Abstract
Objective
This research was to establish a mitochondrial-related Drp1 gene and a lung cancer-related Erbb4 gene to participate in the regulatory network of lung cancer cell apoptosis, and to provide theoretical support for mitochondria to participate in tumor regulation.
Method
The GO and KEGG methods were used to construct the regulatory networks of lung cancer related Drp1 and Erbb4 proteins that involved in the apoptosis of tumor cells, and to combine with the Bayesian network theory to screen out the largest possible action path acting on this network; The information about Drp1 in Oncomine database was collected, and the data in current database were analyzed twice. The role of Drp1 in lung cancer was meta-analyzed.
Result
A regulatory network of Drp1 and Erbb4 involved in the apoptosis of tumor cells was successfully constructed; the optimal pathway was optimized using Bayesian theory; a total of 446 different types of research results were collected in the Oncomine database, of which there were 18 studies with statistical differences in Drp1 expression, 13 studies with increased Drp1’s expression, and 5 studies with decreased expression. Compared with the control group, Drp1 was expressed in lung cancer tissues highly (P < 0.05).
Conclusion
Establishment and optimization of mitochondrial-related Drp1 and tumor-related Erbb4 genes involved in the regulation of apoptosis of cancer cells. It was proposed that Drp1 was expressed in lung cancer tissues highly through in-depth excavation of tumor-associated gene information in the Oncomine gene chip database.
Keywords: Lung cancer, Drp1, Erbb4, Regulation pathway, Oncomine
1. Introduction
The development of tumors is a complex biological process involving multiple genes and multiple signaling pathways. Compared with normal cells, tumor cells have infinite proliferation, abnormal energy metabolism, elevated reactive oxygen species, infiltration, and metastasis and resistance to cell death (Shen et al., 2019). In recent years, with the deepening of research, people have gradually realized that mitochondria is of significance in the development of tumors, and proposed mitochondria as a targeted new strategies for tumor treatment (Gao et al., 2017a, Gao et al., 2017b, Liu, 2016, Zhang et al., 2019a, Zhang et al., 2019b). The Drp1 protein structural domain is homologous to dynein and also contains a unique proline-rich domain (Yang, 2018). Although the recent researches have demonstrated that Drp1 is of great importance in maintaining proper mitochondrial function, apoptosis and necrosis in various important cellular processes (Xu and Liu, 2013), the potential Drp1 involvement in the formation of cancer has not yet been resolved completely.
In our previous study, we devoted ourselves to the research on Erbb4 and tumors, and constructed a regulatory network of lung cancer cells involved by Erbb4 (Meng and Li, 2016). In order to further elucidate whether mitochondria plays a role in the above-mentioned regulatory networks, a regulatory network of lung cancer cells involved by mitochondrial related Drp1 and Erbb4 genes was constructed.
2. Materials and methods
2.1. GO analysis
GO analysis of the gene Drp1 was conducted with Gene Ontology Consortium (http://geneontology.org/). Firstly, entering the website, the screening conditions were “Drp1”, “Homo sapiens”.
2.2. KEGG network analysis
In the process of KEGG analysis, the KEGG PATHWAY database was registered. The screening condition was that “hsa” was an organism and the key word was “Drp1”. The regulatory network of the gene in the apoptosis of cancer cells was queried (Hussain et al., 2016). With the protein as the starting point and the “avoidance of apoptosis” as the ending point, the pathways and nodes of each reference protein were collected, and there were intersections among these pathways.
2.3. Bayesian regulation network construction and optimization
In the Bayesian regulation network model, in the case of given a parent node, and no parent node was using the prior probability to express information. In this network, by applying the conditional probability knowledge and based on the occurrence probability of the parent node and the conditional possibility, whose occurrence of each node was calculated as shown in formula (1):
| (1) |
2.4. Data extraction from Oncomine database
The Oncomine database is a database based on a gene chips and a integrated platform of data mining. In this data, the conditions of screening and mining data can be set up according to own requirements. In this study, the screening conditions were set as: (1) “Cancer Type: Lung cancer”; (2) “Gene: Drp1”; (3) “Analysis Type: Cancer vs Normal Analysis”; (4) Threshold setting conditions (P value < 0.05, fold change > 2, gene rank = top 10%).
3. Results
3.1. GO analysis results
The function of Drp1 gene is shown in Table 1. It can be seen that mitochondrial fission and release of cytochrome c from mitochondria is involved in biological processes, also included dynamin family protein polymerization involved in mitochondrial fission. It's involved Golgi membrane in cellular_component and GTPase activity in molecular_function.
Table 1.
GO analysis results for the Drp1 gene.
| Gene | Name | Ontology | Accession |
|---|---|---|---|
| Drp1 | Golgi membrane | cellular_component | GO:0000139 |
| mitochondrial fission | biological_process | GO:0000266 | |
| release of cytochrome c from mitochondria | biological_process | GO:0001836 | |
| dynamin family protein polymerization involved in mitochondrial fission | biological_process | GO:0003374 | |
| GTPase activity | molecular_function | GO:0003924 |
3.2. Lung cancer control network that Erbb4 and Drp1 involved in
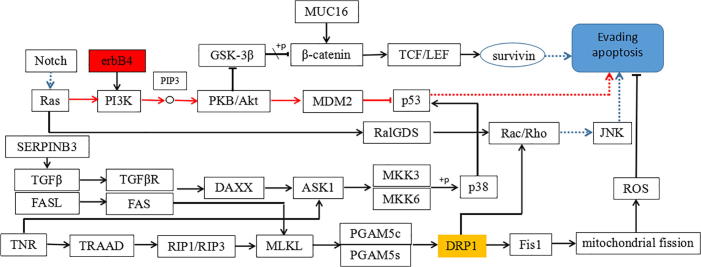
The KEGG analysis was performed on the Erbb4 and Drp1 genes using the KEGG signaling pathway database. At the same time, the literature on the regulation of the two genes was improved. The aforementioned genes involved regulation network of the lung cancer tumor cells evading apoptosis was successfully established, shown in Fig. 1.
Fig. 1.
Regulatory network of lung cancer tumor cell evading apoptosis.
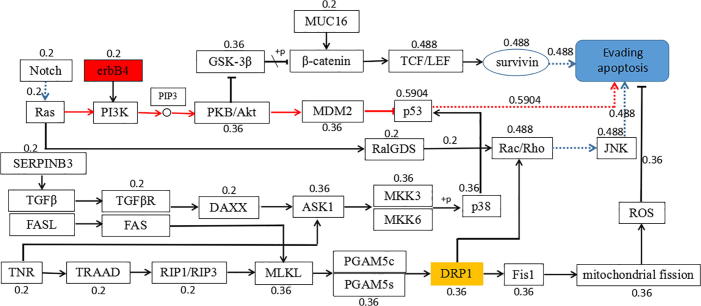
3.3. Bayesian optimization based regulation network
Calculate the probability of each path based on Bayesian theorem. The results are shown in Fig. 2. In the figure, the red path was the most likely path for tumor cell evading apoptosis.
Fig. 2.
The possible optimal path for cancer tumor cell evading apoptosis of Bayesian theorem optimization.
3.4. Expression results of Drp1 in common tumor types
In order to further verify the relationship between Drp1 and lung cancer, this study used the Oncomine database for the following analysis (Rhodes et al., 2004). Oncomine database collected 446 different types of research results (Fig. 3), of which there were 18 studies about the statistical differences in Drp1 expression, 13 studies about the increased Drp1 expression, and 5 studies with about the decreased expression.
Fig. 3.
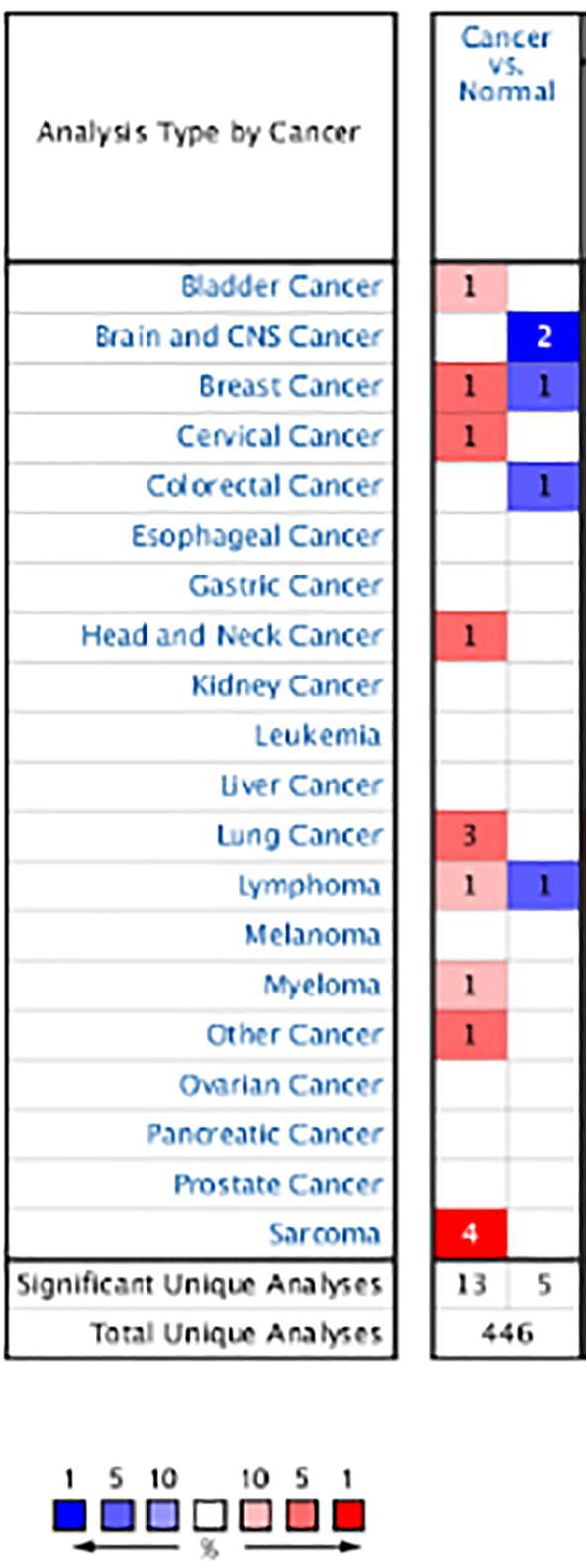
Expression data for Drp1 in a variety of normal and cancerous human tissues in the Oncomine database.
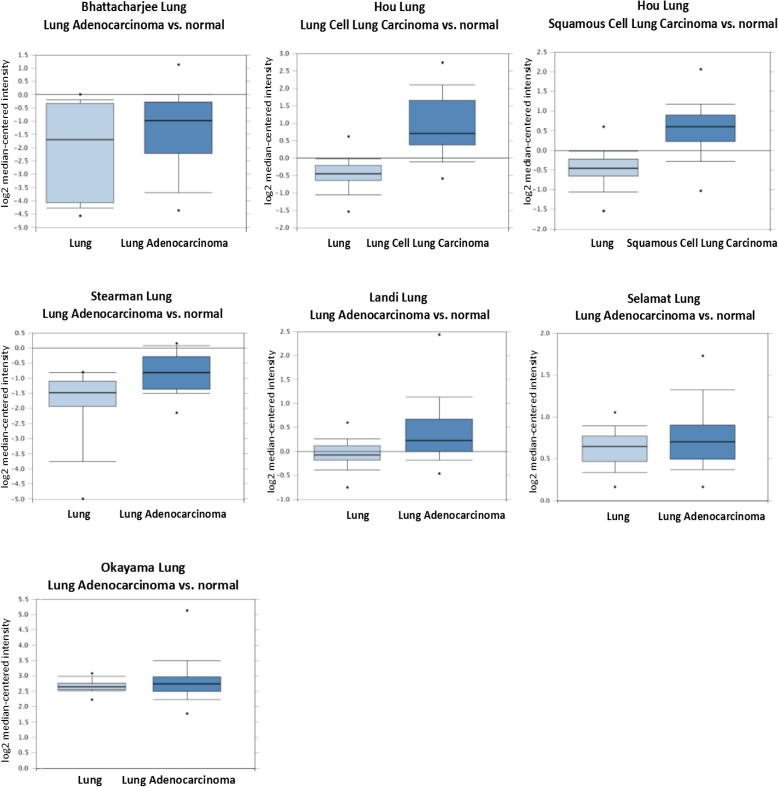
3.5. Expression results of Drp1 in lung cancer
In the Oncomine database, we found that since 2000, there have been 19 studies involving the expression of Drp1 in lung cancer tissues and normal tissues (Fig. 4). There were 1205 samples including squamous cell carcinoma, lung adenocarcinoma, cell lung cancer with large size and that with small size to compare with normal tissue. In the Oncomine database, a meta-analysis of 19 studies was found that as compared with the control group, Drp1 gene had a median of 1053.0 in all differentially expressed genes, P = 0.025, suggesting that Drp1 was highly expressed in lung cancer.
Fig. 4.
Drp1’s expression in lung cancer studies identified in the Oncomine database. 1–19 represents the 19 studies on the expressions of Drp1 in lung cancer, respectively. The darker the red, the higher the Drp1’s expression gene in the chips.
3.6. Expression differences of Drp1 in different lung cancer research chips
Fig. 5 shows the expression results of Drp1 in the Oncomine database in different lung cancer research chips. The Drp1’s expression the normal group was lower than that in the lung cancer (P all < 0.05).
Fig. 5.
Drp1’s expression in different studies of lung cancer identified in the Oncomine database. *: P < 0.05.
4. Discussion
According to the survey, there were approximately 4.292 million new cancer cases in China in 2015, and approximately 2.841 million cases of cancer deaths. Currently EGFR-TKI gene targeted therapy presented great limitations. Therefore, the platinum-based combination chemotherapy remains the primary treatment for most lung adenocarcinoma patients. However, with the popularization of chemotherapy, the problem of chemotherapy resistance has become increasingly prominent (Zhou and Fen, 2015).
Mitochondria can be used as targets to remove tumor cells, such as inhibiting glycolysis and anti-apoptotic protein expression, inducing increased mitochondrial permeability and regulating mitochondrial respiratory chain activity, etc. (Jiang et al., 2018, Zhang, 2013). In recent years, the regulation of mitochondrial fission and fusion has become a hotspot of tumor therapy. Mitochondrial division was driven by the dynamic-related proteins DRP1 (mammal) and DNM1 (yeast) as the main regulatory role (Deng et al., 2019). The Drp1 protein was a protein-driven GTPase molecules with a GTPase domain at its N-terminus (head), C-terminus (stem) as the effector domain of GTP enzyme (GED), the middle (stem) was the Dynamin homeodomain (also known as the middle region) and the variable region (Insert B) and the tail (Chan, 2012). The Drp1 protein was distributed in the cytoplasm and mitochondria, and generally existed in the form of a dimer/tetramer structure. When it binded onto the mitochondrial membrane, it formed a more advanced structure. Studies have shown that cisplatin resistance in cholangiocarcinoma was associated with mitochondrial dynamics and mitochondrial division induces apoptosis (Yu et al., 2018). In xenograft models, inhibition of Drp1 expression promotes tumor growth. It is speculated that impaired fusion and enhanced fission have fundamentally lead to the imbalance of proliferation/apoptosis of tumors and become a promising new therapeutic target. In a xenograft tumor model, knockout Drp1 has increased the migration activity and tumor formation of human lung cancer cells (Gao et al., 2017a, Gao et al., 2017b). The results obtained by oncomine analysis in this study show that Drp1 is expressed in lung cancer highly, which is different from the above results (Kim, Yun and Yun, 2018). We hypothesize that the Drp1 gene is expressed highly in most lung cancer tissues, and after mutation in another tissue, it becomes low.
The escape of cells from apoptosis involved almost all aspects of cancer progression, including the treatment of drug resistance. In our previous study, we constructed the regulatory pathway for Erb4-involved lung cancer cells to escape apoptosis (Ma et al., 2017). Based on this study, the role of mitochondria in this regulatory network was studied. Mitochondrial morphology determines mitochondrial function. This process is regulated by mitochondrial dynamics. The fusion of two mitochondria inhibited apoptosis, and mitochondrial division initiated apoptosis. The increased escape of apoptosis in A549 cells is associated with impediments to mitochondrial division and mitosis. The study concluded that mitochondrial disruption contributes to the apoptotic resistance of A549 cells (Shen et al., 2017). This is consistent with the study of the pathway we have established. This study calculates the optimal path of regulatory pathways, but at present this part still needs experimental verification.
In summary, this study used GO and KEGG database and Bayesian algorithm to construct the regulatory pathways of Drp1 and our previously studied lung cancer-related protein Erbb4 involved in lung cancer cell evasion apoptosis. This experiment also used the Oncomine database to analyze the relationship between Drp1 and lung cancer. Our study provides a new supplement to the pathogenesis of lung cancer. Drp1 gene and Erbb4 gene are jointly involved in the formation of lung cancer. In the future, these two genes can be used as tumor markers in clinical studies to provide guiding recommendations for the prognosis of patients. In addition, Drp1 gene and Erbb4 gene can be used as targets of lung cancer drugs in clinical treatment, greatly promoting the development of cancer drugs. In the follow-up work, the regulatory pathways need to be experimentally verified to further explore the potential molecular mechanisms of Drp1, mitochondrial division, tumor-associated protein Erbb4, and tumor development.
Acknowledgement
The work was financially supported by The Natural Science Foundation of Henan Province (Grant No.: 162300410290) and The National Key Research and Development Program of China (Grant No.: 2016YFC1303204).
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Chuanliang Chen, Email: henanccl@163.com.
Jianhua Zhang, Email: petermails@zzu.edu.cn.
References
- Chan D.C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- Deng W.J., Wang L., Peng W. Effect of Astragaloside IV on mitochondrial fission induced by high glucose in podocyte. Chinese J. Cli. Pharmacol. 2019;35(8):804–807. [Google Scholar]
- Gao W., Wang Y., Basavanagoud B., Jamil M.K. Characteristics studies of molecular structures in drugs. Saudi Pharm. J. 2017;25:580–586. doi: 10.1016/j.jsps.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Wang Y., Wang W., Shi L. The first multiplication atom-bond connectivity index of molecular structures in drugs. Saudi Pharm. J. 2017;25:548–555. doi: 10.1016/j.jsps.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Atif M.A., Ghafoor M.B. Beneficial effects of sitagliptin and metformin in non-diabetic hypertensive and dyslipidemic patients. Pakistan J. Pharm. Sci. 2016;29S:2385–2389. [PubMed] [Google Scholar]
- Jiang Z.Z., Xu D.Q., Yang H. Progress in research of drug targets. J. Exp. Hematol. 2018;42(21):838–859. [Google Scholar]
- Kim Y.Y., Yun S.H., Yun J. Downregulation of Drp1, a fission regulator, is associated with human lung and colon cancers. Acta Bioch. Bioph. Sin. 2018;50(2) doi: 10.1093/abbs/gmx137. [DOI] [PubMed] [Google Scholar]
- Liu Q.L. Abnormality of mitochondrial DNA and cancer. Chem. Life. 2016;36(6):862–867. [Google Scholar]
- Ma X., Li L., Tian T. Study of lung cancer regulatory network that involves erbB4 and tumor marker gene. Saudi J. Biol. Sci. 2017;24(3):649–657. doi: 10.1016/j.sjbs.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L.P., Li W.X. The curative effect analysis and nursing measures of the transvaginal myomectomy and transabdominal myomectomy. Pakistan J. Pharm. Sci. 2016;29S:2281–2285. [PubMed] [Google Scholar]
- Rhodes D.R., Yu J., Shanker K. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Gai J., Xing J. Dynasore suppresses proliferation and induces apoptosis of the non-small-cell lung cancer cell line A549. Biochem. Biophys. Res. Commun. 2017;2017 doi: 10.1016/j.bbrc.2017.11.109. S0006291X1732288X. [DOI] [PubMed] [Google Scholar]
- Shen W.S., Han Q.J., Zhang J. Advances in research on the effects of immune microenvironment on cancer stem cells. Prog. Biochem. Biophys. 2019;46(7):454–662. [Google Scholar]
- Xu F.F., Liu X.H. Physiological and pathophysiological function of myocardial autophagy. Prog. Biochem. Biophys. 2013;44(3):193–199. [PubMed] [Google Scholar]
- Yang Q.F. Zhejiang University; 2018. The Mechanism Study of Mitochondrial Dynamin-Related Protein drpl in Hepatocellular Carcinoma. [Google Scholar]
- Yu Y., Xu L., Liu S.B. Induction effect of myricetin on apoptosis of human ovarian cancer SKOV3 cells by promoting DRP1-dependent mitochondrial fission. J. Jilin Univ. (Medicine Edition) 2018;44(5):903–907. [Google Scholar]
- Zhang L.T., Yang L.J., Yang T. The role of mitochondrial ribosomal proteins in human malignant tumors. Chinese J. Biochem. Mol. Biol. 2019;35(6):620–624. [Google Scholar]
- Zhang Y. Zhejiang University; 2013. Azurin and its Combination with Chemotherapy Drugs induced Apoptosis of Human Osteosarcoma Cell by P53 mediated ROS and Mitochondrial Pathway. [Google Scholar]
- Zhang Z., Dong Q.Z., Qin L.X. Research progress of mitochondrial dynamics and cancer. Fudan Univ. J. Med. Sci. 2019;46(1):128–134. [Google Scholar]
- Zhou J., Fen S.Q. The clinical effects of pemetrexed or docetaxel combined with cisplatin as the first line treatment for advanced lung adenocarcinoma, 2016. China Oncol. 2015;25(9):671–676. [Google Scholar]