Summary
Secretion principles are conserved from yeast to humans and many yeast orthologs have established roles in synaptic vesicle exocytosis in the mammalian brain. Surprisingly, SEC4 orthologues and their effectors, the exocyst, are dispensable for synaptic vesicle exocytosis. Here, we identify the SEC4 orthologue RAB3 and its neuronal effector RIM1 as essential molecules for neuropeptide/neurotrophin release from dense-core vesicles (DCVs) in mammalian neurons. Inactivation of all four RAB3 genes nearly ablated DCV exocytosis, and re-expression of RAB3A restored this deficit. In RIM1/2-deficient neurons, DCV exocytosis was undetectable. Full-length RIM1, but not mutants that lack RAB3 or MUNC13 binding, restored release. Strikingly, a short N-terminal RIM1 fragment only harboring RAB3- and MUNC13-interacting domains was sufficient to support DCV exocytosis. We propose that RIM/MUNC13 emerged as mammalian alternative to the yeast exocyst complex as essential RAB3/SEC4 effectors and organizers of DCV fusion sites by recruiting DCVs via RAB3.
Introduction
Secretion mechanisms are highly conserved across species and rely on ancient principles. In yeast Saccharomyces cerevisiae, 23 proteins were initially identified to drive the secretory pathway (Novick et al., 1981; Novick et al., 1980; Novick and Schekman, 1979), of which ten, SEC1–6, 8–10 and 15, act in the last steps of secretion (Novick et al., 1981), together with SNC1/2 (Protopopov et al., 1993) and SSO1/2 (Aalto et al., 1993). Subsequent research revealed that orthologs for many of these proteins drive regulated secretion of synaptic vesicles (SVs) in mammalian neurons (Jahn and Scheller, 2006; Kaeser and Regehr, 2014; Sudhof, 2013; Sudhof and Rothman, 2009). Strikingly, among the ten SEC genes that act in the last steps, only two have established roles in mammalian SV fusion, SEC1 (MUNC18) and SEC9 (SNAP25). Orthologs of the other eight, encoding the GTPase SEC4 and its effectors: SEC2, a guanine exchange factor for SEC4 (Walch-Solimena et al., 1997), and 6 subunits of the SEC4 effector complex called the exocyst complex, SEC3, 5, 6, 8, 10 and 15 (Bowser et al., 1992; Guo et al., 1999; TerBush et al., 1996; TerBush and Novick, 1995) are largely dispensable for SV fusion (Mehta et al., 2005; Murthy et al., 2003; Schlüter et al., 2006; Schlüter et al., 2004; Schwenger and Kuner, 2010). The role of these orthologs in regulated secretion in mammalian neurons remains poorly understood.
RAB3 proteins, orthologs of yeast SEC4p (Zahraoui et al., 1989), are highly expressed in brain (Fischer von Mollard et al., 1990; Schlüter et al., 2002) and dynamically associate with SVs (Fischer von Mollard et al., 1990; Takamori et al., 2006). Null mutant mice, lacking all four mammalian RAB3 paralogs (RAB3A-D, RAB3 QKO from hereon), show perinatal lethality, but only subtle changes in synaptic transmission (Schlüter et al., 2006; Schlüter et al., 2004), in contrast to the essential function of SEC4p in vesicle secretion in yeast (Novick et al., 1980; Salminen and Novick, 1987). RAB3A null mice, which are homozygous viable and have minor changes in synaptic transmission (Geppert et al., 1997), show several altered behaviors, including circadian rhythmicity (Kapfhamer et al., 2002), reversal learning and exploration (D’Adamo et al., 2004), memory precision (Ruediger et al., 2011) and ethanol responses (Kapfhamer et al., 2002), although other mnemonic capabilities were normal (Hensbroek et al., 2003). These data suggest important, as yet unidentified, roles of mammalian RAB3 proteins.
Neuropeptides, neurotrophins and other signaling molecules, together referred to as neuromodulators, are secreted by dense-core vesicles (DCVs) and control diverse physiological functions such as brain development, synaptic plasticity, circadian rhythm, many behaviors and emotions (Cheng et al., 2011; Malva et al., 2012; Mertens et al., 2007; Meyer-Lindenberg et al., 2011; Pang et al., 2004). Defects in neuromodulator signaling are associated with psychiatric disorders, obesity and diabetes (Meyer-Lindenberg et al., 2011; Vahatalo et al., 2015). While SV fusion principles are well-characterized, many fundamental questions remain unanswered for neuromodulator secretion.
Here, we identify SEC4-ortholog RAB3 and its mammalian effector RIM as essential molecules for regulated secretion of neuromodulators from DCVs in mammalian neurons. We used hippocampal excitatory neurons to describe essential components of the DCV secretory pathway. We show that, unlike the previously reported (mild) effects on SV fusion (Schlüter et al., 2006), DCV fusion was reduced by >90% in RAB3 QKO neurons. Furthermore, in RIM-deficient conditional double knockout (cDKO) neurons DCV fusion was completely lost. N-terminal RAB3 and MUNC13-interacting domains of RIM co-trafficked with DCVs in a RAB3-dependent manner and were sufficient to fully restore DCV fusion. We propose that RIMs and MUNC13 emerged as mammalian alternative to the yeast exocyst complex as essential RAB3/SEC4 effectors and organizers of DCV fusion sites by positioning MUNC13 and recruiting DCVs via RAB3.
Results
Deletion of all RAB3 proteins severely impairs DCV fusion
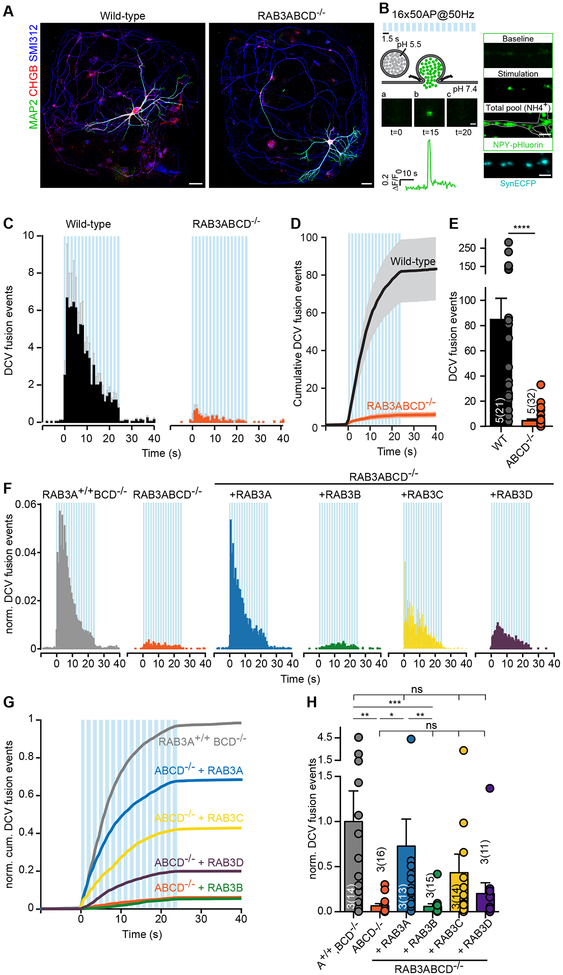
The involvement of mammalian RAB3 proteins in release of neuromodulators was assessed by recording DCV fusion in single hippocampal neurons on glia micro-islands from RAB3ABCD−/− (RAB3 QKO) and wild-type mice at days in vitro 14 (DIV 14; Fig. 1A). Expression of Neuropeptide Y (NPY) fused to pH-sensitive EGFP (pHluorin) using lentivirus, targets this reporter to virtually all DCVs, with >90% overlap with endogenous neurotrophin BDNF and neuropeptide co-factors chromogranins A/B and without altering the total number of DCVs per neuron (Dominguez et al., 2018; Persoon et al., 2018). Hence, this reporter labels DCVs irrespective of their endogenous neuropeptide/neurotrophin content and can be used to study their general secretion principles applicable to many cargo types. The NPY-pHluorin reporter detects single DCV fusion events by an instant increase of fluorescence due to rapid deacidification of the vesicle’s interior when the fusion pores opens (Fig. 1B; Arora et al., 2017; Emperador Melero et al., 2017; Farina et al., 2015; Persoon et al., 2018; van Keimpema et al., 2017). Upon calcium influx induced by action potential trains (16 bursts of 50 action potentials (APs) at 50 Hz; Balkowiec and Katz, 2002; de Wit et al., 2009; Gartner and Staiger, 2002; Hartmann et al., 2001; Matsuda et al., 2009; van de Bospoort et al., 2012), DCV fusion in RAB3 QKO neurons was almost absent (Fig. 1C–E) showing an ~20-fold reduction as compared to wild-type neurons (Fig. 1C–E). The loss of DCV fusion in RAB3QKO neurons was confirmed using BDNF as an independent reporter for neuromodulator release (Fig. S1A–E). Taken together, these data indicate that RAB3 is a key factor in neuromodulator release from DCVs.
Figure 1. RAB3 deletion impairs DCV fusion.
(A) Representative composite confocal image of single cultured hippocampal neurons from wild-type (left) or RAB3ABCD−/− (right) mice. Dendrites (MAP2, green), axons (SMI312, blue) and DCVs (CHGB, red) were labeled. Scale bars: 40 μm.
(B) NPY-pHluorin as optical reporter for DCV fusion. Repetitive electrical stimulation (16 trains of 50APs at 50 Hz) is represented by blue bars. Middle panels a-c show a single DCV fusion event reported by NPY-pHluorin, with ΔF/F0 (inset below). Scale bar: 1 μm. NPY-pHluorin is quenched in the acidic environment of the DCV lumen (a). Upon depolarization-induced Ca2+-influx, the DCV fusion pore opens indicated by a rapid increase in fluorescence (b), followed by a rapid decline upon cargo release or fusion pore closure and vesicle reacidification (c). Right panels: DCV fusion events during stimulation (Stimulation), total number of DCVs upon NH4 superfusion (Total pool), Synapsin-ECFP labeled synapses (SynECFP; Fig. S2A–C). Scale bar: 5 μm.
(C-H) DCV fusion analysis using NPY-pHluorin in single wild-type or RAB3ABCD−/− hippocampal neurons. (C) Histograms, (D) cumulative plot and (E) summary graph of DCV fusion events per cell. Mann-Whitney U test: **** p < 0.0001.
(F) Histograms, (G) cumulative plot and (H) summary graph of DCV fusion events per cell for RAB3A+/+BCD−/−, RAB3ABCD−/− or RAB3ABCD−/− expressing RAB3A, -B, -C or -D neurons, normalized to RAB3A+/+ BCD−/−. Kruskal-Wallis with Dunn’s correction: * p < 0.05, ** p < 0.01, ***p < 0.001. ns = non-significant, p > 0.05.
In all figures: repetitive electrical stimulation (16 trains of 50APs at 50 Hz) is represented by blue bars. All data shown as mean ± SEM. N represents number of experiments and number of single neuron observations in brackets. Individual neurons are represented as dots. Detailed information (average, SEM, n and detailed statistics) is shown in STAR Methods.
We excluded several potentially confounding factors that could in principle contribute to this major reduction in DCV fusion: (i) the number of NPY-pHluorin labeled DCVs per neuron were similar between genotypes (Fig. S1F), (ii) the temporal distribution of fusion before and during stimulation (Fig. S1G–H) and (iii) calcium dynamics (Fig. S1I) were also similar; (iv) the location of fusion events, synaptic or extra-synaptic, did not differ between wild-type and RAB3 QKO neurons (Fig. S1K, S2A–C); (v) the number of synapses and the total dendritic length of wild-type, RAB3A+/+, BCD−/− (RAB3 Triple knockout (TKO)) and RAB3 QKO neurons were similar (Fig. S2D–G), as observed before (Schlüter et al., 2006); (vi) the fluorescence intensity of VGLUT1 (Fig. S2H) at synapses was not altered in RAB3 QKO neurons; (vii) the number and distribution of puncta labeled with the endogenous DCV marker Chromogranin B (CHGB) was similar between genotypes (Fig. S2I–L, O). While the number of CHGB puncta and total CHGB protein levels (Fig. S2N) were similar between RAB3 QKO and control neurons, the intensity of CHGB puncta (Fig. S2M) and intensity of individual DCV fusion events (Fig. S1J) were slightly but significantly reduced (<20%), indicating a possible mild impairment in DCV loading. Together, these data exclude several potentially confounding effects and strengthen the conclusion that RAB3 proteins are important regulators of DCV fusion.
To test for functional redundancy between RAB3A-D (Schlüter et al., 2002), we expressed each paralog in RAB3 QKO neurons and analyzed DCV fusion events (Fig. 1F–H). Re-expression of RAB3A, RAB3C and RAB3D, but not RAB3B, restored the number of DCV fusion events to levels that were not significantly different from RAB3 TKO neurons (Fig. 1F–H). However, only re-expression of RAB3A significantly increased DCV fusion compared to RAB3 QKO neurons (Fig. 1F–H). The expression levels of rescue constructs (Fig. S1L–M) and number of NPY-pHluorin labeled DCVs per neuron (Fig. S1N) was similar between each condition. Together these data suggest that RAB3C and RAB3D partially restore DCV release but less efficiently than RAB3A, while RAB3B does not rescue DCV release.
RIM1 is essential for DCV fusion
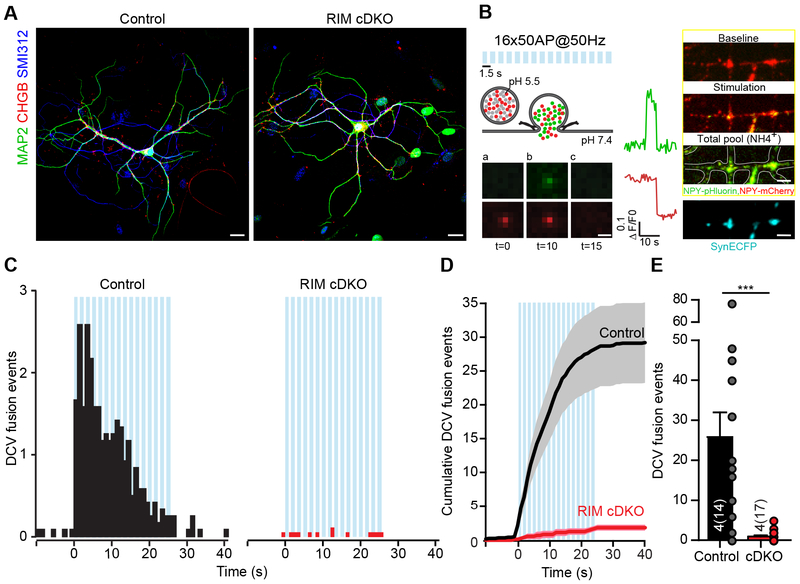
RIM1/2 proteins, multi-domain scaffolding proteins enriched at presynaptic active zones, are established mammalian RAB3A/C effectors at the target membrane (Wang et al., 1997). To test if RIM1/2 are important in DCV fusion, we used single isolated hippocampal neurons from conditional RIM1/RIM2 DKO mice in which Cre-recombinase deletes expression of all RIM 1(α & β) and RIM 2(α, β & γ) isoforms (Kaeser et al., 2011). Hippocampal neurons (Fig. 2A) were infected at DIV 0 with lentiviral constructs expressing active, EGFP-tagged Cre-recombinase (RIM cDKO), resulting in the absence of RIM protein expression from DIV 8 (Fig. S3A), or inactive, EGFP-tagged mutant Cre-recombinase (control). To test if RIMs function in neuromodulator release, DCVs were co-labeled with NPY-pHluorin and NPY fused to red fluorescent mCherry (NPY-mCherry), which upon DCV fusion shows a rapid decrease in fluorescence due to cargo diffusion (Fig. 2B). NPY-mCherry allows for analysis of DCV transport and behavior prior to fusion as mCherry does not quench in the low pH of the DCV lumen (Fig. 2B). RIM cDKO neurons showed a more than 95% reduction in NPY-mCherry labeled DCV fusion events upon stimulation compared to controls (Fig. 2C–E, Fig. S3B). The few remaining events in RIM cDKO neurons occurred mostly outside synapses (Fig. S3C) of which the fluorescence disappeared within 1 second (Fig. S3D–E), and were not detected as NPY-pHluorin-labeled fusion events (Fig. S3F–G). The total number of NPY-mCherry puncta was not altered (Fig. S3H). The loss of DCV fusion in RIM cDKO neurons was confirmed using BDNF-pHluorin as an independent reporter for neuromodulator release (Fig. S3J–L).
Figure 2. RIM depletion blocks DCV fusion.
(A) Representative composite confocal image of single hippocampal control (left) or RIM 1/2 cDKO (right) neurons. Dendrites (MAP2, green), axons (SMI312, blue) and DCVs (CHGB, red) were labeled. Scale bars: 20 μm..
(B) NPY-pHluorin and NPY-mCherry as dual-color optical reporters for DCV fusion. Lower panels a-c show a single DCV fusion event reported by NPY-pHluorin and NPY-mCherry, with ΔF/F0 traces (green, NPY-pHluorin; red, NPY-mCherry). Right panels: DCV fusion events during stimulation (Stimulation), total number of DCVs upon NH4 superfusion (Total pool), Synapsin-ECFP labeled synapses (SynECFP). Scale bar (left): 1 μm; scale bare (right): 5 μm..
(C-E) DCV fusion analysis using NPY-pHluorin and NPY-mCherry in single hippocampal control (black) or RIM1/2 cDKO (red) neurons. (C) Histograms, (D) cumulative plot and (E) summary graph of DCV fusion events per cell. Mann-Whitney U test: *** p = < 0.0002.
Morphological characterization at DIV 14 showed a modestly reduced number of synapses and MUNC13 intensity when Cre-recombinase was expressed at DIV 0 (Fig. S4A–G). No differences were found in the number or intensity of CHGB puncta (Fig. S4A, H–L). CHGB puncta co-localized slightly more with VGLUT1-positive synapses compared to control (Fig. S4M–N) and synaptic electron micrograph sections showed a trend towards accumulation of DCVs at the pre-synapse (Fig. S4O–P). DCVs are actively transported throughout the neuron by microtubule-based motor proteins (Lo et al., 2011; Stucchi et al., 2018; Zahn et al., 2004), but DCV transport parameters (speed, distance moved) were not altered in RIM cDKO neurons (Fig. S5A–H). Hence, RIM cDKO neurons have less synapses when cre-infected at DIV 0 with a trend for DCV accumulation, but have a normal DCV population indicating that RIMs are not required for DCV biogenesis, loading or transport.
To test if reduced synapse numbers could explain the DCV fusion phenotype, we expressed Cre-recombinase at DIV 5 instead of DIV 0, which does not affect synapse numbers (Fig. S5I–L), and observed a similar block in DCV fusion in RIM deficient neurons at DIV 18 (Fig. S5M–O), showing that changes in synapse number does not correlate with a reduction in DCV fusion in RIM-deficient neurons. Cre-recombinase expression in wild-type neurons did not negatively affect DCV fusion (Fig. S5P–R). Together we conclude that RIMs are essential for DCV fusion.
To study whether RIM1 or RIM2 is required for DCV fusion, single hippocampal neurons from RIM1 cKO mice or RIM2 cKO mice were infected with Cre-recombinase or control virus at DIV 0. Upon stimulation RIM2 deficient neurons (RIM1+/+, RIM2−/−) showed similar number of DCV fusion events compared to controls, while DCV fusion was strongly reduced in RIM1 deficient neurons (RIM1−/−, RIM2+/+ and RIM1−/−, RIM2+/−) and RIM2 deficient neurons heterozygous for RIM1 (Fig. S6A–F). Multiple bursts of action potentials were required to trigger DCV fusion in RIM1 deficient neurons (Fig. S6B–C). Total DCV numbers were similar in all genotypes (Fig. S6E). These data show that RIM1 is required for efficient DCV fusion and in absence of RIM1, RIM2-dependent DCV fusion is strongly reduced and delayed.
N-terminal interactions of RIMs with RAB3 and MUNC13 regulate DCV fusion
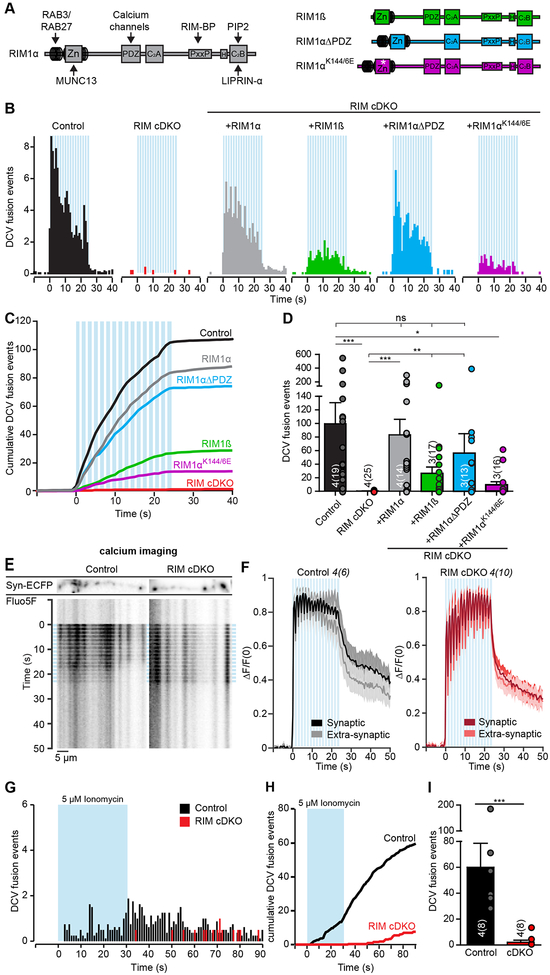
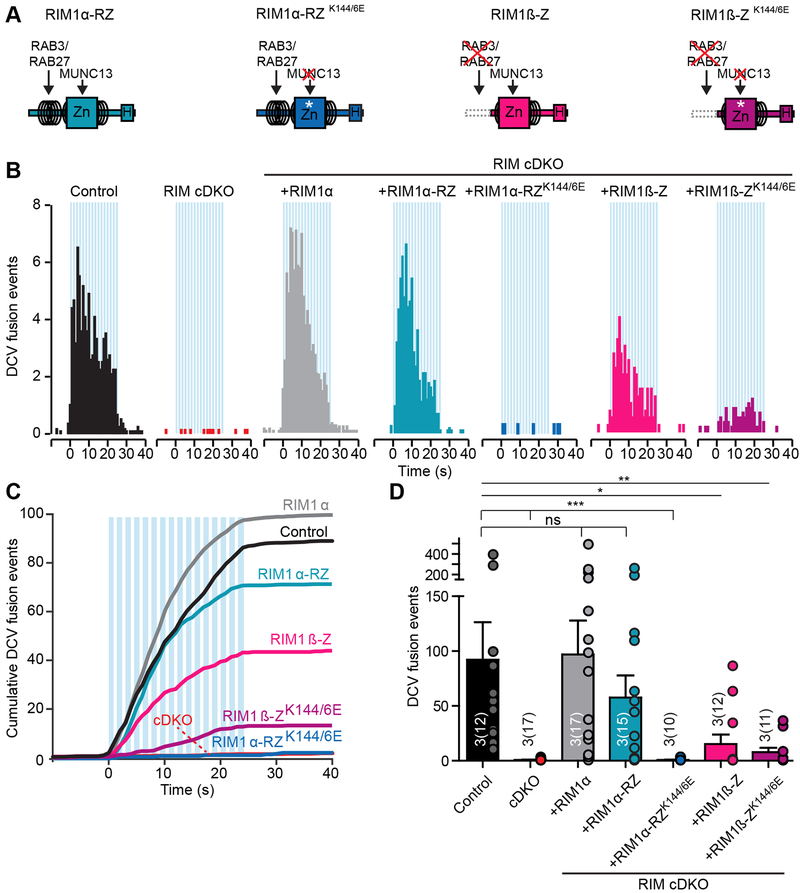
RIMs regulate synaptic vesicle fusion through interactions with MUNC13, voltage-gated calcium channels and PIP2 (Fig. 3A (left); de Jong et al., 2018; Deng et al., 2011; Han et al., 2011; Kaeser et al., 2011). To study if these interactions are required for DCV fusion, we expressed multiple RIM-rescue constructs (Fig. 3A) in RIM cDKO neurons from DIV 0, which were expressed at similar levels and all localized to synaptic regions at DIV 14 (Fig. S6G–H), as reported before (Kaeser et al., 2011). Upon stimulation, full-length RIM1α rescued DCV fusion to control levels (Fig. 3B–D). RIM1β lacks interaction with RAB3 (Kaeser et al., 2008), which binds RIM1α through the N-terminal α-helix (Fig. 3A; Wang et al., 1997). DCV fusion in RIM1β expressing RIM cDKO was reduced by almost 75 % compared to controls (Fig. 3B–D). RIM1α-ΔPDZ, lacking the PDZ-domain that binds to voltage-gated calcium channels (Fig. 3A; Kaeser et al., 2011), almost completely rescued DCV fusion in RIM cDKO neurons (Fig. 3B–D). MUNC13 binds to the N-terminal zinc-finger of RIM (Betz et al., 2001; Dulubova et al., 2005; Schoch et al., 2002), and mutating two lysine residues (K144 and K146) of RIM to glutamates (RIM1α-K144/6E, Fig. 3A) results in a loss of MUNC13 binding (Deng et al., 2011; Dulubova et al., 2005; Lu et al., 2006). Expression of RIM1α-K144/6E in RIM cDKO neurons did not restore DCV fusion (Fig. 3B–D). The onset of fusion or the total pool was not altered (Fig. S6I–J), but the percentage of synaptic fusion events was increased in the different rescue conditions (Fig. S6K), in line with their synaptic expression (Fig. S6G–H). These data show that the N-terminal interactions of RIMs with RAB3 and MUNC13 are essential for efficient DCV fusion.
Figure 3. N-terminus interactions of RIM1/2 regulate DCV fusion.
(A) Domain structure of full-length RIM1α (left) with key interactions. Diagram of RIM1β, RIM1α-ΔPDZ and RIM1α-K144/6E rescue proteins expressed in RIM cDKO neurons (right). Zn, zinc-finger domain with surrounding α-helical regions; PxxP, proline-rich region; H, location of a human influenza hemagglutinin (HA)-tag; asterisk, K144/6E substitution..
(B-D) DCV fusion analysis using NPY-pHluorin in single control or RIM cDKO hippocampal neurons without or with rescue constructs (RIM cDKO + rescue). (B) Histograms, (C) cumulative plot and (D) summary graph of DCV fusion events per cell. Kruskal-Wallis with Dunn’s correction: *p < 0.05, **p < 0.01, ***p < 0.001. ns, p > 0.05..
(E) Kymograph and (F) average normalized ΔF/F0 traces of intracellular calcium (Fluo5-AM) levels upon repetitive electrical stimulation (blue bars) in control or RIM cDKO neurons at synaptic (labeled by Synapsin-ECFP) or extra-synaptic regions.
(G-I) DCV fusion analysis using NPY-pHluorin in single control or RIM cDKO hippocampal neurons upon 30 sec. application of 5 μM Ionomycin (blue bar). (G) Histograms, (H) cumulative plot and (I) summary graph of DCV fusion events per cell. Mann-Whitney U test: *** p = < 0.0007.
See also Figure S6.
To confirm that RIM interaction with voltage-gated calcium channels is not essential for DCV fusion, we first measured calcium influx using Fluo5-AM in synaptic and extra-synaptic regions. Both RIM cDKO neurons and control neurons showed a calcium influx profile corresponding with the bursts of repetitive stimulation (Fig. 3E–F). In RIM cDKO neurons multiple bursts of activity were required to reach maximum fluorescence (Fig. 3E–F). Next, we applied the calcium ionophore Ionomycin to increase intracellular calcium levels independently of voltage-gated calcium channels. Ionomycin (5 μM) elicited robust DCV fusion in control but not in RIM cDKO neurons (Fig. 3G–I, S6L–M). From these data, we conclude that a defect in calcium entry or location of voltage-gated calcium channels cannot explain the lack of DCV fusion in RIM cDKO neurons.
Over-expression of MUNC13 rescues DCV fusion in RIM cDKO neurons
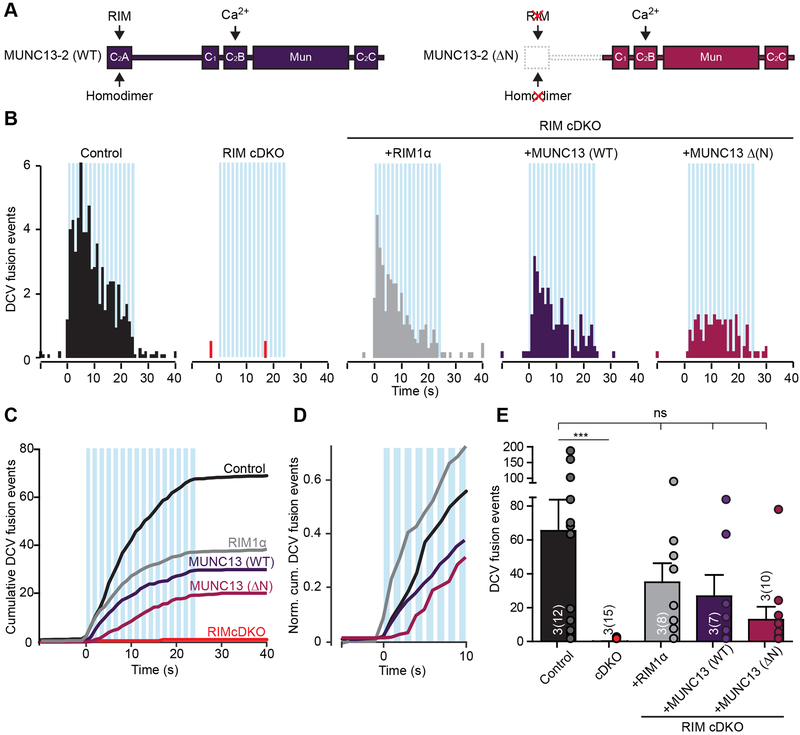
RIMs prime synaptic vesicle fusion by converting an autoinhibitory MUNC13 homodimer into an activated heterodimer by interaction of the RIM Zn2+ finger with the C2A domain of MUNC13 (Camacho et al., 2017). To test whether activation of MUNC13 is required for DCV fusion, we expressed wild-type ubMUNC13–2 (Fig. 4A, left) or N-terminally truncated ubMUNC13–2 (Fig. 4A, right; MUNC13–2 ΔN), which does not interact with RIM1α or forms homodimers (Deng et al., 2011), in RIM cDKO neurons. Over-expression of MUNC13–2 (WT) or MUNC13–2 (ΔN) restored DCV fusion (Fig. 4B–E, S7A-D), although RIM-deficient cDKO neurons expressing MUNC13–2 (ΔN) required multiple bursts of stimulation before DCV fusion to peak (Fig. 4D). These results show that over-expression of MUNC13–2 rescues DCV fusion in RIM cDKO neurons and that the N-terminal C2A domain of MUNC13–2 is not required for DCV fusion. Furthermore, the data suggest that MUNC13 supports DCV fusion independent of RIM when over-expressed.
Figure 4. Expression of MUNC13 rescues DCV fusion in RIM cDKO neurons.
(A) Diagram of MUNC13–2 wild-type (WT) and MUNC13–2 (ΔN) rescue proteins expressed in RIM cDKO neurons. Key domains and interactions are indicated. Constructs were labeled at the C-terminus with mCherry to visualize expression (not indicated)..
(B-E) DCV fusion analysis using NPY-pHluorin in single control or RIM cDKO hippocampal neurons without or with expression of full-length RIM1α (grey), MUNC13–2 WT (purple) or MUNC13–2 (ΔN) (magenta). (B) Histograms, (C) cumulative plot, (D) normalized cumulative plot of first 10 seconds of stimulation and (E) summary graph of DCV fusion events per cell. Kruskal-Wallis with Dunn’s correction: *** p < 0.001. ns, p > 0.05..
See also Figure S7.
N-terminal domain of RIM1α is sufficient to support DCV fusion
To test if the N-terminal interactions of RIMs are sufficient to support DCV fusion, we expressed different N-terminal RIM1 fragments in RIM cDKO neurons (Fig. 5A; Deng et al., 2011). Rescue with the N-terminal RIM1α fragment containing RAB3- and MUNC13-binding sequences (RIM1α-RZ) fully restored DCV fusion (Fig. 5B–D, S7E–I). Expression of RIM1β-Z, which lacks RAB3-binding but still binds MUNC13, rescued DCV fusion in RIM cDKO neurons but significantly lower compared to control (Fig. 5B–D, S7E–I, similar to full-length RIM1β, Fig. 3D). Both the N-terminal RIM1α and RIM1β fragments containing the Zn2+-finger mutations eliminating MUNC13 binding (RIM1α-RZ-K144/6E and RIM1β-Z-K144/6E) did not restore DCV fusion in RIM cDKO neurons (Fig. 5B–D, S7E–I). Together, these data show that the N-terminal fragment of RIM is sufficient to support DCV fusion, and it does so with more efficiency than for synaptic vesicle fusion (Deng et al., 2011). Furthermore, interactions with both RAB3 and MUNC13 are important.
Figure 5. RIM1α N-terminal domain is sufficient to restore DCV fusion in RIM cDKO neurons.
(A) Diagram of N-terminal wild-type and mutant RIM rescue domains expressed in RIM cDKO neurons. Key domains and interactions are indicated. Zn, zinc-finger domain with surrounding α-helical regions; H, location of a HA-tag; asterisk, K144/6E substitution..
(B-D) DCV fusion analysis using NPY-pHluorin in single control or RIM cDKO hippocampal neurons without or with expression of full-length RIM1α (grey) or N-terminal rescue constructs (RIM cDKO + rescue). (B) Histogram, (C) cumulative plot and (D) summary graph of DCV fusion events per cell. Kruskal-Wallis with Dunn’s correction: *p < 0.05, **p <0.01, ***p < 0.001. ns, p > 0.05..
See also Figure S7.
N-terminus of RIM interacts with DCVs through RAB3A.
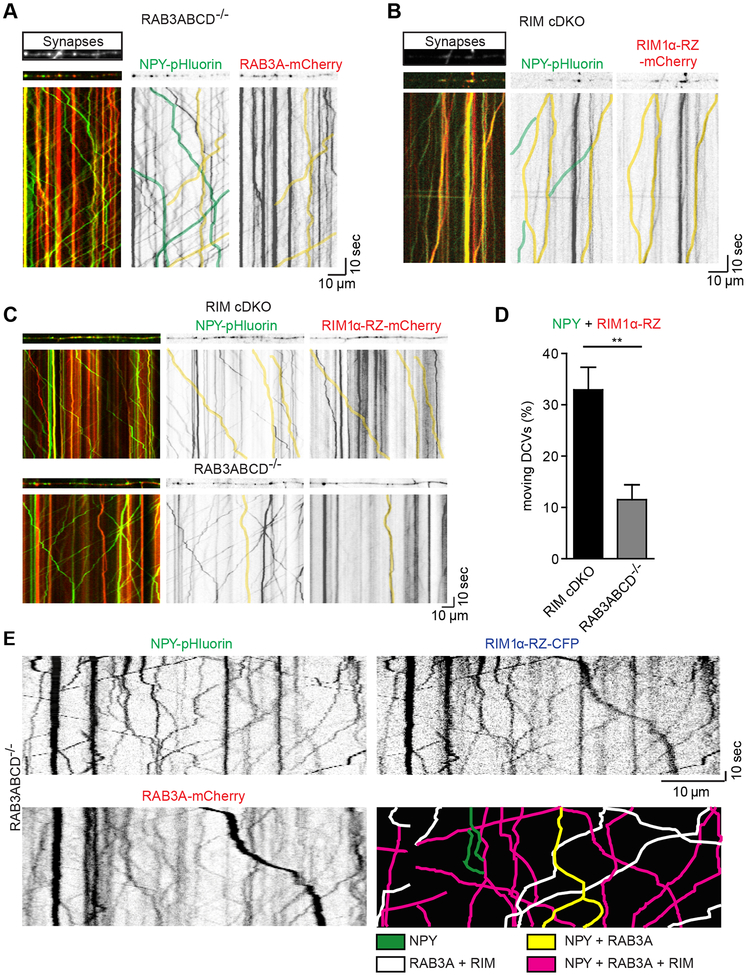
RAB3 binds to SVs (Fischer von Mollard et al., 1990) and to Secretogranin II positive secretory granules in PC12 cells (Handley et al., 2007). To assess whether RAB3A is present on DCVs in hippocampal neurons, colocalization experiments were performed in RAB3 QKO neurons expressing RAB3A-mCherry and NPY-pHluorin (Fig. 6A). RAB3A-mCherry was found in stationary deposits at synapses (Fig. 6A), co-transported with NPY-pHluorin (Fig. 6A; yellow lines) or transported without colocalizing with NPY-pHluorin (Fig. 6A). Also, moving NPY-pHluorin puncta negative for RAB3A-mCherry were found (Fig. 6A; green lines). These data suggest that RAB3A is transported on a subset of DCVs.
Figure 6. RIM N-terminus interaction with DCVs is reduced in RAB3 deficient neurons.
(A) Representative kymograph showing trajectories of DCVs (NPY-pHluorin, green) and RAB3A-mCherry (red) in RAB3ABCD−/− neurons. Synapsin-ECFP labels synapses (above kymograph). NPY only transport (green) and co-transport of NPY and RAB3A (yellow) is indicated by overlays.
(B) Representative kymograph showing trajectories of DCVs (NPY-pHluorin, green) and N-terminal fragment RIM1α-RZ-mCherry (red) in RIM cDKO neurons. Synapsin-ECFP labels synapses (above kymograph). NPY only transport (green) and co-transport of NPY and RIM1α-RZ-mCherry (yellow) is indicated by overlays.
(C) Example kymographs of NPY-pHluorin and RIM1α-RZ-mCherry transport in RIM cDKO (top) and RAB3 QKO (bottom) neurons. Co-transport is indicated by yellow lines.
(D) Quantification of co-transport of NPY-pHluorin with RIM1α-RZ-mCherry in RIM cDKO (black) and RAB3 QKO (grey) neurons. A subset of moving NPY puncta per cell were quantified for trafficking with or without RIM1α-RZ-mCherry. Percentage of moving NPY co-trafficking with RIM1α-RZ-mCherry per cell is shown.
(E) Example kymograph of co-transport of NPY-pHluorin with RIM1α-RZ-ECFP and RAB3A-mCherry in RAB3 QKO neurons. Graphical overlay (bottom right) indicates transport of NPY (green), co-transport of NPY with RAB3A (yellow), NPY with RIM1α-RZ (white) or NPY with RAB3A and RIM1α-RZ (magenta).
Neurons were imaged in the presence of Tyrode’s solution containing 50 mM NH4Cl to visualize all DCVs..
See also Figure S8.
RIMs are expressed at the presynaptic active zone (Wong et al., 2018). To investigate if RIMs also interact with DCVs, we infected neurons with full-length mCherry-tagged RIM1 but did not obtain sufficient expression. However, the mCherry-tagged N-terminal RIM rescue construct (RIM1α-RZ-mCherry) did express well enough to study transport of RIM1α-RZ-mCherry and NPY-pHluorin in RIM cDKO neurons (Fig. 6B). RIM1α-RZ-mCherry was predominantly located at synapses (Fig. 6B), but also co-transported with a subset of NPY-pHluorin labeled vesicles outside synapses (Fig. 6B; yellow lines). We hypothesized that the interaction between DCVs and the N-terminus of RIM1α is mediated by RAB3A. To test this, the percentage of moving NPY-pHluorin labeled DCVs positive for RIM1α-RZ-mCherry was quantified in RAB3 QKO neurons and RIM cDKO neurons (Fig. 6C–D). In RIM cDKO neurons, approximately 33% of moving NPY-pHluorin puncta co-transported RIM1α-RZ-mCherry (Fig. 6D; black), while in RAB3 QKO neurons only 11.5% of moving NPY-pHluorin puncta were positive for RIM1α-RZ-mCherry (Fig. 6D; grey). Hence, in RAB3 QKO neurons the interaction of DCVs with the N-terminus of RIMs is partly lost, suggesting RIMs interact with DCVs mainly through RAB3.
To test if RAB3A and RIMs are transported together on DCVs, NPY-pHluorin, RIM1α-RZ-ECFP and RAB3A-mCherry were co-expressed in RAB3 QKO neurons (Fig. 6E) or wild-type neurons (Fig. S8A). Moving NPY-pHluorin puncta colocalized with RIM1α-RZ (Fig. 6E, S8A; white), with RAB3A (Fig. 6E, S8A; yellow) and co-transport of NPY with RAB3A and RIM1α-RZ (Fig. 6E, S8A; magenta) was observed. These data suggest that RAB3A and RIM1α -RZ are transported together on a subset of DCVs.
To test if MUNC13 and RIMs are transported together on DCVs, NPY-pHluorin, RIM1α-RZ-ECFP and MUNC13–2-mCherry were co-expressed in wild-type neurons (Fig. S8B). Endogenous MUNC13 localizes predominantly to synapses, with little immunoreactivity colocalizing with an endogenous DCV marker outside synaptic regions (Fig. S8C–E). However, MUNC13-mCherry colocalizes with moving NPY-pHluorin puncta positive for RIM1α-RZ (Fig. S8B; magenta). This co-trafficking of MUNC13 and RIM1α with DCVs suggests that MUNC13 and RIMs may form heterodimers on DCVs and RIM activates MUNC13 already while traveling through the axon. However, the limited evidence for colocalization of endogenous MUNC13 and DCVs and the fact that live imaging of full length RIM was not feasible, prevent strong conclusions.
Discussion
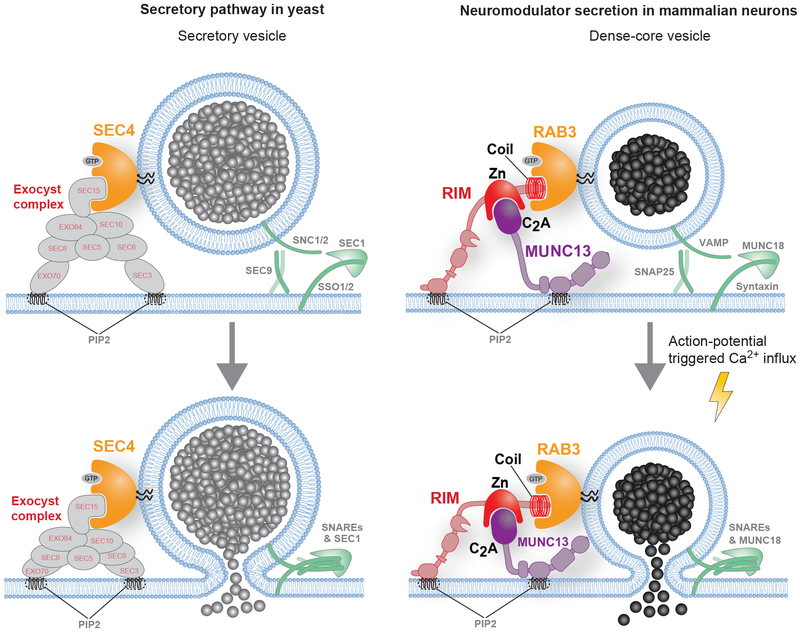
In this study, we identify essential roles for RAB3 and RIM1/2 in neuromodulator release in mammalian CNS neurons. RAB3 QKO neurons showed a 20-fold decrease in DCV fusion and RIM1/2 cDKO neurons a 100-fold decrease. DCV fusion in RIM cDKO neurons was rescued by expression of wild-type RIM1α, but not RAB3- or MUNC13-binding deficient RIM1 mutants. The N-terminal fragment of RIM1 that interacts with RAB3 and MUNC13 was sufficient to fully restore DCV fusion. This N-terminal fragment also co-trafficked with DCVs via RAB3. We conclude that RIMs are essential RAB3 effectors for mammalian neuromodulator release and organize DCV fusion by positioning/activating MUNC13 and recruiting DCVs through RAB3 interactions (Fig. 7), in analogy to the exocyst complex in yeast.
Figure 7. Function of RAB3, RIM and MUNC13 in DCV fusion.
In yeast (left), secretory vesicle binding to the plasma membrane (top, left) relies on the interaction between SEC4 (RAB3) and the Exocyst complex before SNARE-mediated fusion (bottom, left). In mammalian neurons (right), RAB3, RIM and MUNC13 regulate the late steps in DCV fusion. Through N-terminal interactions, RIMs position MUNC13 and recruit DCVs via RAB3, which is located on the vesicle (top, right). After this step, action potential triggered, SNARE-mediated fusion can occur (bottom, right). These interactions are essential for the organization of DCV fusion sites, in analogy to the exocyst complex in yeast. Zn, zinc-finger domain.
To our knowledge, the RIM1/2 null DCV fusion phenotype is stronger than any other null mutation studied so far: In MUNC13–1/2 double knock out neurons, 40% of DCV fusion events remain (van de Bospoort et al., 2012), 10–40% in the CAPS1/2 DKO (Farina et al., 2015; van Keimpema et al., 2017) and 10–20% upon deletion of SNAREs, i.e., SNAP25 knock out/down or TeTx expression (Arora et al., 2017; Shimojo et al., 2015). RIM deficient neurons showed a 100-fold reduction in evoked DCV fusion (approximately 0.75 fusion events per cell (72 events in 96 cells), compared to approximately 70 events per cell in controls (5063 event in 72 cells)). Hence, RIMs are required for neuromodulator release in hippocampal neurons and no redundant pathways exist. Furthermore, it is evident that SNAREs are not sufficient for DCV fusion in living neurons. Finally, the almost complete loss of NPY-pHluorin or BDNF-pHluorin events in the absence of RIM1/2 also confirms the specificity of the DCV-fusion reporters, because synaptic vesicle exocytosis is less strongly impaired in RIM1/2 cDKO neurons (de Jong et al., 2018; Kaeser et al., 2011).
RAB3A regulates the mammalian DCV secretory pathway at a late step, analogous to SEC4p in yeast
Since the original discovery that the ras-like GTP-binding protein SEC4p is one of the essential components in the last step of the secretory pathway in yeast (Goud et al., 1988), members of the RAB protein family have been found to regulate many intracellular fusion reactions (see for a review: Galvez et al., 2012). RAB3A has been considered to serve a similar role as SEC4p in the mammalian brain, due to the high homology to SEC4p, high expression levels in brain and dynamic association to synaptic vesicles (Fischer von Mollard et al., 1990; Takamori et al., 2006; Zahraoui et al., 1989). Furthermore, RAB3A regulates secretory granule fusion in pancreatic beta-cells (Yaekura et al., 2003), PC12 cells (Tsuboi and Fukuda, 2006) and sperm (Bustos et al., 2012), but synaptic transmission in RAB3A knock out mice is only mildly affected (Geppert et al., 1997) and deficiency for all four RAB3 paralogs hardly affects synaptic transmission (Schlüter et al., 2006). Also in C. elegans neurons and mouse chromaffin cells, RAB3 deficiency produces partial effects on membrane fusion, largely/partially explained by other defects (impaired vesicle biogenesis; Nonet et al., 1997; Schonn et al., 2010). The current data indicate that although RAB3s may not be the (only) unequivocal SEC4p ortholog for synaptic vesicle fusion, RAB3s are crucial in a late step of the DCV secretory pathway, analogous to its ortholog SEC4p.
To our knowledge, the current study describes the first major phenotype for RAB3 deficiency in the mammalian brain. Whereas synaptic vesicle fusion is hardly affected (Schlüter et al., 2006), DCV fusion is reduced 20-fold, while the number of DCVs and their transport were unaffected. RAB3 function may have become redundant for synaptic transmission altogether, or robust regulation of synaptic vesicle fusion is secured by emergence of additional paralogs such as RAB27A/B (Mahoney et al., 2006). For DCV fusion and neuromodulator release, no other (RAB) proteins endogenously expressed in hippocampal neurons compensate for the loss of RAB3 expression.
Interestingly, while synaptic transmission was largely intact, several behaviors were altered in RAB3A null mice, including circadian rhythmicity (Kapfhamer et al., 2002), reversal learning and exploration (D’Adamo et al., 2004), memory precision (Ruediger et al., 2011) and ethanol responses (Kapfhamer et al., 2002), although other mnemonic capabilities were normal (Hensbroek et al., 2003). While such effects have been interpreted in the context of synaptic deficits, for example the loss of mossy fiber LTP in RAB3A KO (Castillo et al., 1997), they are equally consistent with loss of neuromodulator signaling.
RIM1 is an essential RAB3 effector in and outside synapses
The absence of RIM1/2 resulted in a 100-fold reduction in DCV fusion, which was rescued by RIM1α-RZ, harboring only the RAB3- and MUNC13-binding domains. Conversely, DCV fusion was not rescued by re-expression of RIM1beta, which binds all known RIM1 binding partners except RAB3. Hence, RIM1 is an essential RAB3A effector for neuromodulator release in hippocampal neurons. The selective loss of RIM2 (i.e., in the presence of 1 or 2 intact RIM1 alleles) tended to increase DCV fusion compared to control (Fig. S6), suggesting that RIM2 may not have the same role in neuromodulator release as RIM1 in the neurons studied here.
The essential role of RIM1 for all neuromodulator release is unexpected because many DCV fusion events occur outside synapses, albeit with a low release probability (de Wit et al., 2009; Persoon et al., 2018; van de Bospoort et al., 2012), whereas RIM1 is an active zone protein. Fusion events that occur outside synapses or at dendrites were also absent in RIM1/2 deficient neurons. Hence, a small number of RIM molecules may be present at non-synaptic sites to support non-synaptic DCV fusion. However, despite detailed sub-cellular localization studies (Tang et al., 2016; Wong et al., 2018), there is no evidence for such non-synaptic localization of RIM1/2. Therefore, the possibility that RIM1/2 travels on DCVs, probably via RAB3 interaction (Fig. 6), and provides on-board support for DCV fusion, is an alternative and possible scenario. While expression of full length RIM1 produced cellular levels too low to detect unequivocal co-trafficking, the co-trafficking of the N-terminal fragment with DCVs was evident (Fig. 6) and the efficient rescue of DCV fusion with this construct (Fig. 5) supports such a scenario. The fact that labeled RIM1 zinc finger constructs travel with a 3-fold larger fraction of DCVs in RIM1/2-deficient neurons than in RAB3 QKO neurons is also consistent with the idea that endogenous RIM1/2 associates and travels with DCVs by binding RAB3.
Despite substantial co-trafficking, most DCVs appear to travel without detectable RIM1α-RZ (Fig. 6). However, non-synaptic DCV fusion is relatively rare (40%) and requires extreme stimulation intensity/frequency (Persoon et al., 2018; van de Bospoort et al., 2012). The number of RIM1-containing DCVs seems enough to explain these sparse events. DCV fusion depends on the t-SNARE SNAP25 (Arora et al., 2017; Shimojo et al., 2015) and most likely on Syntaxins, which are known to be abundantly expressed in axons, also outside synapses (Garcia et al., 1995). Taken together, these considerations suggest that some DCVs fuse at non-synaptic sites with a low probability using RAB3A and possibly RIM1 on the DCV, requiring only cytosolic molecules and the t-SNAREs at the target membrane, while most DCVs fuse at synapses exploiting the local enrichment of RIM1 and t-SNAREs at the target membrane.
The RIM1-MUNC13 interaction is essential for DCV fusion and neuromodulator release
In RIM1/2 deficient neurons, the loss of neuromodulator release is effectively rescued with full length RIM1, but not a RIM1 mutant with two amino acid mutations that prevent MUNC13–1 binding (Fig. 3). Moreover, a small N-terminal RIM1-fragment, containing only the RAB3 and MUNC13–1 binding sites also rescues release, but not when the same fragment contains the same two mutations (Fig. 5). These four observations suggest that in addition to the essential role of the RAB3-RIM interaction, the interaction between RIM1 and MUNC13–1 is also essential for neuromodulator release.
MUNC13–1 levels are 35% (Fig. S4G) to 67% (Deng et al., 2011) decreased in RIM1/2 deficient hippocampal neurons. The remaining endogenous MUNC13–1 levels apparently are not sufficient to support neuromodulator release in the absence of RIMs and the interaction between the two molecules is required, as recently proposed for synaptic vesicle fusion (Camacho et al., 2017). RIM1 is considered to disinhibit MUNC13s by competitive binding to their N-terminal homo-dimerization domain (Camacho et al., 2017; Deng et al., 2011; Dulubova et al., 2005). The fact that full length MUNC13 is even more efficient in rescuing neuromodulator release in the absence of RIM1/2 compared to a MUNC13 mutant lacking this homo-dimerization domain (Fig. 4), suggests that under physiological conditions, the interaction between RIM1 and MUNC13, and the disinhibition of MUNC13, may not be essential for neuromodulator release. Alternatively, other factors that monomerize MUNC13 could be present for DCV but not synaptic vesicles, or, more trivially, over-expression may have strong gain of function effects on DCV fusion independent of the presence of RIM. While MUNC13 effectively rescued DCV fusion in RIM-deficient neurons, the onset of evoked DCV fusion was slow (Fig. 4D). This may be explained by delayed calcium influx, as calcium responsiveness is not restored by expression of MUNC13 in RIM-deficient neurons (Deng et al., 2011).
MUNC13 is proposed to tether synaptic vesicles by bridging between vesicle and plasma membrane via its C2C-domain and C1/C2B-domains, respectively (Liu et al., 2016; Rizo, 2018). Such a mechanism may explain why MUNC13 over-expression rescues neuromodulator release in the absence of RIMs: high cellular MUNC13 levels may also bridge between DCV- and plasma membrane and partially compensate for the loss of the dominant native RIM-RAB3 tethering mechanism for DCVs. Recruitment of MUNC13 to release sites and MUNC13 activation may both contribute to the essential role of RIM1 in DCV fusion.
RAB3/RIM1 functions define diverging aspects among CNS regulated secretory pathways
Secretion of neurotransmitters from synaptic vesicles, of neuromodulators from DCVs and other forms of regulated secretion most likely emerged from a common ancestral mechanism. This study reveals several robust molecular differences between synaptic vesicle and DCV secretory pathways. First, while deficiency for all four RAB3s produces only a mild effect on synaptic vesicle fusion, DCV fusion is affected by >90%. Second, the role of RIMs PDZ domain and its known role in organizing/clustering Ca2+-channels (Kaeser et al., 2011) is important for synaptic vesicle- but not DCV fusion. This is likely because DCV fusion does not require tight coupling of Ca2+-channels to DCVs before fusion, and is consistent with the observation that DCV fusion relies on Ca2+ buildup during long action potential trains and bypassing Ca2+-channels using Ionomycin still produced a >95% reduction of DCV fusion in RIM1/2 null neurons. Third, RIM’s disinhibition of MUNC13 is important for synaptic vesicle- but not DCV fusion (see above). Fourth, while RIM1’s C2B domain is essential for RIM’s role in synaptic vesicle fusion by interaction with phosphatidylinositol 4,5-bisphosphate (de Jong et al., 2018), this domain, as well as 80% of RIM1 sequence downstream of the Zn-finger domain, which also interacts with ELKS, RIM-binding proteins and α-liprins (Hibino et al., 2002; Schoch et al., 2002; Wang et al., 2002), is dispensable for efficient neuromodulator release. These functional differences suggest that RIM’s two N-terminal domains are the core domains essential for ancestral secretion principles, and that the rest of the protein contains evolutionary adaptations that specifically support the ultra-fast, synchronous fusion of synaptic vesicles in synapses.
In yeast, the six exocyst complex subunits SEC3, 5, 6, 8, 10 and 15 are essential effectors of SEC4/RAB3 in the last steps in the secretory pathway, but their orthologs appear not to be important for regulated secretion in neurons, although available information is still scarce (Schwenger and Kuner, 2010). In invertebrates, the exocyst complex appears to be dispensable for synaptic transmission (Mehta et al., 2005; Murthy et al., 2003), but the RIM orthologs unc10/dRIM are not (Koushika et al., 2001; Liu et al., 2011), similar to vertebrate synapses and striatal dopamine varicosities (Deng et al., 2011; Kaeser et al., 2011; Liu et al., 2018). One possible scenario is that RIMs and MUNC13 have emerged in evolution, between unicellular organisms and nematodes/flies, as an alternative SEC4/RAB3 effector to the exocyst complex in regulated secretion. Consistent with this idea is that the exocyst complex is ubiquitously expressed in multicellular organisms, but RAB3 and RIM are strongly enriched in cells that specialize in regulated secretion.
In conclusion, our data shows that RAB3A and its effector RIM1 are responsible for the regulated secretion of chemical signals from DCVs in mammalian hippocampal neurons. RIM1 organizes neuromodulator vesicle fusion, also outside synapses, by positioning or activating MUNC13 and recruiting DCVs via RAB3s.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthijs Verhage (matthijs@cncr.vu.nl). All unique plasmids generated in this study are available from the Lead Contact without restriction. No other unique reagents were generated in this study.
Experimental model and subject details
Animals
Homozygous double conditional RIM1αβ/RIM2αβγ mutant mice described previously (Kaeser et al., 2011; Kaeser et al., 2008) were used for timed mating of homozygous cDKO mice. All newborn (P1) RIM cDKO mice used for experiments were genotyped by PCR (Kaeser et al., 2011). RAB3ABCD KO mice have been described previously (Schlüter et al., 2004). As RAB3ABCD−/− embryos die shortly after birth, embryonic day 18.5 embryos were obtained by caesarean section of pregnant females from timed matings of RAB3A+/−, BCD−/− mice and genotyped by PCR as described previously (Schlüter et al., 2004). C57BL/6 mice were used for wild-type cultures. For glia preparations newborn pups from female Wistar rats were used. Animals were housed and bred according to institutional and Dutch governmental guidelines (DEC-FGA 11–03 and AVD112002017824).
Method details
Primary neuronal cultures
Dissociated hippocampal neuron cultures were prepared from newborn (P1) RIM cDKO mice, E18.5 C57BL/6 (wild-type) or RAB3A+/−, BCD−/− littermate mouse embryos. Cerebral cortices were dissected free of meninges in Hanks’ balanced salt solution (Sigma, H9394) supplemented with 10 mM HEPES (Gibco, 15630–056). The hippocampi were isolated from the tissue and digested with 0.25% Trypsin (Gibco, 15090–046) in Hanks-HEPES for 20 min. at 37°C. Hippocampi were washed three times with Hanks-HEPES and triturated with fire-polished glass pipettes. Dissociated neurons were counted and plated in neurobasal medium (Gibco, 21103–049) supplemented with 2% B-27 (Gibco, 17504–044), 1.8% HEPES, 0.25% glutamax (Gibco, 35050–038) and 0.1% Penicillin-Streptomycin (Gibco, 15140–122). To obtain single neuron cultures, hippocampal neurons were plated in 12-well plates at a density of 1100–1400 cells/well on 18 mm glass coverslips containing micro-islands of rat glia. Micro-islands were generated as described previously (Meijer et al., 2012) by plating 8000/well rat glia on UV-sterilized agarose (Type II-A; Sigma, A9918)-coated etched glass coverslips stamped with a mixture of 0.1 mg/ml poly-D-lysine (Sigma, P6407), 0.7 mg/ml rat tail collagen (BD Biosciences, 354236) and 10 mM acetic acid (Sigma, 45731). High-density dissociated neuron cultures for protein quantitation and secretion measurements were prepared from cortex tissue and plated on plastic 12-wells or 6-wells plates coated with 0.01% poly-L-ornithine (Sigma, P4957) and 2.5 μg/ml laminin (Sigma, L2020) diluted in Dulbecco’s phosphate-buffered saline (DPBS; Gibco, 14190–250) overnight at RT.
Constructs
EGFP-Cre and EGFP-control (mutant Cre) constructs were created previously (Kaeser et al., 2011) and contained an additional nuclear localization sequence of nucleoplasmin in the N-terminus of EGFP to ensure complete nuclear localization of EGFP. NPY-pHluorin or NPY-mCherry were generated by replacing Venus in NPY-Venus with super-ecliptic pHluorin or red fluorescent mCherry (van de Bospoort et al., 2012). Generation of BDNF-pHluorin was described previously (de Wit et al., 2009). Synapsin-mCherry was a kind gift of Dr A. Jeromin (Allen Brain Institute, Seattle, USA) and synapsin-ECFP was obtained by replacing mCherry with ECFP. The RIM1 rescue constructs RIM1α, RIM1β, RIM1α-ΔPDZ, RIM1α-K144/6E, RIM1α-RZ, RIM1α-RZ-K144/6E, RIM1β-Z, RIM1β-Z-K144/6E were described previously (Deng et al., 2011; Kaeser et al., 2011). They were generated from a rat RIM1α or RIM1β expression plasmid and contained an HA tag. RIM1α-RZ-mCherry or RIM1α-RZ-ECFP were created by adding mCherry or ECFP to the C-terminus. The MUNC13–2(WT) and MUNC13–2(ΔN) constructs were described previously (Deng et al., 2011) and contained mCherry at the C-terminus. RAB3A, B, C and D constructs were obtained from a mouse cDNA library by PCR and labeled with mCherry at the C-terminus. All constructs were cloned to synapsin-promoter driven constructs, sequenced verified and subcloned into pLenti vectors and viral particles were produced. Titration of lentiviral particle batches was performed by assessment of number of fluorescent cells upon infection to ensure 100% infection efficiency.
Lentiviral Infection
Neuronal RIM cultures were infected with EGFP-Cre or EGFP-control lentiviral particles at DIV 0 or DIV 5. To visualize DCV fusion and transport (Fig. 2, S3B–I, S5B–H,M–O) cultures were infected with lentiviral particles encoding for NPY-pHluorin, NPY-mCherry and Syn-ECFP at DIV 9–10. For other DCV fusion experiments cultures were infected at DIV 9–10 with Syn-ECFP and the DCV reporter NPY-pHluorin or BDNF-pHluorin, allowing single color live cell imaging of DCV fusion and/or the addition of other constructs. For rescue experiments cultures were infected with the corresponding rescue construct at DIV 0. Neurons were post-hoc fixed and immunostained for the HA-tag present on all rescue constructs to validate expression of rescue constructs in individual neurons.
Protein quantitation
To characterize protein expression levels of RIM1/2 upon Cre-recombinase expression, high-density dissociated cortical cultures of RIM cDKO mice were infected at DIV 0 with Cre or control virus. At indicated time-points neurons were washed in cold PBS and homogenized in Laemmli sample buffer consisting of 2% SDS (VWR chemicals, M107), 10% glycerol (Merck, 818709), 0.26 M β-mercaptoethanol (Sigma, M3148), 60 mM Tris-HCl (Serva, 37180) pH 6.8, and 0.01% Bromophenol blue (Applichem, A3640). To measure chromogranin B (CHGB) levels in RAB3 KO neurons, cortex tissue of RAB3A+/+, +/− or −/−, BCD−/− animals was homogenized in Laemmli sample buffer. Samples were separated on 7% SDS-polyacrylamide gels with 2,2,2-Trichloroethanol using standard SDS-PAGE technique and scanned in a Gel Doc EZ imager (Bio-Rad). Proteins were transferred to membrane O/N at 150 mA, 4°C. Blots were incubated in 2% BSA (Acros Organics, 268131000) - PBS containing 0.1% Tween-20 (Sigma, P2287) for 1 hour at 4°C. Subsequently blots were incubated with primary antibodies in 2% BSA-PBS-0.1% Tween20 for 4 hours at RT. Primary antibodies included polyclonal rabbit RIM1/2 ZN-finger (1:1000; SySy, 140203), monoclonal mouse β3-Tubulin (1:1000; Cell Signaling, 4466), polyclonal rabbit Chromogranin B (1:500; SySy 259103) and monoclonal mouse Actin (1:10,000; Chemicon, MAB1501). After washing with PBS-0.1%Tween-20, blots were incubated with secondary antibodies (anti-rabbit or anti-mouse IRDye secondary antibodies (1:10,000; LI-COR) or alkaline phosphatase-conjugated secondary antibodies (1:10,000; Jackson) in 2% BSA-PBS-0.1% Tween20 for 45 min. at 4°C. After washing blots were scanned with Odyssey FC dual-mode imaging system (LI-COR) for 2 minutes in each channel (700 and 800 nm laser). When labeled with alkaline phosphatase-conjugated secondary antibodies, blots were incubated with AttoPhos substrate for 5 min, and scanned on a Fujifilm FLA-5000 Reader.
ELISA
High-density cultures (DIV 8) were washed ones with Tyrode’s solution (119 mM NaCl, 2.5 mM KCl, 2 mM CaCl2*2H20, 2 mM MgCl2*6H20, 25 mM HEPES and 30 mM Glucose*H20, pH 7.4, mOsmol 280) containing protease inhibitor cocktail (Sigma, S8830), and subsequently incubated for 1 min. with 200 μl Tyrode’s solution and supernatant was collected. Cultures were then incubated for 1 min. with 200 μl Tyrode’s solution containing 60 mM KCl, which replaced NaCl on an equimolar basis in the solution, and supernatant was collected. Mouse BDNF ELISA (Biosensis, BEK-2003) was used according to the manufacturer’s instructions and measured with a Spectramax I3 plate reader (Molecular Devices) to quantify BDNF levels in supernatant samples.
Immunocytochemistry
Hippocampal cultures were fixed in 3.7% formaldehyde (Electron Microscopies Sciences, 15680) in PBS, pH 7.4, for 20 min at RT. After several washing steps with PBS, cells were permeabilized for 5 min with 0.5% Triton X-100 (Fisher Chemical, T/3751/08)-PBS and subsequently incubated for 30 min. with PBS containing 2% normal goat serum (Gibco, 16210–072) and 0.1% Triton X-100 to block nonspecific binding. Incubations with primary and secondary antibodies were performed for 1 hr at RT with PBS washing steps in between. Primary antibodies used were: Polyclonal rabbit Chromogranin B (1:500; SySy 259103), polyclonal chicken MAP2 (1:10,000; Abcam ab5392), monoclonal mouse SMI‐312 (1:5000; Covance), polyclonal guinea pig VGLUT1 (1:5000; Millipore AB5905), monoclonal mouse HA (12CA5, 1:500; Roche 11583816001), monoclonal mouse MUNC13–1 (1:1000; SySy 126111), polyclonal guinea pig Synaptophysin1 (1:500; SySy 101004), monoclonal mouse Homer1 (1:500; SySy 160011), monoclonal mouse PSD95 (1:200; Abcam ab2723). Alexa Fluor conjugated secondary antibodies were from Invitrogen (1:1000). Coverslips were washed again and mounted with Mowiol 4–88 (Sigma, 81381) and examined on a confocal A1R microscope (Nikon) with LU4A laser unit using a 40x oil immersion objective (NA=1.3). Images were acquired at 1024×1024 pixels as z-stacks (5 steps of 0.5 μm) and resulting maximum projection images were used for analysis. A 60x oil immersion objective (NA 1.4) was used for zooms. Confocal settings were kept constant for all scans within an experiment.
Electron Microscopy
Hippocampal single cultured RIM cDKO neurons (Cre and control infected) were fixed for 60 min. at room temperature with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4), post-fixed for 1h at room temperature with 1% OsO4/1% K4Ru(CN)6 in double distilled water. Following dehydration through a series of increasing ethanol concentrations, cells were embedded in Epon and polymerized for 24 h at 65°C. After polymerization of the Epon, the coverslip was removed by alternately dipping in liquid nitrogen and hot water. Cells were selected by observing the Epon embedded culture under the light microscope, and mounted on pre-polymerized Epon blocks for thin sectioning. Ultrathin sections (approximately 70nm) were cut parallel to the cell monolayer and collected on single-slot, formvar-coated copper grids, and stained in uranyl acetate and lead citrate.
Synapses were selected at low magnification using a JEOL 1010 electron microscope. All analyses were performed on single ultrathin sections of randomly selected synapses. Digital images of synapses were taken at 80,000x magnification using iTEM software (EMSIS, Germany). For all morphological analyses, we selected only synapses with intact synaptic plasma membranes with a recognizable pre- and postsynaptic density and clear synaptic vesicle membranes. DCVs were recognized as an electron dense core surrounded by a vesicular membrane.
Live imaging
Live imaging experiments were performed on a Nikon Ti-E Eclipse inverted microscope system fitted with a Confocal A1R (LU4A Laser) unit and an EMCCD camera (Andor DU-897). The inverted microscope together with the EMCCD were used for live imaging using the LU4A laser unit with a 40x oil objective lens (NA 1.3) and appropriate filter sets. NIS elements software (version 4.30) controlled the microscope and image acquisition. Coverslips were placed in an imaging chamber and perfused with Tyrode’s solution (119 mM NaCl, 2.5 mM KCl, 2 mM CaCl2*2H20, 2 mM MgCl2*6H20, 25 mM HEPES and 30 mM Glucose*H20, pH 7.4, mOsmol 280). Isolated single neurons on glial‐islands were selected for acquisition and Synapsin-ECFP signal was recorded (z-stack, 5 steps of 0.5μm). Time-lapse (2Hz, exposure time dual-color imaging: 80ms per channel, single color imaging: 150 ms) recordings consisted of 30 seconds baseline recordings followed by stimulation. Electrical field stimulation was applied through parallel platinum electrodes powered by a stimulus isolator (WPI A385) delivering 30-mA, 1-ms pulses, regulated by a Master‐8 pulse generator (A.M.P.I.) providing 16 trains of 50 action potentials (Aps) at 50 Hz with a 0.5 sec interval. Chemical stimulation of 5 μM Ionomycin, (Fisher BioReagent) dissolved in normal Tyrode’s solution, was applied through glass capillaries placed in close proximity to the cell by gravity flow. To visualize the total pool of NPY-pHluorin, intracellular pH was neutralized by barrel application of normal Tyrode’s solution containing 50 mM NH4Cl, which replaced NaCl on an equimolar basis in the solution. To define calcium influx profiles upon stimulation, neurons were incubated for 15 min with 1 μM Fluo-5F-AM (Molecular Probes, F14222; stock in DMSO). Co-trafficking experiments with NPY-pHluorin were conducted in the presence of normal Tyrode’s solution containing 50 mM NH4Cl, to visualize the total pool of NPY-pHluorin during the recording. All experiments were performed at RT (20–24°C). To ensure expression of HA-tagged rescue constructs, coverslips were fixed in 4% PFA-PBS after imaging for immunocytochemistry.
Imaging analysis
Synapse number, DCV poolsize, neuronal morphology
Neuronal morphology and synapse or DCV numbers were analyzed using automated image analysis software synD (Schmitz et al., 2011). Synapse detection settings were optimized to measure VGLUT1, CHGB puncta or NPY-pHluorin signal and kept constant for the corresponding dataset. For co-localization analysis of different markers, morphological masks were drawn using SynD and imported in ImageJ to remove background fluorescence. Co-localization was measured in ImageJ with JACoP. Thresholds were set manually to correct for background.
DCV fusion
DCV fusion events were analyzed in stacks of time-lapse recordings (2 Hz, 512×512 pixels). In ImageJ DCV fusion events were manually selected and fluorescent traces were measured in a circular 4×4 pixel ROI (1.56×1.56 μm). NPY-pHluorin or BDNF-pHluorin events were defined by a sudden increase in fluorescence, NPY-mCherry events as a sudden decrease in fluorescence. Resulting fluorescent traces were loaded in a custom-built Matlab plugin where the traces were expressed as fluorescence change (ΔF) compared to initial fluorescence (F0) obtained by averaging the first 10 frames of the time-lapse recording. Fusion events were automatically detected and included when fluorescence showed a sudden increase (NPY-pHluorin/BDNF-pHluorin) or a sudden decrease (NPY-mCherry) two standard deviations above or below F0. Start of a fusion event was defined as the first frame above/below 2xSD of F0 and end of the fusion event as the first frame below this threshold. Total pool measurements were performed in SynD on recordings of NPY-pHluorin after application of 50 mM NH4Cl containing Tyrode’s solution to neutralize the intracellular pH and defined as the number of NPY-pHluorin puncta. For normalization of cumulative number of DCV fusion events over time, each condition was normalized to 1. The number of DCV fusion events in RAB3 QKO neurons rescued with RAB3A,B,C or D (Fig. 1F–H) was normalized by dividing the number of fusion events by the cumulative average of RAB3 TKO as control. Co-localization of the area of a fusion event with synapsin-ECFP (z-stacks) was defined as a synaptic event, remaining events were classified as extra-synaptic events. Using ImageJ software binary images of maximum projections of the z-stacks were created and events were defined synaptic if the fluorescent region of the fusion event co-localized with Synapsin-ECFP signal. Immunostainings with a panel of endogenous pre- and post-synaptic markers (Synaptophysin, VGLUT1 and Homer, PSD95, respectively) on neurons expressing Synapsin-ECFP (Fig. S2A) were used to quantify co-localization of Synapsin-ECFP with these markers, which showed 90% colocalization with pre-synaptic, and 80% with post-synaptic markers (Fig. S2B–C).
Calcium imaging
Calcium measurements were performed in ImageJ. Maximum projections of Synapsin-ECFP were used to define synaptic and extra-synaptic regions. Five neurite-located ROIs (4×4 pixels) and a background ROI were measured per neuron. Normalized ΔF/F0 data was calculated per cell after background subtraction.
DCV transport
Transport of DCVs in RIM cDKO neurons and controls was measured in time-lapse recordings (2Hz) consisting of 30 s baseline imaging, 24 s stimulation (16×50AP at 50 Hz) and 30 s imaging after stimulation. Stacks were divided in 20×20 regions (ImageJ, Grid) and transport was measured in three random regions (coordinates generated by random number generation in Matlab). NPY-mCherry labeled vesicles were manually tracked and vesicle trajectories were obtained using the imageJ plugin MtrackJ. If no vesicles were present in a selected region, the closest region containing DCVs was analyzed. Resulting velocity and distance measurements were analyzed. A vesicle was regarded as moving when the minimum distance between two consecutive frames was 400 nm (1 pixel) or more and if the total distance moved from start was at least 800 nm (2 pixels). Kymographs were created in ImageJ (MultipleKymograph, line width 3) to assess colocalization of NPY-pHluorin and RAB3A-mCherry or RIM1α-RZ-mCherry. Colocalization was only measured in moving NPY-pHluorin puncta. The soma was always excluded from analysis.
Quantification and statistical analysis
Shapiro-Wilk test was used to assess distribution normality. When assumptions of normality or homogeneity of variances were met, parametric tests were used: Student’s t-test or one-way ANOVA (Tukey as post-hoc test). Otherwise, non-parametric tests used were: Mann–Whitney U-test for 2 independent groups, or Kruskal–Wallis with Dunn’s correction for multiple groups. Data are plotted as mean with standard error of the mean; N represents number of independent experiments, n the number of cells and are indicated in figures and/or figure legends. Detailed information per data set (average, SEM, n and detailed statistics) is shown in table below.
Table:
detailed data and statistics:
| Dataset | Condition | Value (mean ± SEMs) | n1 | p-value | Statistical test |
|---|---|---|---|---|---|
| RAB3ABCD: DCV fusion experiments | |||||
| DCV fusion events (#) –NPY | Wild-type | 84.76 ± 16.88 | 5(21) | **** p < 0.0001 | Mann-Whitney U test |
| Total DCV pool (#) | Wild-type | 1260 ± 103.5 | 5(21) | ns2, p=0.1049 | Student’s t-test |
| Spontaneous DCV fusion (#/30 sec) | Wild-type | 0.67 ± 0.24 | 5(21) | ns, p = 0.1783 | Mann-Whitney U test |
| Peak fusion intensity (F/F0) | Wild-type | 1.809 ± 0.01 | 5(1780) | **** p<0.0001 | Mann-Whitney U test |
| DCV fusion events (#) – NPY | (1) A+/+, BCD−/− | 427.4 ± 145.4 | 3(14) | ns, p>0.05: 1 vs 3, 1 vs 5, 1 vs 6, 2 vs 4, 2 vs 5, 2 vs 6, 3 vs 5, 3 vs 6, 4 vs 5, 4 vs 6, 5 vs 6 * p < 0.05: 2 vs 3 ** p<0.01: 1 vs 2, 3 vs 4 ***p<0.001: 1 vs 4 |
Kruskal-Wallis with Dunn’s correction |
| Normalized DCV fusion events (#) | (1) A+/+, BCD−/− | 1.00 ± 0.34 | 3(14) | ns, p>0.05: 1 vs 3, 1 vs 5, 1 vs 6, 2 vs 4, 2 vs 5, 2 vs 6, 3 vs 5, 3 vs 6, 4 vs 5, 4 vs 6, 5 vs 6 * p < 0.05: 2 vs 3 ** p<0.01: 1 vs 2, 3 vs 4 ***p<0.001: 1 vs 4 |
Kruskal-Wallis with Dunn’s correction |
| Total DCV pool (#) | (1) A+/+, BCD−/− | 4441 ± 372.0 | 3(14) | ns, p > 0.05 | One-way ANOVA |
| Spontaneous DCV fusion (#/30 sec) | (1) A+/+, BCD−/− | 4.29 ± 1.91 | 3(14) | ns, p>0.05: 1 vs 2, 1 vs 3, 1 vs 4, 1 vs 5, 1 vs 6, 2 vs 4, 2 vs 5, 2 vs 6, 3 vs 5, 4 vs 5, 4 vs 6, 5 vs 6 * p < 0.05: 2 vs 3, 3 vs 4, 3 vs 6 |
Kruskal-Wallis with Dunn’s correction |
| Fluorescent intensity RAB3A-D constructs | (1) Endogenous RAB3A – synaptic extra-synaptic |
0.86 ± 0.08 1.99 ± 1.76 |
15 15 |
1: synaptic vs extra-synaptic: **** p<0.0001 | 1+4: Mann-Whitney U test 2,3,5: Student’s t-test |
| DCV fusion events (#) –BDNF | Wild-type | 59.85 ± 13.97 | 5(20) | ** p < 0.0054 | Mann-Whitney U test |
| ELISA – BDNF | (1) A+/+, BCD−/−; control | 21.92 ± 6.17 | 6(12) | ns, p>0.05 | Kruskal-Wallis with Dunn’s correction |
| ELISA – (corrected for baseline secretion) | A+/+, BCD−/− | 10.84 ± 14.01 | 6(12) | ns, p>0.05 | Mann-Whitney U test |
| Pearson’s correlation SynapsinECFP | SynECFP::SYP | 0.853 ± 0.02 | 1(10) | ns, p >0.05: 1 vs 2, 3 vs 4 | Mann-Whitney U test |
| Manders’ coefficients | (1) M1; SynECFP::SYP | 0.916 ± 0.02 | 1(10) | ns, p>0.05: 3 vs 4 **p <0.0073: 1 vs 2 |
Mann-Whitney U test |
| RAB3ABCD: morphological and protein level analysis | |||||
| Synapse number | (1) Wild-type | 710.2 ± 58.41 | 3(28) | ns, p>0.05 | Kruskal-Wallis with Dunn’s correction |
| Dendritic length (mm) | (1) Wild-type | 2147 ± 132.9 | 3(28) | ns, p>0.05 | One-way ANOVA |
| Synapse number per μm dendrite | (1) Wild-type | 0.32 ± 0.01 | 3(28) | ns, p>0.05: 1 vs 3 | One-way ANOVA |
| VGLUT1 intensity (F) | (1) Wild-type | 2883 ± 123.5 | 3(28) | ns, p>0.05 | Kruskal-Wallis with Dunn’s correction |
| CHGB puncta (#) | (1) Wild-type | 6358 ± 558.9 | 3(20) | ns, p>0.05: 1 vs 3 | Kruskal-Wallis with Dunn’s correction |
| Neurite length (mm) | (1) Wild-type | 12598 ± 1328 | 3(20) | ns, p>0.05: 1 vs 2, 1 vs 3 * p < 0.05: 2 vs 3 |
Kruskal-Wallis with Dunn’s correction |
| CHGB puncta per μm neurite | (1) Wild-type | 0.54 ± 0.03 | 3(20) | ns, p>0.05: 1 vs 3, 2 vs 3 * p < 0.05: 1 vs 2 |
One-way ANOVA |
| CHGB intensity (F) | (1) Wild-type | 2172 ± 162.2 | 3(20) | ns, p>0.05: 1 vs 2 *** p < 0.001: 1 vs 3, 2 vs 3 |
One-way ANOVA |
| ChgB levels (westernblot) | (1) Wild-type | 1.0 ± 0.0 | 5 | ns, p = 0.5231 | Kruskal-Wallis with Dunn’s correction |
| RIM 1/2 experiments: DCV fusion experiments | |||||
| DCV fusion events (#) – NPY | Control | 25.8 ± 6.2 | 4(14) | ***p < 0.0002 | Mann-Whitney U test |
| NPY-pHluorin DCV fusion events | Control | 13.9 ± 4.4 | 4(14) | **** p = < 0.0001 | Mann-Whitney U test |
| Total DCV pool (#) | Control | 1300 ± 138.6 | 4(14) | ns, p= 0.9551 | Student’s t-test |
| Spontaneous DCV fusion (#/30 sec) | Control | 0.50 ± 0.17 | 4(14) | ns, p= 0.0508 | Mann-Whitney U test |
| DCV fusion events (#) – BDNF | Control | 75.68 ± 12.68 | 3(31) | ****p < 0.0001 | Mann-Whitney U test |
| DCV fusion events (#) | (1) Control | 99.6 ± 31.1 | 4(19) | ns, p > 0.05: 1 vs 3, 1 vs 4, 1 vs 5, 2 vs 6, 3 vs 4, 3 vs 5, 4 vs 5, 4 vs 6, 5 vs 6 *p < 0.05: 1 vs 6, 3 vs 6 **p < 0.01: 2 vs 4, 2 vs 5 ***p < 0.001: 1 vs 2, 2 vs 3 |
Kruskal-Wallis with Dunn’s correction |
| Total DCV pool (#) | (1) Control | 778.7 ± 66.63 | 4(19) | ns, p>0.05 | Kruskal-Wallis with Dunn’s correction |
| Fluorescent intensity RIM constructs | (1) cDKO+RIM1α: synaptic extra-synaptic |
2.65 ± 0.14 0.47 ± 0.27 |
24 24 |
1: synaptic vs extra-synaptic: **** p<0.0001 | Mann-Whitney U test |
| DCV fusion events (#) | Control (5 μM Ionomycin) | 60.0 ± 18.6 | 4(8) | *** p = < 0.0007 | Mann-Whitney U test |
| Total DCV pool (#) | Control (5 μM Ionomycin) | 611.9 ± 74.72 | 4(8) | ns, p=0.3229 | Student’s t-test |
| DCV fusion events (#) | (1) Control | 65.3 ± 18.4 | 3(12) | ns, p > 0.05: 1 vs 3, 1 vs 4, 1 vs 5, 2 vs 5, 3 vs 4, 3 vs 5, 4 vs 5 *p < 0.05: 2 vs 4 ** p < 0.01: 2 vs 3 *** p < 0.001: 1 vs 2 |
Kruskal-Wallis with Dunn’s correction |
| Total DCV pool (#) | (1) Control | 842.3 ± 118.0 | 3(12) | ns, p>0.05 | Kruskal-Wallis with Dunn’s correction |
| Fluorescent intensity MUNC13–2 constructs | (1) cDKO+ Munc13–2 (WT):Synaptic Extra-synaptic |
2.50 ± 0.42 1.59 ± 0.31 1.40 ± 0.18 |
18 18 21 |
1: ns, p=0.0556 2: ns, |
1: Mann-Whitney U test |
| DCV fusion events (#) | (1) Control | 92.1 ± 34.4 | 3(12) | ns, p > 0.05: 1 vs 3, 1 vs 4, 2 vs 4, 2 vs 5, 2 vs 6, 2 vs 7, 3 vs 4, 3 vs 7, 4 vs 5, 4 vs 6, 4 vs 7, 5 vs 6, 5 vs 7, 6 vs 7 *p < 0.05: 1 vs 7 ** p < 0.01: 1 vs 5, 3 vs 5 *** p < 0.001: 1 vs 2, 1 vs 6, 2 vs 3, 3 vs 6 |
Kruskal-Wallis with Dunn’s correction |
| Total DCV pool (#) | (1) Control | 741.5 ± 108.7 | 3(12) | ns, p > 0.05: 1 vs 2, 1 vs 3, 1 vs 4, 1 vs 5, 1 vs 6, 1 vs 7, 2 vs 4, 2 vs 5, 2 vs 6, 2 vs 7, 3 vs 4, 3 vs 6, 3 vs 7, 4 vs 5, 4 vs 6, 4 vs 7, 5 vs 6, 5 vs 7, 6 vs 7 *p < 0.05: 3 vs 5 *** p < 0.001: 2 vs 3 |
Kruskal-Wallis with Dunn’s correction |
| Fluorescent intensity RIM constructs | (1) cDKO+ RIM1α: synaptic extra-synaptic |
1.20 ± 0.23 0.32 ± 0.12 |
18 18 |
1: synaptic vs extra-synaptic: *** p<0.0002 | 1, 2, 4: Mann-Whitney U test 3, 5: Student’s t-test |
| DCV fusion events (#) | (1) Control | 63.67 ± 14.84 | 3(15) | ns, p > 0.05: 1 vs 2, 1 vs 3, 1 vs 4, 2 vs 3, 2 vs 4, 3 vs 4, 3 vs 5, 4 vs 5 *p < 0.05: 2 vs 5 **p < 0.01: 1 vs 5 |
Kruskal-Wallis with Dunn’s correction |
| Total DCV pool (#) | (1) Control | 577.9 ± 62.12 | 3(15) | ns, p > 0.05 | Kruskal-Wallis with Dunn’s correction |
| Spontaneous DCV fusion (#/30 sec) | (1) Control | 0.53 ± 0.24 | 3(15) | ns, p > 0.5027 | Kruskal-Wallis with Dunn’s correction |
| DCV fusion events (#) -Cre at DIV 5 | Control | 58.6 ± 20.91 | 2(5) | ** p < 0.01 | Mann Whitney U test |
| DCV fusion events (#) Wild-type Cre/control | Wild-type + control | 17.83 ± 3.71 | 1(6) | ns, p= 0.667 | Mann-Whitney U test |
| RIM 1/2 experiments: DCV transport | |||||
| Co-trafficking NPY + RIM1α-RZ (%) | RIM cDKO | 32.92 ± 4.40 | 1(14) | **, p = 0.0011 | Mann-Whitney U test |
| Average velocity (μm/s) | Control | 0.343 ± 0.03 | 6(18) | ns, p= 0.438 | Mann-Whitney U test |
| Velocity (μm/s) | (1) Control – before stimulation | 0.397 ± 0.039 | 6(18) | ns, p=0.2424 (1 vs 2) ns, p=0.1991 (3 vs 4) ns, p=0.6050 (5 vs 6) |
Mann-Whitney U test |
| Distance (μm) | Control | 2.55 ± 0.34 | 6(18) | ns, p=0.502 | Mann-Whitney U test |
| RIM 1/2 experiments: morphology, protein level and EM analysis | |||||
| Synapse number | Control | 670.1 ± 47.37 | 5(59) | ** p = 0.0015 | Mann–Whitney U test |
| Dendritic length (mm) | Control | 2.343 ± 0.129 | 5(59) | ** p = 0.0024 | Student’s t-test |
| Synapse number per μm dendrite | Control | 0.278 ± 0.01 | 5(59) | * p = 0.0172 | Student’s t-test |
| Synapse area (μm2) | Control | 1.470 ± 0.05 | 5(59) | ns, p = 0.46 | Mann-Whitney U test |
| VGLUT1 intensity (F) | Control | 2.955 ± 0.087 | 5(59) | * p = 0.233 | Mann-Whitney U test |
| MUNC13 intensity (F) | Control | 1.921 ± 0.170 | 2(20) | ** p=0.007 | Student’s t-test |
| CHGB puncta (#) | Control | 2442 ± 184.6 | 4(50) | ns, p = 0.50 | Mann–Whitney U test |
| Neurite length (mm) | Control | 5.80 ± 0.34 | 4(50) | ns, p = 0.25 | Mann–Whitney U test |
| CHGB puncta per μm neurite | Control | 0.422 ± 0.02 | 4(50) | ns, p = 0.23 | Student’s t-test |
| CHGB intensity (F) | Control | 1.329 ± 0.105 | 4(50) | ns, p = 0.44 | Student’s t-test |
| Pearson’s correlation VGLUT1::CHGB | Control | 0.631 ± 0.02 | 3(28) | * p < 0.025 | Mann–Whitney U test |
| Manders’ coefficients | Control (M1: VGLUT1::CHGB) | 0.678 ± 0.03 | 3(28) | ns, p > 0.05 | Student’s t-test |
| DCVs per synapse section | Control | 1.58 ± 0.19 | 3(60 synapses) | ns, p=0.0571 | Mann–Whitney U test |
| Synapse number - Cre at DIV 5 | Control | 386.3 ± 60.56 | 3(25) | ns, p = 0.67 | Mann–Whitney U test |
| Dendritic length (mm) - Cre at DIV 5 | Control | 1718 ± 167.0 | 3(25) | ns, p = 0.92 | Mann–Whitney U test |
| Synapse number per μm dendrite - Cre at DIV 5 | Control | 0.204 ± 0.02 | 3(25) | ns, p = 0.457 | Student’s t-test |
| VGLUT1 intensity (F)- Cre at DIV 5 | Control | 2.08 ± 0.185 | 3(25) | ns, p = 0.7432 | Student’s t-test |
| Manders’ coefficients | (1) MUNC13::VGLUT1 | 0.803 ± 0.03 | 8 | * p<0.05: 1 vs 2 ns, p>0.05: 1 vs 3, 2 vs 3 |
Kruskal-Wallis with Dunn’s correction |
n= number of experiments (cells); unless otherwise indicated
ns=non-significant
Data and code availability
The data that support the findings of this study are available from the Lead Contact upon reasonable request.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal mouse Homer1 | SYSY | Cat#160011; RRID:AB_2120992 |
| Monoclonal mouse anti-Pan-Axonal Neurofilament marker (SMI-312R) | Covance | Cat# SMI-312R; RRID:AB_2314906 |
| Monoclonal mouse β3-tubulin | Cell Signaling | Cat# 4466 |
| Monoclonal mouse Actin | Chemicon | Cat# MAB1501; RRID:AB_2223041 |
| Monoclonal mouse HA (12CA5) | Roche | Cat#11583816001; RRID:AB_514505 |
| Monoclonal mouse MUNC13–1 | SYSY | Cat# 126111; RRID:AB_887735 |
| Monoclonal mouse PSD95 | Abcam | Cat# ab2723; RRID:AB_303248 |
| Polyclonal chicken MAP2 | Abcam | Cat# Ab5392; RRID:AB_2138153 |
| Polyclonal guinea pig Synaptophysin1 | SYSY | Cat# 101 004; RRID:AB_1210382 |
| Polyclonal guinea pig VGLUT1 | Millipore | Cat# AB5905; RRID:AB_2301751 |
| Polyclonal rabbit RIM1/2 ZN-finger | SYSY | Cat# 140 203; RRID:AB_887775 |
| Polyclonal rabbit Chromogranin B | SYSY | Cat# 259103; RRID:AB_2619973 |
| Alexa Fluor conjugated secondary antibodies | Invitrogen | Cat# A21244, A11001, A21450 A11040, A11010, A11003; RRID:AB_141663, AB_2534069, AB_141882, AB_1500590, AB_2534077, AB_141370 |
| anti-rabbit or anti-mouse IRDye secondary antibodies | LI-COR | Cat# LI 925–32210, 926–68073; RRID:AB_2687825, AB_10954442 |
| alkaline phosphatase-conjugated secondary antibodies | Jackson | Cat#111-055-003, 115-055-003; RRID:AB_2337947, AB_2338528 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Agarose type II-A | Sigma | Cat# A9918; CAS: 9012-36-6 |
| Laminin | Sigma | Cat# L2020 |
| Poly-D-lysine | Sigma | Cat# P6407 |
| Rat tail collagen | BD Biosciences | Cat# 354236 |
| B-27 | Gibco | Cat# 17504–044 |
| poly-L-ornithine | Sigma | Cat# P4957 |
| normal goat serum | Gibco | Cat# 16210–072 |
| Mowiol 4–88 | Sigma | Cat# 81381 |
| Glutaraldehyde | Merck | Cat# 1042390250 |
| Cacodylate | Merck | Cat# 820670 |
| OsO4 | EMS | Cat# 19172 |
| K4Ru(CN)6 | Sigma | Cat# 378232.2 |
| Glycid Ether (Epon) | Serva | Cat# 21045.02 |
| Dodecenyl succinic anhydride (Epon) | Serva | Cat# 20755.02 |
| Methyl nadic anhydride (Epon) | Serva | Cat# 29452.03 |
| benzyldimethylamine (Epon) | EMS | Cat# 1140025 |
| Uranyl acetate | Polyscience | Cat# 21447 |
| Lead nitrate (lead citrate) | Merck | Cat# 1.07398.0100 |
| Sodium citrate (lead citrate) | VWR | Cat# 27831.297 |
| Ionomycin | Fisher BioReagent | Cat# 10429883 |
| Fluo-5F-AM | Molecular Probes | Cat# F14222 |
| Critical Commercial Assays | ||
| Mouse BDNF ELISA | Biosensis | Cat# BEK-2003 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: Rab3ABCD null | Schlüter et al., 2004 | N/A |
| Mouse: Rim1/2 conditional knock-out | Kaeser et al., 2011; Kaeser et al., 2008 | N/A |
| Rat: Wistar (Crl:WI) | Charles River | Strain code: 003 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| pFSW nclcre | Kaeser et al., 2011 | N/A |
| pFSW nclDeltacre | ||
| pSyn(pr) hNPYpHluorin-N1 | van de Bospoort et al., 2012 | N/A |
| pSyn(pr) hNPYmCherry | ||
| pSyn(pr) rBDNFpHluorin | De Wit et al., 2009 | N/A |
| pSyn(pr) Synapsin-1ECFP | Modified gift from: Dr A. Jeromin (Allen Brain Institute, Seattle, USA) | N/A |
| pSyn(pr) RIM1alfa-HA | Modified from: Deng et al., 2011 Kaeser et al., 201 |
N/A |
| pSyn(pr) RIM1alfa-K144–6E-HA | ||
| pSyn(pr) RIM1alfa-dPDZ-HA | ||
| pSyn(pr) RIM1beta-HA | ||
| pSyn(pr) RIM1alfa-Zn-HA | Modified from: Deng et al., 2011 Kaeser et al., 201 |
N/A |
| pSyn(pr) RIM1alfa-Zn-K144–6E-HA | ||
| pSyn(pr) RIM1beta-Zn-HA | ||
| pSyn(pr) RIM1beta-ZN-HA(K144/6E) | ||
| pSyn(pr) RIM1alfa-Zn-HA-ECFP | Modified from: Deng et al., 2011 Kaeser et al., 201 |
N/A |
| pSyn(pr) RIM1alfa-Zn-HA-mCherry | ||
| pSIN-TRE-rUnc-13 (WT)mCherryN1-Syn-rtTA2 | Modified from: Deng et al., 2011 |
N/A |
| pSyn(pr) rUnc-13 (delta N term)mCherry | ||
| pSyn(pr) mCherry-RAB3A(mus) | Created in this study | N/A |
| pSyn(pr) RAB3B(mus)mCherry | ||
| pSyn(pr) RAB3C(mus)mCherry | ||
| pSyn(pr) RAB3D(mus)mCherry | ||
| Software and Algorithms | ||
| MATLAB R2018a | MathWorks | https://mathworks.com |
| SynD – Synapse and neurite detection | Schmitz et al., 2011 | https://www.johanneshjorth.se/SynD/SynD.html |
| ImageJ |
https://imagej.net/ RRID:SCR_003070 |
|
| GraphPad Prism | GraphPad Software |
http://www.graphpad.com/ RRID:SCR_002798 |
| NIS-Elements | Nikon Instruments |
https://www.nikoninstruments.com/Products/Software RRID:SCR_014329 |
| Other | ||
Acknowledgements
The authors thank Tobias Moser (University Medical Center Goettingen) for providing the conditional RIM1/2 mouse line. Robbert Zalm for cloning and producing viral particles, Frank den Oudsten and Desiree Schut for producing glia feeders and primary culture assistance, Joke Wortel for animal breeding and Electron Microscopy, Frank den Oudsten, Erik Ceelen and Joost Hoetjes for genotyping, Ingrid Saarloos for protein chemistry and members of the CNCR DCV project team for fruitful discussions. EM analysis was performed at the VU/VUmc EM facility (ZonMW 91111009). This work is supported by an ERC Advanced Grant (322966) of the European Union (to M.V.) and by the NIH (R01MH113349 and R01NS103484 to P.S.K.).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Aalto MK, Ronne H, and Keränen S (1993). Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. The EMBO Journal 12, 4095–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S, Saarloos I, Kooistra R, van de Bospoort R, Verhage M, and Toonen RF (2017). SNAP-25 gene family members differentially support secretory vesicle fusion. J Cell Sci 130, 1877–1889. [DOI] [PubMed] [Google Scholar]
- Balkowiec A, and Katz DM (2002). Cellular mechanisms regulating activity-dependent release of native brain-derived neurotrophic factor from hippocampal neurons. J Neurosci 22, 10399–10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A, Thakur P, Junge HJ, Ashery U, Rhee JS, Scheuss V, Rosenmund C, Rettig J, and Brose N (2001). Functional interaction of the active zone proteins Munc13–1 and RIM1 in synaptic vesicle priming. Neuron 30, 183–196. [DOI] [PubMed] [Google Scholar]
- Bowser R, Muller H, Govindan B, and Novick P (1992). Sec8p and Sec15p are components of a plasma membrane-associated 19.5S particle that may function downstream of Sec4p to control exocytosis. Journal of Cell Biology 118, 1041–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos MA, Lucchesi O, Ruete MC, Mayorga LS, and Tomes CN (2012). Rab27 and Rab3 sequentially regulate human sperm dense-core granule exocytosis. Proc Natl Acad Sci U S A 109, E2057–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho M, Basu J, Trimbuch T, Chang S, Pulido-Lozano C, Chang SS, Duluvova I, Abo-Rady M, Rizo J, and Rosenmund C (2017). Heterodimerization of Munc13 C2A domain with RIM regulates synaptic vesicle docking and priming. Nat Commun 8, 15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Janz R, Südhof TC, Tzounopoulos T, Malenka RC, and Nicoll RA (1997). Rab3A is essential for mossy fibre long-term potentiation in the hippocampus. Nature 388, 590–593. [DOI] [PubMed] [Google Scholar]
- Cheng PL, Song AH, Wong YH, Wang S, Zhang X, and Poo MM (2011). Self-amplifying autocrine actions of BDNF in axon development. Proceedings of the National Academy of Sciences 108, 18430–18435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Adamo P, Wolfer DP, Kopp C, Tobler I, Toniolo D, and Lipp HP (2004). Mice deficient for the synaptic vesicle protein Rab3a show impaired spatial reversal learning and increased explorative activity but none of the behavioral changes shown by mice deficient for the Rab3a regulator Gdi1. Eur J Neurosci 19, 1895–1905. [DOI] [PubMed] [Google Scholar]
- de Jong APH, Roggero CM, Ho MR, Wong MY, Brautigam CA, Rizo J, and Kaeser PS (2018). RIM C2B Domains Target Presynaptic Active Zone Functions to PIP2-Containing Membranes. Neuron 98, 335–349 e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, Toonen RF, and Verhage M (2009). Matrix-dependent Local Retention of Secretory Vesicle Cargo in Cortical Neurons. Journal of Neuroscience 29, 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Kaeser PS, Xu W, and Sudhof TC (2011). RIM proteins activate vesicle priming by reversing autoinhibitory homodimerization of Munc13. Neuron 69, 317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez N, van Weering JRT, Borges R, Toonen RFG, and Verhage M (2018). Dense-core vesicle biogenesis and exocytosis in neurons lacking chromogranins A and B. J Neurochem 144, 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Lou X, Lu J, Huryeva I, Alam A, Schneggenburger R, Sudhof TC, and Rizo J (2005). A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity? EMBO J 24, 2839–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emperador Melero J, Nadadhur AG, Schut D, Weering JV, Heine VM, Toonen RF, and Verhage M (2017). Differential Maturation of the Two Regulated Secretory Pathways in Human iPSC-Derived Neurons. Stem Cell Reports 8, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina M, van de Bospoort R, He E, Persoon CM, van Weering JR, Broeke JH, Verhage M, and Toonen RF (2015). CAPS-1 promotes fusion competence of stationary dense-core vesicles in presynaptic terminals of mammalian neurons. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Mignery GA, Baumert M, Perin MS, Hanson TJ, Burger PM, Jahn R, and Südhof TC (1990). rab3 is a small GTP-binding protein exclusively localized to synaptic vesicles. Proceedings of the National Academy of Sciences of the United States of America 87, 1988–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez T, Gilleron J, Zerial M, and O’Sullivan GA (2012). SnapShot: Mammalian Rab proteins in endocytic trafficking. Cell 151, 234–234 e232. [DOI] [PubMed] [Google Scholar]
- Garcia EP, McPherson PS, Chilcote TJ, Takei K, and De Camilli P (1995). rbSec1A and B colocalize with syntaxin 1 and SNAP-25 throughout the axon, but are not in a stable complex with syntaxin. The Journal of Cell Biology 129, 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, and Staiger V (2002). Neurotrophin secretion from hippocampal neurons evoked by long-term-potentiation-inducing electrical stimulation patterns. Proc Natl Acad Sci U S A 99, 6386–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geppert M, Goda Y, Stevens CF, and Sudhof TC (1997). The small GTP-binding protein Rab3A regulates a late step in synaptic vesicle fusion. Nature 387, 810–814. [DOI] [PubMed] [Google Scholar]
- Goud B, Salminen A, Walworth NC, and Novick PJ (1988). A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell 53, 753–768. [DOI] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena C, and Novick P (1999). The exocyst is an effector for Sec4P, targeting secretory vesicles to sites of exocytosis. The EMBO Journal 18, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Kaeser PS, Südhof TC, and Schneggenburger R (2011). RIM Determines Ca2+ Channel Density and Vesicle Docking at the Presynaptic Active Zone. Neuron 69, 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley MT, Haynes LP, and Burgoyne RD (2007). Differential dynamics of Rab3A and Rab27A on secretory granules. J Cell Sci 120, 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Heumann R, and Lessmann V (2001). Synaptic secretion of BDNF after high-frequency stimulation of glutamatergic synapses. EMBO J 20, 5887–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensbroek RA, Kamal A, Baars AM, Verhage M, and Spruijt BM (2003). Spatial, contextual and working memory are not affected by the absence of mossy fiber long-term potentiation and depression. Behav Brain Res 138, 215–223. [DOI] [PubMed] [Google Scholar]
- Hibino H, Pironkova R, Onwumere O, Vologodskaia M, Hudspeth a.J., and Lesage F (2002). RIM binding proteins (RBPs) couple Rab3-interacting molecules (RIMs) to voltage-gated Ca2+ channels. Neuron 34, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, and Scheller RH (2006). SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol 7, 631–643. [DOI] [PubMed] [Google Scholar]
- Kaeser PS, Deng L, Wang Y, Dulubova I, Liu X, Rizo J, and Sudhof TC (2011). RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell 144, 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser PS, Kwon HB, Chiu CQ, Deng L, Castillo PE, and Sudhof TC (2008). RIM1alpha and RIM1beta are synthesized from distinct promoters of the RIM1 gene to mediate differential but overlapping synaptic functions. J Neurosci 28, 13435–13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser PS, and Regehr WG (2014). Molecular mechanisms for synchronous, asynchronous, and spontaneous neurotransmitter release. Annu Rev Physiol 76, 333–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfhamer D, Valladares O, Sun Y, Nolan PM, Rux JJ, Arnold SE, Veasey SC, and Bucan M (2002). Mutations in Rab3a alter circadian period and homeostatic response to sleep loss in the mouse. Nat Genet 32, 290–295. [DOI] [PubMed] [Google Scholar]
- Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, and Nonet ML (2001). A post-docking role for active zone protein Rim. Nat Neurosci 4, 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kershberg L, Wang J, Schneeberger S, and Kaeser PS (2018). Dopamine Secretion Is Mediated by Sparse Active Zone-like Release Sites. Cell 172, 706–718 e715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KS, Siebert M, Mertel S, Knoche E, Wegener S, Wichmann C, Matkovic T, Muhammad K, Depner H, Mettke C, et al. (2011). RIM-binding protein, a central part of the active zone, is essential for neurotransmitter release. Science 334, 1565–1569. [DOI] [PubMed] [Google Scholar]
- Liu X, Seven AB, Camacho M, Esser V, Xu J, Trimbuch T, Quade B, Su L, Ma C, Rosenmund C, et al. (2016). Functional synergy between the Munc13 C-terminal C1 and C2 domains. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Kuzmin A, Unger SM, Petersen JD, and Silverman MA (2011). KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett 491, 168–173. [DOI] [PubMed] [Google Scholar]
- Lu J, Machius M, Dulubova I, Dai H, Sudhof TC, Tomchick DR, and Rizo J (2006). Structural basis for a Munc13–1 homodimer to Munc13–1/RIM heterodimer switch. PLoS Biol 4, e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney TR, Liu Q, Itoh T, Luo S, Hadwiger G, Vincent R, Wang ZW, Fukuda M, and Nonet ML (2006). Regulation of synaptic transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol Biol Cell 17, 2617–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malva JO, Xapelli S, Baptista S, Valero J, Agasse F, Ferreira R, and Silva AP (2012). Multifaces of neuropeptide Y in the brain--neuroprotection, neurogenesis and neuroinflammation. Neuropeptides 46, 299–308. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Lu H, Fukata Y, Noritake J, Gao H, Mukherjee S, Nemoto T, Fukata M, and Poo M.m. (2009). Differential Activity-Dependent Secretion of Brain-Derived Neurotrophic Factor from Axon and Dendrite. Journal of Neuroscience 29, 14185–14198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SQ, Hiesinger PR, Beronja S, Zhai RG, Schulze KL, Verstreken P, Cao Y, Zhou Y, Tepass U, Crair MC, et al. (2005). Mutations in Drosophila sec15 reveal a function in neuronal targeting for a subset of exocyst components. Neuron 46, 219–232. [DOI] [PubMed] [Google Scholar]
- Meijer M, Burkhardt P, de Wit H, Toonen RF, Fasshauer D, and Verhage M (2012). Munc18–1 mutations that strongly impair SNARE-complex binding support normal synaptic transmission. EMBO J 31, 2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens I, Husson SJ, Janssen T, Lindemans M, and Schoofs L (2007). PACAP and PDF signaling in the regulation of mammalian and insect circadian rhythms. Peptides 28, 1775–1783. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Domes G, Kirsch P, and Heinrichs M (2011). Oxytocin and vasopressin in the human brain: social neuropeptides for translational medicine. Nat Rev Neurosci 12, 524–538. [DOI] [PubMed] [Google Scholar]
- Murthy M, Garza D, Scheller RH, and Schwarz TL (2003). Mutations in the Exocyst component Sec5 disrupt neuronal membrane traffic, but neurotransmitter release persists. Neuron 37, 433–447. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz R, Jorgensen EM, and Meyer BJ (1997). Caenorhabditis elegans rab-3 Mutant Synapses Exhibit Impaired Function and Are Partially Depleted of Vesicles. The Journal of Neuroscience 17, 8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Ferro S, and Schekman R (1981). Order of events in the yeast secretory pathway. Cell 25, 461–469. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, and Schekman R (1980). Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21, 205–215. [DOI] [PubMed] [Google Scholar]
- Novick P, and Schekman R (1979). Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Nati Acad Sci USA 76, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang PT, Teng HK, Zaitsev E, Woo NT, Sakata K, Zhen S, Teng KK, Yung WH, Hempstead BL, and Lu B (2004). Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science 306, 487–491. [DOI] [PubMed] [Google Scholar]
- Persoon CM, Moro A, Nassal JP, Farina M, Broeke JH, Arora S, Dominguez N, van Weering JR, Toonen RF, and Verhage M (2018). Pool size estimations for dense-core vesicles in mammalian CNS neurons. The EMBO journal 37, e99672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopov V, Govindan B, Novick P, and Gerst JE (1993). Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell 74, 855–861. [DOI] [PubMed] [Google Scholar]
- Rizo J (2018). Mechanism of neurotransmitter release coming into focus. Protein Sci 27, 1364–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruediger S, Vittori C, Bednarek E, Genoud C, Strata P, Sacchetti B, and Caroni P (2011). Learning-related feedforward inhibitory connectivity growth required for memory precision. Nature 473, 514–518. [DOI] [PubMed] [Google Scholar]
- Salminen A, and Novick PJ (1987). A ras-like protein is required for a post-Golgi event in yeast secretion. Cell 49, 527–538. [DOI] [PubMed] [Google Scholar]
- Schlüter OM, Basu J, Südhof TC, and Rosenmund C (2006). Rab3 superprimes synaptic vesicles for release: implications for short-term synaptic plasticity. J Neurosci 26, 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter OM, Khvotchev M, Jahn R, and Südhof TC (2002). Localization versus function of Rab3 proteins. Evidence for a common regulatory role in controlling fusion. J Biol Chem 277, 40919–40929. [DOI] [PubMed] [Google Scholar]
- Schlüter OM, Schmitz F, Jahn R, Rosenmund C, and Südhof TC (2004). A complete genetic analysis of neuronal Rab3 function. J Neurosci 24, 6629–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz SK, Hjorth JJ, Joemai RM, Wijntjes R, Eijgenraam S, de Bruijn P, Georgiou C, de Jong AP, van Ooyen A, Verhage M, et al. (2011). Automated analysis of neuronal morphology, synapse number and synaptic recruitment. J Neurosci Methods 195, 185–193. [DOI] [PubMed] [Google Scholar]
- Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, and Südhof TC (2002). RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature 415, 321–326. [DOI] [PubMed] [Google Scholar]
- Schonn JS, van Weering JR, Mohrmann R, Schluter OM, Sudhof TC, de Wit H, Verhage M, and Sorensen JB (2010). Rab3 proteins involved in vesicle biogenesis and priming in embryonic mouse chromaffin cells. Traffic 11, 1415–1428. [DOI] [PubMed] [Google Scholar]
- Schwenger DB, and Kuner T (2010). Acute genetic perturbation of exocyst function in the rat calyx of Held impedes structural maturation, but spares synaptic transmission. Eur J Neurosci 32, 974–984. [DOI] [PubMed] [Google Scholar]
- Shimojo M, Courchet J, Pieraut S, Torabi-Rander N, Sando R 3rd, Polleux F, and Maximov A (2015). SNAREs Controlling Vesicular Release of BDNF and Development of Callosal Axons. Cell Rep 11, 1054–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucchi R, Plucinska G, Hummel JJA, Zahavi EE, Guerra San Juan I, Klykov O, Scheltema RA, Altelaar AFM, and Hoogenraad CC (2018). Regulation of KIF1A-Driven Dense Core Vesicle Transport: Ca(2+)/CaM Controls DCV Binding and Liprin-alpha/TANC2 Recruits DCVs to Postsynaptic Sites. Cell Rep 24, 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC (2013). Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron 80, 675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC, and Rothman JE (2009). Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, et al. (2006). Molecular anatomy of a trafficking organelle. Cell 127, 831–846. [DOI] [PubMed] [Google Scholar]
- Tang AH, Chen H, Li TP, Metzbower SR, MacGillavry HD, and Blanpied TA (2016). A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, and Novick P (1996). The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. The EMBO Journal 15, 6483–6494. [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, and Novick P (1995). Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. The Journal of Cell Biology 130, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, and Fukuda M (2006). Rab3A and Rab27A cooperatively regulate the docking step of dense-core vesicle exocytosis in PC12 cells. J Cell Sci 119, 2196–2203. [DOI] [PubMed] [Google Scholar]
- Vahatalo LH, Ruohonen ST, Makela S, Kovalainen M, Huotari A, Makela KA, Maatta JA, Miinalainen I, Gilsbach R, Hein L, et al. (2015). Neuropeptide Y in the noradrenergic neurones induces obesity and inhibits sympathetic tone in mice. Acta Physiol (Oxf) 213, 902–919. [DOI] [PubMed] [Google Scholar]
- van de Bospoort R, Farina M, Schmitz SK, de Jong A, de Wit H, Verhage M, and Toonen RF (2012). Munc13 controls the location and efficiency of dense-core vesicle release in neurons. J Cell Biol 199, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Keimpema L, Kooistra R, Toonen RF, and Verhage M (2017). CAPS-1 requires its C2, PH, MHD1 and DCV domains for dense core vesicle exocytosis in mammalian CNS neurons. Sci Rep 7, 10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, and Novick PJ (1997). Sec2p Mediates Nucleotide Exchange on Sec4p and Is Involved in Polarized Delivery of Post-Golgi Vesicles. The Journal of Cell Biology 137, 1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu X, Biederer T, and Südhof TC (2002). A family of RIM-binding proteins regulated by alternative splicing: Implications for the genesis of synaptic active zones. PNAS 99, 14464–14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, and Sudhof TC (1997). Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature 388, 593–598. [DOI] [PubMed] [Google Scholar]
- Wong MY, Liu C, Wang SSH, Roquas ACF, Fowler SC, and Kaeser PS (2018). Liprin-alpha3 controls vesicle docking and exocytosis at the active zone of hippocampal synapses. Proc Natl Acad Sci U S A 115, 2234–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaekura K, Julyan R, Wicksteed BL, Hays LB, Alarcon C, Sommers S, Poitout V, Baskin DG, Wang Y, Philipson LH, et al. (2003). Insulin secretory deficiency and glucose intolerance in Rab3A null mice. J Biol Chem 278, 9715–9721. [DOI] [PubMed] [Google Scholar]
- Zahn TR, Angleson JK, MacMorris MA, Domke E, Hutton JF, Schwartz C, and Hutton JC (2004). Dense core vesicle dynamics in Caenorhabditis elegans neurons and the role of kinesin UNC-104. Traffic 5, 544–559. [DOI] [PubMed] [Google Scholar]
- Zahraoui A, Touchot N, Chardin P, and Tavitian A (1989). The human rab genes encode a family of GTP-binding proteins related to yeast YPT1 and SEC4 products involved in secretion. Journal of Biological Chemistry 264, 12394–12401. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the Lead Contact upon reasonable request.