Abstract
Background: The relationship between antisense non-coding RNA (ncRNA) in the INK4 locus (ANRIL) polymorphisms and coronary artery disease (CAD) remains inconclusive. Thus, we conducted this meta-analysis to better evaluate the roles of ANRIL polymorphisms in CAD. Methods: Systematic literature search of PubMed, Medline, and Embase was performed to identify potential relevant articles. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to estimate the strength of association. Results: Fifteen studies were finally enrolled for analyses. Overall analyses suggested that rs1333040 (dominant model: P<0.0001; recessive model: P<0.0001; allele model: P<0.0001), rs1333049 (dominant model: P=0.02; allele model: P=0.02) and rs2383207 (additive model: P=0.004) polymorphisms were significantly associated with the risk of CAD. Further subgroup analyses showed that rs1333040, rs1333049, and rs2383207 polymorphisms were significantly correlated with the risk of CAD in East Asians, rs2383206 and rs10757274 polymorphisms were significantly correlated with the risk of CAD in West Asians, while rs2383206, rs10757274, and rs10757278 polymorphisms were significantly correlated with the risk of CAD in Caucasians. Conclusion: Our findings indicated that rs1333040, rs1333049, rs2383206, rs2383207, rs10757274, and rs10757278 polymorphisms might serve as genetic biomarkers of CAD in certain ethnicities.
Keywords: Antisense non-coding RNA in the INK4 locus (ANRIL), Coronary artery disease (CAD), Gene polymorphisms, Meta-analysis
Introduction
Coronary artery disease (CAD), featured by narrowing or even occlusion of coronary arteries, is the primary cause of death and disability worldwide [1,2]. Until now, the exact pathogenic mechanism of CAD remains unclear. Nevertheless, mounting evidence supports that genetic factors are crucial for its occurrence and development. In the first place, family aggregation of CAD was not uncommon, and past twin studies showed that the heredity grade of CHD was over 50% [3,4]. In the second place, numerous genetic variants were found to be associated with an increased risk of CAD by previous genetic association studies, and screening of common causal variants was also shown to be a cost-efficient way to predict the individual risk of developing CAD [5,6]. In summary, these findings jointly indicated that genetic predisposition to CAD played a central part in its pathogenesis.
Non-coding RNAs (ncRNAs) make up the vast majority of the human genome. Although ncRNAs do not encode proteins, it was evident that they play eminent roles in regulating expression levels of neighboring protein-encoding genes [7,8]. Therefore, dysregulation of ncRNAs may cause abnormal gene expression and give rise to the development of multiple diseases. Antisense ncRNA in the INK4 locus (ANRIL) is located on human chromosome 9p21, a region that has been repeated found to be associated with atherosclerosis and its related ischemia cardiovascular diseases like CAD [9]. Although the exact function of ANRIL is still unknown, it was shown that the expression levels of several neighboring protein-encoding genes like cyclin-dependent kinase inhibitors 2A (CDKN2A), CDKN2B, and methylthioadenosine phosphorylase (MTAP) were modulated by ANRIL [10]. Previous investigations demonstrated that the abovementioned proteins were abundantly expressed in atherosclerotic lesions, and they could promote atherosclerosis by impacting vascular remodeling, thrombogenesis, and plaque stability [9,10]. Hence, it is biologically plausible that these ANRIL polymorphisms may also be involved in atherosclerotic-related diseases like CAD. So far, some pilot studies already investigated potential correlations between ANRIL gene polymorphisms and the risk of CAD. But the results of these studies were controversial. Thus, we performed the present meta-analysis to better evaluate the roles of ANRIL polymorphisms in CAD.
Materials and methods
Literature search and inclusion criteria
This meta-analysis complied with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines [11]. Potentially related literatures that were published before September 2018 were retrieved from PubMed, Medline, and Embase using the following combination of keywords: (antisense noncoding RNA in the INK4 locus OR CDKN2B antisense RNA OR ANRIL OR CDKN2B-AS long non-coding RNA) AND (polymorphism OR variant OR mutation OR genotype OR allele) AND (coronary heart disease OR coronary artery disease OR angina pectoris OR acute coronary syndrome OR myocardial infarction).
The references of retrieved articles were also screened to identify other potentially relevant studies.
To test the research hypothesis of this meta-analysis, included studies must meet all the following criteria: (i) case–control study on correlations between ANRIL polymorphisms and the risk of CAD; (ii) provide genotypic frequencies of investigated ANRIL polymorphisms in cases and controls; and (iii) full text in English available.
Studies were excluded if one of the following criteria was fulfilled: (i) not relevant to ANRIL polymorphisms and CAD; (ii) case reports or case series; and (iii) abstracts, reviews, comments, letters, and conference presentations.
For duplicate publications, we only included the study with the largest sample size for analyses.
Data extraction and quality assessment
The following data were extracted from included studies: (i) name of the first author; (ii) publication time; (iii) country and ethnicity; (iv) sample size; and (v) genotypic distributions of ANRIL polymorphisms in cases and controls. Additionally, the probability value (P-value) of Hardy–Weinberg equilibrium (HWE) was also calculated. When necessary, we wrote to the corresponding authors for raw data. We used the Newcastle–Ottawa scale (NOS) to assess the quality of eligible studies [12]. This scale has a score range of 0–9, and studies with a score of more than 7 were thought to be of high quality. Two reviewers conducted data extraction and quality assessment independently. Any disagreement between two reviewers was solved by discussion until a consensus was reached.
Statistical analyses
All statistical analyses were conducted with Review Manager Version 5.3.3 (The Cochrane Collaboration, Software Update, Oxford, United Kingdom). Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to estimate strength of associations between ANRIL polymorphisms and the risk of CAD in all possible genetic models, and P-values ≤0.05 were considered to be statistically significant. Between-study heterogeneities were evaluated with I2 statistic. If I2 was greater than 50%, random-effect models (REMs) would be used to pool the data. Otherwise, fixed-effect models (FEMs) would be employed for synthetic analyses. Subgroup analyses by ethnicity of participants were subsequently performed. Sensitivity analyses were conducted to examine the stability of synthetic results. Funnel plots were used to assess publication bias.
Results
Characteristics of included studies
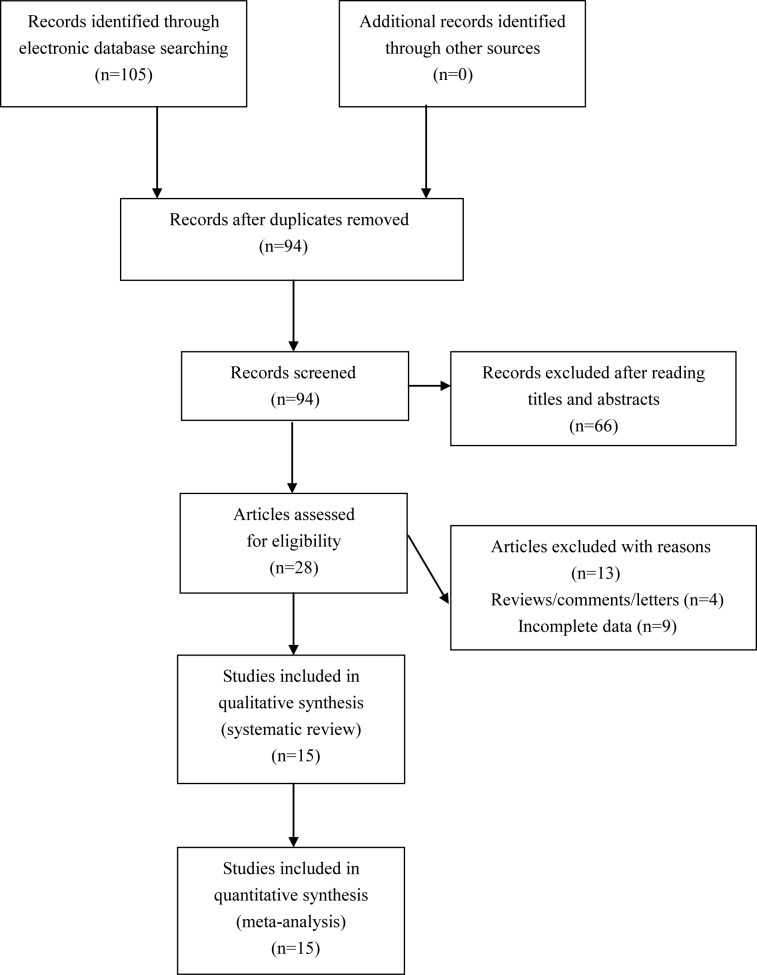
We found 105 potential relevant articles. Amongst these articles, a total of 15 eligible studies were finally included for synthetic analyses (see Figure 1). The NOS score of eligible articles ranged from 7 to 8, which indicated that all included studies were of high quality. Baseline characteristics of included studies are shown in Table 1.
Figure 1. Flowchart of study selection for the present study.
Table 1. The characteristics of included studies for ANRIL polymorphisms and CAD.
| First author, year | Country | Ethnicity | Type of disease | Sample size | Genotype distribution | P-value for HWE | NOS score | |
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||
| rs1333040 | TT/TC/CC | |||||||
| Kumar (2011) | India | West Asian | CAD | 300/423 | 147/116/37 | 175/182/66 | 0.107 | 7 |
| Lin (2011) | Taiwan | East Asian | MI | 411/1352 | 228/156/27 | 649/573/130 | 0.829 | 7 |
| Qi (2012) | China | East Asian | MI | 142/192 | 91/44/7 | 90/93/9 | 0.013 | 8 |
| Scheffold (2011) | Germany | Caucasian | MI | 976/3532 | 292/503/181 | 889/1750/893 | 0.590 | 7 |
| Zhao (2016) | China | East Asian | CAD | 456/683 | 236/184/36 | 336/293/54 | 0.370 | 7 |
| rs1333049 | GG/GC/CC | |||||||
| Ahmed (2013) | Pakistan | West Asian | MI | 294/290 | 65/166/63 | 87/180/23 | <0.001 | 8 |
| Çakmak (2015) | Turkey | Caucasian | CAD | 220/240 | 54/120/46 | 85/115/40 | 0.917 | 7 |
| Haslacher (2016) | Austria | Caucasian | MI | 493/431 | 139/236/118 | 112/222/97 | 0.514 | 7 |
| Lin (2011) | Taiwan | East Asian | MI | 423/1361 | 100/218/105 | 395/655/311 | 0.213 | 7 |
| Qi (2012) | China | East Asian | MI | 142/192 | 42/79/21 | 50/99/43 | 0.651 | 8 |
| Scheffold (2011) | Germany | Caucasian | MI | 976/999 | 212/518/246 | 292/502/205 | 0.688 | 7 |
| Yang (2018) | China | East Asian | CAD | 542/549 | 162/269/111 | 176/273/100 | 0.743 | 8 |
| rs2383206 | AA/AG/GG | |||||||
| Abdullah (2008) | U.S.A. | Caucasian | CAD | 310/560 | 56/124/130 | 143/279/138 | 0.934 | 8 |
| Kumar (2011) | India | West Asian | CAD | 309/434 | 112/143/54 | 116/224/94 | 0.467 | 7 |
| Scheffold (2011) | Germany | Caucasian | MI | 976/9053 | 183/521/272 | 2489/4583/1981 | 0.017 | 7 |
| Shendy (2017) | Egypt | West Asian | CAD | 100/50 | 51/42/7 | 23/15/12 | 0.009 | 8 |
| Zhou (2008) | China | East Asian | CAD | 1360/1360 | 373/600/387 | 382/669/309 | 0.623 | 8 |
| rs2383207 | GG/GA/AA | |||||||
| Abdullah (2008) | U.S.A. | Caucasian | CAD | 310/560 | 139/121/50 | 147/277/136 | 0.807 | 8 |
| Çakmak (2015) | Turkey | Caucasian | CAD | 220/240 | 83/101/36 | 102/118/20 | 0.079 | 7 |
| Chen (2009) | China | East Asian | CAD | 212/232 | 107/69/36 | 71/114/47 | 0.920 | 8 |
| El-Menyar (2015) | Egypt | West Asian | CAD | 236/152 | 146/77/12 | 84/58/10 | 0.998 | 7 |
| Kumar (2011) | India | West Asian | CAD | 301/424 | 137/124/40 | 174/190/60 | 0.485 | 7 |
| Lin (2011) | Taiwan | East Asian | MI | 415/1349 | 214/162/39 | 568/609/172 | 0.660 | 7 |
| Qi (2012) | China | East Asian | MI | 142/192 | 78/55/9 | 82/93/17 | 0.192 | 8 |
| Scheffold (2011) | Germany | Caucasian | MI | 976/3532 | 183/519/274 | 1016/1770/746 | 0.628 | 7 |
| Yang (2018) | China | East Asian | CAD | 540/548 | 247/236/57 | 244/251/53 | 0.317 | 8 |
| Zhou (2008) | China | East Asian | CAD | 1360/1360 | 702/520/138 | 592/605/163 | 0.659 | 8 |
| rs10757274 | GG/GA/AA | |||||||
| Abdullah (2008) | U.S.A. | Caucasian | CAD | 310/560 | 61/119/130 | 156/283/121 | 0.728 | 8 |
| El-Menyar (2015) | Egypt | West Asian | CAD | 309/434 | 122/133/54 | 143/206/85 | 0.486 | 7 |
| Kumar (2011) | India | West Asian | CAD | 310/439 | 116/135/59 | 144/210/85 | 0.591 | 7 |
| Mafi Golchin (2017) | Iran | West Asian | CAD | 103/102 | 40/47/16 | 22/51/29 | 0.962 | 8 |
| Scheffold (2011) | Germany | Caucasian | MI | 976/9053 | 208/515/253 | 2752/4543/1758 | 0.131 | 7 |
| Zhao (2016) | China | East Asian | CAD | 291/385 | 82/137/72 | 130/188/67 | 0.945 | 7 |
| rs10757278 | GG/GA/AA | |||||||
| Abdullah (2008) | U.S.A. | Caucasian | CAD | 310/560 | 59/135/116 | 159/294/107 | 0.162 | 8 |
| Chen (2009) | China | East Asian | CAD | 212/232 | 107/69/36 | 71/114/47 | 0.920 | 8 |
| El-Menyar (2015) | Egypt | West Asian | CAD | 309/427 | 89/142/78 | 103/214/110 | 0.957 | 7 |
| Scheffold (2011) | Germany | Caucasian | MI | 976/718 | 211/522/243 | 224/366/128 | 0.308 | 7 |
| Shendy (2017) | Egypt | West Asian | CAD | 100/50 | 37/45/18 | 13/18/19 | 0.057 | 8 |
P-values ≤0.05 were considered to be statistically significant. Abbreviation: MI, myocardial infarction.
Overall and subgroup analyses
To investigate potential correlations between ANRIL polymorphisms and the risk of CAD, five studies about rs1333040 polymorphism, seven studies about rs1333049 polymorphism, five studies about rs2383206 polymorphism, ten studies about rs2383207 polymorphism, six studies about rs10757274 polymorphism, and five studies about rs10757278 polymorphism were enrolled for analyses.
Significant associations with the risk of CAD were detected for rs1333040 (dominant model: P<0.0001, OR = 1.30, 95% CI: 1.17–1.44; recessive model: P<0.0001, OR = 0.71, 95% CI: 0.62–0.82; allele model: P<0.0001, OR = 1.25, 95% CI: 1.16–1.34), rs1333049 (dominant model: P=0.02, OR = 0.81, 95% CI: 0.67–0.96; allele model: P=0.02, OR = 0.86, 95% CI: 0.76–0.98), and rs2383207 (additive model: P=0.004, OR = 0.80, 95% CI: 0.69–0.93) polymorphisms in overall analyses.
Further subgroup analyses according to ethnicity of participants revealed that rs1333040 (dominant model and allele model), rs1333049 (recessive model), and rs2383207 (all genetic models) polymorphisms were significantly correlated with the risk of CAD in East Asians, rs2383206 (dominant model and allele model) and rs10757274 (dominant model and allele model) polymorphisms were significantly correlated with the risk of CAD in West Asians, while rs2383206 (dominant model, recessive model, and allele model), rs10757274 (dominant model, recessive model, and allele model) and rs10757278 (dominant model, recessive model, and allele model) polymorphisms were significantly correlated with the risk of CAD in Caucasians (see Table 2).
Table 2. Overall and subgroup analyses for ANRIL polymorphisms and CAD.
| Polymorphisms | Population | Sample size | Dominant comparison | Recessive comparison | Additive comparison | Allele comparison | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | |||
| rs1333040 | Overall | 2285/6182 | <0.0001 | 1.30 (1.17–1.44) | <0.0001 | 0.71 (0.62–0.82) | 0.12 | 0.85 (0.69–1.04) | <0.0001 | 1.25 (1.16–1.34) |
| East Asian | 1009/2227 | 0.03 | 1.37 (1.04–1.80) | 0.18 | 0.82 (0.61–1.09) | 0.06 | 0.76 (0.57–1.01) | 0.001 | 1.22 (1.08–1.38) | |
| rs1333049 | Overall | 3090/4062 | 0.02 | 0.81 (0.67–0.96) | 0.08 | 1.24 (0.98–1.57) | 0.35 | 1.05 (0.95–1.15) | 0.02 | 0.86 (0.76–0.98) |
| Caucasian | 1689/1670 | 0.37 | 0.85 (0.60–1.20) | 0.93 | 1.02 (0.69–1.51) | 0.32 | 1.10 (0.91–1.31) | 0.59 | 0.93 (0.72–1.20) | |
| East Asian | 1107/2102 | 0.16 | 0.82 (0.61–1.09) | 0.02 | 1.19 (1.03–1.38) | 0.31 | 1.07 (0.94–1.20) | 0.06 | 0.87 (0.75–1.01) | |
| rs2383206 | Overall | 3055/11457 | 0.60 | 0.91 (0.64–1.29) | 0.33 | 1.18 (0.84–1.65) | 0.36 | 0.90 (0.72–1.13) | 0.45 | 0.91 (0.72–1.16) |
| Caucasian | 1286/9613 | <0.0001 | 0.61 (0.53–0.71) | 0.02 | 1.71 (1.08–2.71) | 0.61 | 0.88 (0.53–1.45) | 0.0002 | 0.68 (0.56–0.84) | |
| West Asian | 409/484 | 0.006 | 1.49 (1.12–1.99) | 0.19 | 0.47 (0.15–1.46) | 0.83 | 1.08 (0.53–2.19) | 0.002 | 1.36 (1.12–1.65) | |
| rs2383207 | Overall | 4712/8589 | 0.09 | 1.29 (0.96–1.72) | 0.66 | 0.94 (0.73–1.22) | 0.004 | 0.80 (0.69–0.93) | 0.15 | 1.17 (0.95–1.45) |
| Caucasian | 1506/4332 | 0.97 | 1.02 (0.42–2.47) | 0.58 | 1.21 (0.62–2.35) | 0.48 | 0.88 (0.61–1.26) | 0.94 | 0.98 (0.55–1.75) | |
| East Asian | 2669/3681 | 0.0003 | 1.45 (1.18–1.79) | 0.03 | 0.83 (0.71–0.98) | <0.0001 | 0.77 (0.69–0.85) | 0.0005 | 1.28 (1.11–1.48) | |
| West Asian | 537/576 | 0.08 | 1.24 (0.97–1.58) | 0.57 | 0.89 (0.61–1.32) | 0.15 | 0.84 (0.65–1.07) | 0.12 | 1.16 (0.96–1.39) | |
| rs10757274 | Overall | 2299/10973 | 0.89 | 0.98 (0.69–1.39) | 0.27 | 1.23 (0.85–1.76) | 0.15 | 0.86 (0.70–1.06) | 0.89 | 0.98 (0.69–1.39) |
| Caucasian | 1286/9613 | <0.0001 | 0.62 (0.54–0.72) | 0.03 | 1.92 (1.08–3.42) | 0.54 | 0.83 (0.46–1.50) | 0.001 | 0.65 (0.50–0.84) | |
| West Asian | 722/975 | 0.003 | 1.36 (1.11–1.67) | 0.17 | 0.84 (0.66–1.08) | 0.08 | 0.84 (0.69–1.02) | 0.04 | 1.28 (1.01–1.62) | |
| rs10757278 | Overall | 1907/1987 | 0.81 | 1.09 (0.63–1.87) | 0.72 | 1.10 (0.67–1.81) | 0.24 | 0.83 (0.61–1.13) | 0.87 | 1.03 (0.70–1.52) |
| Caucasian | 1286/1278 | <0.0001 | 0.60 (0.50–0.73) | 0.008 | 1.95 (1.19–3.19) | 0.61 | 0.89 (0.57–1.39) | <0.0001 | 0.65 (0.52–0.81) | |
| West Asian | 409/477 | 0.06 | 1.33 (0.98–1.80) | 0.35 | 0.63 (0.24–1.66) | 0.54 | 0.92 (0.70–1.20) | 0.22 | 1.37 (0.83–2.27) | |
The values in bold represent that there are statistically significant differences between cases and controls. P-values ≤0.05 were considered to be statistically significant. Abbreviation: MI, myocardial infarction.
Sensitivity analyses
We performed sensitivity analyses by excluding studies that deviated from HWE. No alterations of results were detected in sensitivity analyses, which suggested that our findings were statistically reliable.
Publication biases
Publication biases were evaluated with funnel plots. We did not find obvious asymmetry of funnel plots in any comparisons, which indicated that our findings were unlikely to be impacted by severe publication biases.
Discussion
To the best of our knowledge, this is so far the most comprehensive meta-analysis on correlations between ANRIL polymorphisms and the risk of CAD. The overall analyses suggested that rs1333040, rs1333049, and rs2383207 polymorphisms were significantly associated with CAD risk in overall population. Further subgroup analyses revealed that rs1333040, rs1333049, and rs2383207 polymorphisms were significantly correlated with the risk of CAD in East Asians, rs2383206 and rs10757274 polymorphisms were significantly correlated with the risk of CAD in West Asians, while rs2383206, rs10757274, and rs10757278 polymorphisms were significantly correlated with the risk of CAD in Caucasians.
There are several points that need to be addressed about this meta-analysis. First, the exact function of ANRIL is still unclear, and therefore the underlying mechanisms of our positive findings need to be elucidated by future investigations. Second, the pathogenic mechanism of CAD is highly complex, and therefore it is unlikely that a single gene polymorphism can significantly contribute to its development. As a result, to better illustrate potential correlations of certain gene polymorphisms with CAD, we strongly recommend further studies to perform haplotype analyses and explore potential gene–gene interactions. Thirdly, according to a previous investigation, subjects carrying mutant allele of rs1333049 polymorphism had elevated TC, TG and LDL-C levels, and this may partially explain the observed significant correlation between this polymorphism and the risk of CAD. For other investigated polymorphisms, however, future studies are needed to investigate whether these polymorphisms are correlated with clinical and biochemical parameters of CAD [13].
As with all meta-analysis, the present study certainly has some limitations. First, our results were derived from unadjusted analyses, and lack of further adjusted analyses for age, gender, and co-morbidity conditions may impact the reliability of our findings [14]. Second, obvious heterogeneities were found in several subgroups, which indicated that the controversial results of included studies could not be fully explained by differences in ethnic background and type of disease, and other baseline characteristics of participants may also contribute to between-study heterogeneities [15]. Third, associations between ANRIL polymorphisms and CAD may also be modified by gene-gene and gene-environmental interactions. However, most eligible studies did not consider these potential interactions, which impeded us to perform relevant analyses accordingly [16]. On account of abovementioned limitations, our findings should be cautiously interpreted.
Conclusion
In conclusion, our meta-analysis suggested that rs1333040, rs1333049, rs2383206, rs2383207, rs10757274, and rs10757278 polymorphisms might serve as genetic biomarkers of CAD in certain ethnicities. However, further well-designed studies are still warranted to confirm our findings.
Abbreviations
- ANRIL
antisense non-coding RNA in the INK4 locus
- CAD
coronary artery disease
- CI
confidence interval
- HWE
Hardy–Weinberg equilibrium
- ncRNA
non-coding RNA
- NOS
Newcastle–Ottawa scale
- OR
odds ratio
Author contribution
L.H. conceived the study and participated in its design. L.H. and G.S. conducted the systematic literature review. X.W. performed data analyses. L.H. drafted the manuscript. All authors gave final approval and agreed to be accountable for all aspects of work ensuring integrity and accuracy.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Moran A.E., Forouzanfar M.H., Roth G., Mensah G.A., Ezzati M., Flaxman A. et al. (2014) The Global Burden of Ischemic Heart Disease in 1990 and 2010: The Global Burden of Disease 2010 Study. Circulation 129, 1493–1501 10.1161/CIRCULATIONAHA.113.004046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Study 2013 Collaborators (2015) Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386, 743–800 10.1016/S0140-6736(15)60692-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayer B., Erdmann J. and Schunkert H. (2007) Genetics and heritability of coronary artery disease and myocardial infarction. Clin. Res. Cardiol. 96, 1–7 10.1007/s00392-006-0447-y [DOI] [PubMed] [Google Scholar]
- 4.Evans A., Van Baal G.C., McCarron P., DeLange M., Soerensen T.I., De Geus E.J. et al. (2003) The genetics of coronary heart disease: the contribution of twin studies. Twin Res. 6, 432–441 10.1375/136905203770326439 [DOI] [PubMed] [Google Scholar]
- 5.Sayols-Baixeras S., Lluís-Ganella C., Lucas G. and Elosua R. (2014) Pathogenesis of coronary artery disease: focus on genetic risk factors and identification of genetic variants. Appl. Clin. Genet. 7, 15–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai X., Wiernek S., Evans J.P. and Runge M.S. (2016) Genetics of coronary artery disease and myocardial infarction. World J. Cardiol. 8, 1–23 10.4330/wjc.v8.i1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peschansky V.J. and Wahlestedt C. (2014) Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 9, 3–12 10.4161/epi.27473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M. (2011) Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 9.Congrains A., Kamide K., Oguro R., Yasuda O., Miyata K., Yamamoto E. et al. (2012) Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 220, 449–455 10.1016/j.atherosclerosis.2011.11.017 [DOI] [PubMed] [Google Scholar]
- 10.Cunnington M.S. and Keavney B. (2011) Genetic mechanisms mediating atherosclerosis susceptibility at the chromosome 9p21 locus. Curr. Atheroscler. Rep. 13, 193–201 10.1007/s11883-011-0178-z [DOI] [PubMed] [Google Scholar]
- 11.Moher D., Liberati A., Tetzlaff J., Altman D.G. and PRISMA Group (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann. Intern. Med. 151, 264–269 [DOI] [PubMed] [Google Scholar]
- 12.Stang A. (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 13.Ahmed W., Ali I.S., Riaz M., Younas A., Sadeque A., Niazi A.K. et al. (2013) Association of ANRIL polymorphism (rs1333049:C>G) with myocardial infarction and its pharmacogenomic role in hypercholesterolemia. Gene 515, 416–420 10.1016/j.gene.2012.12.044 [DOI] [PubMed] [Google Scholar]
- 14.Xie X., Shi X. and Liu M. (2017) The roles of TLR gene polymorphisms in atherosclerosis: a systematic review and meta-analysis of 35317 subjects. Scand. J. Immunol. 86, 50–58 10.1111/sji.12560 [DOI] [PubMed] [Google Scholar]
- 15.Shi X., Xie X., Jia Y. and Li S. (2016) Associations of insulin receptor and insulin receptor substrates genetic polymorphisms with polycystic ovary syndrome: a systematic review and meta-analysis. J. Obstet. Gynaecol. Res. 42, 844–854 10.1111/jog.13002 [DOI] [PubMed] [Google Scholar]
- 16.Xie X., Shi X., Xun X. and Rao L. (2017) Endothelial nitric oxide synthase gene single nucleotide polymorphisms and the risk of hypertension: a meta-analysis involving 63,258 subjects. Clin. Exp. Hypertens. 39, 175–182 10.1080/10641963.2016.1235177 [DOI] [PubMed] [Google Scholar]