Abstract
Background
Current guidelines recommend surgical resection as the first-line option for patients with solitary hepatocellular carcinoma (HCC); unfortunately, postoperative recurrence rate remains high and there is no reliable prediction tool. We explored the potential of radiomics coupled with machine-learning algorithms to improve the predictive accuracy for HCC recurrence.
Methods
A total of 470 patients who underwent contrast-enhanced CT and curative resection for solitary HCC were recruited from 3 independent institutions. In the training phase of 210 patients from Institution 1, a radiomics-derived signature was generated based on 3384 engineered features extracted from primary tumor and its periphery using aggregated machine-learning framework. We employed Cox modeling to build predictive models. The models were then validated using an internal dataset of 107 patients and an external dataset of 153 patients from Institution 2 and 3.
Findings
Using the machine-learning framework, we identified a three-feature signature that demonstrated favorable prediction of HCC recurrence across all datasets, with C-index of 0.633–0.699. Serum alpha-fetoprotein, albumin-bilirubin grade, liver cirrhosis, tumor margin, and radiomics signature were selected for preoperative model; postoperative model incorporated satellite nodules into above-mentioned predictors. The two models showed superior prognostic performance, with C-index of 0.733–0.801 and integrated Brier score of 0.147–0.165, compared with rival models without radiomics and widely used staging systems (all P < 0.05); they also gave three risk strata for recurrence with distinct recurrence patterns.
Interpretation
When integrated with clinical data sources, our three-feature radiomics signature promises to accurately predict individual recurrence risk that may facilitate personalized HCC management.
Keywords: Hepatocellular carcinoma, Radiomics, Machine learning, Recurrence, Prediction model
Abbreviations: HCC, hepatocellular carcinoma; LT, liver transplantation; BCLC, Barcelona Clinic Liver Cancer; HKLC, Hong Kong Liver Cancer; CLIP, Cancer of the Liver Italian Program; AJCC, American Joint Committee on Cancer; TNM, tumor-node-metastasis; ERASL, Early Recurrence After Surgery for Liver tumor; ML, machine learning; CT, computed tomography; ROI, region of interest; ICC, intraclass correlation coefficient; MRMR, maximum relevance minimum redundancy; MI, mutual information; RSF, random survival forest; VIMP, variable importance; LASSO, least absolute shrinkage and selection operator; AIC, Akaike information criteria; AFP, alpha-fetoprotein; MRI, magnetic resonance imaging; RFS, recurrence-free survival; C-index, concordance index; ROC, receiver operating characteristic; IBS, integrated Brier score; IQR, interquartile range
Graphical abstract
Highlights
-
•
We identified a three-feature fusion signature using machine-learning framework.
-
•
The signature coupled with clinical sources accurately predicted HCC recurrence.
-
•
This signature may serve as an early detector of aggressive disease.
-
•
We highlight the complementary nature of radiomics and existing variables.
Research in context.
Evidence before this study
The ideal candidates for resection are patients with a solitary hepatocellular carcinoma (HCC) at an early stage, regardless of tumor size, and preserved liver function; unfortunately, disease recurrence rate remains high and there is no reliable prediction tool. Radiomics is a nascent technique that quantifies imaging phenotypes using automated and high-throughput method. Only a few studies have employed radiomics to predict the likelihood of HCC recurrence, with most solely extracting engineered features from tumor; however, peritumoral area harbors highly invasive tumor cells. In addition, to the best of our knowledge, no multi-institutional studies on the use of radiomics to predict recurrence after surgical resection of early-stage HCC have been reported to date.
Added value of this study
We identified a three-feature radiomics signature using aggregated machine-learning framework. The signature coupled with clinical sources accurately predicted HCC recurrence either before or after surgery and showed superior prognostic performance compared with rival models without radiomics as well as widely used staging systems. Moreover, the two models could give three risk strata with low, intermediate, or high risks of recurrence and distinct recurrence patterns, backed by internal and external validations.
Implications of all the available evidence
This study highlights the complementary nature of radiomics and existing variables. Our results suggest that the three-feature signature may serve as an early detector of aggressive disease in patients with solitary HCC. When integrated with clinical data sources, the radiomics signature promises to accurately predict individual recurrence risk that may facilitate personalized HCC management.
CRediT authorship contribution statement
Ji Gu-Wei: Writing - original draft, Writing - review & editing. Zhu Fei-Peng: . Xu Qing: . Wang Ke: Data curation. Wu Ming-Yu: Data curation. Tang Wei-Wei: Writing - review & editing. Li Xiang-Cheng: Writing - review & editing. Wang Xue-Hao: Writing - review & editing.
Alt-text: Unlabelled box
1. Introduction
Hepatocellular carcinoma (HCC) ranks number four among the most common causes of cancer-related death globally, with prognosis predominantly driven by tumor burden and liver dysfunction [1,2]. In selected patients diagnosed at early stages, surgical resection and liver transplantation (LT) are potentially curative and can achieve 5-year survival rates of 60–80% [2,3]. LT offers definite advantages of oncologic cure but demand for organs far exceeds supply. Surgical resection is accepted as the first-line treatment option for a solitary HCC at an early stage, regardless of tumor size, in patients with preserved liver function; unfortunately, 50%−70% of cases present with disease recurrence within 5 years, reflecting either disseminative or de novo recurrence [1], [2], [3], [4].
Patients with HCC do not recur through the evolutionary stages of this disease after curative therapies [1]. Although several staging systems - such as Barcelona Clinic Liver Cancer (BCLC), Hong Kong Liver Cancer (HKLC), Cancer of the Liver Italian Program (CLIP), and American Joint Committee on Cancer (AJCC) tumor-node-metastasis (TNM) systems - have been proposed for prognostic prediction and treatment allocation, they are not derived from surgically treated patients and therefore inadequate for predicting recurrence after resection [1–5]. Recently, two Early Recurrence After Surgery for Liver tumor (ERASL) models have been developed specifically to predict HCC recurrence; however, their discriminatory ability is barely satisfactory and none of them has been specifically validated in patients with solitary lesion, who are the ideal candidates for resection [5]. Genomic investigation may provide prognostic information, but this approach has not been used in routine clinical care [2,6]. By contrast, medical imaging is an indispensable tool in oncology.
A rapidly evolving field named “Radiomics” that quantifies imaging phenotypes using automated and high-throughput method promises to make great strides in precision medicine [7], [8], [9]. This new paradigm, as an extension of traditional imaging semantics, is mined with machine-learning (ML) algorithms to select robust features and develop models that may potentially improve outcome prediction. Recently, a few studies have employed radiomics to predict early recurrence of HCC after resection; however, the outcome of interest was transformed to a dichotomized endpoint (early versus late or no recurrence) that incurs the risk of biasing prediction accuracy in these studies while few attempts have been made to assess recurrence risk based on continuous time-to-event survival data [10], [11], [12]. In addition, most previous studies solely extracted engineered features from within tumor annotations; however, peritumoral area harbors highly invasive tumor cells on pathology and peritumoral changes captured by radiomic analysis of HCC may hold prognostic information [11], [12], [13], [14], [15]. To our knowledge, no multi-institutional studies on the use of radiomics to predict recurrence after surgical resection of early-stage HCC have been reported to date.
This study aimed to investigate whether radiomic analysis of contrast-enhanced computed tomography (CT) coupled with ML-based algorithms could improve the prediction of HCC recurrence for patients with solitary lesion in clinical settings, backed by internal and external validations. We benchmarked the performance of established models against that achieved by rival models and staging systems to determine the added value of radiomics.
2. Materials and methods
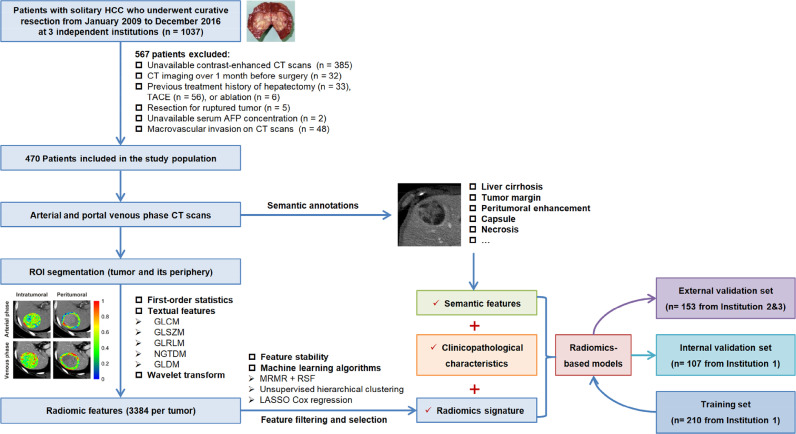
2.1. Study population
The institutional review boards of all participating institutions approved this retrospective study, and all boards waived patient written informed consent. A total of 1037 patients with pathologically-confirmed solitary HCC treated by curative resection between January 2009 and December 2016 were received from 3 independent institutions. Among these, 470 patients were included in the final analysis according to the study criteria shown in Fig 1. We randomly assigned the 317 patients recruited from Institution 1 (The First Affiliated Hospital of Nanjing Medical University, Nanjing, China) in a two-to-one ratio into training (n = 210) and internal validation (n = 107) datasets. The external validation set consisted of 153 patients treated at 2 independent institutions [Institution 2 (n = 94): Wuxi People's Hospital, Wuxi, China; Institution 3 (n = 59): Nanjing First Hospital, Nanjing, China].
Fig. 1.
Flowchart of study design. HCC, hepatocellular carcinoma; CT, computed tomography; TACE, transcatheter arterial chemoembolization; AFP, alpha-fetoprotein; ROI, region of interest.
2.2. Image analysis
CT protocols are detailed in Table S1. Two abdominal radiologists with 10 years (reader 1: F.P.Z) and 20 years (reader 2: Q.X) of experience in liver imaging blinded to all clinical data independently reviewed the baseline CT images to evaluate the following traits: (a) tumor size; (b) liver cirrhosis; (c) arterial rim enhancement; (d) arterial peritumoral enhancement; (e) tumor margin; (f) radiological capsule; (g) intratumoral necrosis; (h) radiogenomic venous invasion. Diagnostic criteria are introduced in Supplementary Methods. Tumor size was recorded as mean value. Discordant annotations were resolved through consensus review in a third session and inter-reader variation was measured by Kappa statistics.
2.3. Radiomic analysis
The radiomics workflow is summarized in Fig 1. Regions of interest (ROIs) were semiautomatically delineated on each transverse slice at arterial and portal venous phases using Fast GrowCut algorithm implemented in 3D Slicer (version 4.9.0; www. slicer.org). Annotated lesion within tumor was termed intratumoral ROI; peritumoral ROI was generated with automated dilatation and shrinkage of tumor boundary by 2 mm on each side, resulting in a 4mm-wide band. All images were resampled to a voxel size of 1 × 1 × 1 mm and voxel intensities were discretized using a bin-width of 25 Hounsfield units [16,17]. We extracted 846 radiomic features from each three-dimensional ROI using open-source Pyradiomics package (version: 2.12; https://pyradiomics.readthedocs.io/en/2.1.2/), including 19 first-order statistics, 75 textual features, and 752 wavelet features. Accounting for biphasic radiomics from the tumor and its periphery analyzed, a total of 3384 features were obtained per patient. Feature extraction algorithms are provided in Supplementary Methods and feature values were standardized with Z-scores derived from the training set. All segmentations were completed by reader 1. To test feature stability, reader 1 and reader 2 repeated feature extraction independently in a one-week period on 30 randomly chosen patients. The stability was calculated by using intraclass correlation coefficient (ICC). Features with excellent stability (ICC>0.90) in both test-retest and inter-reader settings were included in subsequent analysis.
2.4. Machine learning framework
Two most commonly used ML algorithms were implemented for feature filtering and selection without involvement of the model: (ⅰ) maximum relevance minimum redundancy (MRMR) and (ⅱ) random survival forest (RSF) consist in computing feature importance associated with time-to-event outcomes. MRMR ranks the input features by maximizing the mutual information (MI) with outcome and minimizing the average MI with all higher ranked features [18]. RSF ranks candidate features based on the variable importance (VIMP), which is calculated by comparing out-of-bag prediction performance for the permuted feature to the original feature [19]. A forest of 1000 trees was grown using log-rank splitting, and VIMP for each feature was recorded. The analysis was repeated 100 times independently, and VIMP was averaged over the runs.
We then aggregated the top 20 engineered features from either MRMR or RSF algorithm, and employed least absolute shrinkage and selection operator (LASSO) Cox regression algorithm [20] with penalty parameter tuning conducted by 10-fold cross-validation to compile a radiomics signature. Unsupervised hierarchical clustering was done to identify similar expression patterns of candidate features using Pearson correlation-based distance and complete linkage.
2.5. Model development and validation
Predictors of HCC recurrence that achieved statistical significance in univariate analysis were included in the multivariate analysis. The final model was formulated based on the results of multivariate Cox regression by using backward step wise elimination with Akaike information criteria (AIC) as the stopping rule, and Cox regression coefficients were utilized to generate the nomogram. The proportional hazards assumption was tested by scaled Schoenfeld residuals. Two radiomics-based models were constructed: the preoperative model (model-pre) included radiomics-derived signature and parameters available before surgery; the post-operative model (model-post) included aforementioned parameters plus pathological results. Correspondingly, two clinical models were generated without radiomics. Proposed models were validated in independent internal and external datasets.
2.6. Follow-up surveillance
All patients underwent surveillance after resection with alpha-fetoprotein (AFP), liver function, contrast-enhanced CT or magnetic resonance imaging (MRI) of abdomen and chest every 3 months in the first 2 years and every 6 months thereafter. The primary endpoint was recurrence-free survival (RFS), which was calculated from the date of surgery to the date of first detected disease recurrence or metastasis by dynamic CT or MRI studies, censoring recurrence-free patients at the date of last follow-up and those who died of other causes. We also recorded the details at the time of first recurrence, including site of recurrence (intrahepatic only vs extrahepatic only vs intra-and extrahepatic), number of recurrent tumor (single vs multiple), and primary treatment modality. This study was censored on January 15, 2019.
2.7. Statistical analysis
Recurrence and survival probabilities were estimated using the Kaplan-Meier method and compared by the log-rank test. Serum AFP level was normalized with a natural logarithm transformation to reduce the effect of small differences. Model discrimination was measured by the concordance index (C-index) and compared using the method previously described [21]. Model fit was assessed by calibration plots via 1000 bootstrap resamples. Time-dependent receiver operating characteristic (ROC) curve and corresponding AUC were employed to investigate the performance at different time points. The integrated Brier score (IBS) that represents the differences between actual events and predicted probabilities was evaluated using “Boot632plus” split method with 1000 iterations [22]. Clinical utility of models was evaluated by decision curve analysis. Statistical analysis was undertaken using R software (version 3.4.4; www.r-project.org) with R packages listed in Supplementary Methods. We used X-tile software (version 3.6.1; Yale University School of Medicine, New Haven, CT, USA) in the training dataset to determine the optimal cutoff points for risk scores outputted from the prediction model against RFS; the selected thresholds were used to separate patients into low-, intermediate-, and high-risk groups [23]. The predictive ability of prediction model was further evaluated in subgroups of all patients defined by three well-established prognostic factors: tumor size (≤5 vs > 5 cm), serum AFP level (≤400 vs > 400 ng/mL), and microvascular invasion (absent vs present) [3], [4], [5]. Statistical significance was set at P<0.05.
2.8. Data sharing statement
Research data are not available for public access due to patient privacy concerns but can be obtained from the corresponding author on reasonable request approved by the institutional review boards of all participating institutions.
3. Results
3.1. Patient characteristics
Baseline characteristics of the training, internal validation, and external validation sets are summarized in Table S2. The median follow-up was 56.0 months (interquartile range [IQR], 39.0–74.4) for the training set, 41.6 months (IQR, 33.5–53.1) for the internal validation set, and 59.5 months (IQR, 37.0–79.8) for the external validation set. Compared with the training set, the external validation set had significantly longer prothrombin times; other characteristics and RSF were comparable between the training and validation sets.
3.2. Radiomic analysis
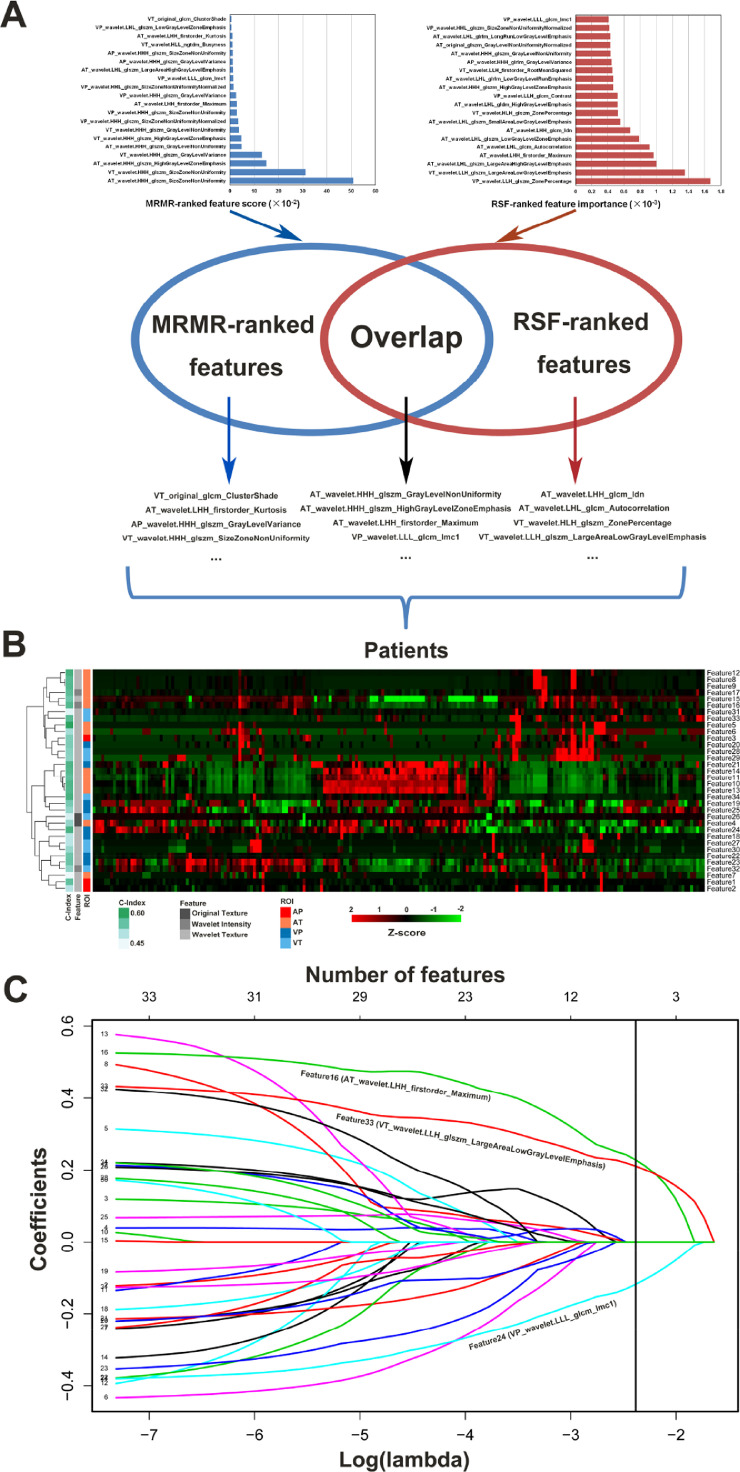
Among the 3384 extracted radiomic features, a total of 2422 with high stability (ICC > 0.90) in both test-retest and inter-reader settings were preliminarily selected. After combining the top 20 engineered features ranked by MRMR and RSF algorithms, 34 features were identified from the training set, with 6 features selected simultaneously by two algorithms (Fig 2A). Unsupervised clustering of filtered features highlighted clusters of highly correlated features with comparable performance (Fig 2B). LASSO Cox regression algorithm further narrowed down a fusion signature that retained three archetypal features (Fig 2C, Fig S1, and Table S3). The resulting signature was formulated as follows:
Fig. 2.
Integrated machine-learning framework for radiomic feature selection. (A) MRMR feature ranking and RSF permutation importance. (B) Unsupervised clustering analysis of candidate features. (C) LASSO regression analysis for prognostic signature-building. A vertical line is drawn at the optimal value chosen by 10-fold cross-validation via minimum criteria. MRMR, maximum relevance minimum redundancy; RSF, random survival forest; LASSO, least absolute shrinkage and selection operator.
This fusion signature indicated favorable prediction of HCC recurrence, with C-index values of 0.633, 0.699 and 0.645 in the training, internal validation and external validation sets, respectively.
3.3. Model development and validation
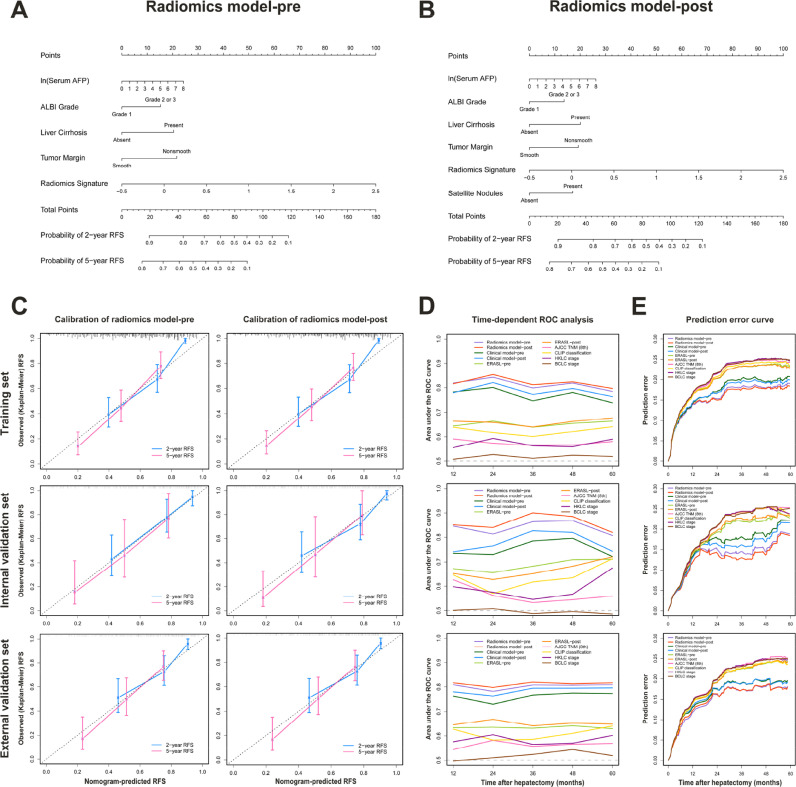
Among the 25 candidate variables, 12 significant predictors of HCC recurrence were identified by univariate analysis in the training set (Fig S2). Stepwise multivariate analysis with the lowest AIC score retained independent predictors for radiomics model-pre and model-post; their formulas and nomograms are shown in Table 1 and Fig 3. The validity of proportional hazards assumption for radiomics models was verified by Schoenfeld residual plots (Fig S3). Similarly, clinical model-pre and model-post were built according to the formulas shown in Table S4.
Table 1.
Multivariate Cox regression analysis of predictors of HCC recurrence using stepwise backward selection method in the training set.
| Variable | Radiomics model - pre |
Radiomics model - post |
||||
|---|---|---|---|---|---|---|
| β | HR (95% CI) | P value | β | HR (95% CI) | P value | |
| ln (Serum AFP) | 0.095 | 1.099 (1.015–1.191) | 0.020 | 0.099 | 1.104 (1.019–1.196) | 0.016 |
| ALBI grade | 0.475 | 1.608 (1.102–2.347) | 0.014 | 0.412 | 1.511 (1.026–2.224) | 0.037 |
| Liver cirrhosis | 0.639 | 1.894 (1.281–2.801) | 0.001 | 0.606 | 1.833 (1.236–2.718) | 0.003 |
| Tumor margin | 0.676 | 1.967 (1.339–2.889) | < 0.001 | 0.582 | 1.789 (1.203–2.662) | 0.004 |
| Radiomics signature | 1.043 | 2.837 (1.875–4.293) | < 0.001 | 1.004 | 2.729 (1.805–4.126) | < 0.001 |
| Satellite nodules | NA | NA | NA | 0.515 | 1.674 (1.046–2.679) | 0.032 |
| C-index (SE) | 0.748 (0.028) | 0.752 (0.028) | ||||
| AIC | 1075.78 | 1073.46 | ||||
| Radiomics model - pre: risk score = 0.095 × ln (Serum AFP) + 0.475 × ALBI grade (0: Grade 1; 1: Grade 2 or 3) + 0.639 × Liver cirrhosis (0: Absent; 1: Present) + 0.676 × Tumor margin (0: Smooth; 1: Nonsmooth) + 0.887 × Radiomics signature | ||||||
| Radiomics model - post: risk score = 0.099 × ln (Serum AFP) + 0.412 × ALBI grade (0: Grade 1; 1: Grade 2 or 3) + 0.606 × Liver cirrhosis (0: Absent; 1: Present) + 0.582 × Tumor margin (0: Smooth; 1: Nonsmooth) + 1.004 × Radiomics signature + 0.515 × Satellite nodules (0: Absent; 1: Present) | ||||||
Abbreviations: HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; ALBI, albumin-bilirubin; HR, hazard ratio; CI, confidence interval; C-index, concordance index; SE, standard error; AIC, Akaike information criteria; NA, not applicable.
Fig. 3.
Calibration and performance of predictive models in the training, internal validation, and external validation sets. Two radiomics models were developed and presented as nomograms to predict the risk of recurrence before (A) and after (B) resection. (C) Plots depict the calibration of each model in each dataset. (D) Time-dependent AUCs for predictive models and staging systems. (E) Comparison of prediction error estimates for established models and staging systems. AFP, alpha-fetoprotein; ALBI, albumin-bilirubin; RFS, recurrence-free survival; ROC, receiver operating characteristic; ERASL, Early Recurrence After Surgery for Liver tumor; AJCC, American Joint Committee on Cancer; TNM, tumor-node-metastasis; CLIP, Cancer of the Liver Italian Program; HKLC, Hong Kong Liver Cancer; BCLC, Barcelona Clinic Liver Cancer.
The resulting radiomics model-pre and model-post yielded respective C-indexes of 0.748 and 0.752 in the training set, 0.781 and 0.801 in the internal validation set, 0.733 and 0.741 in the external validation set; their performance was clearly superior (all P<0.05) to that of clinical models, ERASL models, and commonly used staging systems (Table 2). The radiomics model-predicted RFS was well calibrated with the Kaplan-Meier-observed RFS at 2 and 5 years (Fig 3C). Time-dependent ROC analysis also confirmed that radiomics models improved prediction of HCC recurrence compared with rival models and staging systems at various time points (Fig 3D). Detailed data of time-dependent AUCs are reported in Table S5. Using the prediction error, we found that radiomics model-pre and model-post yielded respective IBSs of 0.165 and 0.162 in the training set, 0.152 and 0.147 in the internal validation set, 0.162 and 0.162 in the external validation set, indicating better performance than rival models and staging systems (Table 2 and Fig 3E). By decision curve analysis, radiomics models provided larger net benefit across a reasonable range of threshold probabilities compared with rival models, staging systems, and simple strategies (ie, follow-up of all patients or no patients) across all datasets (Fig S4).
Table 2.
Prognostic performance of radiomics models compared with rival models and staging systems.
| Model | Training set |
Internal validation set |
External validation set |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C-index (SE) | tdAUC | IBS | P value | C-index (SE) | tdAUC | IBS | P value | C-index (SE) | tdAUC | IBS | P value | |
| Radiomics model-pre | 0.748 (0.019) | 0.813 | 0.165 | Ref | 0.781 (0.028) | 0.840 | 0.152 | Ref | 0.733 (0.026) | 0.803 | 0.162 | Ref |
| Radiomics model-post | 0.752 (0.019) | 0.821 | 0.162 | Ref | 0.801 (0.027) | 0.859 | 0.147 | Ref | 0.741 (0.025) | 0.813 | 0.161 | Ref |
| Clinical model-pre | 0.716 (0.022) | 0.770 | 0.180 | < 0.001* | 0.707 (0.036) | 0.754 | 0.180 | < 0.001* | 0.696 (0.028) | 0.762 | 0.173 | < 0.001* |
| Clinical model-post | 0.727 (0.022) | 0.787 | 0.176 | < 0.001† | 0.739 (0.036) | 0.779 | 0.172 | < 0.001† | 0.716 (0.025) | 0.786 | 0.172 | < 0.001† |
| ERASL-pre model | 0.622 (0.027) | 0.653 | 0.205 | < 0.001* | 0.647 (0.040) | 0.686 | 0.195 | < 0.001* | 0.609 (0.033) | 0.635 | 0.206 | < 0.001* |
| ERASL-post model | 0.622 (0.027) | 0.661 | 0.204 | < 0.001† | 0.624 (0.041) | 0.667 | 0.198 | < 0.001† | 0.621 (0.032) | 0.652 | 0.207 | < 0.001† |
| BCLC stage | 0.582 (0.085) | 0.518 | 0.219 | 0.024* | 0.617 (0.072) | 0.496 | 0.215 | 0.015* | 0.568 (0.097) | 0.519 | 0.213 | 0.039* |
| HKLC stage | 0.599 (0.047) | 0.572 | 0.216 | < 0.001* | 0.626 (0.070) | 0.592 | 0.209 | 0.018* | 0.632 (0.056) | 0.584 | 0.210 | 0.038* |
| CLIP classification | 0.668 (0.039) | 0.624 | 0.207 | 0.012* | 0.659 (0.059) | 0.637 | 0.209 | 0.016* | 0.650 (0.049) | 0.610 | 0.204 | 0.033* |
| AJCC TNM (8th) | 0.649 (0.055) | 0.575 | 0.215 | 0.027† | 0.652 (0.092) | 0.565 | 0.214 | 0.048† | 0.641 (0.060) | 0.564 | 0.215 | 0.048† |
NOTE. The tdAUC represented the median value of AUCs at various time points and all P values were obtained from analyses comparing the C-indices of various models using the “survcomp” package in R software. * P value vs radiomics model-pre; † P value vs radiomics model-post.
Abbreviations: C-index, concordance index; SE, standard error; tdAUC, time-dependent area under the receiver operating characteristic curve; IBS, integrated Brier score; ERASL, Early Recurrence After Surgery for Liver tumor; BCLC, Barcelona Clinic Liver Cancer; HKLC, Hong Kong Liver Cancer; CLIP, Cancer of the Liver Italian Program; AJCC, American Joint Committee on Cancer; TNM, tumor-node-metastasis.
Inter-reader agreement for the two semantic features that were incorporated into predictive models was excellent (κ=0.906 for liver cirrhosis and 0.880 for tumor margin).
3.4. Radiomics models for prognostic stratification
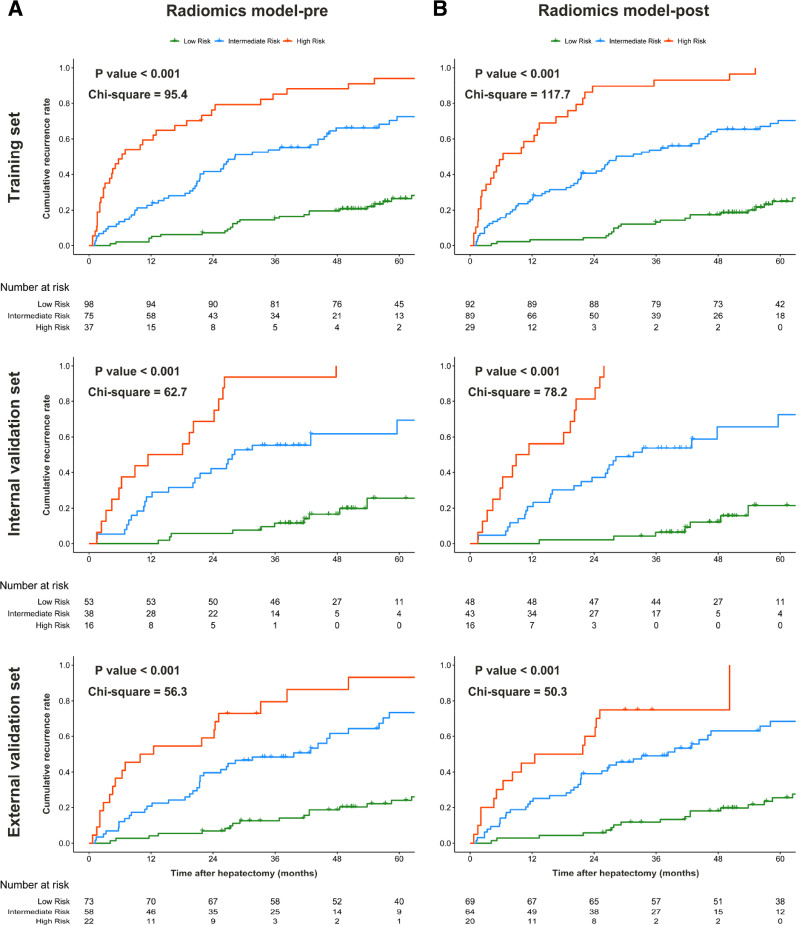
By using X-tile plots of the training set (Fig S5), risk scores of 1.1 and 2.0 (correspond to total points of 52 and 83 in nomogram, respectively) were selected as the optimal cut-points for radiomics model-pre that stratified patients into three risk categories of recurrence (median time to recurrence, 98.7 months, 28.3 months, and 6.4 months for low-risk vs intermediate-risk vs high-risk patients in the training set, respectively; P<0.001). Similar results were achieved for radiomics model-post using 1.0 (51 points) and 2.1 (91 points) as cutoff values derived from X-tile analysis. Powerful prognostic stratification by both radiomics models (P < 0.001 for all) was confirmed in the internal and external validation sets (Table 3 and Fig 4).
Table 3.
Median TTR and cumulative recurrence rate according to each risk group defined by radiomics models.
| Model | Set | Risk group | N | Median TTR, months (95% CI) | 2-year TRR,% (95% CI) | 5-year TRR,% (95% CI) | HR (95% CI) | P value |
|---|---|---|---|---|---|---|---|---|
| Radiomics model-pre | Training | Low | 98 | 98.7 (95.1-NA) | 7.1 (1.9–12.1) | 26.5 (16.6–35.2) | 1 | |
| Intermediate | 75 | 28.3 (21.5–46.6) | 41.6 (29.3–51.8) | 72.5 (58.8–81.6) | 3.64 (2.32–5.71) | < 0.001 | ||
| High | 37 | 6.4 (4.0–18.9) | 76.2 (57.3–86.7) | 94.1 (77.4–98.4) | 9.75 (5.88–16.16) | < 0.001 | ||
| Internal validation | Low | 53 | NA (82.4-NA) | 5.7 (0.0–11.7) | 25.5 (8.3–39.6) | 1 | ||
| Intermediate | 38 | 27.9 (20.5-NA) | 42.1 (24.1–55.9) | 69.4 (42.0–83.9) | 4.63 (2.28–9.41) | < 0.001 | ||
| High | 16 | 14.8 (5.8–25.9) | 68.8 (35.4–84.9) | 100.0 (NA-NA) | 16.56 (7.40–37.07) | < 0.001 | ||
| External validation | Low | 73 | NA (95.1-NA) | 6.8 (0.9–12.5) | 24.0 (12.9–33.7) | 1 | ||
| Intermediate | 58 | 39.6 (22.2–56.9) | 39.7 (25.7–51.0) | 73.2 (55.5–83.9) | 3.71 (2.17–6.32) | < 0.001 | ||
| High | 22 | 11.2 (5.1–33.2) | 59.1 (32.4–75.2) | 93.2 (57.5–98.9) | 9.25 (4.87–17.60) | < 0.001 | ||
| Radiomics model-post | Training | Low | 92 | 98.7 (95.1-NA) | 4.3 (0.1–8.4) | 24.9 (14.8–33.8) | 1 | |
| Intermediate | 89 | 28.3 (21.5–46.0) | 40.6 (29.5–50.1) | 70.4 (51.9–79.0) | 4.07 (2.57–6.44) | < 0.001 | ||
| High | 29 | 6.4 (4.0–16.6) | 89.7 (69.8–96.5) | 100.0 (NA-NA) | 15.53 (8.86–27.24) | < 0.001 | ||
| Internal validation | Low | 48 | NA (95.1-NA) | 2.1 (0.0–6.0) | 21.5 (4.1–35.8) | 1 | ||
| Intermediate | 43 | 31.6 (23.6-NA) | 37.2 (21.0–50.1) | 72.6 (45.2–86.3) | 4.63 (2.28–9.41) | < 0.001 | ||
| High | 16 | 10.2 (5.8–24.2) | 81.3 (48.0–93.2) | 100.0 (NA-NA) | 16.56 (7.40–37.07) | < 0.001 | ||
| External validation | Low | 69 | NA (95.1-NA) | 5.8 (0.1–11.2) | 25.4 (13.7–35.6) | 1 | ||
| Intermediate | 64 | 38.3 (25.5–58.1) | 39.1 (25.9–49.9) | 68.3 (52.0–79.1) | 3.71 (2.17–6.32) | < 0.001 | ||
| High | 20 | 17.1 (6.4-NA) | 89.1 (31.6–76.6) | 100.0 (NA-NA) | 9.25 (4.87–17.60) | < 0.001 |
Abbreviations: TTR, time to recurrence; TRR, tumor recurrence rate; HR, hazard ratio; CI, confidence interval; NA, not applicable.
Fig. 4.
Cumulative rates of tumor recurrence according to three risk strata defined by radiomics model-pre (A) and model-post (B) in the training, internal validation, and external validation sets.
3.5. Subgroup analysis of radiomics models
Subgroup analysis according to predefined factors (tumor size, serum AFP level, and microvascular invasion) suggested that radiomics model-pre and model-post remained statistically significant prognostic tools for prediction of RFS (all P < 0.001), and achieved better prognostic accuracy compared with rival models and staging systems (all P < 0.05) across all subgroups (Table S6 and Fig S6). Kaplan-Meier curves for HCC recurrence also revealed three distinct prognostic strata across all subgroups by using the cutoff values established in the training set (P < 0.001 for all).
3.6. Recurrence pattern and treatment
Of the 470 patients, 247 (52.6%) developed documented recurrence. Recurrence patterns and corresponding treatments were significantly different among three risk categories predicted by radiomics models (Table S7). Briefly, multiple and extrahepatic recurrences were more frequently detected in intermediate-and high-risk groups compared with low-risk group; a higher proportion of low-risk patients received potentially curative therapy (LT, repeat resection, or ablation) for recurrent HCC compared with intermediate-to high-risk patients.
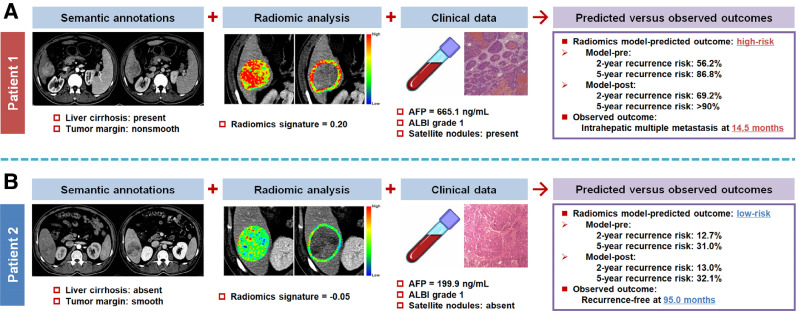
Fig 5 provides two representative cases with similar tumor size, where the proposed radiomics models correctly predicted their recurrence risk and pattern.
Fig. 5.
Two representative cases to show the clinical translation of radiomics-derived models. (A) A 52-year-old man with hepatitis B-related liver cirrhosis and a 4.8-cm liver mass was at a high predicted risk of recurrence. Multiple metastases occurred in the liver 14.5 months after curative resection. (B) A 48-year-old man with non-cirrhotic liver and a 5.7-cm liver mass was at a low predicted risk of recurrence. He remained recurrence-free during 95.0 months of follow-up period after curative resection. AFP, alpha-fetoprotein; ALBI, albumin-bilirubin.
4. Discussion
This study demonstrated that three archetypal features on contrast-enhanced CT radiomics, selected via integrated ML framework and converted into a fusion signature, can complement existing prognostic sources and improve HCC recurrence prediction. Specifically, we developed and validated two new models that incorporate radiomics to predict HCC recurrence before and after resection. The radiomics models exhibited superior prognostic performance, with C-index of 0.733–0.801 and IBS of 0.147–0.165, compared with rival models and widely used staging systems. Both models could stratify surgery patients into three categories of distinct recurrence risk and pattern, suggesting that our findings potentially offer clinical value in HCC management.
Radiomics is a nascent technique that quantifies tumor heterogeneity by the spatial arrangement of imaging voxels with signal-intensity variations and has major implications for personalized oncology [7], [8], [9]. Nevertheless, the pros and cons of radiomics warrant mention. First, although the reproducibility has been addressed from the inter-observer standpoint, engineered features are critically dependent on image acquisition settings that may vary across institutions and operators. We therefore employed voxel intensity discretization and voxel size resampling for feature extraction to reduce the dependency of differences in image specifications and validated the radiomic panel in a multi-institutional dataset. Second, most engineered hard-coded features are difficult to comprehend by clinicians. Our radiomic signature was compiled with three key features that describe voxel intensity information (statistics) and patterns (textures) within tumor and its adjacent region. An intuitive interpretation of this signature is that higher intratumoral peak attenuation value (firstorder_Maximum) in arterial phase and larger proportion of intratumoral necrosis (glszm_LargeAreaLowGrayLevelEmphasis) with relatively heterogeneous peripheral tissue (glcm_InformationalMeasureofCorrelation 1) in portal venous phase are associated with high risk of HCC recurrence after curative resection, backed by the evidence that HCC hemodynamics and microenvironment implicate in aggressive biological behavior [24]. Note that all three features relate to discrete wavelet filters, which decompose the original image in three different directions using a coiflet wavelet transformation and may further reflect the spatial heterogeneity of tumor and its periphery at multiple scales [17]. Third, radiomics-derived data is not a panacea for computerized clinical decision-support system. In addition to radiomics, increased AFP, nonsmooth tumor margin, and satellite lesions that reflect tumor burden were incorporated to achieve holistic models. Our models also highlighted the impact of liver cirrhosis on HCC recurrence through the integration of cirrhosis imaging and albumin-bilirubin grade, which echoes previous investigations [[3], [4], [5],25]. Interestingly, in subgroup analysis, we found that radiomics models exhibited superior performance in patients with favorable characteristics, such as tumor diameter ≤5 cm, AFP value ≤400 ng/mL, and no microvascular invasion, compared to those with established risk factors. These results suggest a possible role for our models as an early detector of aggressive disease.
In the field of radiomics data mining, different ML-based dimensionality reduction techniques have distinct mathematical senses and inherent limitations; therefore, multiple learning algorithms should be combined to select robust features [8]. Two of three features in the signature were selected by the union of two filter methods, which increases the reliability of each component. The Cox model, despite its simplicity, was implemented as the baseline predictive model in our study to sustain its reproducibility and generalizability. Leger et al. [10] reported that Cox model could achieve comparable performance to complex ML models for time-to-event survival data. Although advances in ML-based modeling offer promise in numerous clinical predictions, the interpretability of complex statistical algorithms represents a major bottleneck in attempting to learn any black-box-like model in the clinic [26]. Unlike the Cox model, sophisticated learning algorithms involve hyper-parameter tuning to optimize model performance and may be irreproducible by independent researchers without raw data. Besides, recent studies have addressed that feature selection is more vital for radiomic analysis compared with modeling methodology [10,27].
Although no accepted adjuvant therapies have been demonstrated to reduce recurrence, patients at high risk of recurrence are potential candidates for clinical trials of adjuvant therapy, such as adjuvant chemotherapy, molecular targeted therapy, and immunotherapy [1], [2], [3], [4], [5]. The most effective treatment to prevent HCC recurrence is LT while enlistment of patients at high risk of recurrence after resection prior to the appearance of recurrence permits an optimal use of scarce organs and provides excellent long-term outcomes [1,28,29]. Promisingly, our radiomics models provide individualized estimation of recurrence risk as well as three risk profiles that may affect both the use of adjuvant treatment and the LT strategy. Additionally, choice of surveillance program should balance sensitivity to optimize early detection of recurrence, specificity to minimize harms from follow-up tests, and costs to remain cost-effective. Our radiomics models may therefore facilitate individualized surveillance policy. Specifically, low-risk patients may receive a less intensive surveillance regimen, even within the first 2 years after surgery, given their 2-year cumulative recurrence rate of less than 10%, whereas intermediate-to high-risk patients may need intensive surveillance lasting for 5 years; high-risk patients should also receive intensive screening for distant metastasis since up to 30% of recurrent tumors involved extrahepatic sites in this study.
Several limitations should be noted. First, this study was based on data from China and most patients had hepatitis B-related HCC. Second, this retrospective study suffers from inherent biases. Third, evaluation of solitary HCC by gadoxetate-enhanced MRI has recently been reported to detect additional lesions and therefore improve long-term outcomes [30]. However, recent meta-analysis has precluded support for exclusive use of MRI over CT [31,32]. Fourth, deep learning is a promising method that can automatically learn feature representations from images according to clinical goals and has been widely used in oncology researches [7], but was not explored in this study. Finally, the association between genomic profiles and radiomic phenotypes was not studied.
In conclusion, we demonstrated the complementary nature of engineered radiomics and existing prognostic parameters. When integrated with clinical data sources, a three-feature fusion signature generated by aggregated ML-based framework promises to accurately predict individual recurrence risk that enables appropriate management and surveillance of HCC.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Funding sources
This study was supported by the Key Program of the National Natural Science Foundation of China (31930020), the National Natural Science Foundation of China (81530048, 81470901, 81670570), and the Key Research and Development Program of Jiangsu Province (BE2016789). The funding agency played no role in this study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.10.057.
Contributor Information
Gu-Wei Ji, Email: drjgw@njmu.edu.cn.
Qing Xu, Email: 13776683209@163.com.
Xiang-Cheng Li, Email: drxcli@njmu.edu.cn.
Xue-Hao Wang, Email: wangxh@njmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Forner A., Reig M., Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 2.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 3.European Association for the Study of the Liver EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Vogel A., Cervantes A., Chau I. Hepatocellular carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv238–iv255. doi: 10.1093/annonc/mdy308. [DOI] [PubMed] [Google Scholar]
- 5.Chan A.W.H., Zhong J., Berhane S. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J Hepatol. 2018;69:1284–1293. doi: 10.1016/j.jhep.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Qiu J., Peng B., Tang Y. CpG methylation signature predicts recurrence in early-stage hepatocellular carcinoma: results from a multicenter study. J Clin Oncol. 2017;35:734–742. doi: 10.1200/JCO.2016.68.2153. [DOI] [PubMed] [Google Scholar]
- 7.Bi W.L., Hosny A., Schabath M.B. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. 2019;69:127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambin P., Leijenaar R.T.H., Deist T.M. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 9.Ji G.W., Zhang Y.D., Zhang H. Biliary tract cancer at CT: a radiomics-based model to predict lymph node metastasis and survival outcomes. Radiology. 2019;290:90–98. doi: 10.1148/radiol.2018181408. [DOI] [PubMed] [Google Scholar]
- 10.Leger S., Zwanenburg A., Pilz K. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep. 2017;7:13206. doi: 10.1038/s41598-017-13448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y., He L., Huang Y. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY) 2017;42:1695–1704. doi: 10.1007/s00261-017-1072-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z., Jiang H., Chen J. Hepatocellular carcinoma: radiomics nomogram on gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imaging. 2019;19:22. doi: 10.1186/s40644-019-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S., Shin J., Kim D.Y., Choi G.H., Kim M.J., Choi J.Y. Radiomics on gadoxetic acid-enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin Cancer Res. 2019;25:3847–3855. doi: 10.1158/1078-0432.CCR-18-2861. [DOI] [PubMed] [Google Scholar]
- 14.Zheng B.H., Liu L.Z., Zhang Z.Z. Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer. 2018;18:1148. doi: 10.1186/s12885-018-5024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cong W.M., Bu H., Chen J. Practice guidelines for the pathological diagnosis of primary liver cancer: 2015 update. World J Gastroenterol. 2016;22:9279–9287. doi: 10.3748/wjg.v22.i42.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun R., Limkin E.J., Vakalopoulou M. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 17.van Griethuysen J.J.M., Fedorov A., Parmar C. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Jay N., Papillon-Cavanagh S., Olsen C., El-Hachem N., Bontempi G., Haibe-Kains B. mRMRe: an r package for parallelized mRMR ensemble feature selection. Bioinformatics. 2013;29:2365–2368. doi: 10.1093/bioinformatics/btt383. [DOI] [PubMed] [Google Scholar]
- 19.Grömping U. Variable importance assessment in regression: linear regression versus random forest. Am Stat. 2009;63:308–319. [Google Scholar]
- 20.Tibshirani R. The lasso method for variable selection in the COX model. Stat Med. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Schröder M.S., Culhane A.C., Quackenbush J., Haibe-Kains B. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27:3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen M.E., Ko C.Y., Bilimoria K.Y. Optimizing acs nsqip modeling for evaluation of surgical quality and risk: patient risk adjustment, procedure mix adjustment, shrinkage adjustment, and surgical focus. J Am Coll Surg. 2013;217:336–346. doi: 10.1016/j.jamcollsurg.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 23.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 24.Choi J.Y., Lee J.M., Sirlin C.B. CT and mr imaging diagnosis and staging of hepatocellular carcinoma: part I. development, growth, and spread: key pathologic and imaging aspects. Radiology. 2014;272:635–654. doi: 10.1148/radiol.14132361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson P.J., Berhane S., Kagebayashi C. Assessment of liver function in patients with hepatocellular carcinoma: a new evidence-based approach-the Albi grade. J Clin Oncol. 2015;33:550–558. doi: 10.1200/JCO.2014.57.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choy G., Khalilzadeh O., Michalski M. Current applications and future impact of machine learning in radiology. Radiology. 2018;288:318–328. doi: 10.1148/radiol.2018171820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parmar C., Grossmann P., Bussink J., Lambin P., Aerts H.J.W.L. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrer-Fàbrega J., Forner A., Liccioni A. Prospective validation of ab initio liver transplantation in hepatocellular carcinoma upon detection of risk factors for recurrence after resection. Hepatology. 2016;63:839–849. doi: 10.1002/hep.28339. [DOI] [PubMed] [Google Scholar]
- 29.Kulik L., El-Serag H.B. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156:477–491. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H.D., Lim Y.S., Han S. Evaluation of early-stage hepatocellular carcinoma by magnetic resonance imaging with gadoxetic acid detects additional lesions and increases overall survival. Gastroenterology. 2015;148:1371–1382. doi: 10.1053/j.gastro.2015.02.051. [DOI] [PubMed] [Google Scholar]
- 31.Roberts L.R., Sirlin C.B., Zaiem F. Imaging for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. Hepatology. 2018;67:401–421. doi: 10.1002/hep.29487. [DOI] [PubMed] [Google Scholar]
- 32.Chernyak V., Fowler K.J., Kamaya A. Liver imaging reporting and data system (LI-RADS) version 2018: imaging of hepatocellular carcinoma in at-risk patients. Radiology. 2018;289:816–830. doi: 10.1148/radiol.2018181494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.