Abstract
We delivered plasmid DNA encoding therapeutic genes to the muscles of mouse models of limb girdle muscular dystrophy (LGMD) 2A, 2B, and 2D, deficient in calpain3, dysferlin, and alpha-sarcoglycan, respectively. We also delivered the human follistatin gene, which has the potential to increase therapeutic benefit. After intramuscular injection of DNA, electroporation was applied to enhance delivery to muscle fibers. When plasmids encoding the human calpain3 or dysferlin cDNA sequences were injected into quadriceps muscles of LGMD2A and LGMD2B mouse models, respectively, in 3-month studies, robust levels of calpain3 and dysferlin proteins were detected. We observed a statistically significant decrease in Evans blue dye penetration in LGMD2B mouse muscles after delivery of the dysferlin gene, consistent with repair of the muscle membrane defect in these mice. The therapeutic value of delivery of the genes for alpha-sarcoglycan and follistatin was documented by significant drops in Evans blue dye penetration in gastrocnemius muscles of LGMD2D mice. These results indicated for the first time that a combined gene therapy involving both alpha-sarcoglycan and follistatin would be valuable for LGMD2D patients. We suggest that this non-viral gene delivery method should be explored for its translational potential in patients.
Keywords: electroporation, alpha-sarcoglycan, calpain3, dysferlin, limb girdle muscular dystrophy, non-viral gene therapy, plasmid DNA, follistatin
Introduction
Limb girdle muscular dystrophies (LGMD) are characterized by degeneration in the muscles of the pelvic and shoulder girdles, leading to related mobility problems.1 These disorders currently lack treatments and are potential subjects for gene therapy. Among the 25 forms of LGMD, the 17 disorders classified as type 2 are recessive and amenable to gene replacement strategies.1,2 LGMD2A and LGMD2B result from deficiencies in the intracellular calpain3 protease3,4 and the membrane vesicle-associated protein dysferlin,5,6 respectively. In the case of LGMD2B, dysferlin is known to be involved in repair of the sarcolemmal membrane.7,8 LGMD2D reflects deficiency of the membrane-bound alpha-sarcoglycan protein, part of a complex that associates with dystrophin to stabilize its attachment to the basement membrane.9 The LGMD disorders have characteristic ages of onset, which is in young childhood for LGMD2D and late teens or adulthood for LGMD2A and 2B. Mouse models for each of these disorders3,9,10 make gene therapy studies feasible in these models.
Extensive efforts have been undertaken to develop gene therapies for muscular dystrophies, including LGMD. Due to their excellent delivery characteristics in mouse muscle,11 vectors based on adeno-associated virus (AAV) have been the focus of most recent efforts.10, 11, 12, 13, 14, 15, 16, 17 However, the AAV approach still faces considerable limitations as a gene therapy strategy in LGMD patients. The <5-kb size limit of the vector makes delivery of large genes or combinations of genes difficult.10,14,15 Furthermore, the immunogenicity of the AAV capsid raises further problems, since a good percentage of the human population possesses pre-existing immunity to AAV, eliminating many patients’ access to AAV-mediated gene therapy and hampering re-delivery.18, 19, 20 Because of the large size of the tissue target in muscular dystrophies, anticipated effective doses are high, creating challenges for manufacturing and costs, as well as increasing the chances of toxicity.21
Therefore, alternative delivery methods may be valuable in developing gene therapies for LGMD. Use of non-viral plasmid DNA presents a number of advantages by lessening issues of size limit, immunogenicity, and cost. Plasmid DNA has impressive longevity when introduced into skeletal muscle,22,23 so that if paired with efficient delivery methods, plasmid delivery could produce long-term expression of therapeutic genes. To make delivery more efficient, vascular delivery methods that introduce DNA in a large volume of fluid, promoting extravasation of the DNA and uptake by muscle fibers, can distribute plasmid DNA widely to the muscles of rodent and primate limbs.24, 25, 26, 27 We verified the effectiveness of vascular delivery in a mouse model of LGMD2B,28 and the safety and feasibility of a similar vascular delivery approach was verified in human muscular dystrophy patients.29,30 A different approach for increasing delivery of plasmid DNA in muscle is the use of electroporation to promote the entry of DNA through transient openings in the sarcolemmal membrane stimulated by brief charge pulses.31,32 This type of delivery has been studied intensively to enhance the delivery of DNA vaccines, leading to the development of special electroporation devices designed for easy and relatively non-invasive delivery of DNA as vaccines.33 These devices have been tested in animals and patients, opening a potential translational pathway for their use in gene therapy of LGMD. In this study, we explored the potential of one such electroporation device to deliver DNA to mouse models of LGMD.
Gene therapy of LGMD should supply the therapeutic protein that is deficient in these disorders. In addition, further benefit may accrue from delivery of additional genes that could strengthen diseased muscles. In this regard, the gene for follistatin, a naturally-occurring protein that counteracts the effect of myostatin in suppressing muscle growth, has potential to increase muscle health,34,35 especially when delivered in combination with the therapeutic gene that is mutated in these disorders. For example, follistatin has been found to be beneficial in the mdx mouse model of Duchenne muscular dystrophy,36 and benefit was claimed when an AAV vector expressing follistatin was injected in patients with Becker muscular dystrophy.37 Utilizing the flexibility of the plasmid DNA delivery approach, we introduced follistatin, either in combination with the therapeutic gene or on its own. Since LGMD2B and 2D confer a defect on the muscle membrane that can be easily measured using an assay based on penetration of Evans blue dye,38 we used this assay to show that non-viral gene therapy produced a histological benefit in treated muscles.
Results
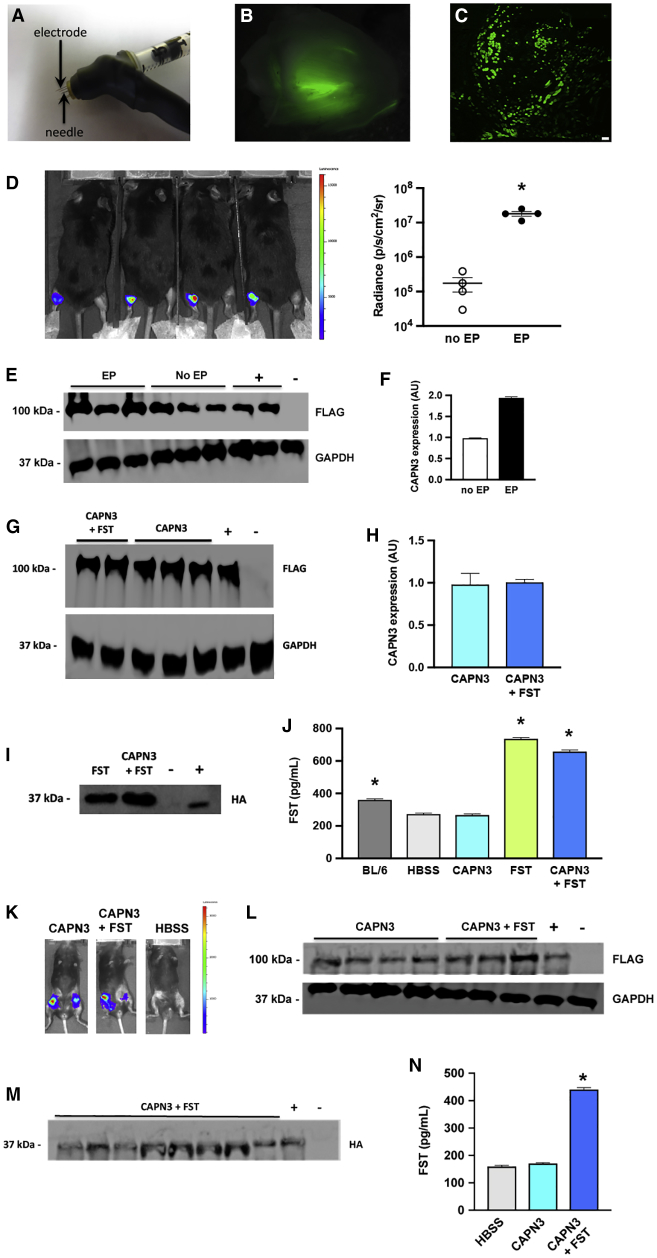
We tested the ability of electroporation to enhance DNA delivery to skeletal muscle by utilizing the TriGrid electroporation system developed by Ichor Medical Systems33 and leased to the Calos lab. The apparatus included the TSD-IM Pulse Stimulator power supply as the source of electric charge, producing a series of rectangular wave electrical pulses that induce electroporation in tissues. To deliver the electric charge to muscle tissue, we utilized the TriGrid electrode array consisting of four slender needles arranged in a diamond pattern that can be directly inserted into the muscle tissue through the skin. DNA was loaded into a syringe whose needle was placed through the center of the TriGrid array, such that the electrodes surrounded the needle (Figure 1A; see Materials and Methods). Unless otherwise indicated, electroporation immediately followed each DNA injection.
Figure 1.
DNA Delivery of GFP, Luciferase, and CAPN3 Genes to Mouse Muscle
(A) Electroporation device. Photograph of syringe attached to Ichor TriGrid for DNA delivery and electroporation. The syringe needle, which delivers the DNA, is surrounded by 4 small electrodes. (B) GFP delivery: Whole mount of GFP fluorescence seen in quadriceps muscle from C57BL/6 mouse injected with GFP plasmid, followed by electroporation. (C) Cross-section of quadriceps muscle seen in (B), showing GFP fluorescence in transfected muscle fibers. Scale bar, 100 um. (D) Comparison of delivery with and without electroporation: luciferase live imaging of Capn3 null mice that received CAPN3 plasmid DNA injected in the gastrocnemius muscle of Capn3 null mice. Left panel, left legs received electroporation, while right legs did not. Right panel, luciferase radiance values revealed that electroporation was associated with an increase of approximately 2 orders of magnitude compared to non-electroporated muscles. Each dot represented one mouse, n = 4 per group. (E) Western blot analysis of protein extracted from treated gastrocnemius muscles from the mice in (D). Upper row, levels of 100 kDa CAPN3 protein made. Lower row, 37 kDa GAPDH was used as a loading control. +, positive control, the CAPN3 plasmid injected in Capn3 null mouse from a previous experiment; −, negative control, the FST plasmid, lacking CAPN3, injected in a Capn3 null mouse. (F) Densitometry of the western blot pictured in (E). The non-electroporated samples were set to 1. Data are mean ± SEM with n = 3 and *p < 0.05. (G) Delivery of CAPN3 with or without follistatin: either CAPN3 + FST or CAPN3 alone were injected into the quadriceps muscles of Capn3 null mice. Muscles were harvested after 1 month. The western blot shows the 100 kDa band representing CAPN3 from two animals that received both plasmids and three animals that received CAPN3 alone. +, positive control, protein extract from human HEK293 cells into which the CAPN3 plasmid had been transiently transfected; −, negative control, extract from a Capn3 null mouse into which the FST plasmid alone, lacking CAPN3, had been injected. (H) Densitometry of the western blot shown in (G). There was no significant difference in CAPN3 level between the muscles treated with both plasmids or the CAPN3 plasmid alone. Data are mean ± SEM with n = 2–3. (I) Follistatin western blot. To verify expression of the 37 kDa follistatin protein in the experiment described in (G), an antibody against the HA tag present on the follistatin-344 gene was used as a probe. The HA tag was detected in all mice that received the FST plasmid. The CAPN3 plasmid alone was used as a negative control (lane 3), and a liver from a mouse that had been injected with the FST plasmid was used as a positive control (lane 4). (J) Follistatin ELISA. The ELISA for human follistatin also detects via cross-reaction a background level of mouse follistatin in untreated animals. Samples from mice that received FST either alone or in combination with CAPN3 showed FST elevation. The BL/6 control had a significant amount of FST compared to the HBSS-injected Capn3 null mouse. Data are mean ± SEM with n = 4 and *p < 0.05. (K) Luciferase live imaging of 3-month experiment. Capn3 null mice were injected in the quadriceps muscles with CAPN3 + FST or CAPN3 alone. Representative luciferase live imaging at the end of the experiment demonstrated retention of the luciferase signal throughout the 3-month time course of the experiment. (L) Western blot for calpain3 of the experiment described in (K). CAPN3 expression was retained through the 3-month time course of the experiment. (M) Western blot for follistatin of the experiment described in (K). Expression of FST in all animals that received the FST plasmid was verified. The CAPN3 plasmid alone was used as a negative control (lane 10), and a liver from a mouse that had been injected with the FST plasmid was used as a positive control (lane 9). Lane 8 in the blot is a second sample from the same animal as lane 3. (N) FST ELISA. FST levels were elevated above background only in samples that received the FST plasmid. Data are mean ± SEM with n = 10 and *p < 0.05.
For easy visualization of in vivo DNA delivery, our initial experiments utilized pmaxGFP (Lonza, Basel, Switzerland), a plasmid expressing GFP. In order to distribute the DNA over the relatively large muscle mass of the quadriceps, two doses of 50 μg pmaxGFP DNA in 50 μl Hank’s balanced salt solution (HBSS) were injected at different positions in the quadriceps muscles of C57BL/6 wild-type mice. One week after administration, the muscles were removed and examined under a fluorescence microscope. A whole-mount of a treated quadriceps muscle is shown in Figure 1B. Green fluorescence was seen in a large area of the muscle and extended for much of its length. When this muscle was sectioned (Figure 1C), numerous fluorescent fibers were seen over a large area of the muscle, corresponding to 15%–20% of the muscle fibers, suggesting that we achieved good delivery. This experiment was performed several times, comparing injected muscles with uninjected controls, allowing us to conclude that the GFP signal was due to presence of GFP in the muscle, not to autofluorescence under these conditions.
To measure the effect of electroporation on DNA delivery, we utilized luciferase live imaging and western blots to compare the level of intramuscular delivery with and without electroporation. For this experiment, we used disease model Capn3 null mice that were deficient in calpain3, as a model for LGMD2A.3 We injected the therapeutic plasmid pgWIZCAPN3gl (CAPN3) (Figure S1A), which encodes human calpain3 tagged with a 3× FLAG tag for easy detection, as well as firefly luciferase for live imaging of muscle delivery. A total of 50 μg of CAPN3 DNA in 50 μl HBSS were injected in the gastrocnemius muscle of each leg. Electroporation was administered after injection of each left leg, while right legs received no electroporation. After 1 week, the mice were imaged and radiance was measured. The intensity of the luciferase signal was more than an order of magnitude stronger in the legs that received electroporation (Figure 1D). To measure the quantity of the therapeutic CAPN3 protein that was delivered, we performed western blots on total protein extracted from these muscles, utilizing a monoclonal antibody against the FLAG tag. The FLAG tag was used because antibodies against calpain3 showed significant background. This problem with the calpain3 antibodies also prevented us from obtaining clear immunohistochemistry images for calpain3. As shown in Figure 1E, more CAPN3 protein was detected when electroporation was utilized to boost intra-muscular injection. When this difference was quantified by densitometry, we observed 2-fold more CAPN3 protein present in the muscles when electroporation was used (Figure 1F). The greater advantage of electroporation when analyzed by luciferase live imaging could be due to the presence of non-muscle cells that were transiently transfected, but not present in the muscle used for western blot, or due to some other aspect that could lead to under-reporting the signal in muscle.
Gene therapy experiments were then undertaken in the Capn3 null mice to examine delivery of therapeutic proteins over a longer time period. The quadriceps muscle was chosen as the target for study in the LGMD2A and 2B models, because these forms of muscular dystrophy develop slowly in both patients and in the corresponding mouse models. The limb muscles closer to the limb girdle are affected earlier than lower limb muscles,39 so at the ages when these mice were used, the quadriceps showed more pathology and thus were a better disease model. In the initial study, the CAPN3 plasmid was delivered, with or without a plasmid encoding human follistatin, p2attB-FST344-HA-CPL (abbreviated FST for follistatin gene) (Figure S1B). The FST plasmid carried a hemagglutinin (HA) tag fused to the 3′ end of the FST coding sequence to provide for easy detection by western blot of the FST protein produced by our plasmid, to distinguish it from the background of cross-reacting mouse FST that was also present. Two doses of either 50 μg of CAPN3 in 50 μL HBSS buffer, 50 μg of FST plasmid in 50 μL HBSS buffer, or 50 μg CAPN3 + 50 μg FST in 50 μL HBSS buffer were injected into the quadriceps muscles of groups of 5 Capn3 null mice, aged 3–4 months. Muscles were isolated after 1 month, and protein extracts were prepared to analyze the presence of therapeutic proteins. Figure 1G shows a western blot result using the anti-FLAG antibody to detect the introduced CAPN3 protein. Both groups that received CAPN3 DNA showed easily detectible levels of the therapeutic protein after 1 month. Densitometry indicated that levels of CAPN3 were similar in animals treated with CAPN3 or CAPN3 + FST (Figure 1H). To detect FST, we used two assays, western blotting and ELISA. Western blots, using an antibody against the HA tag, detected the FST protein only in samples in which the FST plasmid had been injected (Figure 1I). The ELISA detected FST above background levels in these same groups, verifying that FST was expressed in the injected muscles (Figure 1J).
We performed a 3-month study in Capn3 null mice to test whether CAPN3 levels were retained in muscle for a longer time period. Two doses of 50 μg in 50 μL of the CAPN3gl plasmid DNA, with or without the same amount of the FST plasmid were injected into groups of 7 Capn3 null mice, while a group of 7 mice received only HBSS. Luciferase live imaging, which was done at 1, 6, and 12 weeks, indicated retention of the luciferase signal above background (Figure 1K). Western blots performed on protein extracts from the injected quadriceps muscles showed strong signals indicating presence of the FLAG-tagged CAPN3 protein, indicating that the delivery method provided for long-term presence of therapeutic protein (Figure 1L). Western blot (Figure 1M) and ELISA (Figure 1N) assays confirmed that follistatin protein made by the FST plasmid was also present in the muscles.
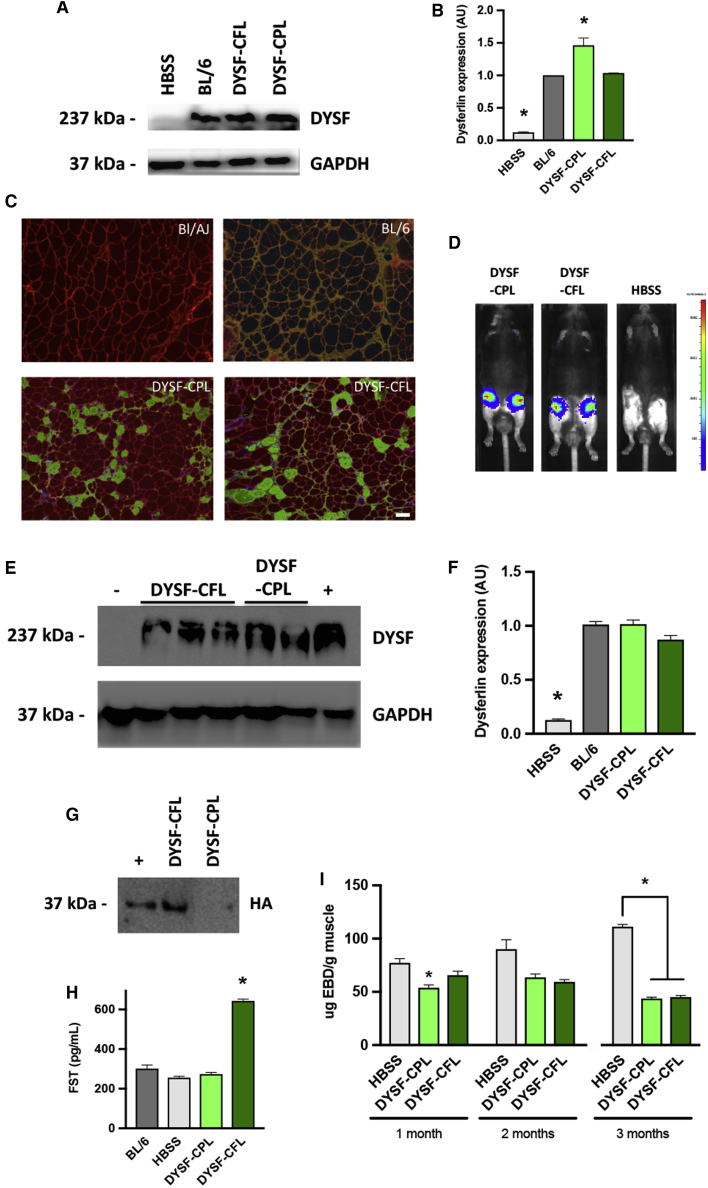
We also performed gene delivery in the BL/AJ mouse model of LGMD2B, deficient in dysferlin.10 Dysferlin deficiency leads to defective membrane repair, rendering the muscle fibers susceptible to damage, which can be measured by the level of penetration of Evans blue dye.38 The level of dye penetration would be expected to decrease if the treatment was effective in repairing the defect in a significant number of muscle fibers. Thus, in the LGMD2B studies, we examined treated muscles for evidence of reduced permeability to Evans blue dye as a measure of histological improvement. Fifteen male BL/AJ mice that were ∼9 months of age were divided into three groups. Five mice were designated negative controls and received HBSS. Five mice received the pKLD-CAG-DSYF-CPL (DYSF-CPL; Figure S1C) plasmid encoding human dysferlin, and five mice received pKLD-CAG-DYSF-CFL (DYSF-CFL; Figure S1D) encoding both dysferlin and follistatin, injected in the quadriceps. In an attempt to maximize DNA delivery, we carried out two injection sessions, with an interval of 3–4 days between sessions. During each session, the quadriceps of each leg received 2 injections of 50 μg of the relevant plasmid in 50 μL HBSS buffer per session, for a total of 200 μg DNA per leg. Three of the mice from each group were harvested at 1 month, and the remaining two were harvested at 2 months.
Figure 2A shows a western blot for dysferlin for representative muscle samples harvested after 1 month. The BL/AJ mouse contained negligible dysferlin, as expected, while the wild-type C57BL/6-positive control illustrated the normal level of dysferlin found in the muscle. Both of the therapeutic plasmids, DYSF-CPL and DYSF-CFL, produced significant levels of dysferlin. Densitometry on blots of all the treated legs (Figure 2B) indicated that the level of dysferlin was similar to the wild-type level for the DYSF-CFL plasmid and was higher than normal for the DYSF-CPL plasmid, perhaps due to better dysferlin expression from that plasmid. Immunohistochemistry was performed to examine the distribution of dysferlin in the muscle fibers. Representative sections for muscles injected with the DYSF-CPL or DYSF-CFL plasmids are shown (Figure 2C), compared to wild-type and BL/AJ controls. By counting the numbers of positive fibers in these and similar sections, we found that the average percent of dysferlin-positive fibers was approximately 15% (Figure S2A). We note that electroporation may cause extra inflammation in the muscle tissue.30,31 While we did not observe inflammation here, any inflammation may have resolved by the time the histology was performed. Near wild-type levels of dysferlin were seen in the western blot, while immunohistochemistry revealed only 15% of the fibers positive. This discrepancy may mean that dysferlin was overproduced in the subset of fibers that stained strongly positive. It is also possible that additional fibers are positive for dysferlin at a lower level that is not detectible next to the more strongly positive subset of fibers seen in the images. We note that when dysferlin is introduced by gene therapy in dysferlin null animals, the entire fiber typically is stained, while in the wild-type situation, dysferlin staining is more prominent near the membrane. This feature might be another indication of overproduction and has also been seen in AAV gene therapy studies.10,14,15 The Evans blue dye assay was performed on muscles harvested at 1 and 2 months, with the result that a downward trend was observed that was statistically significant for CPL at 1 month (included in Figure 2I). The 2-month sample had fewer animals, so statistical significance was not reached.
Figure 2.
Gene Therapy in Dysferlin-Deficient BL/AJ Mice
(A) Western blot from quadriceps muscles of BL/AJ mice injected with plasmids DYSF-CPL and DYSF-CFL, encoding dysferlin with and without follistatin. After one month, muscles were harvested from representative quadriceps receiving DYSF-CPL or DYSF-CFL, as well as samples from untreated BL/AJ (HBSS) and wild-type C57BL/6 controls. (B) Densitometry on the dysferlin bands from western blots of the experiment described in (A). The value for the wild-type C57BL/6 was set at 1.0. Untreated BL/AJ animals (HBSS) had significantly less dysferlin than wild-type, while animals injected with DYSF-CFL had DYSF levels that were similar to those observed in C57BL/6 wild-type mice. Mice injected with DYSF-CPL showed a significantly increased amount of DYSF compared to wild-type. Data are mean ± SEM with n = 3 and *p < 0.05. (C) Immunohistochemistry on cross-sections of muscles treated with DYSF-CPL or DYSF-CFL. We stained for dysferlin (green) to identify protein delivered by gene therapy and laminin (red) to identify all muscle fibers. Representative sections from untreated BL/AJ and wild-type C57BL/6 are shown for comparison. In wild-type muscle, dysferlin co-locates with laminin, whereas in gene therapy, dysferlin appears throughout the fiber. The color in the BL/6 sample is the result of the merge of red laminin and green dysferlin staining. Scale bar, 50 um. (D) Luciferase imaging of a 3-month gene therapy experiment performed in BL/AJ mice, using a lower DNA dose. Mice were injected in the quadriceps with DYSF-CPL, DYSF-CFL, or HBSS alone and were imaged 1 month after injection to monitor expression of the luciferase gene present on the therapeutic plasmids. Representative mice from each of the groups are shown, showing retention of the luciferase signal in animals receiving either of the dysferlin plasmids. (E) Western blot of quadriceps muscles isolated 3 months after treatment. The gel was loaded with protein from untreated BL/AJ, mice injected with DYSF-CPL or DYSF-CFL plasmid, and wild-type C57BL/6, and stained with an antibody against dysferlin. GAPDH was used as a loading control. (F) Densitometry analysis of western blot from the 3-month experiment. The wild-type value was set to 1. Mice treated with either DYSF-CPL or DYSF-CFL showed significantly increased amount of DYSF compared to BL/AJ animals (HBSS), and DYSF levels were similar to those observed in BL/6 mice. Data are mean ± SEM with n = 3 and *p < 0.05. (G) Western blot of follistatin expression in the treated muscles. An antibody against the HA-tag on the follistatin gene on the FST plasmid was used. Representative lanes shown here indicate that muscles injected with DYSF-CPL lack expression, while muscles injected with DYSF-CFL showed a correctly sized band. Protein extracted from livers that had been injected with FST (lane 1) served as a positive control. (H) Follistatin production in the treated muscles monitored by ELISA against human follistatin. Only the protein extracted from muscles injected with DYSF-CFL displayed follistatin levels elevated significantly over background. Data are mean ± SEM with n = 4 and *p < 0.05. (I) Evans blue dye (EBD) assays performed on quadriceps muscles from the 1- and 2-month experiments. DYSF-CPL had a significantly depressed value at 1 month, but not at 2 months, probably due to the lower number of animals assayed at 2 months, which reduced statistical power. In the 3-month experiment, the EBD values were significantly lower for the muscles that received gene therapy with DYSF-CPL and DYSF-CFL. Data are mean ± SEM with n = 6 for the 1-month and 3-month experiments and n = 2 for the 1 month. *p < 0.05.
We performed a 3-month study in order to evaluate the gene therapy over a longer time period. For this experiment, 18 male BL/AJ mice were utilized that were 7 months old at the start of the experiment. The mice were divided into three groups of six, representing untreated, DYSF-CPL, and DYSF-CFL. Because the dysferlin levels for CPL had exceeded wild-type levels in the previous experiment, the DNA dose was reduced in this experiment. A pilot experiment had indicated that there was no benefit from performing two injections a few days apart, perhaps because the DNA dose was already saturated at that time (data not shown). Therefore, only one injection session was performed, consisting of two 50-μl injections of 25 μg DNA each into the quadriceps muscle, for a total of 50 μg DNA per quadriceps muscle, followed by electroporation. Luciferase live imaging was performed periodically to monitor gene delivery. Mice at the 1-month time point of the experiment are shown in Figure 2D.
At the end of three months, the quadriceps muscles were isolated. Some muscles were used for histology, while protein was extracted from others. Western blots were performed on the protein extracts (Figure 2E). Densitometric analysis of the blots revealed that dysferlin levels were robust and not significantly different from normal levels (Figure 2F). The DNA dose had been reduced 4-fold from the previous experiment, with less than 2-fold effect on final protein levels, suggesting that the previous dose was excessive and had reached saturation. The protein extracts were also analyzed by western blot using an antibody to the HA-tag fused to follistatin, revealing that the tagged follistatin was present only in muscles that had been injected with DYSF-CFL, as expected (Figure 2G). An ELISA for follistatin on the same samples indicated that samples that received DYSF-CFL had higher values, as expected (Figure 2H). The Evans blue dye assay revealed that the DYSF-CPL and DYSF-CFL plasmids were both associated with significantly decreased levels of dye penetration at 3 months, consistent with a beneficial change in the muscle as a result of the gene therapy treatment (Figure 2I). We performed H&E staining on some of the muscles in order to examine the levels of centronucleation present. Figure S2B shows representative images of muscles treated with DYSF-CPL or DYSF-CFL, in addition to C57BL/6 and BL/AJ controls. When overall levels of centronucleation were evaluated between the BL/AJ, DYSF-CPL, and DYSF-CFL sections, similar values were seen, with an average centronucleation frequency of ∼60% (Figure S2C). However, when the levels of centronucleation were evaluated only in fibers that were positive for dysferlin, a downward trend to 47% was evident (Figure S2D). This result suggested that the number of overall fibers corrected was too low to cause a measurable shift in centronucleation at this time point. However, when we focused on just the fibers that were corrected, i.e., those that now expressed dysferlin, a decrease in centronucleation was observed.
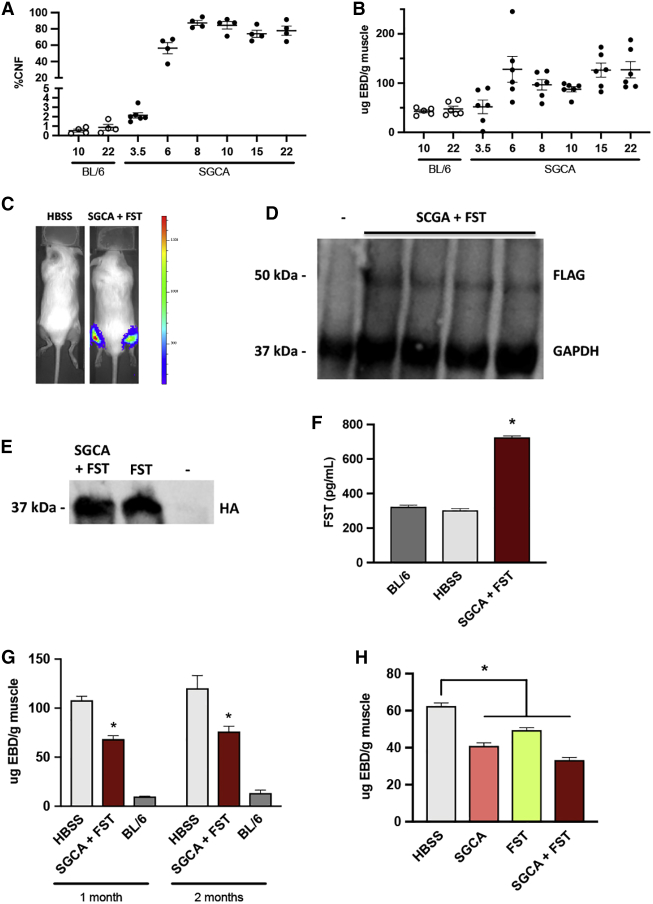
We also pursued gene therapy experiments in the Sgca null mouse model of LGMD2D. This model has advantages for analysis due to faster disease progression compared to LGMD2A and 2B. We had previously crossed an Sgca null mouse model9 with immune-deficient NRG mice40 to generate immune-deficient Sgca/NRG mice for transplantation studies of human stem cells.41 However, in the current study, an immune competent background was preferred, so we backcrossed the Sgca/NRG strain with wild-type C57BL/6 mice. We studied the centronucleation and Evans blue dye phenotype of these mice over a time course of several months to characterize the muscle degeneration present in these mice. As shown in Figure 3A, centronucleation developed quickly in the mice, reaching maximal values of ∼80% by 8 weeks of age. Likewise, the Evans blue dye values in the mice rose quickly to reach a maximum in the first 6 weeks of life (Figure 3B).
Figure 3.
Gene Therapy in Sgca null Mice
(A) Time course of development of centronucleation in gastrocnemius muscles of untreated Sgca null mice. CNF, centronucleation frequency. Each dot represents one mouse, and n = 4–6 per group. (B) Time course of development of Evans blue dye permeability in gastrocnemius muscles of untreated Sgca null mice. Each dot represents one mouse, and n = 6 per group. (C) Luciferase live imaging of representative animals in Sgca null mice. Image was taken 1 month after injection in gastrocnemius muscles of HBSS alone or SGCA + FST plasmids. (D) Western blot of same experiment described in (C). An antibody against the FLAG tag fused to the SGCA gene on the plasmid was used to detect expression of human SGCA. Protein extracted from C57BL/6 mice was used as the negative control for FLAG tag (lane 1). GAPDH was used as a loading control. (E) Western blot of follistatin delivery, from same experiment described in (C). Representative animals received the SGCA and FST plasmids, FST alone positive control (lane 2), or SGCA (negative control, lane 3). (F) ELISA of follistatin delivery, from same experiment described in (C). C57BL/6 and HBSS represent negative controls, while lane 3 animals were treated with the SGCA + FST plasmids. Data are mean ± SEM with n = 12 and *p < 0.05. (G) Evans blue dye analysis of 1- and 2-month studies of SGCA+FST. In both studies, muscles that received the SGCA and FST plasmids had significantly less penetration of Evans blue dye in muscle fibers, compared to HBSS-treated Sgca null mice. Data are mean ± SEM with n = 4–9 for the 1-month and n = 4–12 for the 2-month experiment. *p < 0.05. (H) Evans blue dye analysis of 1-month study in Sgca null mice injected in gastrocnemius muscles with plasmids encoding either SGCA, FST, or SGCA + FST, versus untreated mice. All plasmid-injected mice had a significant drop in EBD 1 month after injection. Data are mean ± SEM, with n = 4 and *p < 0.05.
Using the pgWIZ-SGCAgl plasmid (abbreviated SGCA for alpha-sarcoglycan gene; Figure S1D) encoding a FLAG-tagged version of human SGCA, we performed gene therapy experiments in Sgca null mice, typically using mice 6 weeks to 3 months old. The gastrocnemius muscle was chosen as the target in the LGMD2D model, because in both patients and the corresponding mouse model, LGMD2D develops faster than in LGMD2A and 2B, and at young ages, the gastrocnemius muscle of Sgca null mice already shows profound pathology, as indicated in time course studies (Figures 3A and 3B). Plasmid DNA was injected in the gastrocnemius muscles, using a single dose in 50 μL buffer of either 50 μg SGCA, 50 μg FST, or 50 μg SGCA + 50 μg FST.
Several independent experiments were performed, with lengths of 1 or 2 months. Sets of Sgca null mice ∼6 weeks old were injected with 50 μg SGCA+ 50 μg FST in 50 μL HBSS. Similarly aged C57BL/6 mice served as controls, along with Sgca null mice injected with HBSS buffer alone. Figure 3C shows representative luciferase images of these mice at 1 month after injection. A western blot against the FLAG tag attached to SGCA on the plasmid verified expression of the SGCA gene (Figure 3D). We attempted to perform immunohistochemistry for SGCA on treated muscles, but were unable to obtain high quality images, perhaps due to the extensive degeneration present in the muscles of the Sgca null animals. A western blot for the HA tag fused to follistatin in the FST plasmid documented expression of follistatin in the injected muscles (Figure 3E), as did an ELISA for follistatin in these same samples (Figure 3F). The muscle extracts were subjected to the Evans blue dye assay, which showed that muscles that received the SGCA and FST plasmids absorbed significantly less dye than HBSS-treated Sgca null muscles at 1 month (Figure 3G). In a parallel experiment that was run for 2 months, a similar, statistically significant depression in the Evans blue dye values was seen (Figure 3G). In order to assess the effects of SGCA and FST individually, SGCA and FST plasmids alone, along with the SGCA + FST combination, were injected into Sgca null mice, and the Evans blue dye assay was performed 1 month after injection. As shown in Figure 3H, all of these groups showed a statistically significant drop in Evans blue values, suggesting that the individual SGCA and FST plasmids played a role. The therapeutic effect of the SGCA and FST plasmids was detectible even when electroporation was not used to enhance delivery (Figure S3A). Because elevation in the percentage of fibers showing centronucleation in the Sgca null mouse occurs so quickly, we did not necessarily expect to be able to intervene in this phenotype in short-term experiments. However, by injecting mice at a young age of 4 weeks with the SGCA and FST plasmids and waiting 1 month, we detected a slight downward trend in centronucleation values (Figure S3B). It is possible that this trend would become significant over longer time periods. This experiment also demonstrated that it was feasible to treat 4-week-old animals with this method, and we note that it is generally advantageous to treat earlier, in order to maximize benefit.
Discussion
This study demonstrates that intramuscular injection of plasmid DNA, followed by a brief pulse of electroporation, leads to robust and sustained expression of introduced genes in muscle fibers that can be of value in gene therapy. For example, levels of dysferlin similar to normal values were observed in the mouse quadriceps muscle (Figures 2B and 2F). Expression was documented for up to 3 months in these studies. However, it is likely that expression would continue for much longer and has been documented up to the lifetime of the mouse.27,42 Immunohistochemistry studies of dysferlin delivery here indicated that approximately 15% of fibers were expressing dysferlin after one treatment (Figure S2A). At these levels of expression, we were able to observe statistically significant improvements in muscle histology, as judged by decreased permeability to Evans blue dye, in mouse models of LGMD2B and 2D, detectible by 1 month after treatment (Figures 2I, 3G, and 3H). These results suggest that plasmid delivery to muscle has the potential to produce meaningful, long-term expression of therapeutic genes. In future studies, especially if it were possible to increase the number of corrected fibers, such as with multiple doses of DNA spaced over time, it would be desirable to explore potential benefits more comprehensively, including other measures of muscle histopathology, such as myofiber size, fibrosis, and inflammatory response.
This delivery method, based on DNA injection immediately followed by electroporation of the surrounding area, is being clinically tested for delivery of DNA vaccines.33 In that case, a single injection suffices to stimulate the desired antigenic response. However, for gene therapy, delivery to a more extensive area of muscle mass is needed for distribution of non-secreted therapeutic proteins. A limitation of the electroporation approach is that it is only suitable for local gene delivery and is not currently suitable for systemic delivery. More widespread delivery may be achievable by carrying out multiple injections at various depths. Such a process might be feasible, using regional, deep anesthesia. The next step would be studies in larger animals, such as primates, to develop scale-up strategies. Another viable alternative for more widespread delivery is to make use of higher-volume vascular delivery of DNA, as has been demonstrated in rodent and primate studies.24, 25, 26, 27, 28
Clinical trials have been carried out17 and are underway21 using AAV in local and systemic delivery for muscular dystrophy. We do not yet know whether AAV will provide sufficient delivery to the muscle mass or whether such delivery will be adequate long-term. One strategy would be to focus on use of AAV for coverage of tissues difficult to reach by non-viral methods, such as heart and diaphragm. Conceivably, non-viral delivery could be used to boost gene expression in skeletal muscles, such as limb muscles, if initial delivery was inadequate and/or if expression decreases over time, especially during growth of pediatric patients. Non-viral delivery may also be useful in those patients who possess pre-existing immunity to AAV.
In addition to delivery of therapeutic genes relevant for LGMD2A, 2B, and 2D, we also experimented with simultaneous delivery of the gene expressing human follistatin. We had previously shown a benefit to vascular delivery of dysferlin and follistatin in the hamstring muscle in a mouse model of LGMD2B.28 In the present study in the quadriceps muscle using electroporation, we did not detect an additional benefit when follistatin was present, as opposed to dysferlin alone (Figure 2I). We did not test follistatin alone, because it has been shown to detrimental for LGMD2B.43 In contrast, we did detect a benefit for follistatin for LGMD2D, particularly in combination with the therapeutic alpha-sarcoglycan gene (Figures 3G and 3H). Previously, a benefit of a myostatin inhibitor delivered with AAV was not detectable in Sgca null mice, probably because of the rapid turnover of fibers in this model.44 Our results highlight the advantage of co-delivering SGCA and FST to stabilize transfected fibers. It is likely that combination of CAPN3 and FST would be beneficial in LGMD2A also, because a myostatin inhibitor alone showed benefit in a mouse model of this disorder.44
In addition to introduction of therapeutic genes that remain extrachromosomal as plasmid DNA, non-viral delivery can also be used to introduce gene-editing agents that can carry out permanent additions or corrections to the genome.45 For example, recombinases such as phiC31 integrase have been shown to be effective in bringing about sequence-specific integration in the muscles of 10-day-old mice in a mouse model of Duchenne muscular dystrophy after intramuscular injection and electroporation.42 Use of CRISPR/Cas gene editing systems are currently under intense investigation to correct mutations in the dystrophin gene in muscle cells using AAV for delivery46 and have also been successfully introduced in a mouse model by intramuscular injection and electroporation.47 Because non-viral delivery is relatively non-invasive and lower in cost versus viral delivery, it may also be of interest for delivery of other genes. In situations where the gene product is secreted, as in the case of follistatin and various growth factors, the lower efficiency of this delivery method relative to AAV-mediated delivery may be acceptable and may be a good match for various therapies outside the realm of muscular dystrophy.
Materials and Methods
Plasmids and DNA
The therapeutic plasmids carrying the dysferlin coding sequence, pKLD-CAG-DYSF-CPL and pKLD-CAG-DYSF-CFL, were constructed as described.27,47 Briefly, the plasmid contains the human dysferlin cDNA cloned into the pKLD-CAG backbone, along with a gene expression cassette encoding mCherry, puromycin resistance, and firefly luciferase connected by p2A skipping peptides. To construct a derivative that also encoded follistatin, we replaced the puromycin resistance gene in pKLD-CAG-DYSF-CPL with the coding sequence for the 344-amino acid isoform of human follistatin.28 p2attB-FST344-HA-CPL was constructed by utilizing backbone and functional elements described previously. Briefly, the human FST344-HA cDNA was created by amplifying the FST sequence from plasmid pKLD-CAG-DYSF-CFL28 using primers KpnI-FST317-F: 5′-GAGGAGGGTACCATGGTCCGCGCGAGGC-3′ and FST344-CtHA-MluI-R 5′-CTCCTCACGCGTAGCGTAGTCTGGGACGTCGTATGGGTACCACTCTAGAATAGAAGATA-3′, while also adding the HA-tag through the primer sequence. KpnI and MluI restriction enzyme sites are underlined in the primer sequences, and the HA-tag sequence is italicized in the FST344-CtHA-MluI-R primer sequence. The PCR product of 1,077 bp was digested with restriction enzymes KpnI and MluI and cloned in place of DYSF in plasmid pKLD-CAG-DYSF-CPL. A different plasmid backbone, gWIZgfp (Aldevron, Fargo, ND, USA) was used to express alpha-sarcoglycan and calpain3. To make pgWIZ-SGCAgl, we added the firefly luciferase gene and preceding p2A sequence from pKLD-CAG-DYSF-CPL downstream of the Gfp gene on gWIZgfp, generating pgWIZgl. The human SGCA gene from pKLD-CAG-SGCA was added downstream of the CMV intron, including a gBlock (Integrated DNA Technologies, Coralville, IA, USA) encoding a 3′ 3×FLAG tag and P2A sequence. To make pgWIZ-CAPN3gl, we ligated 2 gBlock sequences (IDT) encoding human calpain3, including a 3′ 3×FLAG tag and P2A sequence into pgWIZgl downstream of the CMV intron. Plasmid DNA used for animal injections was purified with an endotoxin-free maxi- or giga-prep kit (Macherey-Nagel, Düren, Germany), following the manufacturer’s instructions. These plasmids will be made available to researchers through Addgene.
Mice
Calpain3 null mice3 were obtained from the laboratory of Melissa Spencer at UCLA. We utilized dysferlin null A/J mice that had been crossed onto the C57BL/6 background to make the BL/AJ strain.10 BL/AJ mice (B6.A-Dysfprmd/GeneJ, stock number 012767) were provided by the Jackson Laboratory (Bar Harbor, ME, USA) from a colony maintained by the Jain Foundation. The Sgca null mice were derived from our Sgca/NRG strain41 (Jackson Laboratory Stock Number 030443, NOD.Cg-Rag1tm1MomSgcatm1KcamIl2rγtm1Wjl/McalJ) by repeated crossing with C57BL/6 to remove the Rag1 and Il2rg immune-deficient markers, while retaining the homozygous Sgca null allele, as monitored by PCR. C57BL/6 wild-type control mice (labeled as BL/6 in the figures) were purchased from Jackson Laboratory. The Stanford Administrative Panel on Laboratory Animal Care approved all procedures performed on animals. The Stanford Comparative Medicine program is accredited by the Assessment of Laboratory Animal Care International.
Electroporation
Plasmid DNA was injected into mouse hind limb muscles using intramuscular injection with a needle attached to the Ichor TriGrid electrode array (Ichor Medical Systems, San Diego, CA, USA). Briefly, mice were anesthetized with isoflurane, fur was removed, and 25 or 50 μg of plasmid DNA in 50 μl Hank’s balanced salt solution (HBSS; ThermoFisher Scientific, Waltham, MA, USA) or an equal volume of HBSS buffer alone were injected through the skin into the quadriceps or gastrocnemius muscle using a 0.3 mL syringe (Becton Dickinson, Franklin Lakes, NJ, USA) with attached ½” 30G needle (Becton Dickinson Ultra-Fine 328431), inserted through the rodent TriGrid electrode array (Ichor). After injection, the foot pedal on the charge unit was depressed to create a brief electric field surrounding the injection site. Electroporation used the TriGrid Delivery System (TDS-IM) device (Ichor). Electrical stimulation was applied at an amplitude of 250 V/cm of electrode spacing; the total duration of electrical stimulation was 40 ms applied over a 400 ms interval. After electroporation, the needle was withdrawn and mice were returned to their cages for recovery. 100 μl Carprolect (Carprofen, Henry Schein Animal Health, Melville, NY, USA) analgesia per 10 g body weight was administered for pain relief.
Luciferase Live Imaging
Mice to be imaged were anesthetized with 1.5% isoflurane through a nose cone. Luciferin substrate (1 mg/mL PBS; Biosynth, Itasca, IL, USA) was injected into the intraperitoneal cavity (100 μL/10 g body weight). 10 min after luciferin injection, luminescence was detected using an in vivo imaging system (IVIS Spectrum Pre-Clinical In Vivo Imaging System, PerkinElmer, Waltham, MA, USA) and associated software for quantitation. Luminescence images were acquired at exposure times of 60 s or less. Luminescence was quantified within regions of interest in units of photons/s per cm2.
Western Blots
Quadriceps or gastrocnemius muscles were dissected from mouse hind limbs. Muscle lysates were prepared with radioimmunoprecipitation assay (RIPA) Buffer supplemented with HALT Protease Inhibitor Cocktail (ThermoFisher Scientific) according to the manufacturer’s protocol. The supernatant containing protein extract was denatured with Laemmli Sample Buffer (Bio-Rad, Hercules, CA, USA) supplemented with 100 mM dithiothreitol (DTT). In each lane of a 10% TRIS-glycine SDS-PAGE gel (Bio-Rad), 50 μg protein extract was electrophoresed at 120 V in ice cold PAGE running buffer (0.1% SDS, 25 mM Tris, 250 mM glycine). Samples were transferred onto 0.45 μm PVDF membrane (ThermoFisher Scientific) for 60 minutes at 200 mA at 4°C. Membranes were blocked in 3% milk diluted in TBS with 0.2% Tween-20. Detection of calpain3 and alpha-sarcoglycan relied on the rabbit anti-FLAG monoclonal antibody against the 3×FLAG tag used at a 1:1,000 dilution (Sigma Aldrich, St. Louis, MO, USA). Dysferlin detection was achieved with a 1:1,000 dilution of rabbit-anti-dysferlin (Abcam, Cambridge, UK). GAPDH was probed with a 1:10,000 dilution of rabbit-anti-GAPDH (Abcam) in blocking solution. Rabbit antibodies were probed with a 1:5,000 dilution of goat-anti-rabbit immunoglobulin G (IgG) horseradish peroxidase (HRP) secondary antibody (ThermoFisher Scientific). Blots were developed in Clarity Western ECL Substrate according to manufacturer’s protocol (Bio-Rad) and imaged using the ChemiDoc Touch Imaging System (Bio-Rad). In densitometry of the western blots, the values for the target protein were always normalized to the value for the corresponding GAPDH band in the same lane.
Histology
For immunohistochemistry, muscles were harvested and snap frozen in OCT (Satura Finetek, Torrance, CA, USA) in liquid nitrogen. Serial 12 μm cryostat sections were obtained throughout the muscles. Sections were fixed in 4% PFA for 10 min and washed 3 times with PBS for 1–2 min. Sections were then blocked in 10% goat serum in PBS for 1 h. Primary antibodies were prepared in block solution and incubated overnight at 4°C in a humid chamber. The next day, slides were washed 3 times with PBS and stained with secondary antibody for 60 minutes at room temperature. Slides were washed with PBS 3 times and visualized. Dysferlin was detected with the rabbit NCL-Hamlet antibody (1:500, Leica Biosystems, Buffalo Grove, IL, USA), and laminin was detected with a rat anti-laminin antibody (1:1,000, Sigma Aldrich). Goat anti-rabbit conjugated to Alexa 488 (1:1,000, Life Technologies, Carlsbad, CA, USA) and anti-rat conjugated to Alexa 594 (1:1,000, Life Technologies) were used as secondary antibodies.
For H&E staining, quadriceps or gastrocnemius muscles were isolated and flash frozen in OCT mounting media. Cryosections 12 μm-thick were stained using H&E (Sigma Aldrich) following the manufacturer’s specifications. All pictures were taken using a Zeiss Aptiskop microscope. The percentage of centronucleated fibers (CNFs) were counted in the sections. At least 2–3 photos of the muscle were taken per section.
ELISA for Follistatin
Follistatin levels were measured using a follistatin ELISA kit (R&D Systems, London, UK) following instructions provided by the manufacturer. A standard curve was constructed using provided follistatin solution. Protein extracts from each muscle sample were diluted 1:10 in Calibrator Diluent RD5-21. Both standards and samples were pipetted into wells pre-coated with immobilized antibody, followed by incubation with a follistatin-specific enzyme-linked monoclonal antibody. Substrate solution was then added to each well, with color development proportional to amount of follistatin bound. Plates were read with a Tecan Infinite 200Pro microplate reader (Tecan Group, Männedorf, Switzerland) at 450 nm with its reference at 570 nm.
Evans Blue Dye Procedure
To evaluate the membrane permeability of muscle, mice were intraperitoneally injected with Evans Blue dye (EBD; Sigma; 100 μL of 1% EBD in PBS per 10 g body weight). The following day, muscles were flash-frozen and ground with a mortar and pestle. Following an overnight incubation in formamide at room temperature and under shaking, EBD was quantified in the supernatant by measuring light emission at 630 nm.38
Statistical Analysis
To determine statistical significance for two groups, we made comparisons using a Student’s t test; for more than 2 groups, the ANOVA test was used. Statistical analyses were performed using GraphPad Prism v.8 (GraphPad Software, La Jolla, CA, USA). A p value < 0.05 was considered significant.
Author Contributions
T.K.G. and C.P. designed and performed experiments, interpreted results, and contributed to preparing the manuscript; M.P.C. designed experiments, interpreted results, obtained funding, and drafted the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Aaron Xuan Yue and Jon-Michael Knapp for initial work on the delivery method in the Calos lab, Mital Bhakta and Jon-Michael Knapp for help with plasmid constructions, and Aaron Xuan Yue for immunohistochemistry data. We thank Claire Evans of Ichor Medical Systems for helpful discussions. This work was supported by grants to M.P.C. from the Muscular Dystrophy Association, the Jain Foundation, the Coalition to Cure Calpain 3, the James Kanagy Fund, and the Stanford Discovery Innovation Fund.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.10.002.
Supplemental Information
References
- 1.Mercuri E., Muntoni F. Muscular dystrophies. Lancet. 2013;381:845–860. doi: 10.1016/S0140-6736(12)61897-2. [DOI] [PubMed] [Google Scholar]
- 2.Guiraud S., Aartsma-Rus A., Vieira N.M., Davies K.E., van Ommen G.J., Kunkel L.M. The pathogenesis and therapy of muscular dystrophies. Annu. Rev. Genomics Hum. Genet. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 3.Kramerova I., Kudryashova E., Tidball J.G., Spencer M.J. Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet. 2004;13:1373–1388. doi: 10.1093/hmg/ddh153. [DOI] [PubMed] [Google Scholar]
- 4.Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 5.Liu J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J.A., Hentati F., Hamida M.B. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 1998;20:31–36. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- 6.Bashir R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- 7.Bansal D., Miyake K., Vogel S.S., Groh S., Chen C.C., Williamson R., McNeil P.L., Campbell K.P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 8.Ho M., Post C.M., Donahue L.R., Lidov H.G.W., Bronson R.T., Goolsby H., Watkins S.C., Cox G.A., Brown R.H., Jr. Disruption of muscle membrane and phenotype divergence in two novel mouse models of dysferlin deficiency. Hum. Mol. Genet. 2004;13:1999–2010. doi: 10.1093/hmg/ddh212. [DOI] [PubMed] [Google Scholar]
- 9.Duclos F., Straub V., Moore S.A., Venzke D.P., Hrstka R.F., Crosbie R.H., Durbeej M., Lebakken C.S., Ettinger A.J., van der Meulen J. Progressive muscular dystrophy in α-sarcoglycan-deficient mice. J. Cell Biol. 1998;142:1461–1471. doi: 10.1083/jcb.142.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lostal W., Bartoli M., Bourg N., Roudaut C., Bentaïb A., Miyake K., Guerchet N., Fougerousse F., McNeil P., Richard I. Efficient recovery of dysferlin deficiency by dual adeno-associated vector-mediated gene transfer. Hum. Mol. Genet. 2010;19:1897–1907. doi: 10.1093/hmg/ddq065. [DOI] [PubMed] [Google Scholar]
- 11.Gregorevic P., Allen J.M., Minami E., Blankinship M.J., Haraguchi M., Meuse L., Finn E., Adams M.E., Froehner S.C., Murry C.E., Chamberlain J.S. rAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic mice. Nat. Med. 2006;12:787–789. doi: 10.1038/nm1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartoli M., Roudaut C., Martin S., Fougerousse F., Suel L., Poupiot J., Gicquel E., Noulet F., Danos O., Richard I. Safety and efficacy of AAV-mediated calpain 3 gene transfer in a mouse model of limb-girdle muscular dystrophy type 2A. Mol. Ther. 2006;13:250–259. doi: 10.1016/j.ymthe.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 13.Roudaut C., Le Roy F., Suel L., Poupiot J., Charton K., Bartoli M., Richard I. Restriction of calpain3 expression to the skeletal muscle prevents cardiac toxicity and corrects pathology in a murine model of limb-girdle muscular dystrophy. Circulation. 2013;128:1094–1104. doi: 10.1161/CIRCULATIONAHA.113.001340. [DOI] [PubMed] [Google Scholar]
- 14.Sondergaard P.C., Griffin D.A., Pozsgai E.R., Johnson R.W., Grose W.E., Heller K.N., Shontz K.M., Montgomery C.L., Liu J., Clark K.R. AAV.dysferlin overlap vectors restore function in dysferlinopathy animal models. Ann. Clin. Transl. Neurol. 2015;2:256–270. doi: 10.1002/acn3.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llanga T., Nagy N., Conatser L., Dial C., Sutton R.B., Hirsch M.L. Structure-based designed nano-dysferlin significantly improves dysferlinopathy in BLA/J mice. Mol. Ther. 2017;25:2150–2162. doi: 10.1016/j.ymthe.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fougerousse F., Bartoli M., Poupiot J., Arandel L., Durand M., Guerchet N., Gicquel E., Danos O., Richard I. Phenotypic correction of α-sarcoglycan deficiency by intra-arterial injection of a muscle-specific serotype 1 rAAV vector. Mol. Ther. 2007;15:53–61. doi: 10.1038/sj.mt.6300022. [DOI] [PubMed] [Google Scholar]
- 17.Mendell J.R., Rodino-Klapac L.R., Rosales-Quintero X., Kota J., Coley B.D., Galloway G., Craenen J.M., Lewis S., Malik V., Shilling C. Limb-girdle muscular dystrophy type 2D gene therapy restores α-sarcoglycan and associated proteins. Ann. Neurol. 2009;66:290–297. doi: 10.1002/ana.21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 19.Louis Jeune V., Joergensen J.A., Hajjar R.J., Weber T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum. Gene Ther. Methods. 2013;24:59–67. doi: 10.1089/hgtb.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calcedo R., Chichester J.A., Wilson J.M. Assessment of humoral, innate, and T-cell immune responses to adeno-associated virus vectors. Hum. Gene Ther. Methods. 2018;29:86–95. doi: 10.1089/hgtb.2018.038. [DOI] [PubMed] [Google Scholar]
- 21.Duan D. Systemic AAV micro-dystrophin gene therapy for Duchenne muscular dystrophy. Mol. Ther. 2018;26:2337–2356. doi: 10.1016/j.ymthe.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 23.Hagstrom J.E., Hegge J., Zhang G., Noble M., Budker V., Lewis D.L., Herweijer H., Wolff J.A. A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs. Mol. Ther. 2004;10:386–398. doi: 10.1016/j.ymthe.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Zhang G., Wooddell C.I., Hegge J.O., Griffin J.B., Huss T., Braun S., Wolff J.A. Functional efficacy of dystrophin expression from plasmids delivered to mdx mice by hydrodynamic limb vein injection. Hum. Gene Ther. 2010;21:221–237. doi: 10.1089/hum.2009.133. [DOI] [PubMed] [Google Scholar]
- 25.Hegge J.O., Wooddell C.I., Zhang G., Hagstrom J.E., Braun S., Huss T., Sebestyén M.G., Emborg M.E., Wolff J.A. Evaluation of hydrodynamic limb vein injections in nonhuman primates. Hum. Gene Ther. 2010;21:829–842. doi: 10.1089/hum.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wooddell C.I., Hegge J.O., Zhang G., Sebestyén M.G., Noble M., Griffin J.B., Pfannes L.V., Herweijer H., Hagstrom J.E., Braun S. Dose response in rodents and nonhuman primates after hydrodynamic limb vein delivery of naked plasmid DNA. Hum. Gene Ther. 2011;22:889–903. doi: 10.1089/hum.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma J., Pichavant C., du Bois H., Bhakta M., Calos M.P. DNA-mediated gene therapy in a mouse model of limb girdle muscular dystrophy 2B. Mol. Ther. Methods Clin. Dev. 2017;7:123–131. doi: 10.1016/j.omtm.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan Z., Kocis K., Valley R., Howard J.F., Chopra M., An H., Lin W., Muenzer J., Powers W., Powers W. Safety and feasibility of high-pressure transvenous limb perfusion with 0.9% saline in human muscular dystrophy. Mol. Ther. 2012;20:456–461. doi: 10.1038/mt.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan Z., Kocis K., Valley R., Howard J.F., Jr., Chopra M., Chen Y., An H., Lin W., Muenzer J., Powers W. High-pressure transvenous perfusion of the upper extremity in human muscular dystrophy: A safety study with 0.9% saline. Hum. Gene Ther. 2015;26:614–621. doi: 10.1089/hum.2015.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mir L.M., Bureau M.F., Rangara R., Schwartz B., Scherman D. Long-term, high level in vivo gene expression after electric pulse-mediated gene transfer into skeletal muscle. C. R. Acad. Sci. III. 1998;321:893–899. doi: 10.1016/s0764-4469(99)80003-1. [DOI] [PubMed] [Google Scholar]
- 31.Aihara H., Miyazaki J. Gene transfer into muscle by electroporation in vivo. Nat. Biotechnol. 1998;16:867–870. doi: 10.1038/nbt0998-867. [DOI] [PubMed] [Google Scholar]
- 32.Hannaman D. Electroporation based TriGrid™ delivery system (TDS) for DNA vaccine administration. In: Thalhamer J., Weiss R., Scheiblhofer S., editors. Gene Vaccines, Chapter 8. Springer-Verlag; 2012. pp. 163–181. [Google Scholar]
- 33.Lee S.J., McPherron A.C. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S.-J. Quadrupling muscle mass in mice by targeting TGF-β signaling pathways. PLoS ONE. 2007;2:e789. doi: 10.1371/journal.pone.0000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodino-Klapac L.R., Janssen P.M.L., Shontz K.M., Canan B., Montgomery C.L., Griffin D., Heller K., Schmelzer L., Handy C., Clark K.R. Micro-dystrophin and follistatin co-delivery restores muscle function in aged DMD model. Hum. Mol. Genet. 2013;22:4929–4937. doi: 10.1093/hmg/ddt342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendell J.R., Sahenk Z., Malik V., Gomez A.M., Flanigan K.M., Lowes L.P., Alfano L.N., Berry K., Meadows E., Lewis S. A phase 1/2a follistatin gene therapy trial for becker muscular dystrophy. Mol. Ther. 2015;23:192–201. doi: 10.1038/mt.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wooddell C.I., Radley-Crabb H.G., Griffin J.B., Zhang G. Myofiber damage evaluation by Evans blue dye injection. Curr. Protoc. Mouse Biol. 2011;1:463–488. doi: 10.1002/9780470942390.mo110141. [DOI] [PubMed] [Google Scholar]
- 38.Maguire K.K., Lim L., Speedy S., Rando T.A. Assessment of disease activity in muscular dystrophies by noninvasive imaging. J. Clin. Invest. 2013;123:2298–2305. doi: 10.1172/JCI68458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson T., Shultz L.D., Miller D., King M., Laning J., Fodor W., Cuthbert A., Burzenski L., Gott B., Lyons B. Non-obese diabetic-recombination activating gene-1 (NOD-Rag1 null) interleukin (IL)-2 receptor common gamma chain (IL2r gamma null) null mice: a radioresistant model for human lymphohaematopoietic engraftment. Clin. Exp. Immunol. 2008;154:270–284. doi: 10.1111/j.1365-2249.2008.03753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pichavant C., Bjornson C.R.R., du Bois H., Gallagher T.J., Neal T., Calos M.P. Three novel immune-deficient mouse models of muscular dystrophy. PLoS Curr. Musc. Dyst. 2017;1 [Google Scholar]
- 41.Bertoni C., Jarrahian S., Wheeler T.M., Li Y., Olivares E.C., Calos M.P., Rando T.A. Enhancement of plasmid-mediated gene therapy for muscular dystrophy by directed plasmid integration. Proc. Natl. Acad. Sci. USA. 2006;103:419–424. doi: 10.1073/pnas.0504505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee Y.S., Lehar A., Sebald S., Liu M., Swaggart K.A., Talbot C.C., Jr., Pytel P., Barton E.R., McNally E.M., Lee S.J. Muscle hypertrophy induced by myostatin inhibition accelerates degeneration in dysferlinopathy. Hum. Mol. Genet. 2015;24:5711–5719. doi: 10.1093/hmg/ddv288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartoli M., Poupiot J., Vulin A., Fougerousse F., Arandel L., Daniele N., Roudaut C., Noulet F., Garcia L., Danos O., Richard I. AAV-mediated delivery of a mutated myostatin propeptide ameliorates calpain 3 but not alpha-sarcoglycan deficiency. Gene Ther. 2007;14:733–740. doi: 10.1038/sj.gt.3302928. [DOI] [PubMed] [Google Scholar]
- 44.Calos M.P. Genome editing techniques and their therapeutic applications. Clin. Pharmacol. Ther. 2017;101:42–51. doi: 10.1002/cpt.542. [DOI] [PubMed] [Google Scholar]
- 45.Calos M.P. The CRISPR way to think about Duchenne’s. N. Engl. J. Med. 2016;374:1684–1686. doi: 10.1056/NEJMcibr1601383. [DOI] [PubMed] [Google Scholar]
- 46.Xu L., Park K.H., Zhao L., Xu J., El Refaey M., Gao Y., Zhu H., Ma J., Han R. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol. Ther. 2016;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turan S., Farruggio A.P., Srifa W., Day J.W., Calos M.P. Precise correction of disease mutations in induced pluripotent stem cells derived from patients with limb girdle muscular dystrophy. Mol. Ther. 2016;24:685–696. doi: 10.1038/mt.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.