. Recent data have strengthened this analogy by showing that both lncRNAs are able to spread chromatin marks imposed by the Polycomb repressive complexes 1 and 2 (PRC1 and PRC2) across the imprinted cluster, which might be mediated by the HNRNPK RNA-binding protein as for Xist lncRNA [75].
DNA methylation-independent imprinting
Parental allele-specific differences in DNA methylation are key to the regulation of genomic imprinting. Interestingly however, DNA methylation-independent imprinting has been recently described. This new form of genomic imprinting is controlled by the H3K27me3 histone mark, deposited by PRC2 in oocytes [76]. Asymmetric differences in H3K27me3 between gametes persist past fertilisation during preimplantation development and determine paternal-specific expression of a few genes. An interesting example is the silencing of the maternal copy of the Xist gene, which results in specific inactivation of the paternal X chromosome during pre-implantation development in female murine embryos [77]. However, in contrast with canonical imprinting, H3K27me3-dependent genomic imprinting is lost after implantation and only five genes were found to remain imprinted in extra-embryonic lineages [76]. Genomic imprinting remains therefore a classical example of an epigenetic mechanism dictating monoallelic expression of genes, which is predominantly dependent on germline-specified DNA methylation.
X-chromosome inactivation: differential methylation patterns of the active and inactive X chromosomes
XCI is an epigenetic mechanism, which allows dosage compensation of X-linked genes between XX females and XY males in mammals. This process leads to chromosome-wide silencing of one the two X chromosomes, chosen at random, during early embryonic development in female cells [78]. The way gene silencing is initiated and maintained has been the focus of intense studies and results from the interplay of multiple epigenetic mechanisms, including chromatin compaction, histone modifications and DNA methylation, which was proposed to play a role in XCI as early as 1975. Indeed, Riggs [79] argued for a role of DNA methylation in XCI based on a processive spreading mechanism along the chromosome and a model of inheritance of methylation patterns during DNA replication. The first experimental evidence demonstrating the link between DNA methylation and XCI came shortly after from studies revealing that promoter regions of selected X-linked genes are differentially methylated on the two X chromosomes in somatic cells [80–84]. Furthermore, treatment of mouse–human somatic cell hybrids or mouse transformed cells with 5-azacytidine, a drug that inhibits the activity of DNMTs, was shown to lead to sporadic reactivation of selected genes on the Xi [85–87]. These early studies were seminal in establishing a role for DNA methylation in the maintenance of gene silencing on the Xi. Moreover, these analyses already pointed towards DNA methylation to likely be a late event during XCI, as silencing precedes methylation [88].
Despite its clear importance in XCI, genome-wide studies of DNA methylation with a focus on the X chromosome in female cells came later. A first study in human somatic cells using immunoprecipitation of methylated DNA combined with microarrays confirmed the overall hypermethylation of promoter associated-CGI on the Xi in female versus male cells [89]. In contrast, genes escaping XCI that are biallelically expressed remain unmethylated on both X chromosomes (Figure 2A). Surprisingly, this analysis also revealed that the level of CpG methylation along the X chromosome, particularly in gene-poor regions, is lower in female cells when compared with male cells [89]. These sex-specific differences reflect the overall reduced methylation of the Xi compared with the active X (Xa) and autosomes, and confirmed earlier cytogenetic analyses on metaphase chromosomes [90,91]. Another seminal study using microarrays described the first allele-specific analysis of DNA methylation on the Xi and Xa in human somatic cells, interrogating approximately 1000 informative loci along the chromosome [92]. This analysis also reported an overall excess of monoallelically methylated CpG on the Xa compared with the Xi within gene bodies (Figure 2A). Interestingly, some of these CpG sites are biallelically methylated in human embryonic stem (ES) cells prior to XCI [92]. In addition, bodies of genes escaping XCI were shown to be methylated on both X chromosomes [92,93], suggesting a correlation between gene body methylation and expression (Figure 2A). Subsequent genome-wide methylation analysis revealed that the bodies of active genes are indeed heavily methylated and represent the most conserved target of DNA methylation across eukaryote genomes [94,95].
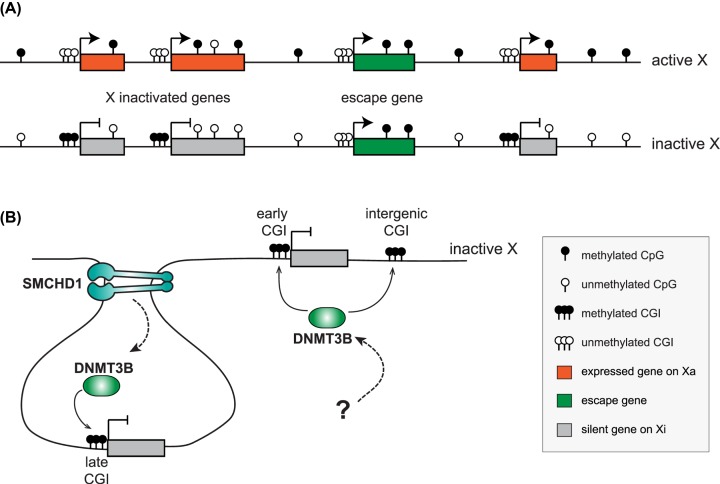
Figure 2. DNA methylation during XCI.
(A) DNA methylation profiles of the active and inactive X chromosomes illustrating the hypermethylation of promoter-CGIs linked to genes undergoing XCI on the Xi (orange/grey boxes), hypomethylation of promoter-CGI associated with escape genes (green boxes), gene-body methylation associated with gene expression on both Xa and Xi and hypomethylation of CpG sites on the Xi compared with Xa. (B) Illustration of the mechanisms involved in methylation of the Xi. DNMT3B establishes methylation of all CGI on the Xi (fast and slow methylating, as well as intergenic CGI). The mechanism of DNMT3 recruitment to the Xi remains unknown, but it is believed to be partially dependent on SMCHD1 function in chromatin architecture. Slow methylating islands are dependent on SMCHD1 for their methylation, whereas fast islands acquire methylation mostly independently of SMCHD1, defining a parallel unknown pathway. Abbreviation: SMCHD1, structural maintenance of chromosomes hinge domain-containing 1.
One study analysed the dynamics of Xi promoter-CGI methylation in a developmental context. This analysis revealed two modes of DNA methylation on the Xi. While most CGI acquire methylation slowly and late throughout development, a subset of CGI show fast methylation kinetics [96]. Interestingly, these CGI differ by their CpG composition, immediate genomic environment and expression levels prior to XCI [96].
Overall, these analyses revealed that differential methylation of the two X chromosomes in female cells is found at CGI associated with inactive gene promoters that are hypermethylated on the Xi and at both intergenic and intragenic sequences that are hypermethylated on the Xa (Figure 2A) [89,97]. While promoter-CGI methylation on the Xi is associated with monoallelic expression, intragenic methylation is associated with transcription and does not have any impact on allelic expression.
Mechanisms of methylation of the inactive X chromosome
Treatment of somatic cells with 5-azacytidine provided the first experimental evidence for the importance of DNA methylation in the maintenance of gene silencing on the Xi [85]. This opened the door to investigate which enzymes are necessary for XCI-induced DNA methylation and in dissecting the interplay between methylation and transcription in XCI. Analysis of mouse post-implantation embryos and differentiating ES cells mutant for Dnmt1 indicated that Xist is ectopically expressed from the Xa in a small proportion of cells in both males and females, which led to occasional aberrant silencing of X-linked genes [98]. This indicates that DNMT1 is essential for the stable maintenance of Xist monoallelic repression in differentiated cells, but is dispensable for the initiation of gene silencing on the Xi [98]. In another study, it was found that an X-linked LacZ transgene actually becomes reactivated in mutant embryos at later post-implantation stages after initial silencing, consistent with a role of DNA methylation in maintenance of silencing [99]. In contrast with the embryonic lineage, absence of DNMT1 does not majorly affect the Xi state in murine extraembryonic tissues, where an imprinted form of XCI with exclusive silencing of the paternal X chromosome takes place [99].
Analysis of embryos mutant for both de novo methyltransferases, Dnmt3a and Dnmt3b, reported hypomethylation of the Xist promoter, which was nevertheless associated with repression of the gene on the Xa [100]. Moreover, promoter-CGI on the Xi were shown to be extensively hypomethylated in double mutant embryos [100,101], however without derepression of the silent alleles. This indicates that the Xist-coated Xi undergoes XCI in mutant embryos and that the absence of both DNMT3A and DNMT3B does not impair neither initiation, nor propagation of XCI [100]. Analysis of single KO embryos revealed that methylation of CGI is actually dependent on the DNMT3B enzyme only, while DNMT3A and DNMT3L are dispensable [96]. This was the case for both fast and slow methylating CGI associated with promoters, but also intra- and intergenic CGI, indicating that DNMT3B establishes methylation of all CGI on the Xi (Figure 2B) [96,102]. This is in agreement with the observation showing reduced Xi CGI methylation in cells from patients with ICF (immunodeficiency centromeric-instability facial anomalies) syndrome, which is caused by mutations in the DNMT3B gene [103]. In agreement with previous observations, monoallelic expression appears to be stably maintained in Dnmt3b−/−females embryos, at least for the few genes tested [96,104]. However, chromosome-wide allele-specific expression analysis would be needed to determine whether some genes, such as those linked to fast methylating CGI [96], may be more sensitive, even sporadically, to the absence of methylation.
The structural maintenance of chromosomes hinge domain-containing 1 (SMCHD1) protein, a non-canonical SMC protein, was also shown to play a role in the maintenance of XCI and methylation of CGI on the Xi. Characterisation of female mutant post-implantation embryos indicated a widespread hypomethylation of most promoter-CGI on the Xi, associated with reactivation of a subset of genes, unlike in Dnmt3b−/− embryos [96,104,105]. Allele-specific analysis of hybrid Smchd1 null XX MEF cell lines confirmed these observations demonstrating extensive promoter-CGI hypomethylation in both gene-poor and gene-dense regions. This is accompanied by depletion of H3K27me3 and expression from the Xi for a large proportion of genes [97]. Direct comparison of Dnmt3b and Smchd1 KO embryos at similar stages indicated that CGI hypomethylation is not sufficient to explain the Smchd1-specific loss of silencing on the Xi [104], suggesting that this protein might act upstream of DNA methylation. Interestingly, recent work showed that SMCHD1 is involved in the establishment of the unique higher order chromatin architecture of the Xi [97,106,107]. This 3D chromatin organisation might be necessary to facilitate accessibility of de novo methyltransferases and subsequent promoter-CGI methylation to lock the inactive state (Figure 2B). Interestingly, fast methylating islands are methylated, at least in part, in an SMCHD1-independent manner [96], suggesting that other mechanisms are involved (Figure 2B).
In contrast with genomic imprinting that depends on defined parent-of-origin dependent DNA methylation patterns for monoallelic expression, methylation of promoter-CGI on the Xi occurs late during XCI and is not the driving force for initiating gene silencing. In any case, long-term silencing of X-linked genes definitely relies on DNA methylation, although this occurs in combination with other epigenetic marks. While it is clear that DNMT3B methylates all CGI on the Xi, it is currently unknown how this enzyme is recruited to the Xi during XCI. This could occur via recognition of specific features of Xi chromatin. It also remains unclear why X-linked gene reactivation following loss of promoter-CGI methylation only occurs in particular developmental or cellular contexts.
DNA methylation of random monoallelically expressed genes
Monoallelic expression also concerns autosomal genes that can be expressed randomly from either the paternal or the maternal allele, in a stable manner, and independently of DNA sequence polymorphisms. RME affects a wide variety of gene functions, from genes encoding cell surface-associated proteins to developmental transcription factors and may be involved in promoting not only cellular specificity, but also diversity in gene expression patterns [1]. An important class of RME genes comprises large clustered gene families, such as AgRs, ORs or Pcdh, that presumably play roles in generating specificity and identity at the cellular surface of highly specialised cells. RME of AgR loci, which undergo genetic recombination in developing B or T cells to generate one expressed functional allele, appears to be predetermined during early development. However, RME of Pcdh or OR genes occurs through a stochastic process. Pcdh genes show monoallelic and combinatorial expression of exons in individual Purkinje neurons in the cortex, while OR genes are expressed in a monogenic and monoallelic manner in olfactory neurons [2]. RME also occurs at the level of single genes, for which monoallelic expression is more labile and less well understood. RME for these genes is established during development or differentiation into particular lineages, presumably through a stochastic and independent regulation of the two alleles. They are thus usually expressed in a cell-type or tissue-specific manner and their expression can vary among individuals [1]. These characteristics make their study rather challenging.
Analysis of clonal cell populations in vitro revealed that single RME genes can be expressed monoallelically from either allele, but also biallelically or not expressed at all in independent clones, in contrast with imprinted and X-inactivated genes (Figure 3A) [5,6]. Moreover, these expression patterns were found to be remarkably stable during cell passaging and differentiation of neural progenitor cell (NPC) clones [5], raising the question of what epigenetic mechanisms could account for this stability. Again, studies in clonal cell populations allowed testing whether promoter DNA methylation could be involved in monoallelic silencing. Bisulfite-based methods were used to measure the methylation levels of CGI associated with the promoters of a few RME genes in NPC clones [5,6]. This analysis revealed that approximately half of the genes analysed show good correlation between the methylation levels (absent, intermediate or full) and the expression status (biallelic, monoallelic or not expressed) in a given NPC clone (Figure 3A) [5,6]. However, for other RME genes, there was no clear correlation between promoter-CGI methylation levels and expression [5,6]. In another study, the identification of sequences with dual methylation patterns, i.e. showing both methylated and unmethylated states, was used to identify new genes showing monoallelic expression in the central nervous system, of which 12% were confirmed to show RME in clonal cell populations [108]. Additionally, a genome-wide analysis of DNA methylation in human clonal neural stem cell lines using Illumina Infinium methylation beadchip reported a significant correlation between monoallelic expression and intermediate levels of DNA methylation, whereas biallelic genes were hypomethylated [109]. In contrast, no evidence for DNA methylation imbalances associated with allele-specific expression was found in a separate genome-wide study in humans, except when linked to variations in DNA sequences (see last paragraph) [110]. It should be noted however that this analysis was not performed on clonal cell populations unlike other studies, thus differences in DNA methylation could potentially be masked.
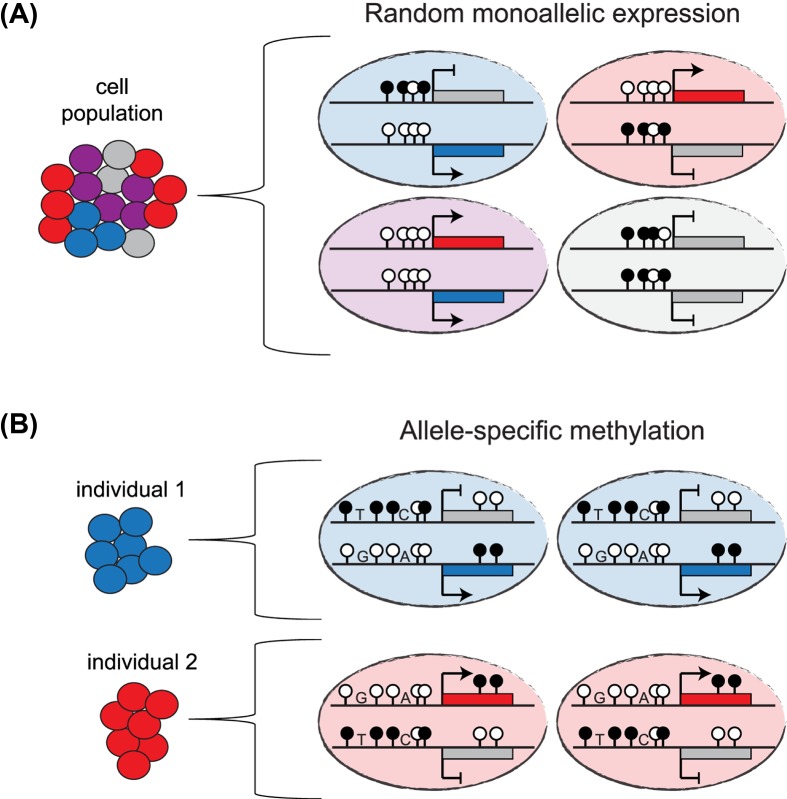
Figure 3. Allele-specific DNA methylation imbalances associated with RME or with DNA sequence polymorphisms.
(A) Schematic representation of the possible allele-specific expression states for an RME gene whose expression shows good correlation with DNA methylation levels. A cell population with four representative cells are shown. The RME gene is expressed from the unmethylated paternal allele in blue cells, from the unmethylated maternal in red cells, biallelically expressed in purple cells and not expressed in grey cells. (B) Schematic representation of allele-specific DNA methylation (ASM) associated with DNA sequence polymorphisms and allele-specific expression imbalance in two individuals. Here, the methylation status and the allelic expression are dependent on cis-acting polymorphisms.
DNA methylation patterns were also analysed for clustered gene families. Stochastically expressed Pcdh exons display mosaic methylation patterns, while constitutively expressed exons are hypomethylated [111]. Interestingly, these methylation patterns are established during early embryonic stages long before lineage specification and expression [104,111]. On the other hand, OR gene clusters are located in regions devoid of DNA methylation and repression appears to be mediated rather through nuclear localisation and histone modifications [112,113].
Mechanisms controlling methylation of random monoallelically expressed genes
The specific role of DNMTs in the methylation of RME genes has been investigated so far only for clustered gene families. It was shown that DNMT3B is required for de novo methylation at promoters of the three Pcdh gene clusters, which is established early during development, after the blastocyst stage when the enzyme is highly expressed in epiblast cells [111,114]. DNMT3A is on the other hand dispensable for this methylation [111]. Dnmt3b-deficient Purkinje cells express an increased number of Pcdh isoforms per cell, indicating that the DNA methylation by DNMT3B regulates the expression frequency of each Pcdh isoforms in neurons. Smchd1-deficient embryos also show hypomethylation of Pcdh-α and Pcdh-β clusters promoter regions, correlated with increased expression of many different isoforms in mutant embryos and adult brain [104]. These observations mirror the phenotype observed in Dnmt3b-deficient embryos reinforcing the idea that DNA methylation plays a role in the regulation of stochastic and monoallelic expression of Pcdh isoforms. Additionally, the chromatin state of OR genes was studied quite extensively in the mouse olfactory epithelium, in particular in mice deficient for Dnmt3a. While this enzyme is important for the regulation of global gene expression in olfactory sensory neurons, it does not appear to regulate choice and monoallelic expression of OR genes [113,115].
Few studies have yet explored the molecular mechanisms involved in methylation of single RME genes due to their random, transient and variable nature within a cell population or a tissue. Treatment of cells with 5-azacytidine allowed testing whether DNA methylation is necessary for the maintenance of RME patterns. The erasure of DNA methylation at 5′ CGI of RME genes did not induce biallelic expression in NPC clonal cell lines, although this was analysed only for a small number of loci [5,6]. Besides these examples, the dependence of monoallelic expression of single RME genes towards DNA methylation is unknown. It is likely that the cellular memory at different RME loci rely on a variety and combination of epigenetic mechanisms, including DNA methylation, which might be relevant for some genes [5,6,108,109]. As RME appears somewhat less stable and more transient than the monoallelic expression of imprinted and X-linked genes, this could account for some of the differences observed. Interestingly, RME is often associated with random monoallelically accessible promoter regions in NPC, suggestive of regulation through transcription factor binding [116]. It has been hypothesised that RME could represent a way of fine-tuning the expression of genes in specific cell types during development or differentiation. In this case, repression of the silent allele through histone modifications or activation by transcription factors would allow easy reversal of the expression states when needed. In contrast, RME genes associated with DNA methylation may indicate a need for stronger stability at these loci.
Importantly, genome-wide studies of DNA methylation in human and mice [117–120] revealed a strong correlation between allele-specific DNA methylation (ASM) and the presence of DNA sequence polymorphisms in the vicinity. This phenomenon is widespread in the human genome and sometimes associated with allele-specific expression imbalances [121] (Figure 3B). These studies demonstrate how allele-specific genetic variations in cis can influence CpG methylation in both mouse and human and occasionally lead to allelic differences in expression. These findings, which connect genetic polymorphisms and phenotypic variability, may help to understand how some of these loci that may be associated with dosage-sensitive genes, could contribute to human diseases.
Concluding remarks
Although the three classes of genes showing monoallelic expression in mammalian genome share multiple properties, they also display specific features. DNA methylation is an epigenetic modification essential for the establishment and maintenance of imprinting by differential marking of parentally inherited alleles at ICRs in the germline. X-inactivated genes are characterised by promoter hypermethylation, which is established after transcriptional silencing. Although X-linked gene promoter methylation is considered to be important for the maintenance of silencing, it appears only essential upon certain cellular or developmental contexts. Finally, whereas DNA methylation is sometimes associated with RME loci, it does not seem to be a common epigenetic signature of this class of genes. This is likely linked to the nature of RME genes, whose expression often arises during differentiation, is highly cell-type specific, often more variable and potentially transient, unlike genes regulated by imprinting and XCI.
In the future, methods combining RNA analysis, methylomes, chromatin signatures, accessibility and lineage tracing will help to understand the extent to which some RME loci share similar properties to imprinted and X-inactivated genes, in particular in vivo. Moreover, the manipulation of the methylation machinery in vitro or in vivo using CRISPR-Cas9 and the use of conditional KOs will allow to determine the extent to which DNA methylation is instrumental for the maintenance of monoallelic expression states in specific developmental or tissue contexts. Furthermore, the ability to use human induced pluripotent stem cells and 3D organoids will allow further investigation on the role of DNA methylation and monoallelic expression in human cells, which could have important implications not only for development, but also for disease.
Summary
Three main classes of genes display monoallelic expression independently of DNA sequence polymorphisms in mammalian genomes: imprinted genes, X inactivated genes and RME genes on autosomes.
DNA methylation is essential for the establishment and maintenance of imprinting.
DNA methylation of X-linked gene promoter regions occurs late during XCI and participates in the long-term silencing of genes on the Xi.
DNA methylation can be associated with RME loci, but it does not appear to be a general feature of this class of genes.
Abbreviations
- AgR
antigen receptor
- AID
activation-induced deaminase
- CGI
CpG island
- CpG
5′-cytosine-phosphate-guanine-3′
- DMR
differentially methylated region
- DNMT
DNA methyltransferase
- DNMT3L
DNMT3-like
- ES
embryonic stem
- H3K27me3
trimethylation of lysine 27 on histone H3
- ICR
imprinting control region
- KO
knock-out
- NPC
neural progenitor cell
- OR
olfactory receptor
- Pcdh
protocadherin
- PRC
Polycomb repressive complex
- RME
random monoallelic expression
- SMCHD1
structural maintenance of chromosomes hinge domain-containing 1
- TET
ten-eleven translocation
- UHRF1
ubiquitin-like containing PHD and RING finger domains 1
- Xa
active X chromosome
- XCI
X-chromosome inactivation
- Xi
inactive X chromosome
- ZFP
zinc finger protein
- ZNF
zinc finger
- 5mC
5-methylcytosine
Contributor Information
Simão Teixeira da Rocha, Email: simaoteixeiradarocha@medicina.ulisboa.pt.
Anne-Valerie Gendrel, Email: anne-valerie.gendrel@curie.fr.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by Fundação para a Ciência e Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Ensino Superior (MCTES), Portugal, through the project grants PTDC/BEX-BCM/2612/2014 and PTDC/BIA-MOL/29320/2017 IC&DT. S.T.d.R. has a CEECUIND/01234/207 assistant research contract from FCT. A.-V.G. is supported by an INSERM investigator position. Publications costs were supported by UID/BIM/50005/2019, project funded by FCT/MCTES through Fundos do Orçamento de Estado.
Author Contribution
S.T.d.R. and A.-V.G. both prepared and wrote the manuscript.
References
- 1.Gendrel A.V., Marion-Poll L., Katoh K. and Heard E. (2016) Random monoallelic expression of genes on autosomes: parallels with X-chromosome inactivation. Semin. Cell Dev. Biol. 56, 100–110 10.1016/j.semcdb.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 2.Khamlichi A.A. and Feil R. (2018) Parallels between mammalian mechanisms of monoallelic gene expression. Trends Genet. 34, 954–971 10.1016/j.tig.2018.08.005 [DOI] [PubMed] [Google Scholar]
- 3.Tucci V., Isles A.R., Kelsey G., Ferguson-Smith A.C. and Erice Imprinting G. (2019) Genomic imprinting and physiological processes in mammals. Cell 176, 952–965 10.1016/j.cell.2019.01.043 [DOI] [PubMed] [Google Scholar]
- 4.Galupa R. and Heard E. (2018) X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 52, 535–566 10.1146/annurev-genet-120116-024611 [DOI] [PubMed] [Google Scholar]
- 5.Gendrel A.V., Attia M., Chen C.J., Diabangouaya P., Servant N., Barillot E. et al. (2014) Developmental dynamics and disease potential of random monoallelic gene expression. Dev. Cell 28, 366–380 10.1016/j.devcel.2014.01.016 [DOI] [PubMed] [Google Scholar]
- 6.Eckersley-Maslin M.A., Thybert D., Bergmann J.H., Marioni J.C., Flicek P. and Spector D.L. (2014) Random monoallelic gene expression increases upon embryonic stem cell differentiation. Dev. Cell 28, 351–365 10.1016/j.devcel.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg M.V.C. and Bourc’his D. (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 10.1038/s41580-019-0159-6 [DOI] [PubMed] [Google Scholar]
- 8.Iurlaro M., von Meyenn F. and Reik W. (2017) DNA methylation homeostasis in human and mouse development. Curr. Opin. Genet. Dev. 43, 101–109 10.1016/j.gde.2017.02.003 [DOI] [PubMed] [Google Scholar]
- 9.Deaton A.M., Webb S., Kerr A.R., Illingworth R.S., Guy J., Andrews R. et al. (2011) Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 21, 1074–1086 10.1101/gr.118703.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Issa J.P. (2004) CpG island methylator phenotype in cancer. Nat. Rev. Cancer 4, 988–993 10.1038/nrc1507 [DOI] [PubMed] [Google Scholar]
- 11.Luo C., Hajkova P. and Ecker J.R. (2018) Dynamic DNA methylation: in the right place at the right time. Science 361, 1336–1340 10.1126/science.aat6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massah S., Beischlag T.V. and Prefontaine G.G. (2015) Epigenetic events regulating monoallelic gene expression. Crit. Rev. Biochem. Mol. Biol. 50, 337–358 10.3109/10409238.2015.1064350 [DOI] [PubMed] [Google Scholar]
- 13.Barau J., Teissandier A., Zamudio N., Roy S., Nalesso V., Herault Y. et al. (2016) The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354, 909–912 10.1126/science.aah5143 [DOI] [PubMed] [Google Scholar]
- 14.Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S. and Jacobsen S.E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 10.1126/science.1147939 [DOI] [PubMed] [Google Scholar]
- 15.Bourc’his D., Xu G.L., Lin C.S., Bollman B. and Bestor T.H. (2001) Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 10.1126/science.1065848 [DOI] [PubMed] [Google Scholar]
- 16.Maenohara S., Unoki M., Toh H., Ohishi H., Sharif J., Koseki H. et al. (2017) Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLoS Genet. 13, e1007042 10.1371/journal.pgen.1007042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyko F. (2018) The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92 10.1038/nrg.2017.80 [DOI] [PubMed] [Google Scholar]
- 18.Wu X. and Zhang Y. (2017) TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 10.1038/nrg.2017.33 [DOI] [PubMed] [Google Scholar]
- 19.Feil R. and Fraga M.F. (2012) Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109 10.1038/nrg3142 [DOI] [PubMed] [Google Scholar]
- 20.Levine M.E., Lu A.T., Quach A., Chen B.H., Assimes T.L., Bandinelli S. et al. (2018) An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackett J.A. and Surani M.A. (2013) DNA methylation dynamics during the mammalian life cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110328 10.1098/rstb.2011.0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charalambous M., da Rocha S.T. and Ferguson-Smith A.C. (2007) Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr. Opin. Endocrinol. Diabetes Obes. 14, 3–12 10.1097/MED.0b013e328013daa2 [DOI] [PubMed] [Google Scholar]
- 23.Soellner L., Begemann M., Mackay D.J., Gronskov K., Tumer Z., Maher E.R. et al. (2017) Recent advances in imprinting disorders. Clin. Genet. 91, 3–13 10.1111/cge.12827 [DOI] [PubMed] [Google Scholar]
- 24.Lin S.P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M. et al. (2003) Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35, 97–102 10.1038/ng1233 [DOI] [PubMed] [Google Scholar]
- 25.Zwart R., Sleutels F., Wutz A., Schinkel A.H. and Barlow D.P. (2001) Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 15, 2361–2366 10.1101/gad.206201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowak K., Stein G., Powell E., He L.M., Naik S., Morris J. et al. (2011) Establishment of paternal allele-specific DNA methylation at the imprinted mouse Gtl2 locus. Epigenetics 6, 1012–1020 10.4161/epi.6.8.16075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie W., Barr C.L., Kim A., Yue F., Lee A.Y., Eubanks J. et al. (2012) Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831 10.1016/j.cell.2011.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartolomei M.S., Webber A.L., Brunkow M.E. and Tilghman S.M. (1993) Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7, 1663–1673 10.1101/gad.7.9.1663 [DOI] [PubMed] [Google Scholar]
- 29.Ferguson-Smith A.C., Sasaki H., Cattanach B.M. and Surani M.A. (1993) Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362, 751–755 10.1038/362751a0 [DOI] [PubMed] [Google Scholar]
- 30.Li E., Beard C. and Jaenisch R. (1993) Role for DNA methylation in genomic imprinting. Nature 366, 362–365 10.1038/366362a0 [DOI] [PubMed] [Google Scholar]
- 31.Stoger R., Kubicka P., Liu C.G., Kafri T., Razin A., Cedar H. et al. (1993) Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 73, 61–71 10.1016/0092-8674(93)90160-R [DOI] [PubMed] [Google Scholar]
- 32.Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E. et al. (2004) Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 10.1038/nature02633 [DOI] [PubMed] [Google Scholar]
- 33.Bourc’his D. and Bestor T.H. (2004) Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 10.1038/nature02886 [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi H., Sakurai T., Imai M., Takahashi N., Fukuda A., Yayoi O. et al. (2012) Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 8, e1002440 10.1371/journal.pgen.1002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chotalia M., Smallwood S.A., Ruf N., Dawson C., Lucifero D., Frontera M. et al. (2009) Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 23, 105–117 10.1101/gad.495809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smallwood S.A., Tomizawa S., Krueger F., Ruf N., Carli N., Segonds-Pichon A. et al. (2011) Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814 10.1038/ng.864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veselovska L., Smallwood S.A., Saadeh H., Stewart K.R., Krueger F., Maupetit-Mehouas S. et al. (2015) Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 16, 209 10.1186/s13059-015-0769-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y. and Sasaki H. (2011) Genomic imprinting in mammals: its life cycle, molecular mechanisms and reprogramming. Cell Res. 21, 466–473 10.1038/cr.2011.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirasawa R., Chiba H., Kaneda M., Tajima S., Li E., Jaenisch R. et al. (2008) Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 22, 1607–1616 10.1101/gad.1667008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee C.C., Peng S.H., Shen L., Lee C.F., Du T.H., Kang M.L. et al. (2017) The role of N-alpha-acetyltransferase 10 protein in DNA methylation and genomic imprinting. Mol Cell. 68, 89–103.e107 10.1016/j.molcel.2017.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P. et al. (2008) A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547–557 10.1016/j.devcel.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Messerschmidt D.M., de Vries W., Ito M., Solter D., Ferguson-Smith A. and Knowles B.B. (2012) Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335, 1499–1502 10.1126/science.1216154 [DOI] [PubMed] [Google Scholar]
- 43.Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H. et al. (2007) PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9, 64–71 10.1038/ncb1519 [DOI] [PubMed] [Google Scholar]
- 44.Takahashi N., Coluccio A., Thorball C.W., Planet E., Shi H., Offner S. et al. (2019) ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 33, 49–54 10.1101/gad.320069.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S. et al. (2011) In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 44, 361–372 10.1016/j.molcel.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuo X., Sheng J., Lau H.T., McDonald C.M., Andrade M., Cullen D.E. et al. (2012) Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J. Biol. Chem. 287, 2107–2118 10.1074/jbc.M111.322644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proudhon C., Duffie R., Ajjan S., Cowley M., Iranzo J., Carbajosa G. et al. (2012) Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol. Cell 47, 909–920 10.1016/j.molcel.2012.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duffie R., Ajjan S., Greenberg M.V., Zamudio N., Escamilla del Arenal M., Iranzo J. et al. (2014) The Gpr1/Zdbf2 locus provides new paradigms for transient and dynamic genomic imprinting in mammals. Genes Dev. 28, 463–478 10.1101/gad.232058.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenberg M.V., Glaser J., Borsos M., Marjou F.E., Walter M., Teissandier A. et al. (2017) Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat. Genet. 49, 110–118 10.1038/ng.3718 [DOI] [PubMed] [Google Scholar]
- 50.Schoenherr C.J., Levorse J.M. and Tilghman S.M. (2003) CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33, 66–69 10.1038/ng1057 [DOI] [PubMed] [Google Scholar]
- 51.Fedoriw A.M., Stein P., Svoboda P., Schultz R.M. and Bartolomei M.S. (2004) Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303, 238–240 10.1126/science.1090934 [DOI] [PubMed] [Google Scholar]
- 52.Zimmerman D.L., Boddy C.S. and Schoenherr C.S. (2013) Oct4/Sox2 binding sites contribute to maintaining hypomethylation of the maternal igf2/h19 imprinting control region. PLoS ONE 8, e81962 10.1371/journal.pone.0081962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodfine K., Huddleston J.E. and Murrell A. (2011) Quantitative analysis of DNA methylation at all human imprinted regions reveals preservation of epigenetic stability in adult somatic tissue. Epigenetics Chromatin 4, 1 10.1186/1756-8935-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baran Y., Subramaniam M., Biton A., Tukiainen T., Tsang E.K., Rivas M.A. et al. (2015) The landscape of genomic imprinting across diverse adult human tissues. Genome Res. 25, 927–936 10.1101/gr.192278.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goovaerts T., Steyaert S., Vandenbussche C.A., Galle J., Thas O., Van Criekinge W. et al. (2018) A comprehensive overview of genomic imprinting in breast and its deregulation in cancer. Nat. Commun. 9, 4120 10.1038/s41467-018-06566-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim J., Bretz C.L. and Lee S. (2015) Epigenetic instability of imprinted genes in human cancers. Nucleic Acids Res. 43, 10689–10699 10.1093/nar/gkv867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin-Trujillo A., Vidal E., Monteagudo-Sanchez A., Sanchez-Delgado M., Moran S., Hernandez Mora J.R. et al. (2017) Copy number rather than epigenetic alterations are the major dictator of imprinted methylation in tumors. Nat. Commun. 8, 467 10.1038/s41467-017-00639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Radford E.J., Isganaitis E., Jimenez-Chillaron J., Schroeder J., Molla M., Andrews S. et al. (2012) An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS Genet. 8, e1002605 10.1371/journal.pgen.1002605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guibert S., Forne T. and Weber M. (2012) Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 22, 633–641 10.1101/gr.130997.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W. et al. (2002) Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 117, 15–23 10.1016/S0925-4773(02)00181-8 [DOI] [PubMed] [Google Scholar]
- 61.Lee J., Inoue K., Ono R., Ogonuki N., Kohda T., Kaneko-Ishino T. et al. (2002) Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 129, 1807–1817 [DOI] [PubMed] [Google Scholar]
- 62.Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F. et al. (2012) The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reik W. and Surani M.A. (2015) Germline and pluripotent stem cells. Cold Spring Harb. Perspect. Biol. 7, 1–23 10.1101/cshperspect.a019422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barlow D.P. and Bartolomei M.S. (2014) Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6, 1–20 10.1101/cshperspect.a018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phillips J.E. and Corces V.G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurukuti S., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z. et al. (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. U.S.A. 103, 10684–10689 10.1073/pnas.0600326103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murrell A., Heeson S. and Reik W. (2004) Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36, 889–893 10.1038/ng1402 [DOI] [PubMed] [Google Scholar]
- 68.Mancini-Dinardo D., Steele S.J., Levorse J.M., Ingram R.S. and Tilghman S.M. (2006) Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 20, 1268–1282 10.1101/gad.1416906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sleutels F., Zwart R. and Barlow D.P. (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813 10.1038/415810a [DOI] [PubMed] [Google Scholar]
- 70.Williamson C.M., Ball S.T., Dawson C., Mehta S., Beechey C.V., Fray M. et al. (2011) Uncoupling antisense-mediated silencing and DNA methylation in the imprinted Gnas cluster. PLoS Genet. 7, e1001347 10.1371/journal.pgen.1001347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Latos P.A., Pauler F.M., Koerner M.V., Senergin H.B., Hudson Q.J., Stocsits R.R. et al. (2012) Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472 10.1126/science.1228110 [DOI] [PubMed] [Google Scholar]
- 72.Chaumeil J., Le Baccon P., Wutz A. and Heard E. (2006) A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20, 2223–2237 10.1101/gad.380906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nagano T., Mitchell J.A., Sanz L.A., Pauler F.M., Ferguson-Smith A.C., Feil R. et al. (2008) The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720 10.1126/science.1163802 [DOI] [PubMed] [Google Scholar]
- 74.Terranova R., Yokobayashi S., Stadler M.B., Otte A.P., van Lohuizen M., Orkin S.H. et al. (2008) Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell 15, 668–679 10.1016/j.devcel.2008.08.015 [DOI] [PubMed] [Google Scholar]
- 75.Schertzer M.D., Braceros K. C.A., Starmer J., Cherney R.E., Lee D.M., Salazar G. et al. (2019) lncRNA-induced spread of polycomb controlled by genome architecture, RNA abundance, and CpG island DNA. Mol. Cell 75, 523–537 10.1016/j.molcel.2019.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inoue A., Jiang L., Lu F., Suzuki T. and Zhang Y. (2017) Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 10.1038/nature23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inoue A., Jiang L., Lu F. and Zhang Y. (2017) Genomic imprinting of Xist by maternal H3K27me3. Genes Dev. 31, 1927–1932 10.1101/gad.304113.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gendrel A.V. and Heard E. (2014) Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu. Rev. Cell Dev. Biol. 30, 561–580 10.1146/annurev-cellbio-101512-122415 [DOI] [PubMed] [Google Scholar]
- 79.Riggs A.D. (1975) X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 14, 9–25 10.1159/000130315 [DOI] [PubMed] [Google Scholar]
- 80.Norris D.P., Brockdorff N. and Rastan S. (1991) Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm. Genome 1, 78–83 10.1007/BF02443782 [DOI] [PubMed] [Google Scholar]
- 81.Grant M., Zuccotti M. and Monk M. (1992) Methylation of CpG sites of two X-linked genes coincides with X-inactivation in the female mouse embryo but not in the germ line. Nat. Genet. 2, 161–166 10.1038/ng1092-161 [DOI] [PubMed] [Google Scholar]
- 82.Lock L.F., Melton D.W., Caskey C.T. and Martin G.R. (1986) Methylation of the mouse hprt gene differs on the active and inactive X chromosomes. Mol. Cell. Biol. 6, 914–924 10.1128/MCB.6.3.914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wolf S.F., Jolly D.J., Lunnen K.D., Friedmann T. and Migeon B.R. (1984) Methylation of the hypoxanthine phosphoribosyltransferase locus on the human X chromosome: implications for X-chromosome inactivation. Proc. Natl. Acad. Sci. U.S.A. 81, 2806–2810 10.1073/pnas.81.9.2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Singer-Sam J., Grant M., LeBon J.M., Okuyama K., Chapman V., Monk M. et al. (1990) Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol. Cell. Biol. 10, 4987–4989 10.1128/MCB.10.9.4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohandas T., Sparkes R.S. and Shapiro L.J. (1981) Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science 211, 393–396 10.1126/science.6164095 [DOI] [PubMed] [Google Scholar]
- 86.Graves J.A. (1982) 5-azacytidine-induced re-expression of alleles on the inactive X chromosome in a hybrid mouse cell line. Exp. Cell Res. 141, 99–105 10.1016/0014-4827(82)90072-6 [DOI] [PubMed] [Google Scholar]
- 87.Paterno G.D., Adra C.N. and McBurney M.W. (1985) X chromosome reactivation in mouse embryonal carcinoma cells. Mol. Cell. Biol. 5, 2705–2712 10.1128/MCB.5.10.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lock L.F., Takagi N. and Martin G.R. (1987) Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell 48, 39–46 10.1016/0092-8674(87)90353-9 [DOI] [PubMed] [Google Scholar]
- 89.Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L. et al. (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37, 853–862 10.1038/ng1598 [DOI] [PubMed] [Google Scholar]
- 90.Viegas-Pequignot E., Dutrillaux B. and Thomas G. (1988) Inactive X chromosome has the highest concentration of unmethylated Hha I sites. Proc. Natl. Acad. Sci. U.S.A. 85, 7657–7660 10.1073/pnas.85.20.7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bernardino J., Lombard M., Niveleau A. and Dutrillaux B. (2000) Common methylation characteristics of sex chromosomes in somatic and germ cells from mouse, lemur and human. Chromosome Res. 8, 513–525 10.1023/A:1009271706488 [DOI] [PubMed] [Google Scholar]
- 92.Hellman A. and Chess A. (2007) Gene body-specific methylation on the active X chromosome. Science 315, 1141–1143 10.1126/science.1136352 [DOI] [PubMed] [Google Scholar]
- 93.Cotton A.M., Price E.M., Jones M.J., Balaton B.P., Kobor M.S. and Brown C.J. (2015) Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 24, 1528–1539 10.1093/hmg/ddu564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lister R. and Ecker J.R. (2009) Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 19, 959–966 10.1101/gr.083451.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zemach A., McDaniel I.E., Silva P. and Zilberman D. (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 10.1126/science.1186366 [DOI] [PubMed] [Google Scholar]
- 96.Gendrel A.V., Apedaile A., Coker H., Termanis A., Zvetkova I., Godwin J. et al. (2012) Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev. Cell 23, 265–279 10.1016/j.devcel.2012.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gdula M.R., Nesterova T.B., Pintacuda G., Godwin J., Zhan Y., Ozadam H. et al. (2019) The non-canonical SMC protein SmcHD1 antagonises TAD formation and compartmentalisation on the inactive X chromosome. Nat. Commun. 10, 30 10.1038/s41467-018-07907-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Panning B. and Jaenisch R. (1996) DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 10, 1991–2002 10.1101/gad.10.16.1991 [DOI] [PubMed] [Google Scholar]
- 99.Sado T., Fenner M.H., Tan S.S., Tam P., Shioda T. and Li E. (2000) X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev. Biol. 225, 294–303 10.1006/dbio.2000.9823 [DOI] [PubMed] [Google Scholar]
- 100.Sado T., Okano M., Li E. and Sasaki H. (2004) De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development 131, 975–982 10.1242/dev.00995 [DOI] [PubMed] [Google Scholar]
- 101.Oda M., Yamagiwa A., Yamamoto S., Nakayama T., Tsumura A., Sasaki H. et al. (2006) DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev. 20, 3382–3394 10.1101/gad.1470906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Auclair G., Guibert S., Bender A. and Weber M. (2014) Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol. 15, 545 10.1186/s13059-014-0545-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hansen R.S., Stoger R., Wijmenga C., Stanek A.M., Canfield T.K., Luo P. et al. (2000) Escape from gene silencing in ICF syndrome: evidence for advanced replication time as a major determinant. Hum. Mol. Genet. 9, 2575–2587 10.1093/hmg/9.18.2575 [DOI] [PubMed] [Google Scholar]
- 104.Gendrel A.V., Tang Y.A., Suzuki M., Godwin J., Nesterova T.B., Greally J.M. et al. (2013) Epigenetic functions of smchd1 repress gene clusters on the inactive x chromosome and on autosomes. Mol. Cell. Biol. 33, 3150–3165 10.1128/MCB.00145-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blewitt M.E., Gendrel A.V., Pang Z., Sparrow D.B., Whitelaw N., Craig J.M. et al. (2008) SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 40, 663–669 10.1038/ng.142 [DOI] [PubMed] [Google Scholar]
- 106.Wang C.Y., Jegu T., Chu H.P., Oh H.J. and Lee J.T. (2018) SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 174, 406–421.e425 10.1016/j.cell.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jansz N., Keniry A., Trussart M., Bildsoe H., Beck T., Tonks I.D. et al. (2018) Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nat. Struct. Mol. Biol. 25, 766–777 [DOI] [PubMed] [Google Scholar]
- 108.Wang J., Valo Z., Bowers C.W., Smith D.D., Liu Z. and Singer-Sam J. (2010) Dual DNA methylation patterns in the CNS reveal developmentally poised chromatin and monoallelic expression of critical genes. PLoS ONE 5, e13843 10.1371/journal.pone.0013843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jeffries A.R., Perfect L.W., Ledderose J., Schalkwyk L.C., Bray N.J., Mill J. et al. (2012) Stochastic choice of allelic expression in human neural stem cells. Stem Cells 30, 1938–1947 10.1002/stem.1155 [DOI] [PubMed] [Google Scholar]
- 110.Gutierrez-Arcelus M., Lappalainen T., Montgomery S.B., Buil A., Ongen H., Yurovsky A. et al. (2013) Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2, e00523 10.7554/eLife.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Toyoda S., Kawaguchi M., Kobayashi T., Tarusawa E., Toyama T., Okano M. et al. (2014) Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron 82, 94–108 10.1016/j.neuron.2014.02.005 [DOI] [PubMed] [Google Scholar]
- 112.Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D. et al. (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 10.1126/science.1237905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Monahan K. and Lomvardas S. (2015) Monoallelic expression of olfactory receptors. Annu. Rev. Cell Dev. Biol. 31, 721–740 10.1146/annurev-cellbio-100814-125308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Borgel J., Guibert S., Li Y., Chiba H., Schubeler D., Sasaki H. et al. (2010) Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42, 1093–1100 10.1038/ng.708 [DOI] [PubMed] [Google Scholar]
- 115.Colquitt B.M., Markenscoff-Papadimitriou E., Duffie R. and Lomvardas S. (2014) Dnmt3a regulates global gene expression in olfactory sensory neurons and enables odorant-induced transcription. Neuron 83, 823–838 10.1016/j.neuron.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu J., Carter A.C., Gendrel A.V., Attia M., Loftus J., Greenleaf W.J. et al. (2017) Landscape of monoallelic DNA accessibility in mouse embryonic stem cells and neural progenitor cells. Nat. Genet. 49, 377–386 10.1038/ng.3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kerkel K., Spadola A., Yuan E., Kosek J., Jiang L., Hod E. et al. (2008) Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 40, 904–908 10.1038/ng.174 [DOI] [PubMed] [Google Scholar]
- 118.Zhang Y., Rohde C., Reinhardt R., Voelcker-Rehage C. and Jeltsch A. (2009) Non-imprinted allele-specific DNA methylation on human autosomes. Genome Biol. 10, R138 10.1186/gb-2009-10-12-r138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schilling E., El Chartouni C. and Rehli M. (2009) Allele-specific DNA methylation in mouse strains is mainly determined by cis-acting sequences. Genome Res. 19, 2028–2035 10.1101/gr.095562.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schalkwyk L.C., Meaburn E.L., Smith R., Dempster E.L., Jeffries A.R., Davies M.N. et al. (2010) Allelic skewing of DNA methylation is widespread across the genome. Am. J. Hum. Genet. 86, 196–212 10.1016/j.ajhg.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Onuchic V., Lurie E., Carrero I., Pawliczek P., Patel R.Y., Rozowsky J. et al. (2018) Allele-specific epigenome maps reveal sequence-dependent stochastic switching at regulatory loci. Science 361, 10.1126/science.aar3146 [DOI] [PMC free article] [PubMed] [Google Scholar]