Abstract
We conducted a feasibility study to predict malignant glioma grades via radiomic analysis using contrast-enhanced T1-weighted magnetic resonance images (CE-T1WIs) and T2-weighted magnetic resonance images (T2WIs). We proposed a framework and applied it to CE-T1WIs and T2WIs (with tumor region data) acquired preoperatively from 157 patients with malignant glioma (grade III: 55, grade IV: 102) as the primary dataset and 67 patients with malignant glioma (grade III: 22, grade IV: 45) as the validation dataset. Radiomic features such as size/shape, intensity, histogram, and texture features were extracted from the tumor regions on the CE-T1WIs and T2WIs. The Wilcoxon–Mann–Whitney (WMW) test and least absolute shrinkage and selection operator logistic regression (LASSO-LR) were employed to select the radiomic features. Various machine learning (ML) algorithms were used to construct prediction models for the malignant glioma grades using the selected radiomic features. Leave-one-out cross-validation (LOOCV) was implemented to evaluate the performance of the prediction models in the primary dataset. The selected radiomic features for all folds in the LOOCV of the primary dataset were used to perform an independent validation. As evaluation indices, accuracies, sensitivities, specificities, and values for the area under receiver operating characteristic curve (or simply the area under the curve (AUC)) for all prediction models were calculated. The mean AUC value for all prediction models constructed by the ML algorithms in the LOOCV of the primary dataset was 0.902 ± 0.024 (95% CI (confidence interval), 0.873–0.932). In the independent validation, the mean AUC value for all prediction models was 0.747 ± 0.034 (95% CI, 0.705–0.790). The results of this study suggest that the malignant glioma grades could be sufficiently and easily predicted by preparing the CE-T1WIs, T2WIs, and tumor delineations for each patient. Our proposed framework may be an effective tool for preoperatively grading malignant gliomas.
Subject terms: Diagnostic markers, CNS cancer
Introduction
Gliomas are primary brain tumors caused by glial cell mutations. The latest reports from the brain tumor registry of Japan indicate that 27% of brain tumor patients in Japan suffered from gliomas between 2005–20081. Gliomas are classified into four grades in accordance with the pathology and genotypic figures issued by the World Health Organization (WHO)2. A surgical approach of removing the visible tumor tissue is typically applied to all glioma grades after imaging diagnosis based on computed tomography (CT), magnetic resonance (MR), and positron emission tomography (PET) images. Adjuvant therapy (namely chemotherapy, radiotherapy, or chemoradiotherapy) after surgery is used to treat high-grade gliomas (HGGs) to address the inevitable extension of tumors beyond margins suggested by imaging3. The glioma grade is determined based on pathological and genetic features of the tissues. Although an imaging diagnosis is preoperatively performed to approximate the malignancy of the tumor, the grade is usually determined based on the tissue obtained from a biopsy or resection during surgery. Glioma grading using medical imaging should be performed prior to surgery for increasing treatment effects while decreasing adverse events. In addition, predicting glioma grades using preoperative images is useful for patient education before surgery.
Methodologies for predicting glioma grades using MR or CT images have been described in previous studies4–11. One concept for predicting the glioma grade is to construct statistical models using some tumor appearance features or imaging indices. A more comprehensive analysis using more quantitative imaging features may provide better accuracy in predicting glioma grades. For this reason, we investigated the feasibility of radiomics in predicting glioma grades.
Radiomics is a comprehensive analysis for describing tumor phenotypes based on high-dimensional quantitative features extracted from the large quantity of medical images collected12–14. It has the potential to be an effective tool for personalized medicine based on phenotypic descriptions of tumors from medical images12, allowing for noninvasive analysis of tumor characteristics comparable with molecular biological approaches such as genomics, epigenomics, transcriptomics, and proteomics12. Some studies for predicting glioma grades based on radiomics using MR images have been conducted15–22. Qin et al., Cho et al., Chen et al., and Vamvakas et al. proposed frameworks for classifying low-grade gliomas (LGGs) and HGGs using images acquired by multiple MR imaging (MRI) sequences15–19. Predicting LGGs and HGGs could be made possible by constructing radiomics-based classifiers using machine learning (ML) algorithms in those frameworks. Zacharaki et al. and Tian et al. investigated the prediction of grade III and IV gliomas as well as the classification of LGGs and HGGs using images acquired via multiple MRI sequences20,21. Zhang et al. investigated both the classification of LGGs and HGGs and the prediction of grade II, III, and IV gliomas22. However, in previous studies, all of which used multiple MRI sequences, tumors needed to be contoured on each MR image for radiomic analysis of each patient, indicating that radiomic analysis for grading gliomas could not be performed unless all images acquired by the multiple MRI sequences were prepared in this manner. Considerable time and effort would be required to prepare tumor contours on multiple MRI sequences images for all the patients in the database. In addition, if the images acquired by a special MRI sequence were used for a framework for glioma grading based on radiomics, the framework would not have versatility for use in other institutions. Therefore, predicting the glioma grade before surgery in a straightforward manner using a few structural MRI sequences images usually acquired by the majority of institutions and volumes of interest of the tumor regions in each patient is crucial. Reza et al. verified the effect of three structural MRI sequences images (contrast-enhanced T1-weighted MR images (CE-T1WIs), T2-weighted MR images (T2WIs), and fluid attenuated inversion recovery (FLAIR) images) for classifying the LGGs and HGGs, and LGGs and grade IV gliomas using a few datasets23. However, there would be no radiomic study for verifying the effect of a few structural MRI sequences images for predicting malignant glioma grades (namely grades III and IV) using various ML algorithms.
Therefore, the purpose of this study was to investigate the feasibility of predicting malignant glioma grades based on radiomic analysis using the CE-T1WIs and T2WIs acquired before surgery.
Materials and Methods
Overall study design
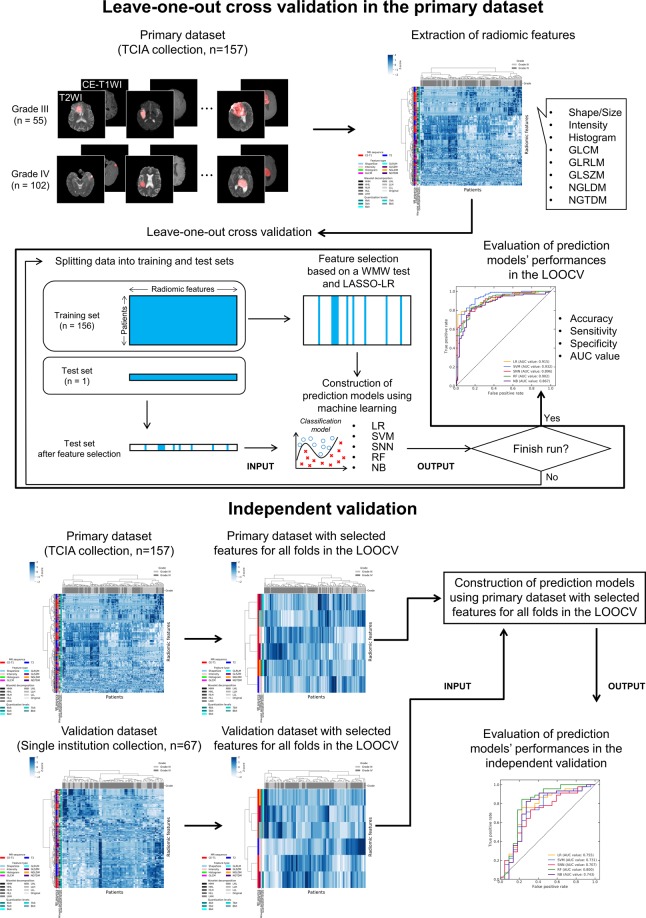
Figure 1 shows a conceptual design for predicting glioma grades based on radiomic features. The database in this study consisted of primary dataset collected in public database and validation dataset collected in our hospital. The high-dimensional radiomic features were extracted from tumor regions on the CE-T1WIs and T2WIs for all patients in the primary and validation datasets. A Wilcoxon–Mann–Whitney (WMW) test and least absolute shrinkage and selection operator logistic regression (LASSO-LR) were employed for selecting the extracted radiomic features to construct prediction models using features potentially related to glioma grades. The prediction models were constructed using the LR, a support vector machine (SVM), a standard neural network (SNN), a random forest (RF), and a naïve Bayes (NB). A leave-one-out cross-validation (LOOCV) was undertaken for evaluating the performance of the prediction models in the primary dataset. Finally, an independent validation was performed using the primary and validation datasets with selected radiomic features for all folds in the LOOCV of the primary dataset.
Figure 1.
A conceptual design for predicting glioma grades based on radiomic features.
Database and equipment
This study was performed in accordance with relevant guidelines and regulations approved by the institutional review board at the University of Tokyo hospital. Ethical approval for the study was also provided by the review board (reference number: 11770-[1]). Written informed consent was obtained from all subjects within the validation dataset collected in our hospital.
The brain CE-T1WIs and T2WIs archived in the cancer genome atlas glioblastoma multiforme (TCGA-GBM)24 and low-grade glioma (TCGA-LGG)25 collections of the cancer imaging archive (TCIA)26 were used in this study. Specifically, 157 malignant glioma patients’ preoperative CE-T1WIs and T2WIs (grade III: 55, grade IV: 102) with tumor segmentations, which were distributed via a third-party analysis using TCGA-GBM and TCGA-LGG collections27–29, were used as the primary dataset. The CE-T1WIs and T2WIs distributed by the third-party analysis using these collections have been transformed into the same coordinate system and interpolated to 1-mm3 isotropic voxels29. The tumor segmentations were delineated using a computerized framework and corrected by a neuroradiologist29. In the segmentations, there were three types of labels: (i) non-enhanced tumor and necrosis, (ii) enhanced tumor, and (iii) edema region29. Cho et al. verified that in accordance with their results, the enhanced and non-enhanced regions should be taken into account for grading the LGGs and HGGs17. Therefore, the tumor segmentations excluding the edema regions were used in this study. TCGA-LGG and TCGA-GBM are multicentered collections. Then, the imaging information and patients’ characteristic have been mentioned in the cited articles24,25,27–29.
The validation dataset comprised brain CE-T1WIs and T2WIs (with tumor region data) acquired preoperatively from 67 malignant glioma patients in our hospital. The mean number of days between image acquisition and surgery for all patients was 13.7 (range: 1–67). None of the patients underwent any treatment prior to the image acquisition that could influence the intensity of the MR images. Table 1 lists the patients’ characteristics in the validation dataset for this study. There were 22 grade III (anaplastic astrocytoma (AA): 8, anaplastic oligodendroglioma (AO): 9, anaplastic oligoastrocytoma (AOA): 5) and 45 glioblastoma (GBM) grade IV patients. The isocitrate dehydrogenase (IDH) mutation and O6-methylguanine-DNA methyltransferase (MGMT) methylation statuses for the GBM patients are listed in Table 1. The CE-T1WIs and T2WIs were acquired using 3.0-T MR scanners (Signa® HDx and HDxt, GE Healthcare, Chicago, IL, USA). The CE-T1WIs were acquired after bolus injection of gadolinium-based contrast agents. The ranges of the repetition time (TR)/echo time (TE) for all CE-T1WIs were 380–640 ms/8–12 ms. The matrix size, pixel size, slice thickness, and spacing between the slices of the CE-T1WIs were 256 × 256, 0.82 × 0.82 mm2, 5.0 mm, and 6.0 mm, respectively. In the T2WIs, the range of TR/TE, matrix size, pixel size, slice thickness, and spacing between slices were 4320–4640 ms/80.77–89.28 ms, 512 × 512, 0.41 × 0.41 mm2, 3.0 mm, and 3.0 mm, respectively. The bit depth of the MR images was 16 bits per pixel (bpp). The CE-T1WIs and T2WIs were transformed into the same coordinate system using ITK-SNAP (ver. 3.6). A radiation technologist (T.N.) manually delineated the tumors excluding the edema regions on the MR images for all patients to extract the radiomic features; this delineation was performed under the supervision of a radiation oncologist (W.T.) and a radiologist (S.K.) for quality assurance. A commercial radiation treatment planning system (Monaco® ver. 5.11, Elekta, Stockholm, Sweden) was used for the tumor delineations.
Table 1.
Patients’ characteristics in the validation dataset for this study.
| Characteristic | Value |
|---|---|
| Total number of patients | 67 |
| Gender | Male: 45 (67.2%) |
| Female: 22 (32.8%) | |
| Mean age | 55.2 ± 16.2 (range: 11–83) |
| Grade | III: 22 (32.8%) |
| IV: 45 (67.2%) | |
| Histological type | GBM: 45 (67.2%) |
| AA: 8 (11.9%) | |
| AO: 9 (13.4%) | |
| AOA: 5 (7.5%) | |
| IDH mutation status in GBM (n = 45) | Mutated: 2 (4.4%) |
| Wild type: 19 (42.2%) | |
| Unknown: 24 (53.3%) | |
| MGMT methylation status in GBM (n = 45) | Methylated: 7 (15.6%) |
| Unmethylated: 13 (28.9%) | |
| Unknown: 25 (55.6%) |
GBM: glioblastoma, AA: anaplastic astrocytoma, AO: anaplastic oligodendroglioma, AOA: anaplastic oligoastrocytoma, IDH: isocitrate dehydrogenase, MGMT: O6-methylguanine-DNA methyltransferase.
The radiomic analysis was performed using a commercial numerical programming language (MATLAB® ver. R2017a and R2017b, MathWorks, Natick, MA, USA) and an open-source numerical programming language (Python® ver. 3.6). There were accessed on two workstations, one with a single 2.26 GHz quad-core central processing unit (CPU) (Intel® Xeon® E5607, Intel Corp., Santa Clara, CA, USA) and the other with double 2.67 GHz quad-core CPUs (Intel® Xeon® X5550). Both workstations had 16 GB of RAM.
Radiomic features
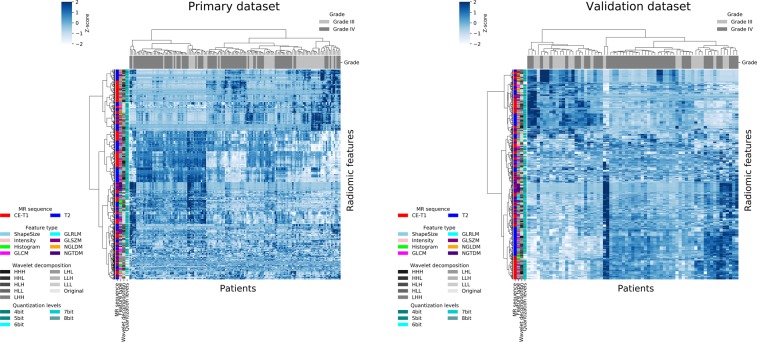
The radiomic features were extracted from the glioma regions on the CE-T1WIs and T2WIs using open-source MATLAB code developed by Vallières et al.30,31 (https://github.com/mvallieres/radiomics and https://github.com/mvallieres/radiomics-develop). Intensity normalization was performed for whole brain regions of the MR images in the primary and validation datasets using Z-score transformation32. The voxels of the MR images in the validation dataset were converted to 1-mm3 isotropic voxels using cubic interpolation before extracting the radiomic features. The interpolation for binary images proposed by Herman et al.33 was employed to isotropically resample the voxels of tumor mask images derived from the tumor delineation data in the validation dataset. The quantitative image features described in the image biomarker standardization initiative (IBSI)34 were used in this radiomic analysis. In this study, 8 shape/size features, 18 intensity features, 20 histogram features, 11 gray-level co-occurrence matrix (GLCM) features, 13 gray-level run length matrix (GLRLM) features, 13 gray-level size zone matrix (GLSZM) features, 16 neighboring gray-level dependence matrix (NGLDM), and 5 neighborhood gray-tone difference matrix (NGTDM) features within the IBSI, which have been widely used in radiomic analyses, were adopted as the radiomic features. The details of the radiomic features are provided in Supplement 1. A three-dimensional (3D) Coiflet wavelet transform35 was applied to the MR images in order to extract the intensity features, histogram features, and GLCM, GLRLM, GLSZM, NGLDM, and NGTDM features known as texture features in frequency decomposed images. The frequency components were HHH, HHL, HLH, HLL, LHH, LHL, LLH, and LLL, where “H” and “L” denote high-pass and low-pass filters, respectively. Thus, the intensity, histogram, and texture features were extracted from the tumor region on the original MR images and eight frequency component-filtered images. Figure 2 shows transverse images of a tumor on the original MR image (T2WI) and on eight frequency component-filtered images to which the 3D Coiflet wavelet transform had been applied. The number of bins for the histogram features was set to 6 bit. The tumor regions on the original MR images and filtered images were quantized to calculate the texture features. The quantization was performed range of μ ± 3σ, where μ and σ denote the mean and standard deviation (SD) of the voxel values in the tumor regions, respectively36. The quantization levels were set to 4, 5, 6, 7, and 8 bit. Figure 3 shows the heat maps of the radiomic features in the primary and validation datasets. The total number of radiomic features was 5912. The radiomic features were normalized by Z-score transformation and clustered using Ward’s method37 in these heat maps.
Figure 2.
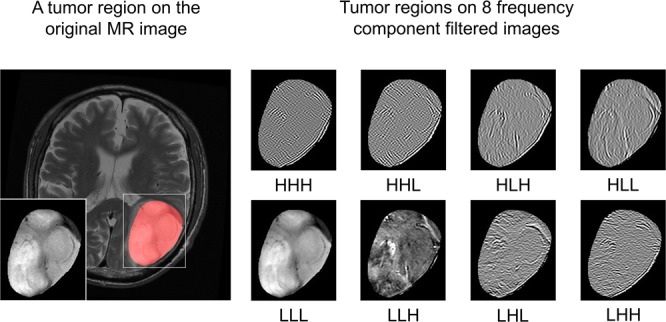
Transverse images of a tumor on original magnetic resonance (MR) image (T2-weighted MR image (T2WI)) and on eight frequency component-filtered images to which a three-dimensional (3D) Coiflet wavelet transform had been applied.
Figure 3.
Heat maps of radiomic features in primary and validation datasets.
Feature selection
Among the extracted radiomic features, some features would not correlate with the malignant glioma grading. Overfitted models for glioma grading would be constructed owing to these uncorrelated radiomic features. Therefore, radiomic features were selected using the WMW test and LASSO-LR38,39 to construct robust prediction models of the glioma grades. The two-tail WMW test was performed for all extracted radiomic features to obtain significant radiomic features (P < 0.001) for grading gliomas. Then, the significant radiomic features were utilize to select features using the LASSO-LR. A scikit-learn (ver. 0.19), open ML library for Python40 was used for the LASSO-based feature selection. The LASSO-LR can construct a classification model with sparse explanatory variables by solving an L1-norm regularized objective function expressed as follows:
| 1 |
where
| 2 |
| 3 |
| 4 |
where is an optimal coefficient vector, n is the number of patients, y is a label for the glioma grades, and λ is a hyper-parameter of the regularization. x, β, and p are explanatory vectors comprising the significant radiomic features, coefficient vector, and number of the significant radiomic features, respectively. The optimization problem was solved using a coordinate descent algorithm41. would be a sparse vector owing to L1-norm regularization. The features with non-zero coefficients of the were selected in this study. λ, the hyper-parameter determining the regularization effect in the optimization problem42, was tuned in this study by using a grid search technique. In the grid search, five-fold cross-validation (CV) was performed five times in the training set while changing the values of the hyper-parameter, and mean values for the area under receiver operating characteristic (ROC) curve (or simply the area under the curve (AUC)) for the five-times five-fold CV were calculated for each value of the hyper-parameter. The value of the hyper-parameter that maximized the mean AUC value for the five-times five-fold CV was used for the regularization Figure 4 shows the mean AUC values for the five-times five-fold CV for each value of the regularization hyper-parameter. The range of the hyper-parameter values was 10−6–102.
Figure 4.
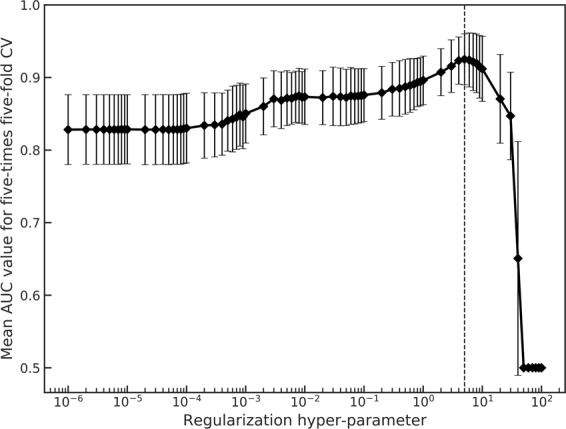
Mean area under the curve (AUC) values for five-times five-fold cross-validation (CV) for each value of a regularization hyper-parameter. The dashed line depicts a hyper-parameter value, which maximizes the mean AUC value for five-times five-fold CV.
Construction of prediction models for glioma grades using machine learning algorithms
The scikit-learn was also used in this procedure. The LR, SVM43, SNN44, RF45, and NB46 were used to construct the prediction models for the malignant glioma grades using the selected radiomic features. Some hyper-parameters of the LR, SVM, SNN, and RF were tuned by the same methodology as that used for feature selection. The ranges for tuning the hyper-parameters by using grid search are provided in Supplement 2. In the SVM, a radial basis function kernel was used to construct nonlinear models43. Almost all hyper-parameters of the SNN and RF were fixed default values provided by scikit-learn40. In the RF, number of trees was fixed to 1000. There was no parameter for tuning in the NB. The LOOCV was conducted to evaluate the performance of prediction models derived from the LR, SVM, SNN, RF, and NB in the primary dataset. Independent validation was also performed to investigate the versatility of the radiomic analysis with a few structural MRI sequences for predicting the malignant glioma grades using the primary and validation datasets. Specifically, the prediction models were constructed using the primary dataset with the selected radiomic features for all folds in the LOOCV; then, the prediction models were evaluated using the validation dataset with the selected radiomic features. Accuracies, sensitivities, specificities, and AUC values for all prediction models were calculated as evaluation indices. Grade III and IV gliomas were defined as negative and positive, respectively, for calculating the evaluation indices.
Results
The range and mode of the number of the significant radiomic features for grading malignant gliomas for the LOOCV were 593–717 and 638, respectively. The range and mode of the number of selected radiomic features for the LOOCV were 21–39 and 30, respectively. The mean percentage of number of selected radiomic features for the LOOCV was 0.53%. The mean ± SD of the value of the hyper-parameter of regularization for the LOOCV was 5.02 ± 0.76 (95% confidence interval (CI), 4.90–5.14). Table 2 lists the selected radiomic features for all folds in the LOOCV of the primary dataset. The number of selected radiomic features for all LOOCV folds in the CE-T1WIs and T2WIs were 5 (intensity: 1, GLRLM: 1, GLSZM: 2, NGLDM: 1), and 1 (intensity: 1), respectively.
Table 2.
Selected radiomic features for all folds in a leave-one-out cross-validation (LOOCV) of the primary dataset.
| MRI sequence | Wavelet | Quantization levels | Feature type | Feature name |
|---|---|---|---|---|
| CE-T1 | LLL | — | Intensity | Median |
| CE-T1 | LHL | 8 bit | GLRLM | Run-length variance |
| CE-T1 | LLL | 5 bit | GLSZM | Gray-level non-uniformity normalized |
| CE-T1 | HLL | 7 bit | GLSZM | Gray-level variance |
| CE-T1 | HLL | 7 bit | NGLDM | High dependence low gray-level emphasis |
| T2 | LLL | — | Intensity | Root mean square |
CE-T1: contrast-enhanced T1, L: low-pass filter, H: high-pass filter, GLRLM: gray-level run length matrix, GLSZM: gray-level size zone matrix, NGLDM: neighboring gray-level dependence matrix.
Figure 5 shows the ROC curves of the prediction models constructed by the five ML algorithms in the LOOCV of the primary dataset. The AUC values of the prediction models constructed by the LR, SVM, SNN, RF, and NB were 0.915, 0.932, 0.896, 0.902, and 0.867, respectively. Table 3 lists the accuracies, sensitivities, specificities, and AUC values of the prediction models in the LOOCV of the primary dataset. The mean ± SD of these four parameters for all prediction models were 0.824 ± 0.027 (95% CI, 0.790–0.858), 0.863 ± 0.033 (95% CI, 0.822–0.903), 0.753 ± 0.065 (95% CI, 0.672–0.833), and 0.902 ± 0.024 (95% CI, 0.873–0.932), respectively. The prediction models using the SVM demonstrated the best performance for classifying the malignant glioma grades in the LOOCV of the primary dataset, based on the resulting AUC value (0.932).
Figure 5.
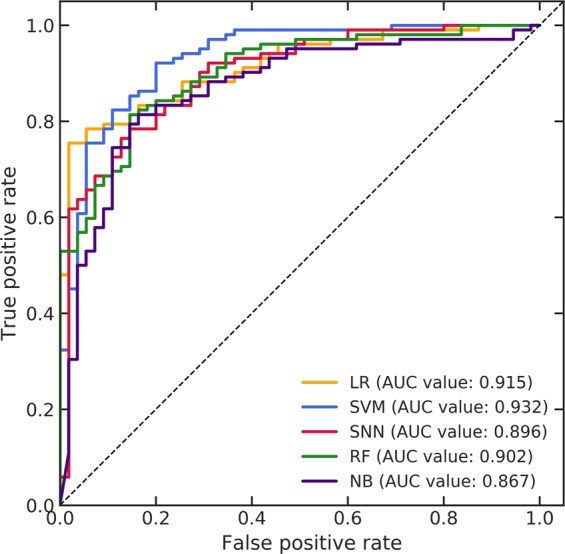
Receiver operating characteristic (ROC) curves of the prediction models constructed by the five machine learning (ML) algorithms in a leave-one-out cross-validation (LOOCV) of the primary dataset.
Table 3.
Accuracies, sensitivities, specificities, and area under the curve (AUC) values of prediction models in a leave-one-out cross-validation (LOOCV) of the primary dataset.
| Machine learning algorithm | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| LR | 0.834 | 0.833 | 0.836 | 0.915 |
| SVM | 0.866 | 0.902 | 0.800 | 0.932 |
| SNN | 0.796 | 0.833 | 0.727 | 0.896 |
| RF | 0.815 | 0.892 | 0.673 | 0.902 |
| NB | 0.809 | 0.853 | 0.727 | 0.867 |
| Mean ± SD | 0.824 ± 0.027 | 0.863 ± 0.033 | 0.753 ± 0.065 | 0.902 ± 0.024 |
| 95% CI | 0.790–0.858 | 0.822–0.903 | 0.672–0.833 | 0.873–0.932 |
LR: logistic regression, SVM: support vector machine, SNN: standard neural network, RF: random forest, NB: naïve Bayes.
Figure 6 shows the ROC curves for all prediction models in the independent validation constructed by using selected radiomic features for all folds in the LOOCV. The AUC values of the prediction models constructed by the LR, SVM, SNN, RF, and NB were 0.755, 0.731, 0.707, 0.800, and 0.743, respectively. Table 4 lists the accuracies, sensitivities, specificities, and AUC values of the prediction models in the independent validation. The mean ± SD of these four parameters for all prediction models were 0.758 ± 0.034 (95% CI, 0.716–0.800), 0.822 ± 0.042 (95% CI, 0.771–0.874), 0.627 ± 0.149 (95% CI, 0.443–0.812), and 0.747 ± 0.034 (95% CI, 0.705–0.790), respectively. The prediction models using the RF demonstrated the best performance in the independent validation, based on the resulting AUC value (0.800).
Figure 6.
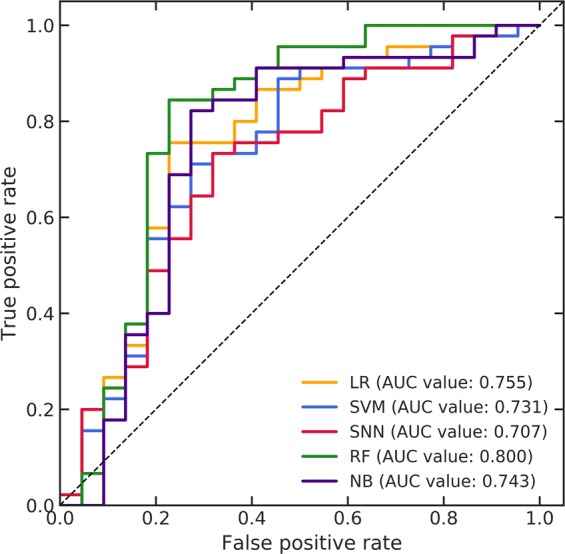
Receiver operating characteristic (ROC) curves for all prediction models in an independent validation constructed by using selected radiomic features for all folds in a leave-one-out cross-validation (LOOCV).
Table 4.
Accuracies, sensitivities, specificities, and area under the curve (AUC) values of prediction models in an independent validation.
| Machine learning algorithm | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| LR | 0.746 | 0.756 | 0.727 | 0.755 |
| SVM | 0.746 | 0.844 | 0.545 | 0.731 |
| SNN | 0.716 | 0.867 | 0.409 | 0.707 |
| RF | 0.806 | 0.822 | 0.773 | 0.800 |
| NB | 0.776 | 0.822 | 0.682 | 0.743 |
| Mean ± SD | 0.758 ± 0.034 | 0.822 ± 0.042 | 0.627 ± 0.149 | 0.747 ± 0.034 |
| 95% CI | 0.716–0.800 | 0.771–0.874 | 0.443–0.812 | 0.705–0.790 |
LR: logistic regression, SVM: support vector machine, SNN: standard neural network, RF: random forest, NB: naïve Bayes.
Discussion
The feasibility of predicting malignant glioma grades based on radiomics by using images acquired with two structural MRI sequences was investigated herein. The classification of LGGs and HGGs using MR-based radiomic frameworks has been investigated and successfully performed in the past15–23. However, this study is focused on only classification of the grade III and IV malignant gliomas because it is also crucial to preoperatively classify the grade IV and the others gliomas for appropriate surgical planning and prognosis prediction. The primary dataset derived from TCIA collection and the validation dataset derived from our institution collection were used to evaluate prediction performances. High-dimensional radiomic features were extracted from both CE-T1WIs and T2WIs in various feature types, wavelet sub-bands, and quantization levels to comprehensively obtain effective features for predicting the malignant glioma grades. The effective features were selected by using combination of the WMW test and LASSO-LR. Five ML algorithms were applied to construct various prediction models using the selected radiomic features for each fold in the LOOCV of the primary dataset. The primary and validation datasets with the selected radiomic features for all folds in the LOOCV of the primary dataset were utilized in the independent validation. The prediction performances of various models were compared using four evaluation indices.
The AUC values of the prediction models constructed by the LR, SVM, and RF in the LOOCV of the primary dataset reached 0.90 and those in the SNN and NB reached 0.80. Moreover, the mean AUC values for all prediction models was 0.902 ± 0.024. In general, classification models with AUC values of 1.00–0.90, and 0.90–0.80 are regarded as excellent and good, respectively47,48. Therefore, the proposed framework could accurately predict malignant glioma grades despite using images acquired with a few structural MRI sequences in the primary dataset. The best prediction performance in the LOOCV of the primary dataset was 0.932 of AUC value using the SVM. Therefore, the SVM was an effective classifier for predicting the grade III and IV gliomas in the primary dataset.
The radiomic features extracted from the CE-T1WIs were dominantly selected for each fold in the LOOCV. In addition, there were five radiomic features extracted from the CE-T1WIs and one radiomic feature extracted from the T2WIs, which were selected for all folds in the LOOCV using the primary dataset. The selected radiomic features for all LOOCV folds comprised almost all texture features extracted from the CE-T1WIs. Tian et al. reported that the texture features extracted from the CE-T1WIs contributed the most to optimal feature subsets for predicting the LGGs and HGGs and grade III and IV gliomas in the multiple MRI sequences images21. They then suggested that the texture features extracted from the CE-T1WIs might lead to high performance while grading the gliomas21. Reza et al. have also reported that in accordance with the results of feature importance ranking in the feature selection, the radiomic features extracted from the CE-T1WIs were more important than those extracted from other structural MRI sequences images23. The result of feature selection for all LOOCV folds in this study was consistent with those reports. Cho et al. and Vamvakas et al. have used 7 and 8 bit of fixed quantization levels, respectively for extracting the texture features17,19. Then, the values of the quantization levels have not been mentioned in almost all previous studies15,16,18,21,22. Few studies have been reported the appropriate values of the quantization levels for grading the gliomas. In this study, five types of values were used to have various combinations of quantization levels in the texture features for achieving high performance. The texture features derived from high quantization levels (7 and 8 bit) were dominantly selected for all folds in the LOOCV. Therefore, the texture features with the high quantization levels might be effective for predicting the malignant glioma grades.
The AUC values of the prediction models were greater than 0.70 but less than 0.80 excluding that of the model constructed by the RF in the independent validation. These results suggested that the performances for predicting the malignant glioma grades in the independent validation were acceptable but not good excluding that of the RF. In addition, the mean AUC values for all prediction models in the independent validation was lower than that in the LOOCV of the primary dataset. The prediction performance degradation in the independent validation could be attributed to the difference in observers for delineating tumors in the primary and validation datasets. The performance for the radiomic analysis varied, depending on the MR scanners, imaging parameters, and tumor delineations49,50. We used MR images acquired by various scanners and imaging parameters in the entire dataset. Therefore, MR intensity normalization was performed as preprocessing for the entire dataset to reduce the influences on the performances caused by those variabilities. However, in terms of delineation, the tumor regions in the primary dataset were delineated by combination of a computerized framework and manual correction by an expert29, while tumor regions in the validation dataset were manually delineated by an observer under the supervision of two experts. Consequently, the selected radiomic features for all folds in the LOOCV of the primary dataset could not have robustness to delineations of the difference observer. The results of independent validation suggested that reproducible radiomic features to the observer delineation variability should be investigated to obtain high prediction performance in case using difference datasets.
Previous studies20,21 had already proposed radiomics-based frameworks for classifying malignant glioma grades using images acquired via multiple MRI sequences. Table 5 lists the prediction performances for malignant glioma grade identification using a radiomic approach in the proposed framework and in previous studies. The best prediction performances of the LOOCV and independent validation using the CE-T1WIs and T2WIs in the proposed framework were listed in Table 5. Prediction performances with more than 0.90 of the AUC values reported by Zacharaki et al. were listed in Table 5 because they investigated various combinations of feature selection methods and classifiers for grading the malignant gliomas20. The AUC values of the previous studies with the multiple MRI sequences were higher than those of our proposed framework with a few structural MRI sequences. The frameworks of previous studies using multiple MRI sequences were indeed effective for classifying malignant glioma grades. However, there might be selection bias in the prediction performances of the previous studies owing to the relatively small datasets used compared with those of this study and using single scanner and unified parameters for acquiring MR images in the datasets. Moreover, an independent validation for investigating versatility to the different datasets was not performed in previous studies. In this study, the AUC values of the best prediction performances in the LOOCV and independent validation using datasets with variety were reached 0.90 and 0.80, respectively. Therefore, we can conclude that our proposed framework with a few structural MRI sequences could sufficiently predict malignant glioma grades despite using datasets comprising MR images acquired by various scanners and imaging parameters.
Table 5.
Prediction performances for malignant glioma grade identification using a radiomic approach in the proposed framework and in previous studies.
| Study | No. of data | MRI sequence | Feature type | Filtering | Feature selection | ML algorithm | Data augmentation | Validation method | Accuracy | Sensitivity | Specificity | AUC value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proposed framework |
Primary dataset: 157 (III: 55, IV: 102) |
•CE-T1 •T2 |
•Shape/size •Intensity •Histogram •GLCM •GLRLM •GLSZM •NGLDM •NGTDM |
Wavelet transform high-pass and low-pass filters for all feature types excluding the shape/size | WMW test & LASSO-LR | SVM (rbf kernel) | No | LOOCV | 0.866 | 0.902 | 0.800 | 0.932 |
|
Entire dataset: 224 (Primary dataset: 157 & Validation dataset: 67 (III: 22, IV: 45)) |
Using the selected radiomic features for all folds in the LOOCV of the primary dataset | RF | No | Independent validation | 0.806 | 0.822 | 0.773 | 0.800 | ||||
| Zacharaki et al.20 | 52 (III: 18, IV: 34) |
•CE-T1 •T1 •T2 •FLAIR •rCBV |
•Shape •Intensity •Rotation invariant texture |
Gabor filter for rotation invariant texture features | SVM-RFE | SVM (rbf kernel) | No | LOOCV | 0.904 | 1.000 | 0.722 | 0.985 |
| t-test with bagging | 0.942 | NR | NR | 1.000 | ||||||||
| Tian et al.21 | 111 (III: 33, IV: 78) |
•CE-T1 •T1 •T2 •Diffusion •3D pCASL |
•GLCM •GLGCM |
No | SVM-RFE | SVM (rbf kernel) | No | 100-times 10-fold CV | 0.937 | 0.942 | 0.927 | 0.982 |
| SMOTE | 0.981 | 0.987 | 0.974 | 0.992 |
CE-T1: contrast-enhanced T1, FLAIR: fluid attenuated inversion recovery, rCBV: relative blood volume, 3D-pCASL: three-dimensional pseudo-continuous arterial spin labeling, GLCM: gray-level co-occurrence matrix, GLRLM: gray-level run length matrix, GLSZM: gray-level size zone matrix, NGLDM: neighboring gray-level dependence matrix, NGTDM: neighborhood gray-tone difference matrix, GLGCM: gray-level gradient co-occurrence matrix, WMW: Wilcoxon-Mann-Whitney, LASSO-LR: least absolute shrinkage and selection operator logistic regression, RFE: recursive feature elimination, SMOTE: synthetic minority over sampling technique, SVM: support vector machine, rbf: radial basis function, RF: random forest, LOOCV: leave-one-out cross validation, AUC: area under the curve, NR: not reported.
There are limitations to our study. Owing to the difficulty of collecting a large number of available malignant glioma cases for a study at our institution, the number of cases in the validation dataset was small. In future, a multi-institutional study would be more helpful. Moreover, some cases lacked several MRI sequences images in the validation dataset owing to retrospective data collection. Therefore, insufficient multiple MRI sequences images were available at our institution for comparison with CE-T1WIs and T2WIs, and the prediction performances using the CE-T1WIs and T2WIs in this study were compared instead with those using multiple MRI sequence images in the previous studies. In addition, the effect of inter-observer tumor delineation variability on the prediction performances of the malignant glioma grades, the reproducible features to the delineation variability, and an appropriate tumor delineation procedure for radiomic analysis should be investigated in future. Finally, although prediction of the glioma grades using preoperative MR images would be useful for planning surgery, the genomic statuses of the gliomas (for example IDH mutation, alpha-thalassemia/mental retardation syndrome X-linked (ATRX) mutation, TP53 mutation, and 1p19q codeletion2) should be identified using radiomics-based analysis (namely radiogenomics) with a few structural MRI sequences for precision medicine. The genomic statuses of the gliomas were difficult to analyze in this study because genomic analyses were not always performed for all cases. In a future study, the proposed framework should be applied to prediction of the genomic features of the gliomas by collecting a large quantity of patients’ preoperative MR images and genomic statuses.
In conclusion, we investigated the feasibility of a framework for predicting malignant glioma grades based on radiomics using CE-T1WIs and T2WIs. Our proposed framework could sufficiently and easily predict malignant glioma grades by preparing images acquired by a few structural MRI sequences. The proposed framework with a few MRI sequences could mitigate the tedious process of tumor contouring on each MRI sequence image compared with the frameworks with multiple MRI sequences. In addition, the best prediction performances of this study indicated that our proposed framework with a few MRI sequences could have versatility to varied datasets. Our proposed framework for noninvasively grading malignant gliomas based on the preoperative images could be an effective tool for selection of appropriate surgery and educating the patients.
Supplementary information
Acknowledgements
We thank Libby Cone, MD, MA, from DMC Corp. (http://www.dmed.co.jp/) for editing drafts of this manuscript. We thank Editage (http://www.editage.jp) for editing revised drafts of this manuscript. This study was founded by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant-in-Aid for Scientific Research), Grant numbers 18J00599 and 18K15625.
Author contributions
T.N. conducted and designed this study. T.N. wrote the manuscript. W.T. and Ke.N. supervised this study. T.N., W.T. and T.O. collected image data. Sa.T., Sh.T., and A.M. collected medical information data and provided knowledge of the neurooncology. T.N. contoured tumor region in images. W.T. and S.K. checked and supervised the tumor contouring. T.N. and A.H. provided programming scripts for analysis. Ka.N., S.O. and Y.N. provided knowledge of the machine learning and data science. All authors read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-55922-0.
References
- 1.The committee of brain tumor registry of Japan. Report of brain tumor registry of Japan (2005–2008) 14th edition. Neurologia Medico-Chirurgica (Tokyo)57, s9–s102 (2017). [DOI] [PMC free article] [PubMed]
- 2.Louis DN, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathologica. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Brada M, van den Bent MJ, Tonn JC, Pentheroudakis G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment, and follow-up. Annals of Oncology. 2014;25(suppl 3):iii93–iii101. doi: 10.1093/annonc/mdu050. [DOI] [PubMed] [Google Scholar]
- 4.Chrity PS, Tervonen O, Scheithauer BW, Forbes GS. Use of a neural network and a multiple regression model to predict histologic grade of astrocytoma from MRI appearances. Neuroradiology. 1995;37:89–93. doi: 10.1007/BF00588619. [DOI] [PubMed] [Google Scholar]
- 5.Lev MH, et al. Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas. American Journal of Neuroradiology. 2004;25:214–221. [PMC free article] [PubMed] [Google Scholar]
- 6.Higano S, et al. Malignant astrocytic tumors: clinical importance of apparent diffusion coefficient in prediction of grade and prognosis. Radiology. 2006;241:839–846. doi: 10.1148/radiol.2413051276. [DOI] [PubMed] [Google Scholar]
- 7.Whitmore RG, et al. Prediction of oligodendroglial tumor subtype and grade using perfusion weighted magnetic resonance imaging. Journal of Neurosurgery. 2007;107:600–609. doi: 10.3171/JNS-07/09/0600. [DOI] [PubMed] [Google Scholar]
- 8.Jakab A, Molár P, Emri M, Berényi E. Glioma grade assessment by using histogram analysis of diffusion tensor imaging-derived maps. Neuroradiology. 2011;53:483–491. doi: 10.1007/s00234-010-0769-3. [DOI] [PubMed] [Google Scholar]
- 9.Beppu T, et al. Prediction of malignancy grading using computed tomography perfusion imaging in nonenhancing supratentorial gliomas. J Neuro-Oncology. 2011;103:619–627. doi: 10.1007/s11060-010-0433-0. [DOI] [PubMed] [Google Scholar]
- 10.Garzón B, et al. Multiparametric analysis of magnetic resonance images for glioma grading and patient survival time prediction. Acta Neuropathologica. 2011;52:1052–1060. doi: 10.1258/ar.2011.100510. [DOI] [PubMed] [Google Scholar]
- 11.Khalid L, et al. Imaging characteristics of oligodendrogliomas that predict grade. American Journal of Neuroradiology. 2012;33:852–857. doi: 10.3174/ajnr.A2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aerts HJ, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature Communications. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peeken JC, et al. Radiomics in radiooncology - challenging the medical physicist. Physica Medica. 2018;48:27–36. doi: 10.1016/j.ejmp.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Qin JB, et al. Grading of gliomas by using radiomic features on multiple magnetic resonance imaging (MRI) sequences. Medical Science Monitor. 2017;23:2168–2178. doi: 10.12659/MSM.901270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho HH, Park H. Classification of low-grade and high-grade glioma using multi-modal image radiomics features. Conference Proceedings of IEEE Engineering in Medicne and Biology Society. 2017;2017:3081–3084. doi: 10.1109/EMBC.2017.8037508. [DOI] [PubMed] [Google Scholar]
- 17.Cho HH, Lee SH, Kim J, Park H. Classification of the glioma grading using radomics analysis. PeerJ. 2018;6:e5982. doi: 10.7717/peerj.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Liu B, Peng S, Sun J, Qiao X. Computer-aided grading of gliomas combining automatic segmentation and radiomics. International Journal of Biomedical Imaging. 2018 doi: 10.1155/2018/2512037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vamvakas A, et al. Imaging biomarker analysis of advanced multiparametric MRI for glioma grading. Physica Medica. 2019;60:188–198. doi: 10.1016/j.ejmp.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Zacharaki EI, et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magnetic Resonance in Medicne. 2009;62:1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Qiang, Yan Lin-Feng, Zhang Xi, Zhang Xin, Hu Yu-Chuan, Han Yu, Liu Zhi-Cheng, Nan Hai-Yan, Sun Qian, Sun Ying-Zhi, Yang Yang, Yu Ying, Zhang Jin, Hu Bo, Xiao Gang, Chen Ping, Tian Shuai, Xu Jie, Wang Wen, Cui Guang-Bin. Radiomics strategy for glioma grading using texture features from multiparametric MRI. Journal of Magnetic Resonance Imaging. 2018;48(6):1518–1528. doi: 10.1002/jmri.26010. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Optimizing a machine learning based glioma grading system using multi-parametric MRI histogram and texture features. Oncotarget. 2017;8:47816–47830. doi: 10.18632/oncotarget.18001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reza SMS, et al. Glioma grading using structural magnetic resonance imaging and molecular data. Journal of Medical Imaging. 2019;6:024501. doi: 10.1117/1.JMI.6.2.024501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scarpace L, et al. Radiology data from the cancer genome atlas glioblastoma multiforme [TCGA-GBM] collection. The Cancer Imaging Archive. 2016 doi: 10.7937/K9/TCIA.2016.RNYFUYE9. [DOI] [Google Scholar]
- 25.Pedano, N. et al. Radiology data from the cancer genome atlas low grade glioma [TCGA-LGG] collection. The Cancer Imaging Archive10.7937/K9/TCIA.2016.L4LTD3TK (2016).
- 26.Clark K, et al. The cancer imaging archive (TCIA): Maintaining and operating a public information repository. Journal of Digital Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakas, S. et al. Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive, 10.7937/K9/TCIA.2017.KLXWJJ1Q (2017).
- 28.Bakas S, et al. Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. The Cancer Imaging Archive. 2017 doi: 10.7937/K9/TCIA.2017.GJQ7R0EF. [DOI] [Google Scholar]
- 29.Bakas S, et al. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Nature Scientific Data. 2017;4:170117. doi: 10.1038/sdata.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Physics in Medicne and Biology. 2015;60:5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
- 31.Vallières M, et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Scientific Reports. 2017;7:10117. doi: 10.1038/s41598-017-10371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loizou, C. P., Pantziaris, M., Seimenis, I. & Pattichis. Brain MR image normalization in texture analysis of multiple sclerosis. In: 9th IEEE International Conference on Information Technology and Applications in Biomedicine, 10.1109/ITAB.2009.5394331 (2009).
- 33.Herman GT, Zheng J, Bucholtz CA. Shape-based interpolation. IEEE Computer Graphics and Applications. 1992;12:69–79. doi: 10.1109/38.135915. [DOI] [Google Scholar]
- 34.Zwanenburg, A., Leger, S., Valliéres, M. & Löck, S. Image biomarker standardization initiative. arXiv: 1612.07003 [cs.CV] (2016).
- 35.Beylkin G, Coifman R, Rokhlin V. Fast wavelet transforms and numerical algorithms I. Communications on Pure and Applied Mathmatics. 1991;XLIV:141–183. doi: 10.1002/cpa.3160440202. [DOI] [Google Scholar]
- 36.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magnetic Resonance Imaging. 2004;22:81–91. doi: 10.1016/j.mri.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Ward JH. Hierarchical grouping to optimize an objective function. Journal of the American Statistical Association. 1963;58:236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 38.Abdollahi H, et al. Cochlea CT radiomics predicts chemoradiotherapy induced sensorineural hearing loss in head and neck cancer patients: a machine learning and multi-variable modelling study. Physica Medica. 2018;45:192–197. doi: 10.1016/j.ejmp.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 39.Wang G, et al. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. European Journal of Radiology. 2018;98:100–106. doi: 10.1016/j.ejrad.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Pedregosa H, et al. Scikit-learn: machine learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 41.Wright SJ. Coordinate descent algorithms. Mathematical Programming Series B. 2015;151:3–34. doi: 10.1007/s10107-015-0892-3. [DOI] [Google Scholar]
- 42.Tibshirani R. Regression shrinkage and selection via the Lasso. Journal of Royal Statistical Society Series B. 1996;58:267–288. [Google Scholar]
- 43.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 44.Jain AK, Mao J, Mohiuddin KM. Artificial neural networks: a tutorial. Computer. 1996;29:31–44. doi: 10.1109/2.485891. [DOI] [Google Scholar]
- 45.Breiman L. Random forests. Machine Learning. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 46.Demichelis F, Magni P, Piergiorgi P, Rubin MA, Bellazi R. A hierarchical naïve Bayes model for handling sample heterogeneity in classification problems: an application to tissue microarrays. BMC Bioinformatics. 2006;7:514. doi: 10.1186/1471-2105-7-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hosmer, D. W. & Lemeshow, S. Applied Logistic Regression, 2nd Edition. New York City: John Wiley & Sons, Inc (2001).
- 48.El Khouli RH, et al. Relationship of temporal resolution to diagnostic performance for dynamic contrast enhanced MRI of the breast. Journal of Magnetic Resonance Imaging. 2009;30:999–1004. doi: 10.1002/jmri.21947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saha A, Yu X, Sahoo D, Mazurowski MA. Effects of MRI scanner parameters on breast cancer radiomics. Expert Systems with Applications. 2017;87:384–391. doi: 10.1016/j.eswa.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haga A, et al. Classification of early stage non-small cell lung cancers on computed tomographic images in to histological types using radiomic features: interobserver delineation variability analysis. Radiological Physics and Technology. 2018;11:27–35. doi: 10.1007/s12194-017-0433-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.