Abstract
Proteinaceous plaques associated with neurodegenerative diseases contain many biopolymers including the polyanions glycosaminoglycans and nucleic acids. Polyanion-induced amyloid fibrillation has been implicated in disease etiology, but structural models for amyloid/nucleic acid co-assemblies remain limited. Here we constrain nucleic acid/peptide interactions with model peptides that exploit electrostatic complementarity and define a novel amyloid/nucleic acid co-assembly. The structure provides a model for nucleic acid/amyloid co-assembly as well as insight into the energetic determinants involved in templating amyloid assembly.
Keywords: Alzheimer’s disease, solid-state NMR, biopolymer co-assembly, nucleic acid/amyloid co-assembly structure
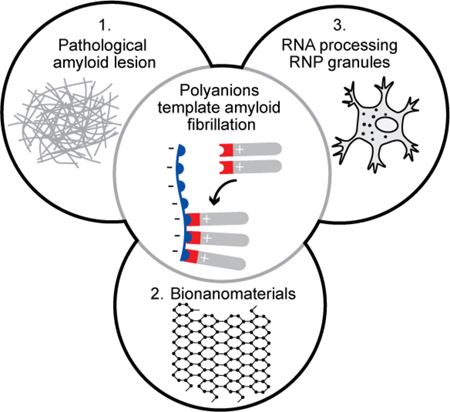
Graphical Abstract
The central role of polyanion-templated amyloid assembly to pathological lesions, bionanomaterials, and RNP granules.
Introduction
The accumulation and deposition of misfolded proteins and peptides as amyloid is associated with the onset and progression of at least 50 disease states[1], [2]. Though these pathogenic proteins have high sequence variability, amyloid assemblies have a common core structure known as cross-β where β-sheets stack perpendicular to the long axis of the fiber with context-dependent differences directed by side chain identity[2]–[10]. These proteinaceous deposits also contain nucleic acids[11], glycosaminoglycans[12]–[16], lipids, metals[17], and small molecules. Polyanionic species at lesions have been implicated in the enhancement of amyloid fibrillation and cytotoxicity[12], [13]–[16], [18], [19]. For example, both sulfated glycosaminoglycans and nucleic acids increase the rate of amyloid assembly[13], [20], and co-assembly of amyloid with the sulfated glycosaminoglycan heparin yields three-fold higher amyloid-specific ThioflavinT fluorescence at three days than amyloid alone[13]. That desulfated heparin does not increase the rate of amyloid fibrillation, suggests a critical role for sulfate moieties in the observed enhancement[13]. For nucleic acids, fibrillation is accelerated through an initial molecular interaction independent of sequence[20], and once assembled, nucleobases of the incorporated nucleic acids are available for hybridization with complementary oligonucleotides[21]. The availability of nucleobases and the sequence-independent enhancement of fibrillation, further supports a critical role for polyanions in amyloid assembly and disease progression through nucleic acid-induced amyloid cytotoxicity[18], [19]. Finally, the identification of nucleic acids in prion protein (PrP) preparations led to the demonstration that both RNA and DNA were able to induce misfolding of recombinant PrP into infectious aggregates in vitro[18], [19], [22], [23].
Since the precise role of electrostatics in the energetics of amyloid-forming polypeptides remains unclear, we sought to evaluate the involvement of polyanionic phosphates as templates. Accordingly, we have investigated simple model peptides derived from the nucleating core of amyloid-β (Aβ), the misfolded peptide of Alzheimer’s disease[24], to assess and define the electrostatic contribution of nucleic acid-influenced amyloid assembly. As outlined in Figure 1, we theorized that nucleic acids template amyloid assembly through complementary electrostatic interactions, with nucleic acids organizing within the amyloid manifold. Here, we provide structural evidence that nucleic acids and peptides co-assemble via electrostatic templating, and characterize the structure of a novel co-assembly. We find specifically that nucleic acid phosphate periodicity contributes to the complementarity, structural integrity, and homogeneity of the final cross-β assembly. The defined structure provides insight into the possible context-dependent folding of amyloid at pathological lesions, and reveals pathways for the design and development of novel nucleic acid/peptide-based nanomaterials. We further propose that these described cooperative interactions may play a critical role in Biology’s more transient nucleic acid/peptide assemblies such as those of the dynamic ribonucleoprotein granules central to cellular RNA processing[25]–[29].
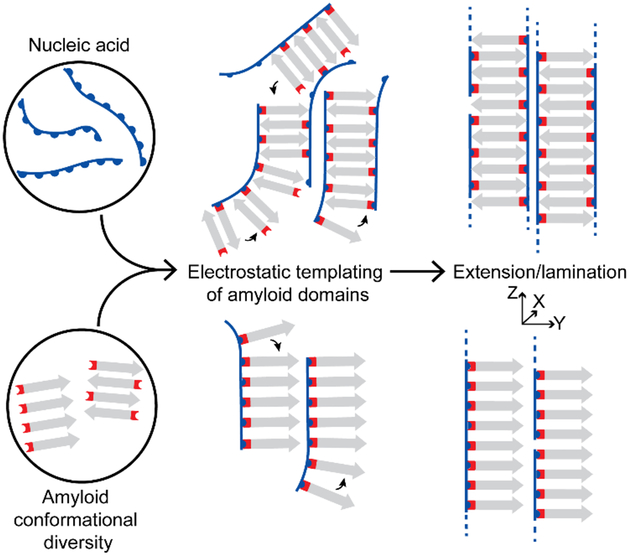
Figure 1. Proposed templating of peptides by nucleic acids.
Positively-charged peptides form cationic phases capable of complementary electrostatic interactions with nucleic acid polyanions. Nucleic acids might selectively template β-sheets into two distinct organized co-assemblies.
Results
Design of peptides and nucleic acids
The sequences we designed to examine the electrostatic contribution of nucleic acid-induced amyloid assembly are based on the nucleating core of Aβ, Ac-KLVFFAE-NH2 (Figure 2a). At neutral pH, Ac-KLVFFAE-NH2 forms fibers with strands oriented anti-parallel and in-register due to the stabilizing influence of salt bridges between terminal lysine and glutamic acid side chains[8]. To release cationic lysine residues from these salt bridges and create greater surface plasticity, the C-terminus was substituted with glycine. To maintain overall hydrophobicity[4], [30], and avoid the potential competing ability of the ‘FF’ dyad to associate with nucleobases, FF was replaced with the ‘II’ dyad. The resulting peptide, Ac-KLVIIAG-NH2 (pep-KG) and its congener Ac-RLVIIAG-NH2 (pep-RG), served as peptide monomers (Figure 2a). The nucleic acids were similarly simplified based on previous studies suggesting nucleic acids and peptides undergo complexation independent of sequence. Sequences rich in guanine or cytosine were avoided based on their inherent ability to fold as g-quadraplexes and i-motifs, respectively. Repeating oligonucleotides of adenine were used in most experiments.
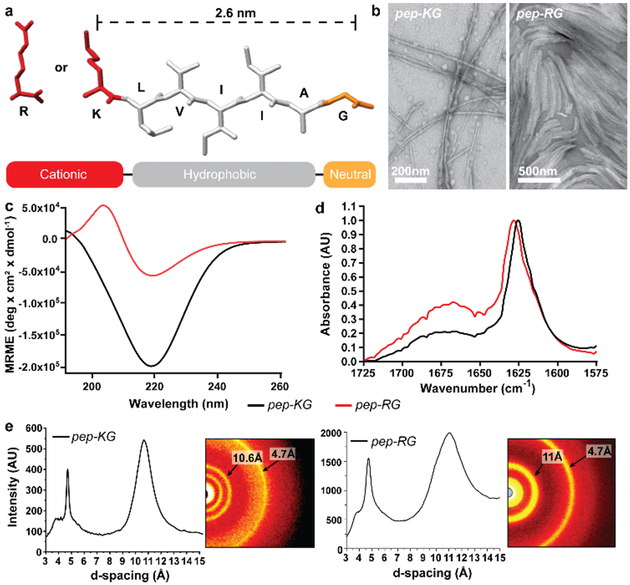
Figure 2. Assembled peptides Ac-KLVIIAG-NH2 (pep-KG) and Ac-RLVIIAG-NH2 (pep-RG).
(a) Primary structure of the peptides. Characterization of the assemblies by (b) transmission electron microscopy, (c) circular dichroism, (d) FT-IR amide I transition normalized to 1, and (e) powder x-ray diffraction. Black lines for CD and FT-IR correspond to pep-KG; Red lines to pep-RG. Scale bars are as shown.
Peptide self-assembly
As shown in Figure 2b and Figure 3a/b, pep-KG and pep-RG self-assembled into fibrillar nanostructures as determined by transmission electron microscopy (TEM). Circular dichroism (CD) identified a negative molar ellipticity at 220nm and Fourier transform infrared analyses (FT-IR) revealed a strongly delocalized amide I stretch at ~1625cm−1, both assigned to β-sheet formation (Figure 2c/d)[6]. The lower molar ellipticity for pep-RG by CD at 220nm was attributed to an overall decrease in β-sheet assembly, consistent with the broad amide I stretch at ~1670cm−1 assigned to unassembled peptide. Powder x-ray diffraction (XRD) of both pep-KG and pep-RG confirmed cross-β assemblies with reflections at 4.7Å and ~11Å assigned to inter-strand and inter-sheet (laminate) distances, respectively (Figure 2e). Given confirmation of amyloid character for peptide self-assembly, we explored mixtures of pep-KG or pep-RG with polyanions.
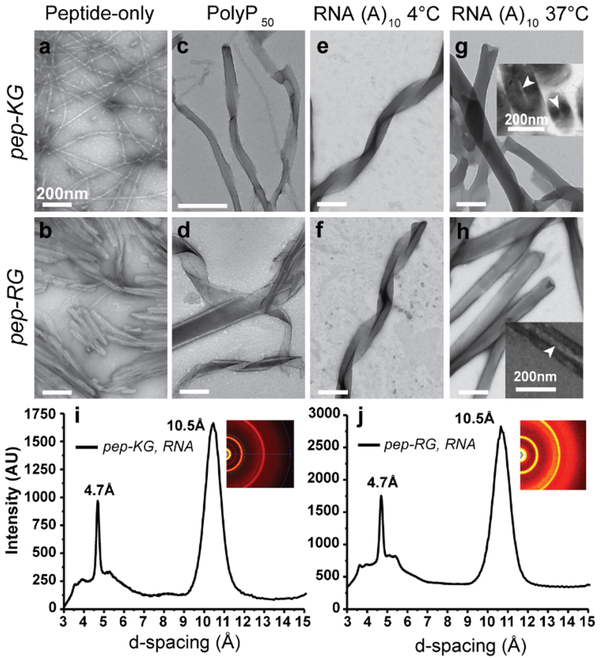
Figure 3. RNA/Peptide co-assembly.
(a/b) TEM micrographs of pep-KG and pep-RG self-assemblies. (c/d) PolyP50 templated assembly of pep-KG and pep-RG. (e/f) RNA (A10)-templated assembly of pep-KG and pep-RG at 4°C. (g/h) At 37°C, the ribbons fuse to form thick-walled nanotubes; TEM Insets show nanotube cross-sections. (i/j) Powder x-ray diffraction of peptide/RNA co-assemblies. Pep-KG refers to Ac-KLVIIAG-NH2 and pep-RG to Ac-RLVIIAG-NH2. Arrowheads indicate hollow nanotube interior. Scale bars are 200nm.
Amyloid/Nucleic acid co-assembly
Polyphosphate 50 (PolyP50), a linear polyanion with approximately 50 phosphate groups, was initially selected for co-assembly with pep-KG and pep-RG to assess specificity of polyanion-induced assembly. Incubation with both peptides gave morphologically heterogeneous assemblies, with the presence of ribbons, fibers, and nanotubes noted by TEM (Figure 3c/d). This heterogeneity was not apparent in co-assemblies with RNA or DNA. As shown in Figure 3e/f, co-assembly of pep-KG and pep-RG at 4°C with RNA(A)10 gave homogeneous positively-stained ribbons by TEM. At 37°C, the co-assemblies appeared as thick-walled nanotubes with multiple well-defined lamellae (Figure 3g/h, Figure S1). Cross-sectioned assemblies confirmed hollow nanotube architectures (Figure 3g/h insets, Figure S1). The lamellae spacing in both the ribbons and thick-walled nanotubes were approximately the lengths of extended peptides, suggesting peptide monolayers made up each lamella (Figure 2a, Figure S1). These morphologically-distinct nanotubes were only observed in preparations containing both peptides (pep-KG or pep-RG) and nucleic acids. XRD analyses of these pep-KG/RNA and pep-RG/RNA co-assemblies identified strong signatures at 4.7Å and 10.5Å, confirming the core cross-β architecture (Figure 3i/j). However, no reflections could be assigned specifically to the nucleic acids. While both DNA and RNA oligonucleotides were effective templates for co-assembly, each required an oligonucleotide length threshold of ≥6 phosphates (Figure S2). Remarkably, DNA strands of at least 7,500 nucleotides gave similar morphologies when co-assembled at concentrations necessary for overall charge balance.
To evaluate the co-assembly of the nucleic acids and peptides further, RNA tagged with Cy3 at the 3’-end was co-assembled with pep-KG and pep-RG and the resulting nanostructures examined by laser scanning confocal fluorescence microscopy. The colocalization of Cy3 and, the amyloid-specific dye, ThioflavinT (ThT) fluorescence supported homogeneous co-assembly (Figure S3). Electrostatic force microscopy (EFM) further supported a homogeneous negatively-charged surface for pep-KG/RNA co-assemblies (Figure S4), consistent with the even distribution of nucleic acids across the outer surface of nanostructures.
Further support for co-assembly, and the electrostatic contribution, was obtained through the addition of 300mM MgCl2 to the assembly conditions. By TEM, the resulting nanotubes had fewer lamellae, with nanotube widths about half those of peptide/RNA nanostructures at 37°C (Figure S5). ThT-positive nanotubes from peptide/RNA co-assemblies in MgCl2 still co-stained with Cy3-tagged RNA (Figure S3). However, in spin-down experiments, where centrifugation at 16,000g enriched for nanostructures in the pellet, significantly higher levels of free nucleic acid were measured in the supernatant of samples containing magnesium ions.
Enrichment of nucleic acid in the supernatant is consistent with the attenuation of electrostatic interactions between nucleic acids and peptides by magnesium ions. Despite these results, nucleation and propagation of amyloid is highly context-dependent[7]–[10]. Therefore, it is also possible that the presence of MgCl2 creates a different propagating amyloid nucleus. Indeed, peptide-alone assembly in the presence and absence of MgCl2 gave nanotubes and fibers by TEM, respectively (Figure S6). These data suggest that added salts may be impacting both peptide and nucleic acid assembly.
Domain architecture of pep-KG and pep-KG/DNA assemblies
With the cross-β architectures assigned for both peptide assemblies by XRD (Figure 2e), precise peptide strand arrangements were determined by measurement of the 13C-15N dipolar recoupling of 13C- and 15N-enriched amino acids between pep-KG β-strands by solid-state NMR (SSNMR)[31], [32]. Pep-KG was selected for analysis by SSNMR because of its higher assembly propensity than pep-RG. Since there were no significant morphological differences between pep-KG/RNA and pep-RG/RNA nanostructures, pep-KG/nucleic acid co-assemblies were also analyzed by SSNMR. Given the amount of materials needed for these NMR experiments, we relied on DNA templates for co-assembly.
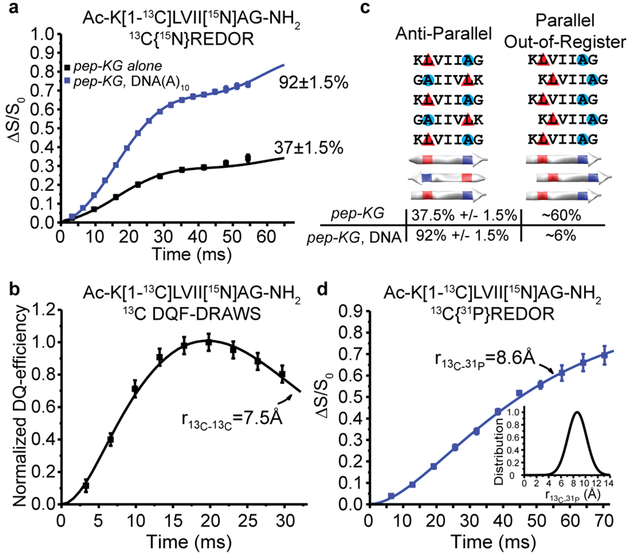
13C{15N} Rotational-Echo Double-Resonance (REDOR)[33] analyses of isotope-enriched pep-KG (Ac-K[1-13C]LVII[15N]AG-NH2) assemblies (Figure 4a) defined an intermolecular 13C-15N distance that positioned leucine and alanine residues as H-bonded between adjacent β-strands. Spin counting constrained 37±1.5% of the leucine amide 13C nuclei as proximal to two alanine amide 15N nuclei from adjacent strands with distances of 4.35Å and 5.2Å, uniquely defining these strands as directly H-bonded in anti-parallel in-register β-sheets (Figure 4a/c). Measurement of 13C-13C homonuclear dipolar recoupling with Double-Quantum Filtered Dipolar Recoupling in a Windowless Sequence (DQF-DRAWS)[34] placed the remaining peptides in the pep-KG self-assemblies with a 13C-13C distance of 7.5Å, consistent with extended H-bonded β-sheets containing parallel strands out-of-register by one residue (Figure 4b/c). The pep-KG-alone assemblies were heterogeneous in β-strand orientation and registry according to these analyses. In contrast, 13C-15N distance measurements of DNA(A)10-templated Ac-K[1-13C]LVII[15N]AG-NH2 co-assemblies assigned 92±1.5% of the peptide as anti-parallel in-register β-strands (Figure 4a/c).
Figure 4. Structural characterization of DNA/Peptide co-assemblies.
(A) 13C{15N}REDOR measurements of 13C-15N distances for pep-KG 13C enriched at the leucine carbonyl ([1-13C]L) and 15N-enriched alanine ([15N]A) in both neat assemblies (black) and templated with DNA(A)10 (blue). Both assemblies have β-sheets with leucine H-bonded to alanine from adjacent strands indicating antiparallel, in-register β-strands with 37±1.5% of the peptides adopting this orientation in pep-KG assemblies, whereas 92±1.5% of the peptides in pep-KG/DNA(A)10 co-assemblies have this orientation. (B) 13C DQF-DRAWS measurement of 13C-13C distances in neat pep-KG assemblies assigned the ~60% of the [1-13C]Leu, not detected in the 13C{15N}REDOR experiment as parallel out-of-register by one amino acid β-sheets. (C) Models of peptide orientation and registry and their associated population from solid-state NMR measurements. Red triangles indicate position of [1-13C] and blue circles indicate 15N. (D) 13C{31P} REDOR of pep-KG/DNA(A)10 show that all of the [1-13C]L nuclei are proximal to at least one 31P nucleus and fit to a single distance of 8.6Å with a Gaussian distribution of 2.2Å (inset). Unless shown, error bars are the size of the data points and represent standard deviation.
Peptide strand registry in amyloid is known to be modulated by sequence and environment, but little structural data is available for nucleic acid-templated peptide amyloid assemblies. To define the arrangement of nucleic acids within the co-assembly, the precise distance of backbone phosphates to the N-terminus of the peptide was measured with 13C{31P}REDOR (Figure 4d). For these analyses, the same isotope-enriched pep-KG/DNA sample was used. The 13C-enrichment of leucine’s carbonyl placed 13C nuclei near the peptide N-terminus in Ac-K[1-13C]LVII[15N]AG-NH2. Measurement of the 13C – 31P distance within the co-assemblies was consistent with the [1-13C]Leu having an average distance to the DNA backbone 31P of 8.6Å. The broad Gaussian distribution of 2.2Å (Figure 4d, inset) suggested that multiple 31P spins contributed to the REDOR dephasing, but with homogeneous and highly-ordered phosphate packing. Notably, these 13C{31P}REDOR data also indicated that all of the leucine 13C-enriched carbonyls were close to a 31P nucleus.
Given that peptide β-strand arrangement, as defined by 13C{15N}REDOR, was constrained to anti-parallel in-register β-sheets within pep-KG/DNA nanostructures (Figure 4a/c), the positively-charged lysine residues must be precisely patterned on each face of the cross-β peptide monolayer. With nucleic acid phosphates pacifying the leaflet surface, nanotubes arise through multilamellar packing of cross-β monolayers (Figure 5). These results lead to a model where the nucleic acids specifically templated anti-parallel in-register β-sheet peptide assembly through complementarity of the phosphate periodicity and the arrangement of positively-charged amino acids at the cross-β leaflet interface. Given the sequence-independence and polymer-length plasticity of the nucleic acids in these co-assemblies, as well as the absence of any diffraction signatures for the nucleic acids, the electrochemical forces between the nucleic acid phosphates and the lysine side chains must dominate templating energetics.
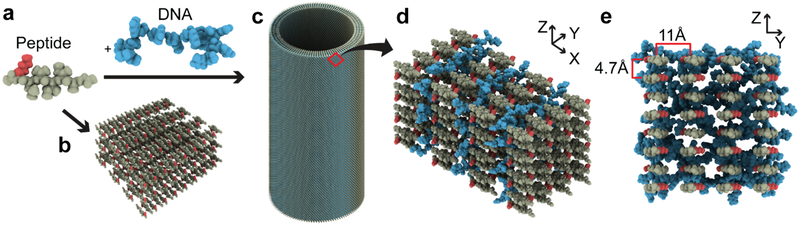
Figure 5. Structural models of peptide-alone and NA/pep co-assemblies.
(A) Peptide and DNA are depicted as globular structures with space filling models. (B) Peptides assemble as parallel and anti-parallel β-sheets and as (C) multi-lamellar nanotubes when templated by NAs. (D) Cross-β monolayers make up the individual lamellae of the peptide/DNA nanotubes with DNA passivating the positively-charged cross-β surface. (E) The peptides in the co-assembly are anti-parallel, in-register containing 4.7Å and 10.5Å d-spacings assigned to inter-strand and inter-sheet distances, respectively. The red N-terminal lysine amine and the blue NA highlight their electrochemical distribution in the co-assemblies.
Discussion
Diseases classified collectively as amyloidoses result from the accumulation and deposition of misfolded proteins or peptides at pathological lesions [1], [2]. The organization of these misfolded proteins and peptides is largely context-dependent, making it difficult to recapitulate disease-associated amyloid structure in vitro[7]–[10]. Polyanions, including glycosaminoglycans and nucleic acids, associated with pathological lesions have been shown to directly influence the rate of amyloid assembly[1], [13], [20], [21], structural dynamics[7]–[10], and cytotoxicity of aggregates[18], [19]. However, a more detailed examination of amyloid/polyanion interactions and their structures has not yet been described. Definition of a high-resolution amyloid/nucleic acid structure will provide a starting point for consideration of the polymer dynamics at these pathological lesions, the inherent cytotoxicity of infectious aggregates, and the accessible domains for the design of new co-assembled bionanomaterials.
In this study, the amyloid-forming peptide Ac-KLVIIAG-NH2, a congener of KLVFFAE, the nucleating core of amyloid-β of Alzheimer’s disease provided the framework for the electrostatic cross-β surface being templated with oligonucleotides. We found that the global structure of amyloid/nucleic acid co-assemblies was morphologically distinct from peptide-alone fibers, and using diffraction and solid state NMR, we were able to assign the location of peptides and nucleic acids at angstrom-level resolution. The phosphate periodicity of the nucleic acid backbone was integral to the electrostatic complementarity and selective templating of antiparallel in-register amyloid domains. This complementarity is notable relative to the heterogeneity of nanostructures observed for PolyP50-templated assemblies. In the literature, amyloid/nucleic acid complexation and aggregation consistently yields beta-sheet-rich nanostructures[12], [18], [20], [21]. Peptides six to ten amino acids in length of alternating hydrophobic and hydrophilic residues have been shown to maintain core amyloid architecture when complexed with single- or double-stranded nucleic acids[21]. A study suggesting cationic amyloid surfaces formed ideal polyelectrolytes for complementary electrostatics with polyanions, found that magnesium or calcium attenuated DNA-, ATP-, or Heparin-induced amyloid fibrillation[12]. Further, nucleic acids convert prion protein to a β-sheet conformation and promote amyloid assembly[18]. These reports are consistent with our comprehensive global and structural analysis of an amyloid/nucleic acid co-assembly.
The plasticity regarding nucleic acid sequence and length may be an important factor required of the stabilizing amyloid domains of the messenger ribonucleoprotein granules[25], [26], [29], [35]. These membraneless organelles guide RNA through the many stages of information processing[25], reversibly form amyloid assemblies[36], [37] for granule maturation[27], [38], interact with RNA Pol II[32], and contribute to nuclear import dynamics[23], [40], all critical events for cellular information flow. These structural insights highlight the dynamic and complementary electrostatic interactions between nucleic acids and amyloid domains that could underlie these diverse processes as well as the transition to infectious and cytotoxic aggregates[18], [41].
Conclusion
Through use of short peptides and oligonucleotides, we defined for the first time the structure of an amyloid/nucleic acid nanostructure with angstrom-level constraints. Analyses of amyloid/nucleic acid co-assemblies and their peptide-alone counterparts by TEM and biophysical methodologies identified critical criteria for the complementarity and electrostatic templating of cationic cross-β surfaces by nucleic acids. These findings provide insight into electrostatic templating of amyloid folding at pathological lesions, and support other crucial studies describing the influence of polyanions on the enhancement of amyloid fibrillation and transition to cytotoxic aggregates. As a reductionist model to disease-relevant amyloid/nucleic acid complexation, these results provide a foundation for in vivo examination of amyloid/nucleic acid dynamics and co-assembly. The complementarity of the nucleic acid backbone with cationic antiparallel in-register amyloid domains introduces new possibilities for the design of novel co-assembled bionanomaterials.
Experimental Section
Peptide and RNA Co-Assembly.
Peptides were desalted using Sep-Pak C18 cartridges (Waters Co., Milford, MA), vortexed briefly in 40% acetonitrile, and sonicated for 5 minutes at pH5. Nucleic acid oligomers were obtained from Integrated DNA Technologies (Skokie, IL). The order of addition for all samples was peptide, solvent, nucleic acid to a final peptide concentration of 1mM. The nucleic acid concentration depended on chain length with all co-assemblies having a 1:1 (peptide: nucleic acid) charge ratio. Further experimental details can be found in the supplementary information.
Circular Dichroism (CD).
Circular Dichroism (CD) spectra were recorded with a Jasco-810 Spectropolarimeter (Easton, MD, USA) at room temperature in a 50μL cell with a 0.1mm path length. Spectra were acquired from 260nm to 190nm with a step size of 0.2nm and a speed of 100nm/s, and are the average of three independent scans. Ellipticity, in mdeg, was converted to Molar ellipticity [θ] with [θ] = θ/(10 × c × l), where ‘c’ is peptide concentration in moles/L and ‘l’ is the pathlength in cm.
Fourier Transform Infrared (FT-IR) Spectroscopy.
FT-IR spectra were acquired using a Jasco FT-IR 4100 (Easton, MD, USA) averaging 750 scans at 2cm−1 resolution. For each sample, 8μl of peptide solution was dried as a thin film on a Pike GaldiATR (Madison, WI, USA) ATR diamond surface using a MCT-M detector with a 5mm aperture and a scan speed of 4mm/sec. Spectra were normalized to the amide-I band at ~1625cm−1.
Powder X-Ray Diffraction.
Assembled samples were flash frozen using liquid nitrogen and lyophilized to yield a white powder and loaded into mylar capillary sample holders. Diffraction patterns were recorded using a Bruker APEX-II diffractometer with graphite monochromatic Cu radiation, K-alpha radiation, λ=1.54184Å, 40kV and 35mA, with a 0.5 pinhole collimator and with exposure times of 900s per frame. Data integration was performed with XRD2SCAN[42] and Bruker AXS software for analysis of the resulting diffraction patterns. Diffraction data was converted from 2θ to d-spacing(Å):
Polyphosphate Synthesis.
Polyphosphate (PolyP50) was synthesized according to the literature[43]. KH2PO4 was melted 450° for 4 hours and the resulting glass purified by stepwise precipitation in Acetone-Water mixtures. The length was determined from 31P NMR peak integration[44].
Electron Microscopy.
Electron microscopy images were obtained using Hitachi H-7500 and JEOL transmission electron microscopes at 75kV. All samples were placed on 200 mesh copper grids with carbon coating and negatively stained using 2% w/w uranyl acetate (Sigma-Aldrich). Cross-section images were obtained by embedding pelleted assemblies in epoxy resin and using a microtome on embedded assemblies to obtain 1μm thick slices that are placed on grids for analysis by TEM.
Solid-State NMR.
NMR spectra were collected using a Bruker Advance AV 600MHz solid-state NMR spectrometer with a Bruker 4mm HCN BioSolids magic-angle spinning (MAS) probe. Boron nitride spacers were used to center samples in 4mm MAS ceramic rotors. All spectra were collected with 1H-13C cross polarization and spin-temperature alternation of the initial 1H (600.133 MHz) 1.9μs π/2 pulse. 1H cross-polarization RF fields were ramped from 50 to 75 kHz and 13C (150.929 MHz) cross-polarization RF field was kept constant at 62.5kHz. Further experimental details can be found in the supplementary information.
Modeling Peptides and RNA Co-Assemblies.
Individual peptide and RNA models were generated in Chimera[45]. Peptide and DNA were further organized to the models of 3D superstructures using Strata 3D (https://www.strata.com/). Peptides were oriented anti-parallel with all amino acids in-register, the inter-strand distance was set to 4.7Å, and the inter-sheet distance set to 10.5Å and 11Å for peptide-only assemblies and peptide/DNA co-assemblies, respectively, based on XRD reflections. DNA was placed at high-density between peptide monolayers in peptide/DNA co-assembly models to account for 100% dipolar recoupling between 13C and 31P in 13C{31P}REDOR experiments.
Supplementary Material
Acknowledgements
We thank Hong Yi and Jeanette Taylor from the Robert P. Apkarian Electron Microscopy Core of Emory University for Electron Microscopy assistance, Dr John Bacsa from the Emory X-ray Center for diffraction analyses, Dr Bing Wang and Dr Shaoxiong Wu from the Emory NMR Center for NMR assistance, and Dr Fred Strobel for mass spectrometry analysis. We acknowledge Pieter Burger for access to MacroModel and the processing power to run minimizations. The research was supported by grants from NSF CHE-1507932 and NSF/DMR-BSF 1610377, and NIH Alzheimer’s Disease Research Center: P50AG025688.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, and Radford SE, Nat. Rev. Mol. Cell Biol 2018, 19, 755–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Greenwald J and Riek R, Structure 2010, 18, 1244–1260. [DOI] [PubMed] [Google Scholar]
- [3].Fändrich M and Dobson CM, EMBO J 2002, 21, 5682–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Maurer-Stroh S et al. , Nat. Methods 2010, 7, 237–242. [DOI] [PubMed] [Google Scholar]
- [5].Toyama BH and Weissman JS, Annu. Rev. Biochem 2011, 80, 557–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nelson R et al. , Nature 2005, 435, 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jucker M and Walker LC, Nature 2013, 501, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mehta AK et al. , J. Am. Chem. Soc 2008, 130, 9829–9835. [DOI] [PubMed] [Google Scholar]
- [9].Childers WS, Anthony NR, Mehta AK, Berland KM, and Lynn DG, Langmuir 2012, 28, 6386–6395. [DOI] [PubMed] [Google Scholar]
- [10].Qiang W, Yau W-M, Lu J-X, Collinge J, and Tycko R, Nature. 2017, 541, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Marcinkiewicz M, J. Neuropathol. Exp. Neurol 2002, 61, 815–829. [DOI] [PubMed] [Google Scholar]
- [12].Calamai M et al. , Biochemistry. 2006, 45, 12806–12815. [DOI] [PubMed] [Google Scholar]
- [13].Castillo GM, Lukito W, Wight TN, and Snow AD, J. Neurochem 2001, 72, 1681–1687. [DOI] [PubMed] [Google Scholar]
- [14].Snow AD, Kisilevsky R, Willmer J, Prusiner SB, and DeArmond SJ, Acta Neuropathol. 1989, 77, 337–342. [DOI] [PubMed] [Google Scholar]
- [15].McLaurin J, Franklin T, Zhang X, Deng J, and Fraser PE, Eur. J. Biochem 1999, 266, 1101–1110. [DOI] [PubMed] [Google Scholar]
- [16].Snow AD, Willmer J, and Kisilevsky R, Lab. Invest 1987, 56, 120–123. [PubMed] [Google Scholar]
- [17].Pratíco D, Uryu K, Sung S, Tang S, Trojanowski JQ, and Lee VM-Y, FASEB J 2002, 16, 1138–1140. [DOI] [PubMed] [Google Scholar]
- [18].Macedo B et al. , Biochemistry. 2012, 51, 5402–5413. [DOI] [PubMed] [Google Scholar]
- [19].Deleault NR, Harris BT, Rees JR, and Supattapone S, Proc. Natl. Acad. Sci. USA 2007, 104, 9741–9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang M, Law M, Duhamel J, and Chen P, Biophys. J 2007, 93, 2477–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Braun S, Humphreys C, Fraser E, Brancale A, Bochtler M, and Dale TC, PLoS One 2011, 6, e19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Giraldo R, Proc. Natl. Acad. Sci 2007, 104, 17388–17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Silva JL and Cordeiro Y, J. Biol. Chem 2016, 291, 15482–15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Serpell LC, Biochim. Biophys. Acta 2000, 1502, 16–30. [DOI] [PubMed] [Google Scholar]
- [25].Erickson SL and Lykke-Andersen J, J. Cell Sci 2011, 124, 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ramaswami M, Taylor JP, and Parker R, Cell 2013, 154, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Calabretta S and Richard S, Trends Biochem. Sci 2015, 40, 662–672. [DOI] [PubMed] [Google Scholar]
- [28].Lin Y, Protter DSW, Rosen MK, and Parker R, Mol. Cell 2015, 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mitrea DM and Kriwacki RW, Cell Commun. Signal 2016, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fauchere J-L and Pliska V, Eur. J. Med. Chem 1983, 18, 369–375. [Google Scholar]
- [31].Balbach JJ et al. , Biochemistry. 2000, 39, 13748–13759. [DOI] [PubMed] [Google Scholar]
- [32].Wälti MA et al. , Proc. Natl. Acad. Sci. USA 2016, 113, E4976–E4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gullion T in Modern Magnetic Resonance, (Dordrecht), Springer, Netherlands, 2008, pp. 713–718. [Google Scholar]
- [34].Gregory DM et al. , Solid State Nucl. Magn. Reson 1998, 13, 149–166. [DOI] [PubMed] [Google Scholar]
- [35].Shin Y and Brangwynne CP, Science 2017, 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- [36].Hayes MH and Weeks DL, Biol. Open 2016, 5, 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Han TW et al. , Cell 2012, 149, 768–779. [DOI] [PubMed] [Google Scholar]
- [38].Lin Y, Protter DSW, Rosen MK, and Parker R, Mol. Cell 2015, 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Feric M et al. , Cell 2016, 165, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Guo L et al. , Cell 2018, 173, 677–692.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Patel A et al. , Cell 2015, 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- [42].Rodriguez-Navarro AB, J. Appl. Cryst 2006, 39, 905–909. [Google Scholar]
- [43].Van Wazer JR, J. Am. Chem. Soc 1950, 72, 647–655. [Google Scholar]
- [44].Smith SA et al. , Blood 2010, 116, 4353–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pettersen EF et al. , J. Comput. Chem 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.