Abstract
Dynamic binding of transcription factors (TFs) to regulatory elements controls transcriptional states throughout organism development. Epigenetics modifications, such as DNA methylation mostly within cytosine-guanine dinucleotides (CpGs), have the potential to modulate TF binding to DNA. Although DNA methylation has long been thought to repress TF binding, a more recent model proposes that TF binding can also inhibit DNA methylation. Here, we review the possible scenarios by which DNA methylation and TF binding affect each other. Further in vivo experiments will be required to generalize these models.
Keywords: DNA methylation, gene regulation, Transcription factor
Introduction
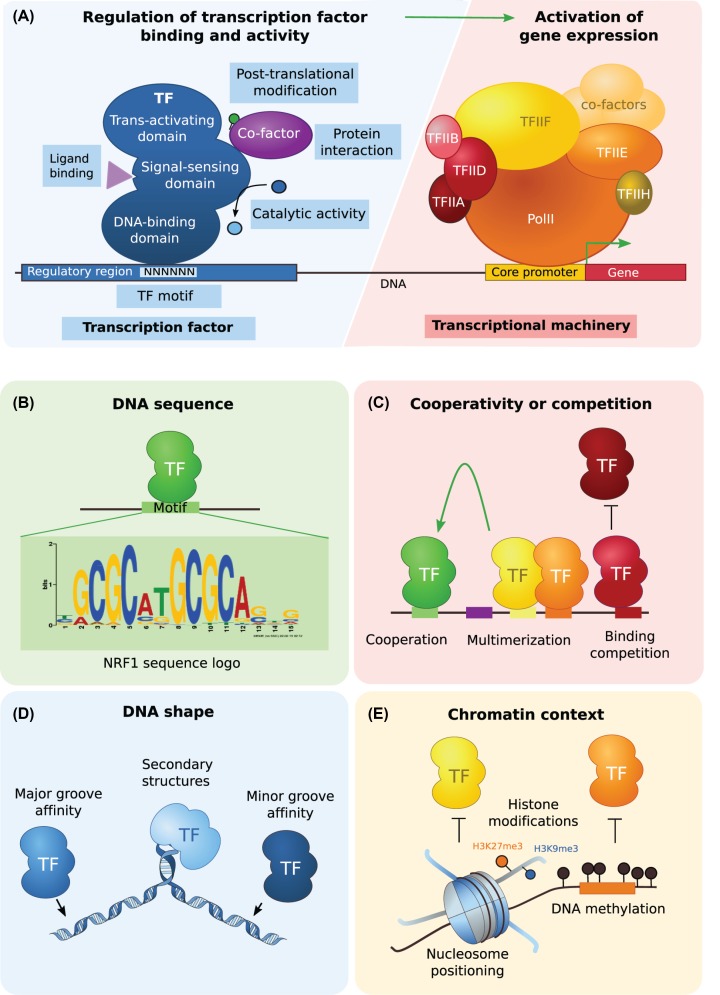
Multicellular organisms establish and maintain different transcriptional states in diverse cell types through dynamic and specific regulation of gene expression. This regulation is mediated by transcription factors (TFs) binding to regulatory elements [1]. TFs are often defined as any protein able to bind DNA and affect gene expression. They are composed of a DNA-binding domain, a trans-activating domain, which binds other proteins and mediates activator or repressor functions and an optional signal-sensing domain, which regulates their activity (Figure 1A). TFs bind to DNA, through the recognition of short specific DNA sequence motifs (Figure 1B). In the past years, much effort has been invested into the identification of TF motifs, using in vitro experiments such as electrophoretic mobility shift assay (EMSA) [2], high-throughput systematic evolution of ligands by exponential enrichment (HT-SELEX) [3] and protein binding microarrays (PBM) [4]. As a result, thousands of TF consensus sequence motifs (represented as sequence logos; Figure 1B) have been stored in databases such as JASPAR [5] and have been used as valuable input for further functional studies. However, while hundreds of thousands of occurrences of specific TF motifs are found within genomes, only a few thousands are bound in vivo in a context-specific manner, as assessed by chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq). Therefore, the specificity of TF binding must be controlled by other means. Combinatorial binding of TFs is a hallmark of regulatory regions, where TF binding relies on the presence of its motif, as well as motifs of other co-expressed TFs [6] (Figure 1C). More recently, the 3D structure of DNA or ‘DNA shape’, characterized by physical features of DNA major or minor grooves, has been thought to affect TF binding [7] (Figure 1D).
Figure 1. TF binding to DNA.
(A) TFs act as regulators of gene expression by binding to regulatory regions and recruiting the transcriptional machinery. They are composed of a DNA-binding domain that recognizes specific DNA sequence motifs, an optional signal-sensing domain that can alter TF activity after integrating signals from ligand binding, catalytic activity, protein interaction or post-translational modification, and a trans-activating domain that translates all these cellular cues into a repression or activation activity to regulate gene expression. (B) TF binding to DNA mainly relies on the recognition of specific DNA sequence motifs, the interaction with other TFs such through cooperativity or competition, the DNA shape (e.g. propeller twist, loops, rolls) and the chromatin context (e.g. nucleosome positioning, histone modifications, DNA methylation). NRF1 motif JASPAR ID: MA0596.1. Abbreviation: NRF1, nuclear respiratory factor 1.
The physical access of TFs to DNA can further be modulated by epigenetic regulation such as nucleosome positioning, histone modifications or DNA methylation [8] (Figure 1E). Early on, DNA methylation of cytosines, mostly within cytosine-guanine dinucleotides (CpGs) (Box 1), emerged as a mechanism to modulate TF affinity. DNA methylation was described as a mark repressing transcription, where the presence of DNA methylation at CpG-rich gene promoters, called CpG islands, would block TF binding leading to gene silencing [11]. With the emergence of high-throughput sequencing and the development of new methods to better profile TF binding and DNA methylation, our views on the sensitivity of TFs to DNA methylation have started to change. DNA methylation was found to be abundant throughout the genome except at active regulatory regions bound by TFs, such as gene promoters and distal enhancers [19–21]. In parallel, pioneer TFs have been described as able to bind non-accessible chromatin and recruit remodeling complexes, which can displace or remove nucleosomes, thus facilitating the context-specific binding of TFs [22]. Together with the observation of a strong anti-correlation between TF binding and DNA methylation patterns genome wide, a new actively discussed hypothesis has emerged where DNA methylation could be a consequence rather than a cause of TF binding and transcriptional activity [23].
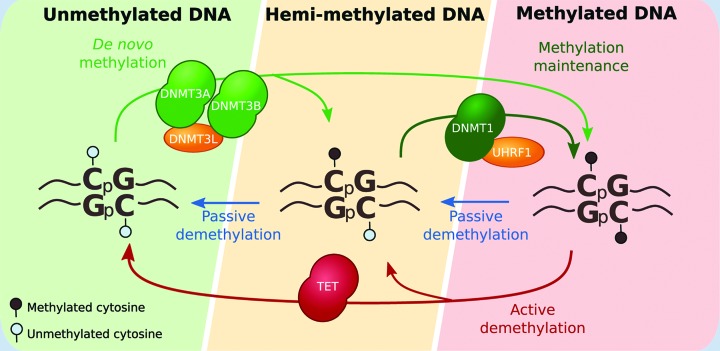
Box 1. DNA methylation under the spotlight.
DNA methylation is a chemical modification conserved across evolution from archeas to higher vertebrates such as human, mouse, zebrafish, plants or sea squirts although sporadic or even absent from invertebrate species such as fruit flies, worms and yeast [9]. DNA methylation affects cytosine residues, where a methyl group is added to the fifth carbon atom, thus forming a 5-methylcytosine (5mC). While in most vertebrates, DNA methylation occurs mainly in a CpG context, in other species such as plants, it also occurs in a CHG or CHH context [9].
DNA methylation is mediated by the DNA methyltransferases (DNMTs): DNMT3A, DNMT3B, and their cofactor DNMT3L, establish 5mC de novo, while DNMT1, with its co-cofactor UHRF1, maintains DNA methylation patterns during replication [10,11]. 5mC is a reversible modification. It can be removed passively during successive rounds of replication through an intermediate state of hemimethylation [12]. Recently, Ten-Eleven Translocation (TET) enzymes have been identified as drivers of active demethylation through an intermediate state of hydroxymethylation (and further formyl- and carboxylmethylation) [13–15].
While the chemical evidence of cytosine methylation was initially discovered in 1948 [16], the first functional evidence that it could regulate physiological processes emerged in 1975 [17]. DNA methylation is now known to be implicated in regulating gene expression in many biological processes such as development, cellular differentiation and cancer [18].
In this review, we summarize the progress and evolution of our understanding of the sensitivity of TFs to DNA methylation, which appears to be factor-specific and condition-dependent. We investigate the determinants of TF sensitivity to DNA methylation, the impact of TF binding on DNA methylation patterns and discuss how these principles can be generalized.
Impact of DNA methylation on TF binding
DNA methylation represses TF binding
The sensitivity of TF to DNA methylation started being investigated in vitro in the late 80s and already different TFs appeared to have different sensitivities [24]. Several TFs were identified as sensitive to DNA methylation by EMSA (Figure 2A). Methylation of a CpG (mCpG) central to the MLTF (also called USF) motif prevented binding of the TFs and inhibited the expression of the adenovirus major late promoter, whereas methylation of a CpG 6 base pairs away had no effect [25]. Methylation of the CREB binding sites also resulted in a loss of TF binding and transcriptional activity [26]. Other TFs, such as AP-2, MYC, E2F, NF-kB and ETS were repressed from binding by mCpG within their binding sites [24,27].
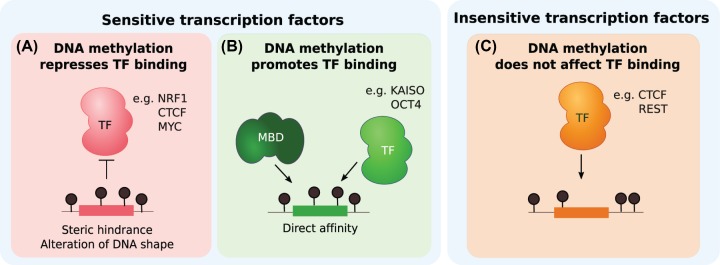
Figure 2. Possible scenarios by which DNA methylation could impact TF binding.
(A) TFs sensitive to DNA methylation are repressed from binding by mCpGs within their motifs, causing steric hindrance or alteration of the DNA shape. (B) Methyl-binding domain proteins (MBDs) recognize mCpGs in a sequence-independent manner. TFs bind sequence motifs containing mCpGs through direct affinity. (C) TFs insensitive to DNA methylation bind their motifs regardless of the DNA methylation status of the surrounding region.
Later in 2000, CTCF was shown to be sensitive to DNA methylation at the imprinted control region of the mouse Igf2-H19 genes both in vitro and later in vivo [28], thus establishing CTCF as a paradigm of TF sensitivity to DNA methylation (Box 2).
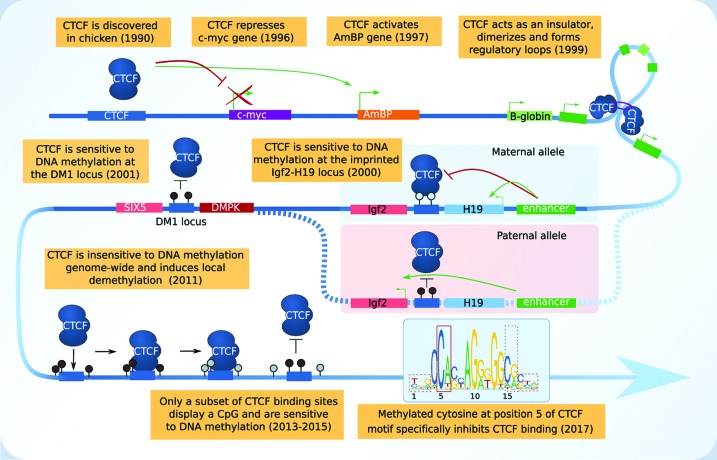
Box 2. CTCF, a paradigm of TF sensitivity to DNA methylation.
The zinc finger CTCF is one of the most studied TFs. It was first identified in chickens [29] and shortly after in humans, as a negative regulator of the c-myc gene [30] and an activator of the amyloid β protein precursor (AmBP) [31]. It was demonstrated to act as an insulator on the chicken β-globin locus, blocking enhancers from activating distal genes [28]. By dimerizing and tethering loop formation, it acts on the genome architecture and regulates gene expression [32,33].
By investigating imprinted genes, CTCF was found to be sensitive to DNA methylation at the Igf2-H19 locus in mouse. When bound to the unmethylated maternal allele, CTCF acts as an insulator, restricting the action of the downstream enhancer to the H19 gene. Whereas, on the methylated paternal allele, CTCF cannot bind and the enhancer activates the Igf2 gene [28]. It was later shown by point mutations of the four CTCF sites in vivo that impaired CTCF binding led to an increase in local DNA methylation in newborn mice although CTCF binding was not required for establishing an unmethylated state during oogenesis [34]. Additionally, methylation of two CTCF motifs at the myotonic dystrophy type 1 (DM1) locus was also shown to abolish CTCF binding thus altering the expression of the DMPK and SIX5 genes [35].
Ten years later, the development of high-throughput sequencing approaches to probe DNA methylation patterns (whole-genome bisulfite sequencing) and TF binding (ChIP-seq), enabled to better study CTCF binding genome wide and challenged traditional views. In mouse embryonic stem (ES) cells, CTCF binding sites were found to be mainly located in regions with no or low levels of DNA methylation [20]. However, CTCF did not bind additional sites in absence of DNA methylation (DNMTs triple knockout), except at known imprinted loci, suggesting that CTCF binding was not repressed by DNA methylation genome wide in vivo. Validations using stable insertions of a reporter construct containing the CTCF motif showed that CTCF could bind methylated DNA and led to local demethylation whereas mutated CTCF motifs were not bound and remained methylated [20]. Therefore, in contrary to what was expected, CTCF appeared to be insensitive to DNA methylation genome wide in vivo [20], which was further confirmed in HCT116 cells [36]. CTCF profiling across 19 different cell types showed that while CTCF binds distinct sites in different cell types, 41% of its variable binding sites are linked to DNA methylation [37]. When looking at CTCF motif occurrences within the genome, 25% do contain CpGs [38] whereas 45% of those located within CTCF ChIP-seq binding sites do. This highlights the fact that only a fraction of putative CTCF binding sites are potentially affected by DNA methylation. Recent reports investigated the contribution of specific CpGs to binding methylated DNA using structural information and a binding affinity assay (Methyl-Spec-seq) and could identify that it is the methylated cytosine at position 5 in the JASPAR motif that specifically inhibit CTCF binding [39,40].
In summary, although CTCF was originally described as a methylation-sensitive factor at the imprinted Igf2-H19 locus, only a limited set of in vivo CTCF binding sites, presumably harboring CpGs in their motifs, will be sensitive to DNA methylation. Its flexible binding sites might help regulate complex developmental and cellular processes. CTCF thus perfectly illustrates the fact that TF sensitivity to DNA methylation, despite its early discovery, remains an open question. CTCF motif JASPAR ID: MA0139.1.
The development of high-throughput technologies later enabled to test the sensitivity of many more TFs in vitro. A quantitative mass-spectrometry approach identified ZBTB2, JUND, CREB1, ATF7 as preferentially bound to unmethylated cytosines over methylated ones [41]. An approach using methylated binding microarrays found that DNA methylation inhibited binding of the basic leucine zipper (BZIP) TFs CREB, ATF4, JUN, JUND, CEBPD and CEBPG [42]. TFs identified as being repressed from binding by DNA methylation were in good agreement in both studies and earlier ones.
NRF1 was the first TF shown to be sensitive to DNA methylation genome-wide in vivo [43]. Sensitive TFs were identified in mouse embryonic stem (ES) cells by profiling open chromatin regions in presence and absence of DNA methylation (using DNMTs triple knockout (TKO) cells) as they were expected to bind new accessible sites in absence of DNA methylation. A motif analysis identified NRF1 as well as MYC/USF/CREB and GABPA/ETS as candidate methylation-sensitive TFs. NRF1 was then validated in vivo by ChIP-seq, where it could bind many new sites in absence of DNA methylation. Further, validations using stable insertions of a reporter construct containing the NRF1 motif showed that it could only bind its unmethylated motif and was not able to bind when methylated [43].
More recent in vitro approaches using methylation-sensitive SELEX expanded the catalog of TFs known to be repressed from binding by DNA methylation. The first study investigated 519 TFs using methyl-SELEX and bisulfite-SELEX and found that 23% (117 out of 519) were inhibited by mCpG (called methyl-minus) [44]. Their global analysis could identify TF families that tend to be inhibited by mCpG such as basic helix–loop–helix (BHLH), BZIP and ETS TFs. The majority (96 TFs, 82%) had CpGs in their original motif and validated known sensitive TFs such as MYC, USF, CREB, ATF, AP (JUN, FOS), E2F, ETS although NRF1 was not mentioned in their study. However, profiling of MYC binding in vivo in cells lacking DNA methylation (ChIP-seq in DNMTs TKO) showed that DNA methylation had only a minimal effect on its binding sites [44].
Another study probed ATF4 sensitivity using methyl-SELEX [45]. Since the motif does not have a prominent CpG, they found that motifs with no CpG showed no preferential binding for methylated or unmethylated DNA, motifs with CpGs in the center were not bound when methylated and motifs with CpGs in the flank were bound when methylated [45]. This confirms that not only an mCpG but also its position within the motif is critical for inhibiting TF binding.
In parallel, an in vitro study in plants identified 234 TFs (72% out of 327 tested) to be inhibited from binding by mCpGs [46] although plants have a different TF repertoire.
Most studies agree that inhibition by an mCpG might be due to steric hindrance of TF binding [26,44]. More recently, DNA shape has been described as an additional feature of TF binding [7] and roll and propeller twist DNA shapes have been found to be strongly affected by mCpGs [47]. Additionally, nucleosome positioning and histone modifications are linked to DNA methylation and might also affect TF binding [48,49].
These different studies identified several TFs to be inhibited from binding by mCpGs and therefore sensitive to DNA methylation, suggesting a widespread mechanism (Figure 2A). Of note, most of these observations came from in vitro studies, and few in vivo studies confirm their sensitivity. Therefore, the functional impact of this sensitivity on a genome-wide scale remains to be further investigated.
DNA methylation promotes TF binding
In parallel to the discovery of TFs that are inhibited by mCpGs, proteins that recognize mCpGs specifically through a methyl-binding domain (MBD) were identified: MeCp2, MBD1, MBD2 and MBD4 [50–52]. However, this recognition can be considered as independent of the underlying DNA sequence, as opposed to TFs that recognize specific DNA sequence motifs [51].
Later, both individual and high-throughput in vitro studies have identified sequence-specific TFs that bind mCpGs (Figure 2B). They are also described as sensitive to DNA methylation since they require an mCpG to bind as opposed to the sensitive TFs that are inhibited by mCpGs. A quantitative mass-spectrometry approach identified 19 proteins as binding preferentially to mCpGs over unmethylated ones (MeCP2, MBD1, MBD4, UHRF1, RFX1/5, ZFHX3,KLF2/4/5) although it does not account for sequence specificity [41]. A competition assay on methylated protein microarrays identified 41 TFs and 6 cofactors (3% of 1321 TFs and 210 cofactors tested) to preferentially bind motifs with mCpGs although in presence of a ten-fold excess of unmethylated sequences [53]. However, the majority recognized several distinct motifs and only 22 recognized fewer than three different motifs. Eight out of eleven were validated by EMSA (including the TFs ARNT2, DIDO1, MEF2A and HOXA9) implying a 27% false-positive rate [53]. NRF1 was found to bind both states, but more strongly to methylated than unmethylated motifs [53], although it was later shown to be inhibited from binding by mCpG in vivo [43]. A study in plants identified 14 TFs (4.3% out of 327 tested), which preferentially bind methylated motifs [46]. More recently, an approach using methyl-SELEX and bisulfite-SELEX found that 34% (175 out of 519) of the tested TFs could bind mCpGs (called methyl-plus) such as KAISO/ZBTB33, CEBPB/E/G, KLF, OCT4, HOX, PAX or SP1 [44]. However, only 49% of these (85 out of 175) had a CpG in their canonical motif whereas the others recognized a weaker methylated site [44]. Another approach called Methyl-Spec-seq, measuring the effect of mCpGs on TF binding affinity at every position within a binding site, could quantify the specific positions that affected ZFP57, CTCF, BATF1, GLI1 and HOXB13 binding, including hemimethylation of one of the two strands [40].
The zinc-finger Cys2His2-like (C2H2) TF KAISO (also called ZBTB33) was first described to bind mCpGs by EMSA in vitro [54] as well as by a structural report that showed the molecular basis for KAISO bi-modal recognition of both unmethylated and methylated CpGs [55]. Re-analysis of KAISO binding sites and DNA methylation patterns in vivo suggests that KAISO does not bind methylated DNA but rather highly active promoters marked with high levels of acetylated histones [56], although this interpretation does not take DNA methylation dynamics into account.
Other zinc-finger proteins were identified as readers of methylated DNA [57], such as KAISO-like ZBTB4 and ZBTB38 in transient transfections in mice [58]. ZFP57 is a well-known example of a TF that binds methylated DNA at imprinted regions in the mouse genome in vivo [59–61] and its mCpG binding preference was shown to be asymmetric [40].
Another zinc-finger family protein KLF4 was identified as binding mCpGs in a proteomics-based approach and by DNA pull-down [41]. Re-analysis of KLF4 binding sites in mouse ES cells in vivo identified 18.5% as methylated [41]. It was also found by a methylated microarray approach although it could bind both methylated and unmethylated sites, but displaying different sequence preferences [53]. Re-analysis of KLF4 binding sites in human ES cells in vivo identified that of the KLF4 binding sites having a CpG, 48% were methylated [53], but represented only 3% of all binding sites [62]. Probing the methylation levels of KLF4 binding sites at four different loci in vivo by ChIP bisulfite sequencing found that it could bind two unmethylated sites (TACpGCC) and two methylated sites (CCmCpGCC) [53].
Several members of the BZIP CEBP TF family were found to bind mCpGs. CEBPA was shown to bind mCpG within the CRE motif by EMSA in vitro [63]. An approach using methylated binding microarrays found that mCpGs promoted binding of CEBPA and CEBPB although CEBPD and CEBPG, which bind similar motifs, were inhibited [42]. Profiling of CEBPB by ChIP-seq in vivo identified only 11% of its methylated motifs as bound, in contrast with 54% of its unmethylated motifs, located in open-chromatin regions [42]. However, TFs are known not to bind all their motif occurrences in a certain context. Further, a similar re-analysis identified 25% of the CEBPB binding sites as methylated [62], which is surprisingly high since most TF binding sites are located in unmethylated open chromatin regions. More recently, a methyl-SELEX approach identified CEBPB only as weakly binding to mCpGs (called methyl-plus) as well as CEBPE and CEBPG [44]. A different methyl-SELEX approach found that CEBPB could bind both methylated and unmethylated sequences suggesting that CEBPB could tolerate DNA methylation [45]. The methyl-SELEX approach identified several other TFs that could bind to mCpGs [44]. OCT4 (also called POU5F1) was classified as a methyl-plus TFs although it does not have a CpG in its canonical motif. They further tested its sensitivity in vivo by profiling OCT4 binding by ChIP-seq in WT and DNMTs TKO mouse ES cells and could identify a few sites that lost OCT4 binding in absence of DNA methylation, suggesting that OCT4 requires DNA methylation at these sites in vivo [44].
Additionally, several HOX TFs were also classified as methyl-plus TFs, with some containing CpGs in their motifs. They showed that HOXC11 could specifically drive luciferase activity of an exogenously inserted construct containing its motif only when methylated [44]. HOXA9 was also previously found to bind mCpGs by EMSA [53]. Structural reports further showed the recognition of HOX TFs to mCpGs, such as HOXB13 [44] and the PBX-HOXA9 complex [45]. However, in the case of HOXB13, only the mCpG on the top strand contributes to binding whereas the other strand does not [40].
Structural reports proposed mechanisms for TF binding to mCpGs. Studies on the HOX TFs suggest that a mCpG in their motif could mimic a thymidine base, which could explain different sensitivities among HOX paralogs, and could be generalized to other TFs [44,45]. Other structural reports suggest that the binding of several TFs to mCpGs such as KAISO, ZFP57 and KLF4 depends on an arginine preceding the first zinc-binding histidine (called the arginine-histidine (RH) motif) [64], although the presence of an RH motif in zinc-finger proteins may not be a good predictor of mCpG binding [65,66].
These different studies identified many TFs as able to bind mCpGs and therefore were sensitive to DNA methylation (Figure 2B), suggesting a widespread mechanism. However, most results report in vitro affinities and some are contradictory. Recent studies have compiled the methylation status of TF binding sites from in vivo datasets [67,68] although those analyses are static and only correlative. Therefore, the functionality of TFs binding to mCpGs remains to be further investigated experimentally.
DNA methylation does not affect TF binding
Alongside the discovery of TFs sensitive to DNA methylation, others appeared not to be affected and are therefore called insensitive to DNA methylation (Figure 2C). In 1988, SP1 was the first TF to be described as insensitive to mCpGs located both at the center and at the periphery of the SP1 motif by EMSA in vitro [69,70]. However, a later study found that an mCpG affected SP1 binding in vitro and that the aberrant methylation of the retinoblastoma gene promoter in cancer was suggested to prevent SP1 binding in vivo [71]. The YY1 TF was then identified as insensitive to DNA methylation at the Surf genes promoter whereas ETS TFs were blocked by mCpGs [27].
More recently high-throughput in vitro approaches identified more TFs as insensitive to DNA methylation. A study in plants identified 79 insensitive TFs (24% out of 327 tested) [46]. An approach using methyl-SELEX and bisulfite-SELEX found that 40% (202 out of 519) of the tested TFs were not affected and the majority (84%, 169 out of 202) did not have CpGs in their motifs [44].
The first evidence for TF insensitivity in vivo came in 2011, when upon removal of DNA methylation (using DNMTs TKO cells), CTCF binding sites were globally unaltered, suggesting that DNA methylation was not preventing CTCF binding in WT mouse ES cells [20]. This was surprising knowing that CTCF was a well-known example of TF sensitivity to DNA methylation (Box 2) and that a similar approach identified NRF1 as sensitive [43]. CTCF as well as REST were then validated using stable insertions of methylated reporters containing their motifs where they could bind methylated regions and lead to local demethylation [20]. In fact, relatively few new regions of open chromatin bound by TFs were identified in absence of DNA methylation in mouse ES cells suggesting that most TFs did not seem to be affected by DNA methylation in vivo in this cell type [43].
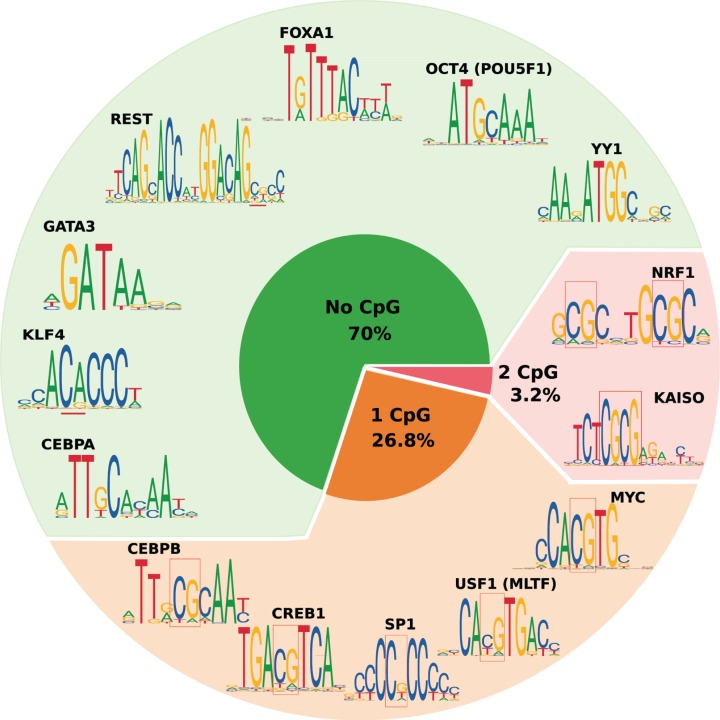
So far, the main feature predicting TF sensitivity appears to be the presence and position of CpGs within each TF motif, where only TFs that recognize a specific CpG within a motif could be affected when this CpG is methylated. Even though some TFs can recognize alternative motifs containing mCpGs (e.g. OCT4), those binding sites are not prevalent in vivo. When analyzing the non-redundant JASPAR CORE 2018 vertebrate motifs [5], and assuming that the 579 motifs reflect well the 1600 human TFs [1], we found that 70% of the TF motifs do not contain prominent CpGs and are therefore more likely not to be affected by DNA methylation. Only 30% of the motifs contain CpGs and might therefore be directly sensitive to DNA methylation including 3% with two CpGs such as NRF1 (Figure 3).
Figure 3. Proportion of TFs having CpGs in their motifs.
The majority of TFs (70%) do not have prominent CpGs in their motifs whereas 26.8% have one CpG and 3.2% have two CpGs. We used 579 position weight matrices from the vertebrates JASPAR CORE 2018 TF motifs [5] assuming that these represent well all 1600 TFs [1]. The presence of prominent CpGs in TF motifs was calculated by counting consecutive C and G positions with more than 0.5 frequency. JASPAR motif IDs: NRF1: MA0596.1; KAISO: MA0527.1; MYC: MA0147.3; USF1: MA0093.3; SP1: MA0079.3; CREB1: MA0018.3; CEBPB: MA0466.2; CEBPA: MA0102.3; KLF4: MA0039.3; GATA3: MA0037.3; REST: MA0138.2; FOXA1: MA0148.3; POU5F1: MA1115.1; YY1: MA0095.2. Abbreviation: USF1, transcription factor CP2-like protein 1.
Impact of TF binding on DNA methylation
TF binding promotes DNA methylation
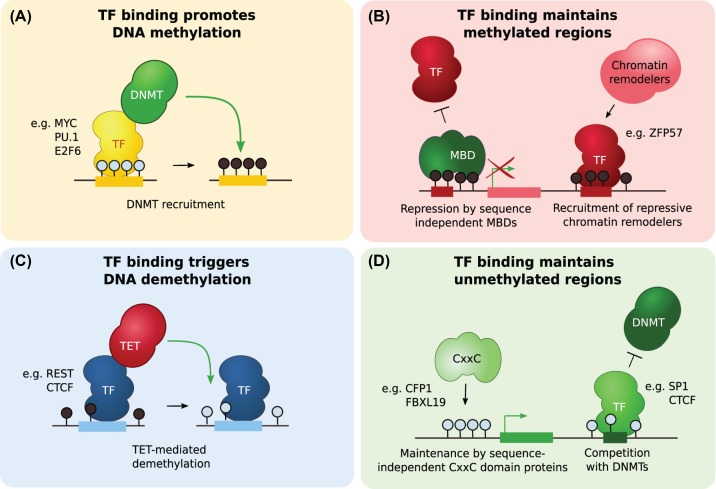
Although the DNMT enzymes have long been identified as writers of DNA methylation, the players involved in the recruitment of DNMT3A and DNMT3B to establish DNA methylation de novo remains elusive. Some TFs have been shown to recruit DNMTs and promote DNA methylation at specific sites (Figure 4A). For example, this was shown for the leukemia-promoting PML-RAR fusion protein at the RARβ2 promoter [72], MYC through association with MIZ1 at the p21Cip1 gene [73], PU.1 in reporter constructs [74], E2F6 at germline-specific genes [75], ZFP354B at the FAS promoter in NIH 3T3 cells [76] and ZNF304 at the INK4-ARF promoter in human ES cells [77]. However, those are few examples by distinct TFs at specific sites, some of which could result from indirect effects. Considering that in most cell types, most of the genome is methylated [19–21], the establishment of DNA methylation de novo could also be unspecific to the DNA sequence and rely on the chromatin context and other regulatory mechanisms or be a default state [10].
Figure 4. Possible scenarios by which TF binding could impact DNA methylation.
(A) TF binding to unmethylated DNA promotes de novo DNA methylation by recruitment of DNMTs. (B) MBDs bind to mCpGs inhibiting TF binding at CpG-rich regions and maintaining transcriptional repression. Direct binding of TFs to motifs containing mCpGs recruit chromatin remodelers to maintain silent transcriptional states. (C) TF binding to methylated regions recruits TET proteins and triggers DNA demethylation. (D) CxxC domain proteins recognize unmethylated CpGs in a sequence-independent manner in CpG-rich regions to maintain unmethylated and transcriptionally active states. TF binding to unmethylated motifs protects from de novo DNA methylation by DNMTs.
TF binding maintains DNA methylation states
Once DNA methylation states are established, mCpG readers, such as the MBD family members, are thought to induce transcriptional repression through chromatin remodeling and maintain DNA methylation states [48] (Figure 4B). However, MBD binding was shown to correlate with CpG content [78] and might therefore be limited to regions with high mCpG densities, often located at CpG-island gene promoters [79]. Accumulation of MBD binding to mCpG is also thought to protect from TF binding and DNA demethylation by steric interference but remains to be shown. For example, the inhibition of NRF1 binding genome-wide by DNA methylation in vivo genome-wide is independent of mCpG and MeCP2 density [43].
Sequence-specific TFs might also be involved in maintaining DNA methylation states. For example, ZFP57, which binds to mCpGs, was found to be necessary for the maintenance of DNA methylation and the histone 3 lysine 9 tri-methylation (H3K9me3) histone mark at these sites [60]. However, more examples of such a mechanism remain to be found and TFs able to bind mCpGs have recently been suggested to induce demethylation.
TF binding triggers demethylation
Recently, a model where TFs would instruct DNA methylation patterns has emerged (Figure 4C). TFs that are insensitive or bind to mCpGs were shown to induce active demethylation through ten-eleven translocations (TETs) recruitment [13,14] (Box 1). This was first shown for CTCF and REST using stable insertions of methylated reporters in mouse ES cells where they could bind and induce local demethylation [20]. It was later demonstrated for PU.1 by motif analyses in differentially methylated regions [80], for RUNX1, RUNX3, GATA2, CEBPB, MAFB, NR4A2, MYOD1, CEBPA and TBX5 by methylation array [81], for CEBPA, KLF4 and TFCP2L1 by profiling DNA hydroxymethylation [82] and for EGR1 by ChIP-seq [83].
The ability of TFs to mediate DNA demethylation was suggested for TFs insensitive to DNA methylation, such as REST, to enable binding of TFs otherwise repressed by DNA methylation, such as NRF1, and speculated to be a useful feature of pioneer TFs [43]. Pioneer TFs, such as GATA4, KLF4 or OCT4, were also proposed to bind mCpGs and then thought to induce DNA demethylation [44,62]. This would explain why TFs that bind mCpGs appear to have unmethylated binding sites in genomic studies. However, most of the TFs described as pioneers or reprogramming do not contain CpGs in their canonical motifs (e.g. OCT4, KLF4, FOXA1, GATA3, CEBPA; Figure 3) and might therefore be insensitive to DNA methylation and able to induce local demethylation of the surrounding region.
TF binding maintains unmethylated states
Once a region is unmethylated, binding of TFs was shown to protect from DNA methylation (Figure 4D). CxxC zinc-finger proteins, such as CXXC1/CFP1, FBXL19 or TET1, were identified to bind clustered unmethylated CpGs within CpG islands, recruit chromatin remodelers and help maintaining unmethylated regions [84–86].
Sequence-specific TFs have also been shown to protect unmethylated regions from de novo methylation. Early on, SP1 was shown to bind unmethylated CpG islands and mutations of its motif resulted in increased DNA methylation at the Aprt gene promoter [87,88] and later at the Gtf2a1l promoter [89]. CTCF was also shown to maintain unmethylated regions at the Igf2-H19 locus [34] (Box 2).
The maintenance of unmethylated regions by TFs was mainly shown at CpG islands that mark most housekeeping genes in vertebrate genomes [90]. In fact, CpG islands are thought to have emerged during evolution due to high rates of methylated cytosine deamination into thymine (5mC to T), which affected most of the methylated genome except at constitutively active unmethylated promoters [91].
In CpG-poor regions, reintroduction of de novo DNA methylation could outcompete NRF1 binding suggesting that binding of TFs sensitive to DNA methylation cannot maintain unmethylated states [43]. However, binding of insensitive TFs should both be able to induce demethylation and maintain unmethylated DNA at active regulatory regions (e.g. CTCF, REST).
Conclusions
DNA methylation patterns are highly dynamic during development, cellular differentiation and disease states [92–95]. Since they strongly anti-correlate with chromatin accessibility and TF binding, regions with no or low DNA methylation levels can be good predictors of TF binding sites [20,96]. However, this mere anti-correlation does not inform us about which of the DNA methylation or TF binding comes first.
The established model proposes that DNA methylation patterns instruct TF binding and act as a transcriptional repressor, although the functions of DNA methylation still remain to be further investigated [97]. Recent studies support this model and several TFs were identified as sensitive to DNA methylation, such as NRF1 in vivo genome wide [43] (Figure 2A). However, although DNA methylation can indeed inhibit TF binding, the extent of this repression in vivo seems limited in mouse ES cells, and remains to be explored in differentiated cell types.
More recently, a new model has emerged where TF binding instructs DNA methylation patterns. Recent studies support this model where TFs, that are insensitive to DNA methylation (e.g. REST or CTCF [20]) or recognize mCpGs (e.g. OCT4 [44]), bind methylated regions, induce demethylation through the recruitment of TET enzymes and maintain regions unmethylated by protecting them from de novo methylation (Figure 4C,D). This ability for TFs to induce demethylation in turn enables binding of sensitive TFs to these sites (e.g. NRF1 [43]), which could explain why DNA methylation seems to have a minimal effect on the binding of some sensitive TFs (e.g. MYC [44]). This feature is suggested to be useful for pioneer TFs, to bind closed methylated DNA inducing chromatin remodeling, leading to dynamic patterns of TF binding and gene expression.
Although the sensitivity of TFs to DNA methylation was mainly assessed in vitro, it appears to be dependent on the presence of mCpGs at specific positions within TF motifs (Figure 3). Therefore, TF sensitivity is factor- and condition-specific, where the same TF could be sensitive at sites with mCpGs and insensitive when CpGs are unmethylated (e.g. CTCF [20] (Box 2), OCT4 [44]).
In order to generalize these models, further in vivo experiments will be required. However, the dynamics of DNA methylation changes driven by TFs cannot be assessed in static conditions where a TF binding site will always appear as unmethylated. Therefore, perturbation or kinetic experiments, facilitated by genome editing tools [94], will be essential to better understand the interplay between TF binding and DNA methylation.
Summary
DNA methylation and TF binding patterns anti-correlate.
Established model: DNA methylation represses TF binding.
Emerging model: TF binding to methylated regions induces demethylation.
TF sensitivity to DNA methylation depends on the position of methylated CpGs within their binding motifs.
Acknowledgments
The authors thank M. Weber, D. Shlyueva and R. Grand for feedback on the review. We apologize to all colleagues whose work could not be cited due to space limitations.
Abbreviations
- ATF4
activating transcription factor 4
- BZIP
basic leucine zipper
- CEBPA/B/E/G/P
CCAAT enhancer binding protein β, η, γ
- ChIP-seq
chromatin immunoprecipitation followed by high throughput sequencing
- CpG
cytosine-guanine dinucleotide
- CREB
cyclic adenosine 3′,5′-monophosphate response element-binding protein
- CTCF
CCCTC-binding factor
- DNMT
DNA methyltransferase
- E2F
E2 transcription factor
- EMSA
electrophoretic mobility shift assay
- ES
embryonic stem
- HOXA9
homeobox A9
- Igf2
insulin-like growth factor 1
- JUND (AP-1)
JunD proto-oncogene, AP-1 transcription factor subunit
- MBD1/2/4
methyl-binding domain 1/2/4
- mCpG
methylation of CpG
- MeCp2
methyl-CpG binding protein 2
- MYC
myc-protooncogene
- NRF1
nuclear respiratory factor 1
- OCT4/POU5F1
octamer binding protein 4/POU class 5 homeobox 1
- PU.1/SPI1
spleen focus integration oncogene 1
- RAR
retinoic acid receptor
- REST
RE1 silencing transcription factor
- RFX1/5
regulatory factor X1/5
- RH
arginine-histidine
- SP1
specificity protein 1
- TET
ten-eleven translocation
- TF
transcription factor
- TKO
DNMT triple knockout
- ZFHX3
zinc finger homeobox
- ZFP57
zinc finger protein 57 homolog
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Systems Biology Cancer Plan grant from the ITMO Cancer AVIESAN (French National Alliance for Life Sciences and Health) [grant number 18CB008-00]; and the Initiative of Excellence IDEX-Unistra from the French national programme ‘Investment for the future’ [grant number ANR-10-IDEX-0002-02].
Author Contribution
E. Héberlé and A.F. Bardet wrote the manuscript and edited the figures.
References
- 1.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M. et al. (2018) The human transcription factors. Cell 172, 650–665 10.1016/j.cell.2018.01.029 [DOI] [PubMed] [Google Scholar]
- 2.Hellman L.M. and Fried M.G. (2007) Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2, 1849–1861 10.1038/nprot.2007.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P. et al. (2013) DNA-binding specificities of human transcription factors. Cell 152, 327–339 10.1016/j.cell.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 4.Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P. et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 10.1016/j.cell.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R. et al. (2018) JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 10.1093/nar/gkx1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spitz F. and Furlong E.E.M. (2012) Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- 7.Slattery M., Zhou T., Yang L., Dantas Machado A.C., Gordân R. and Rohs R. (2014) Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci. 39, 381–399 10.1016/j.tibs.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell O., Tiwari V.K., Thomä N.H. and Schübeler D. (2011) Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 12, 554–564 10.1038/nrg3017 [DOI] [PubMed] [Google Scholar]
- 9.Feng S., Cokus S.J., Zhang X., Chen P.-Y., Bostick M., Goll M.G. et al. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U.S.A. 107, 8689–8694 10.1073/pnas.1002720107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hervouet E., Peixoto P., Delage-Mourroux R., Boyer-Guittaut M. and Cartron P.-F. (2018) Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin. Epigenetics 10, 17 10.1186/s13148-018-0450-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- 12.Xu C. and Corces V.G. (2018) Nascent DNA methylome mapping reveals inheritance of hemimethylation at CTCF/cohesin sites. Science 359, 1166–1170 10.1126/science.aan5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastor W.A., Aravind L. and Rao A. (2013) TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 10.1038/nrm3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen K.D. and Helin K. (2016) Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750 10.1101/gad.276568.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y. et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotchkiss R.D. (1948) The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 175, 315–332 [PubMed] [Google Scholar]
- 17.Holliday R. and Pugh J.E. (1975) DNA modification mechanisms and gene activity during development. Science 187, 226–232 10.1126/science.1111098 [DOI] [PubMed] [Google Scholar]
- 18.Smith Z.D. and Meissner A. (2013) DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- 19.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J. et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 10.1038/nature08514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Schöler A. et al. (2011) DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495 10.1038/nature10716 [DOI] [PubMed] [Google Scholar]
- 21.Tsankov A.M., Gu H., Akopian V., Ziller M.J., Donaghey J., Amit I. et al. (2015) Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344–349 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwafuchi-Doi M. and Zaret K.S. (2014) Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692 10.1101/gad.253443.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blattler A. and Farnham P.J. (2013) Cross-talk between site-specific transcription factors and DNA methylation states. J. Biol. Chem. 288, 34287–34294 10.1074/jbc.R113.512517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tate P.H. and Bird A.P. (1993) Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 3, 226–231 10.1016/0959-437X(93)90027-M [DOI] [PubMed] [Google Scholar]
- 25.Watt F. and Molloy P.L. (1988) Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 2, 1136–1143 10.1101/gad.2.9.1136 [DOI] [PubMed] [Google Scholar]
- 26.Iguchi-Ariga S.M. and Schaffner W. (1989) CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 3, 612–619 10.1101/gad.3.5.612 [DOI] [PubMed] [Google Scholar]
- 27.Gaston K. and Fried M. (1995) CpG methylation has differential effects on the binding of YY1 and ETS proteins to the bi-directional promoter of the Surf-1 and Surf-2 genes. Nucleic Acids Res. 23, 901–909 10.1093/nar/23.6.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell A.C. and Felsenfeld G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 10.1038/35013100 [DOI] [PubMed] [Google Scholar]
- 29.Lobanenkov V.V., Nicolas R.H., Adler V.V., Paterson H., Klenova E.M., Polotskaja A.V. et al. (1990) A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5, 1743–1753 [PubMed] [Google Scholar]
- 30.Filippova G.N., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G. et al. (1996) An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16, 2802–2813 10.1128/MCB.16.6.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vostrov A.A. and Quitschke W.W. (1997) The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 272, 33353–33359 10.1074/jbc.272.52.33353 [DOI] [PubMed] [Google Scholar]
- 32.Braccioli L. and de Wit E. (2019) CTCF: a Swiss-army knife for genome organization and transcription regulation. Essays Biochem. 63, 157–165 10.1042/EBC20180069 [DOI] [PubMed] [Google Scholar]
- 33.Phillips J.E. and Corces V.G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenherr C.J., Levorse J.M. and Tilghman S.M. (2003) CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33, 66–69 10.1038/ng1057 [DOI] [PubMed] [Google Scholar]
- 35.Filippova G.N., Thienes C.P., Penn B.H., Cho D.H., Hu Y.J., Moore J.M. et al. (2001) CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 28, 335–343 10.1038/ng570 [DOI] [PubMed] [Google Scholar]
- 36.Maurano M.T., Wang H., John S., Shafer A., Canfield T., Lee K. et al. (2015) Role of DNA methylation in modulating transcription factor occupancy. Cell Rep. 12, 1184–1195 10.1016/j.celrep.2015.07.024 [DOI] [PubMed] [Google Scholar]
- 37.Wang H., Maurano M.T., Qu H., Varley K.E., Gertz J., Pauli F. et al. (2012) Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 22, 1680–1688 10.1101/gr.136101.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldmann A., Ivanek R., Murr R., Gaidatzis D., Burger L. and Schübeler D. (2013) Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 9, e1003994 10.1371/journal.pgen.1003994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimoto H., Wang D., Horton J.R., Zhang X., Corces V.G. and Cheng X. (2017) Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol. Cell 66, 711–720.e3 10.1016/j.molcel.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo Z., Roy B., Chang Y.K., Granas D. and Stormo G.D. (2017) Measuring quantitative effects of methylation on transcription factor–DNA binding affinity. Sci. Adv. 3, eaao1799 10.1126/sciadv.aao1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W.T.C., Bauer C. et al. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 10.1016/j.cell.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 42.Mann I.K., Chatterjee R., Zhao J., He X., Weirauch M.T., Hughes T.R. et al. (2013) CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 23, 988–997 10.1101/gr.146654.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Domcke S., Bardet A.F., Adrian Ginno P., Hartl D., Burger L. and Schübeler D. (2015) Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528, 575–579 10.1038/nature16462 [DOI] [PubMed] [Google Scholar]
- 44.Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S. et al. (2017) Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 10.1126/science.aaj2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kribelbauer J.F., Laptenko O., Chen S., Martini G.D., Freed-Pastor W.A., Prives C. et al. (2017) Quantitative analysis of the DNA methylation sensitivity of transcription factor complexes. Cell Rep. 19, 2383–2395 10.1016/j.celrep.2017.05.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R. et al. (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 10.1016/j.cell.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao S., Chiu T-P, Kribelbauer J.F., Mann R.S., Bussemaker H.J. and Rohs R. (2018) Systematic prediction of DNA shape changes due to CpG methylation explains epigenetic effects on protein-DNA binding. Epigenetics Chromatin 11, 6 10.1186/s13072-018-0174-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du J., Johnson L.M., Jacobsen S.E. and Patel D.J. (2015) DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532 10.1038/nrm4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C. et al. (2006) The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 10.1038/nature04431 [DOI] [PubMed] [Google Scholar]
- 50.Du Q., Luu P-L, Stirzaker C. and Clark S.J. (2015) Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics 7, 1051–1073 10.2217/epi.15.39 [DOI] [PubMed] [Google Scholar]
- 51.Hendrich B. and Bird A. (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18, 6538–6547 10.1128/MCB.18.11.6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meehan R.R., Lewis J.D., McKay S., Kleiner E.L. and Bird A.P. (1989) Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58, 499–507 10.1016/0092-8674(89)90430-3 [DOI] [PubMed] [Google Scholar]
- 53.Hu S., Wan J., Su Y., Song Q., Zeng Y., Nguyen H.N. et al. (2013) DNA methylation presents distinct binding sites for human transcription factors. eLife 2, e00726 10.7554/eLife.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prokhortchouk A., Hendrich B., Jørgensen H., Ruzov A., Wilm M., Georgiev G. et al. (2001) The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15, 1613–1618 10.1101/gad.198501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniel J.M., Spring C.M., Crawford H.C., Reynolds A.B. and Baig A. (2002) The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 30, 2911–2919 10.1093/nar/gkf398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blattler A., Yao L., Wang Y., Ye Z., Jin V.X. and Farnham P.J. (2013) ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenetics Chromatin 6, 13 10.1186/1756-8935-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hudson N.O., Whitby F.G. and Buck-Koehntop B.A. (2018) Structural insights into methylated DNA recognition by the C-terminal zinc fingers of the DNA reader protein ZBTB38. J. Biol. Chem. 293, 19835–19843 10.1074/jbc.RA118.005147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Filion G.J.P., Zhenilo S., Salozhin S., Yamada D., Prokhortchouk E. and Defossez P-A (2006) A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol. Cell. Biol. 26, 169–181 10.1128/MCB.26.1.169-181.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P. et al. (2008) A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547–557 10.1016/j.devcel.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S. et al. (2011) In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 44, 361–372 10.1016/j.molcel.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Riso V., Cammisa M., Kukreja H., Anvar Z., Verde G., Sparago A. et al. (2016) ZFP57 maintains the parent-of-origin-specific expression of the imprinted genes and differentially affects non-imprinted targets in mouse embryonic stem cells. Nucleic Acids Res. 44, 8165–8178 10.1093/nar/gkw505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu H., Wang G. and Qian J. (2016) Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 17, 551–565 10.1038/nrg.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rishi V., Bhattacharya P., Chatterjee R., Rozenberg J., Zhao J., Glass K. et al. (2010) CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc. Natl. Acad. Sci. U.S.A. 107, 20311–20316 10.1073/pnas.1008688107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y., Zhang X., Blumenthal R.M. and Cheng X. (2013) A common mode of recognition for methylated CpG. Trends Biochem. Sci. 38, 177–183 10.1016/j.tibs.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng X. and Blumenthal R.M. (2013) Response to mackay et Al. Trends Biochem. Sci. 38, 423 10.1016/j.tibs.2013.06.010 [DOI] [PubMed] [Google Scholar]
- 66.Mackay J.P., Segal D.J. and Crossley M. (2013) Is there a telltale RH fingerprint in zinc fingers that recognizes methylated CpG dinucleotides? Trends Biochem. Sci. 38, 421–422 10.1016/j.tibs.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 67.Wang G., Luo X., Wang J., Wan J., Xia S., Zhu H. et al. (2018) MeDReaders: a database for transcription factors that bind to methylated DNA. Nucleic Acids Res. 46, D146–D151 10.1093/nar/gkx1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xuan Lin Q.X., Sian S., An O., Thieffry D., Jha S. and Benoukraf T. (2019) MethMotif: an integrative cell specific database of transcription factor binding motifs coupled with DNA methylation profiles. Nucleic Acids Res. 47, D145–D154 10.1093/nar/gky1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harrington M.A., Jones P.A., Imagawa M. and Karin M. (1988) Cytosine methylation does not affect binding of transcription factor Sp1. Proc. Natl. Acad. Sci. U.S.A. 85, 2066–2070 10.1073/pnas.85.7.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Höller M., Westin G., Jiricny J. and Schaffner W. (1988) Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 2, 1127–1135 10.1101/gad.2.9.1127 [DOI] [PubMed] [Google Scholar]
- 71.Clark S.J., Harrison J. and Molloy P.L. (1997) Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene 195, 67–71 10.1016/S0378-1119(97)00164-9 [DOI] [PubMed] [Google Scholar]
- 72.Di Croce L., Raker V.A., Corsaro M., Fazi F., Fanelli M., Faretta M. et al. (2002) Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295, 1079–1082 10.1126/science.1065173 [DOI] [PubMed] [Google Scholar]
- 73.Brenner C., Deplus R., Didelot C., Loriot A., Viré E., De Smet C. et al. (2005) Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 24, 336–346 10.1038/sj.emboj.7600509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suzuki M., Yamada T., Kihara-Negishi F., Sakurai T., Hara E., Tenen D.G. et al. (2006) Site-specific DNA methylation by a complex of PU.1 and Dnmt3a/b. Oncogene 25, 2477–2488 10.1038/sj.onc.1209272 [DOI] [PubMed] [Google Scholar]
- 75.Velasco G., Hubé F., Rollin J., Neuillet D., Philippe C., Bouzinba-Segard H. et al. (2010) Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. Proc. Natl. Acad. Sci. U.S.A. 107, 9281–9286 10.1073/pnas.1000473107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wajapeyee N., Malonia S.K., Palakurthy R.K. and Green M.R. (2013) Oncogenic RAS directs silencing of tumor suppressor genes through ordered recruitment of transcriptional repressors. Genes Dev. 27, 2221–2226 10.1101/gad.227413.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Serra R.W., Fang M., Park S.M., Hutchinson L. and Green M.R. (2014) A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. eLife 3, e02313 10.7554/eLife.02313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baubec T., Ivánek R., Lienert F. and Schübeler D. (2013) Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell 153, 480–492 10.1016/j.cell.2013.03.011 [DOI] [PubMed] [Google Scholar]
- 79.Boyes J. and Bird A. (1992) Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 11, 327–333 10.1002/j.1460-2075.1992.tb05055.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de la Rica L., Rodríguez-Ubreva J., García M., Islam A.B.M.M.K., Urquiza J.M., Hernando H. et al. (2013) PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 14, R99 10.1186/gb-2013-14-9-r99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suzuki T., Maeda S., Furuhata E., Shimizu Y., Nishimura H., Kishima M. et al. (2017) A screening system to identify transcription factors that induce binding site-directed DNA demethylation. Epigenetics Chromatin 10, 60 10.1186/s13072-017-0169-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sardina J.L., Collombet S., Tian T.V., Gómez A., Stefano B.D., Berenguer C. et al. (2018) Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell 23, 727–741.e9 10.1016/j.stem.2018.08.016 [DOI] [PubMed] [Google Scholar]
- 83.Sun Z., Xu X., He J., Murray A., Sun M-A, Wei X. et al. (2019) EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat. Commun. 10, 3892 10.1038/s41467-019-11905-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boulard M., Edwards J.R. and Bestor T.H. (2015) FBXL10 protects Polycomb-bound genes from hypermethylation. Nat. Genet. 47, 479–485 10.1038/ng.3272 [DOI] [PubMed] [Google Scholar]
- 85.Long H.K., Blackledge N.P. and Klose R.J. (2013) ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem. Soc. Trans. 41, 727–740 10.1042/BST20130028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thomson J.P., Skene P.J., Selfridge J., Clouaire T., Guy J., Webb S. et al. (2010) CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464, 1082–1086 10.1038/nature08924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brandeis M., Frank D., Keshet I., Siegfried Z., Mendelsohn M., Nemes A. et al. (1994) Sp1 elements protect a CpG island from de novo methylation. Nature 371, 435–438 10.1038/371435a0 [DOI] [PubMed] [Google Scholar]
- 88.Macleod D., Charlton J., Mullins J. and Bird A.P. (1994) Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8, 2282–2292 10.1101/gad.8.19.2282 [DOI] [PubMed] [Google Scholar]
- 89.Lienert F., Wirbelauer C., Som I., Dean A., Mohn F. and Schübeler D. (2011) Identification of genetic elements that autonomously determine DNA methylation states. Nat. Genet. 43, 1091–1097 10.1038/ng.946 [DOI] [PubMed] [Google Scholar]
- 90.Deaton A.M. and Bird A. (2011) CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022 10.1101/gad.2037511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bird A.P. (1980) DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8, 1499–1504 10.1093/nar/8.7.1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dor Y. and Cedar H. (2018) Principles of DNA methylation and their implications for biology and medicine. Lancet 392, 777–786 10.1016/S0140-6736(18)31268-6 [DOI] [PubMed] [Google Scholar]
- 93.Greenberg M.V.C. and Bourc’his D. (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 10.1038/s41580-019-0159-6 [DOI] [PubMed] [Google Scholar]
- 94.Luo C., Hajkova P. and Ecker J.R. (2018) Dynamic DNA methylation: in the right place at the right time. Science 361, 1336–1340 10.1126/science.aat6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schübeler D. (2015) Function and information content of DNA methylation. Nature 517, 321–326 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- 96.Ziller M.J., Gu H., Müller F., Donaghey J., Tsai L.T.-Y., Kohlbacher O. et al. (2013) Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481 10.1038/nature12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ambrosi C., Manzo M. and Baubec T. (2017) Dynamics and context-dependent roles of DNA methylation. J. Mol. Biol. 429, 1459–1475 10.1016/j.jmb.2017.02.008 [DOI] [PubMed] [Google Scholar]