Abstract
Objective
To establish an axon regeneration regulatory network for optimal selection, and explore the role of low intensity pulsed ultrasound in the network.
Methods
The axon regeneration regulatory network involving axon regeneration-related proteins NGF, BDNF and PirB was constructed by using GO and KEGG. The maximum possible pathway acting on axon regeneration was screened by Bayesian network theory. The node of low - intensity pulsed ultrasound in NGF - involved axon regeneration network was complemented by combining literature methods.
Results
The NGF, BDNF and PirB-involved axonal regeneration regulatory pathway was successfully constructed. The low intensity pulsed ultrasound played a role in axon regeneration by acting on ERK1/2-CREB pathway and GSK-3β. NGF-TrKA-Rap1-ERK1/2-CREB-Bcl-2 was optimized as optimal pathway by Bayesian theory.
Conclusion
The regulatory pathway of axon regeneration involving nerve growth related factors and low intensity pulsed ultrasound was initially established, which provided a theoretical basis for further study of axon regeneration, and also new ideas for action of low intensity pulsed ultrasound on axon regeneration regulatory pathway.
Keywords: NGF, BDNF, PirB, Axon regeneration, Low intensity pulsed ultrasound, Regulatory network
1. Introduction
Axon regeneration refers to the process of axon budding, growth, extension, and reconstruction of contact with the target cells to achieve nerve reinnervation and function recovery under certain conditions. Axon is often unable to regenerate after damage of the central nervous system (CNNS) of adult mammals, but that of peripheral nervous system (PNS) has some regenerative ability, though there still being problems of slow regeneration rate and poor effect (Ring, 2013). In addition to factors like poor inherent growth capacity of neurons, formation of glial scarring, lack of growth-promoting molecules, nerve regeneration inhibitory factor initiates different intracellular cascade signal transduction pathways, and then affects axon regeneration, which may be the core mechanism for ineffective axon regeneration (Takeuchi et al., 2013). Therefore, understanding and mastering the intracellular signal transduction pathways that promote axon regeneration, and making in-depth study on how to promote nerve regeneration and its mechanism means important significance for improvement of prognosis of nervous system injury.
There are many factors regulating axon regeneration, and intracellular signal transduction mechanism is complex. Where, nerve growth factor (NGF) is one of the most important biological active substances in the nervous system, which plays a decisive role in promoting the proliferation and differentiation of neurons, regulating central and peripheral neuronal survival and accelerating axon regeneration (Aloe et al., 2012). Brain-derived neurophin factor (BDNF), a member of the trophic factors of nerve regeneration microenvironment, is involved in the regulation of nerve fiber regeneration and protection of neurons (Hultman et al., 2014). Pairwise immunoglobulin-like receptor B (PirB) expressed in neuronal cell bodies and growth cone, is closely related to axon growth (Adelson et al., 2012). In recent years, research on intracellular signal transduction pathway influencing axon regeneration has been widely concerned by the medical profession both at home and abroad. However, further studied is needed due to the many factors affecting axon regeneration and the complex intracellular signal transduction mechanism.
Low intensity pulsed ultrasound (LIPUS) is a kind of pressure wave that can be transmitted, which can transfer mechanical energy to tissue and produce beneficial biological reaction (Hersh et al., 2016). LIPUS can promote conduction velocity of motor and sensory nerve after nerve injury, which has been gradually applied to nerve repair therapy, though its mechanism of action is not clear. The aim of this study was to establish NGF, PirB and BDNF-invloved axon regeneration regulatory network, screen the optimal pathway by Bayesian network theory and explore the action mechanism of LIPUS in the regulatory network.
2. Materials and methods
2.1. GO analysis
The NGF gene was analyzed by GO using the online website Gene Ontology Consortium (http://geneontology.org/). For GO analysis, enter the Gene Ontology Consortium home page, set the screening conditions as “NGF”, “Pir-B”, “BDNF” and “Homo sapiens” successively, and make primary analysis of GO function annotation of the axon regeneration genes.
2.2. KEGG network analysis
The network pathways of NGF, Pir-B and BDNF genes in axon regeneration were analyzed by using online network KEGG PATHWAY Database (http://www.kegg.jp/kegg/pathway.html). For KEGG analysis, enter the KEGG PATHWAY Database home page, set the screening conditions organism as “hsa”, key words as “NGF”, “Pir-B”, “BDNF”, to query the regulatory network of the gene in the process of axon regeneration. With the protein as the starting point and the axon regeneration as the end point, collect and sort the acting path and nodes of the reference proteins. The overlapping pathways are then integrated into a primary regulatory network.
2.3. Bayesian regulatory network construction and optimization
The Bayesian network model theory is applied to the network to establish the Bayesian network. In the Bayesian regulatory network model, each node has a conditional probability distribution function in case a parent node is given, and those without parent node are expressed with prior probability. In this network, the starting node (i.e., the node without parent node) is a protein similar to p42.3. Therefore, the similarity between the reference protein and p42.3 is defined as the prior probability of the starting node, and probability of occurrence of each node is calculated based on probability of occurrence and conditional probability of the parent node by using the knowledge of conditional probability, as shown in Eq. (1):
| (1) |
Bayesian network is a directed acyclic graph (DAG), which mainly describes the joint probability distribution in the set of finite variables U = {X1, X2, …, Xn}. The available elements are denoted by B = (G, θ), where G is the directed acyclic graph. The nodes in the graph correspond to the random variables X1, …, Xn, which can represent the expression vector of the gene, while θ represents the conditional probability of each variable. The DAG is specifically interpreted as Markove hypothesis: under the premise of given parent node in G, each variable Xi is independent of its non-child nodes. Taking into account the independent properties of the condition, the only joint probability in the specified set U of G has the following distribution:
where Pa (Xi) represents the parent node of Xi.
It can be seen that the uniqueness of the network lies in application of causal relationship to explain the probability relationship with conditional independence, so that a more advanced causal relationship is developed, that is, the relationship between genes. In the Bayesian network of this study, after probability of occurrence of each node and path is calculated, the probability of action of each node protein is deduced by Bayesian theorem, so that the maximum probability pathway of axon regeneration can be calculated.
3. Results
3.1. GO analysis results
GO analysis, also known as gene ontology, is mainly used to study the specific function of genes which are divided into three main categories: molecular function (MF), cellular component (CC) and biological process (BP). The results of GO analysis of NGF, BDNF and PirB genes are shown in Table 1, Table 2, Table 3. As can be seen from Table 1, NGF has three functions of biological process, molecular function and cellular component. Wherein, the biological process mainly involves the activation of MAPKK activity; molecular function mainly involves binding with its receptor NGFR; cellular component involves extracellular components. As can be seen from Table 2, BDNF has three functions of biological process, molecular function and cellular component. Wherein, the biological process mainly involves angiogenesis, nerve cell migration, protein phosphorylation regulation, neuromuscular junction development; molecular function mainly involves binding with ATP; cellular component involves early endosome, cytoplasmic matrix, etc. As can be seen from Table 3, PirB has three functions of biological process, molecular function and cellular component. Wherein, the biological process mainly involves adaptive immune response, cell surface receptor signal pathway; molecular function mainly involves binding with its receptor, transmembrane signal receptor activity; cellular component involves integral component of cell membrane, plasma membrane, etc.
Table 1.
Results of GO analysis of NGF gene.
| Gene | Name | Ontology | Accession |
|---|---|---|---|
| NGF | activation of MAPKK activity | biological_process | GO:0007255 |
| nerve growth factor receptor binding | molecular_function | GO:0005163 | |
| protein binding | molecular_function | GO:0005515 | |
| extracellular region | cellular_component | GO:0005576 |
Table 2.
Results of GO analysis of BDNF gene.
| Gene | Name | Ontology | Accession |
|---|---|---|---|
| BDNF | vasculogenesis | biological_process | GO:0001570 |
| neuron migration | biological_process | GO:0001764 | |
| positive regulation of protein phosphorylation | biological_process | GO:0001934 | |
| neuromuscular junction development | biological_process | GO:0007528 | |
| circadian rhythm | biological_process | GO:0007623 | |
| ATP binding | molecular_function | GO:0005524 | |
|
cellular_component | GO:0005769 | |
| cytosol | cellular_component | GO:0005829 | |
| integral component of plasma membrane | cellular_component | GO:0005887 |
Table 3.
Results of GO analysis of PirB gene.
| Gene | Name | Ontology | Accession |
|---|---|---|---|
| PirB |
|
biological_process | GO:0002250 |
|
biological_process | GO:0006952 | |
| cell surface receptor signaling pathway | biological_process | GO:0007166 | |
| neutrophil degranulation | biological_process | GO:0043312 | |
| receptor activity | molecular_function | GO:0004872 | |
| transmembrane signaling receptor activity | molecular_function | GO:0004888 | |
| protein binding | molecular_function | GO:0005515 | |
| plasma membrane | cellular_component | GO:0005886 | |
| integral component of plasma membrane | cellular_component | GO:0005887 | |
| secretory granule membrane | cellular_component | GO:0030667 |
3.2. Establishment of axon regeneration regulatory network
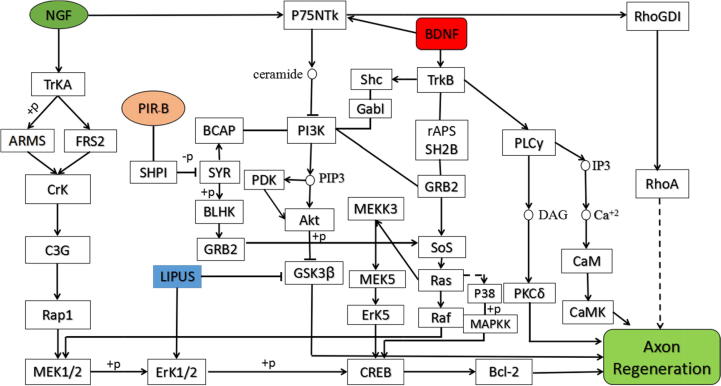
KEGG analysis of NGF, BDNF and PirB genes was carried out by KEGG signal pathway database. The regulation pathway of the three genes was improved by literature method, and the operational nodes of ultrasound in the pathway were supplemented. The axon regeneration regulatory network involving the above three genes and ultrasound was thus successfully established, with the results shown in Fig. 1.
Fig. 1.
Pathway diagram of axon regeneration regulatory network.
3.3. Bayesian-optimized regulatory network
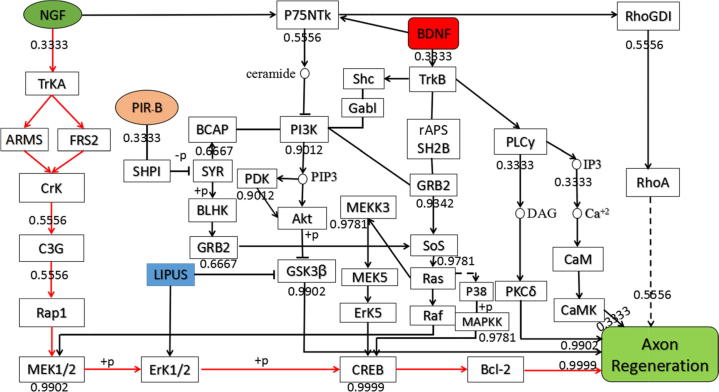
According to the Bayesian theorem, the probability of occurrence of each pathway is calculated. As shown in Fig. 2, the pathway “NGF-TrKA-Rap1-ERK1/2-CREB-Bcl-2” (red pathway in Fig. 2) is the most probable acting pathway of axon regeneration.
Fig. 2.
Bayesian theorem-optimized possible optimal acting pathway of axonal regeneration.
4. Discussions
Neurons are cells difficult to regenerate. After nerve tissue injury, nerve regeneration is mainly manifested as axon extension and synapse remodeling. On the other hand, axon regeneration is affected by glial scar, neurotrophic factor and axon growth inhibitory factor, which is closely related to NGF, BDNF, PirB.
NGF and BDNF are two neurotrophic factors present in mammals that play a role by binding the three receptors (p75NTR, TrkA, TrkB) (Lewin et al., 2014, Qin and Dong, 2000). Both NGF and BDNF bind with p75NTR receptor, but selectively bind with Trk receptor. NGF binds with two transmembrane receptors: tyrosine kinase receptor TrkA and tumor necrosis factor receptor p75NTR. This is consistent with the network pathway constructed in this study. TrkA is a high affinity receptor for mature NGF; p75NTR is a low affinity receptor for mature NGF. At present, studies show that the action mechanism of NGF is mainly to promote the expression of functional proteins like synaptophysin p38, growth-related protein, bind with chondroitin sulfate proteoglycan (CSPGs) and reduce the inhibitory effect of CSPGs on nerve regeneration; control the modification of certain proteins such as phosphorylation process and block the inhibition of regenerated protein produced by central nervous system injury, thus promoting axon regeneration. For instance, NGF binds with cell surface TrkA receptor to activate RAS/ERK and PI3K signal pathway (Skaper, 2012). In this study, NGF -TrKA-Rap1-ERK1/2-CREB-Bcl-2 is calculated as the optimal pathway for axon regeneration according to Bayesian theorem by constructing primary pathway of axon regeneration.
BDNF can prevent the degeneration of damaged neurons. Under certain conditions, it can promote germination of uninjured neurons and reconstruct the damaged neural circuit (Hultman et al., 2014). Gravel et al. showed that BDNF could prevent the motor neurons from death due to axonal cleavage, and that BDNF had a significant effect in promoting regeneration of peripheral nerve axons (Hultman et al., 2014). Recent studies on brain-derived neurotrophic factor found that it can induce the regeneration of axons, protect neurons, prevent nerve injury retrograde degeneration and effector degeneration, and that it helps improve quality of regenerated axons in nerve end-to-side anastomosis. In the early stage of nerve injury, platelets can synthesize and secrete BDNF which will act on the injured motor and sensory neurons to effectively promote nerve regeneration (Qin and Dong, 2000). BDNF plays a role mainly through the binding with TrkB receptor and p75NTR receptor, which is consistent with the pathway in the network constructed in this study.
PirB is mainly expressed in the central nervous system. Recent studies have found that PirB expression is present in cerebral cortical neurons, hippocampal neurons, and olfactory bulb neurons (Akbik et al., 2012, Syken et al., 2006, Omoto et al., 2010), but the expression of PirB has not been found in spinal cord of adult vertebrates (Omoto et al., 2010, Raiker et al., 2010). Studies further found that expression level of PirB was significantly up-regulated after CNS damage (Nakamura et al., 2011, Gou et al., 2013). Adelson et al. (2012) reported in Neuron magazine in 2012 that down-regulation of PirB expression helps remodeling of corticospinal tract and survival of hippocampal neurons following cerebral ischemia-reperfusion injury. Bochner et al. (2014) pointed out in 2014 that closed PirB function is conducive to visual cortical neuronal function recovery. The latest report by Zhao et al. (2016b) in 2016 showed that PirB over-expression could cause apoptosis of in vitro cultured cortical neurons after ischemia and reperfusion. PilB is co-receptor of the central nervous system myelin-associated inhibitory proteins (MAIs): Nogo-A, Myelin-associated glycoprotein (MAG), oligodendrocyte myelin glycoprotein (OMgp), which have high affinity and exhibit a chronic inhibitory effect on axonal growth (Omoto et al., 2010). The network constructed in this study was mainly PirB follow-up pathway. The results showed that PirB inhibits axon regeneration mainly by inhibiting SHPI.
Ultrasound is a sound wave whose frequency is greater than the auditory detection level (>20 kHz) of the human ear and is a mechanical wave that is generated by vibration of the sound source and propagated through the compression and expansion media. Because ultrasound can spread over long distances in specific media and the energy attenuation is very small, it is widely used in medical diagnosis and physical therapy (Hersh et al., 2016). By changing the frequency of ultrasound, pulse repetition frequency, pulse width, duration and intensity of ultrasound, the central nervous system of the stimulated part produces stimulus or inhibitory effect, and thus reversibly two-way adjusts the nerve function (Li et al., 2015). Earlier studies suggested that the mechanism of ultrasound in promoting nerve regeneration is the mechanical effect of ultrasound. The mechanical effect features micro-massage, can promote the proliferation of Schwann cells, and thus promote nerve regeneration (Zhou and Chen, 2003). Lazar et al. (2001) held that it may be associated with the fact that ultrasound accelerates influx of nutrients and elimination of toxic substances in the injured nerve, thereby accelerating denaturation of nerves and regeneration of nerve fibers.
However, these results do not fully elucidate the molecular mechanism in which ultrasound promotes axons. In this study, two pathways on ultrasound action on axon regeneration were collected by literature method, respectively ERA1/2-CREB pathway and GSK-3β. Studies on the former have found that low-intensity pulsed ultrasound (LIPUS) could increase NGF-induced axonal growth. Further studies have found that it played a role mainly through ERK1/2-CREB-Trx-1 pathway (Zhao et al., 2016a). Studies on the latter have shown that LIPUS promotes the growth of axons possibly by inhibiting GSK-3β (Ren et al., 2010). In addition, some studies found that ultrasound can cause increased ne, uronal calcium ion reversibility, and then positively regulates axon regeneration (Ibsen et al., 2015).
To sum up, this study established a regulatory network of axon regeneration involving NGF, BDNF and PirB, and supplemented the acting pathway of low intensity pulsed ultrasound in the network, which provided the theoretical basis for further study on axon regeneration, and also new ideas for action of low-intensity pulsed ultrasound on axon regeneration regulatory pathway.
Acknowledgements
The work was financially supported by The Natural Science Foundation of Henan Province (Grant No.: 162300410290).
Footnotes
Peer review under responsibility of King Saud University.
References
- Adelson J.D., Barreto G.E., Xu L. Neuroprotection from stroke in the absence of MHCI or PirB. Neuron. 2012;73:1100–1107. doi: 10.1016/j.neuron.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbik F., Cafferty W.B., Strittmatter S.M. Myelin associated inhibitors: a link between injury-induced and experience-dependent plasticity. Exp. Neurol. 2012;235:43–52. doi: 10.1016/j.expneurol.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloe L., Rocco M.L., Bianchi P. Nerve growth factor: From the early discoveries to the potential clinical use. J. Transl. Med. 2012;10 doi: 10.1186/1479-5876-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner D.N., Sapp R.W., Adelson J.D. Blocking PirB up-regulates spines and functional synapses to unlock visual cortical plasticity and facilitate recovery from amblyopia. Sci. Transl. Med. 2014;6:140r–258r. doi: 10.1126/scitranslmed.3010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X., Zhang Q., Xu N. Spatio-temporal expression of paired immunoglobulin-like receptor-B in the adult mouse brain after focal cerebral ischaemia. Brain Inj. 2013;27:1311–1315. doi: 10.3109/02699052.2013.812241. [DOI] [PubMed] [Google Scholar]
- Hersh D.S., Kim A.J., Winkles J.A. Emerging applications of therapeutic ultrasound in neuro–oncology: moving beyond tumor ablation. Neurosurgery. 2016;79(5):643–654. doi: 10.1227/NEU.0000000000001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman R., Kumaric U., Micheld N. Gaz regulates BDNF-induction of axon growth in cortical neurons. Mol. Cell Neurosc. 2014;58:53–61. doi: 10.1016/j.mcn.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibsen S., Tong A., Schutt C. Sonogenetics is a non-invasive approach to activating neurons in Caenorhabditis elegans. Nat. Commun. 2015;15(6) doi: 10.1038/ncomms9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar D.A., Curra F.P., Mohr B. Acceleration of recovery after injury to the peripheral nervous system using ultrasound and other therapeutic modalities. Neurosurg. Clin. N. Am. 2001;12(2):353–357. [PubMed] [Google Scholar]
- Lewin G.R., Lechner S.G., Smith E.S. Nerve growth factor and nociception: from experimental embryology to new analgesic therapy. Handb. Exp. Pharmacol. 2014;220:251–282. doi: 10.1007/978-3-642-45106-5_10. [DOI] [PubMed] [Google Scholar]
- Li H.F., Wang Y.R., Huo H.P. Neuroprotective effects of ultrasound-guided nerve growth factor injections after sciatic nerve injury. Neural Regen Res. 2015;10(11):1846–1855. doi: 10.4103/1673-5374.170315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Fujita Y., Ueno M. Paired immunoglobulin-like receptor B knockout does not enhance axonal regeneration or loco-motor recovery after spinal cord injury. J. Biol. Chem. 2011;286:1876–1883. doi: 10.1074/jbc.M110.163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S., Ueno M., Mochio S. Genetic deletion of paired immunoglobulin-like receptor B does not promote axonal plasticity or functional recovery after traumatic brain injury. J. Neurosci. 2010;68 doi: 10.1523/JNEUROSCI.3228-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z.B., Dong M.M. Experimental study on distribution and expression of BDNF and FGF-2 after facial nerve injury. Chin. Arch. Otolaryngol.-Head Neck Surg. 2000;9(1):B39–B41. doi: 10.16066/j.1672-7002.2002.01.020. [DOI] [Google Scholar]
- Raiker S.J., Lee H., Baldwin K.T. Oligodendrocyte-myelin glycoprotein and Nogo negatively regulate activity-dependent synaptic plasticity. J. Neurosci. 2010;30(37):12432–12445. doi: 10.1523/JNEUROSCI.0895-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Li J.M., Lin X. LIPUS enhance elongation of neurites in rat cortical neurons through inhibition of GSK-3β. Biomed. Environ. Sci. 2010;23(3):244–249. doi: 10.1016/S0895-3988(10)60059-1. [DOI] [PubMed] [Google Scholar]
- Ring D. Symptoms and disability after major peripheral nerve injury. Hand Clin. 2013;29(3):421–425. doi: 10.1016/j.hcl.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Skaper S.D. The neurotrophin family of neurotrophic factors: an overview. Methods Mol. Biol. 2012;846:1–12. doi: 10.1007/978-1-61779-536-7_1. [DOI] [PubMed] [Google Scholar]
- Syken J., Grandpre T., Kanold P.O. PirB restricts ocular-dominance plasticity in visual cortex. Science. 2006;313:1795–1800. doi: 10.1126/science.1128232. [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Yoshioka N., Higa O.S. Chondroitin sulphate N-acetylgalactosaminyl-transferase-1 inhibits recovery from neural injury. Nat. Commun. 2013;4 doi: 10.1038/ncomms3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Feng Y., Hu H. Low-intensity pulsed ultrasound enhances nerve growth factor-induced neurite outgrowth through mechanotransduction-mediated ERK1/2-CREB-Trx-1 signaling. Ultrasound Med. Biol. 2016;42(12):2914–2925. doi: 10.1016/j.ultrasmedbio.2016.07.017. [DOI] [PubMed] [Google Scholar]
- Zhao Z.H., Deng B., Xu H. PirB overexpression exacerbates neuronal apoptosis by inhibiting TrkB and mTOR phosphorylation. Cell. Mol. Neurobiol. 2016;37(4):707–715. doi: 10.1007/s10571-016-0406-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Chen W.Z. Research progressing of peripheral nerves regeneration induced by ultrasound. Chin. J. Tissue Eng. Res. 2003;7(16):2342–2343. [Google Scholar]