Abstract
The most frequently mutated oncogene in cancer is KRAS, which utilizes alternative fourth exons to generate two gene products, KRAS4A and KRAS4B, that differ only in their C-terminal membrane-targeting region 1. Because oncogenic mutations occur in exons 2 or 3, when KRAS is activated by mutation two constitutively active KRAS proteins are encoded, each capable of transforming cells 2. No functional distinctions among the splice variants have been established. Oncogenic KRAS alters tumor metabolism 3. Among these alterations is increased glucose uptake and glycolysis, even in the presence of abundant oxygen 4 (the Warburg Effect). Whereas these metabolic effects of oncogenic KRAS have been explained by transcriptional upregulation of glucose transporters and glycolytic enzymes 3–5, direct regulation of metabolic enzymes has not been examined. We report a direct, GTP-dependent interaction between KRAS4A and hexokinase 1 (HK1) that alters the activity of the kinase, establishing HK1 as an effector of KRAS4A. The interaction is unique to KRAS4A because the palmitoylation/depalmitoylation cycle of this RAS isoform permits co-localization with HK1 on the outer mitochondrial membrane (OMM). KRAS4A expression in cancer may drive unique metabolic vulnerabilities that can be exploited therapeutically.
KRAS4A binds to hexokinase
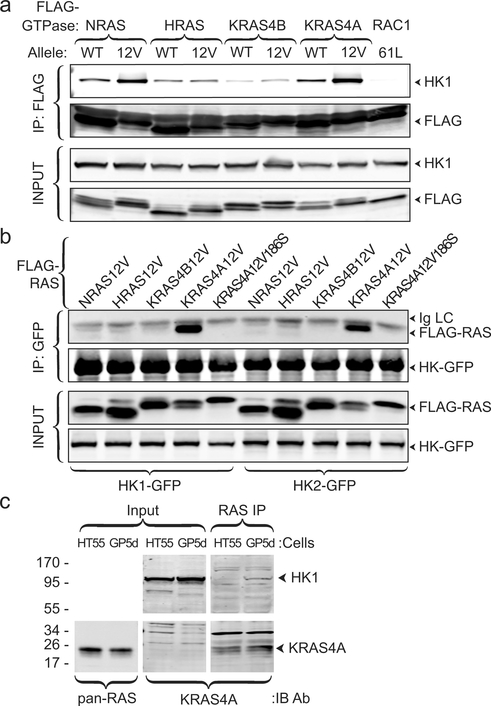
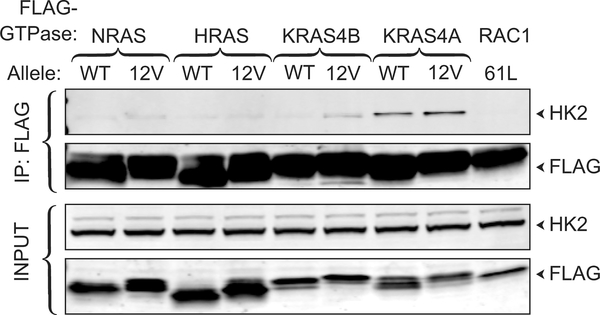
Affinity purification and mass spectroscopic analysis of proteins that interact with NRAS (Supplementary Table 1) identified HK1, the ubiquitously expressed isozyme that initiates glucose metabolism as well as all three isoforms of the mitochondrial voltage-dependent anion channel (VDAC), which form complexes with HK1 on the OMM 6, and the ADP/ATP translocase 1 of the inner mitochondrial membrane, which associates with VDACs 7. Co-immunoprecipitation of FLAG-NRAS and endogenous HK1 validated the interaction and revealed GTP-dependence (Fig. 1a). Interestingly, the ability of RAS proteins to co-immunoprecipitate HK1 was isoform dependent: KRAS4A>NRAS>>HRAS>KRAS4B. Hexokinase 2 (HK2) has 73% sequence identity with HK1, shares all structural features, and is expressed in many cancer cells 8. HK2 associated only with KRAS4A (Extended Data Fig. 1). Reciprocal co-immunoprecipitations pulling down GFP-tagged HK1 or HK2 revealed exquisite isoform specificity with affinity capture of only KRAS4A (Fig. 1b). HT55 and GP5d are human colorectal cancer cell lines, both of which express relatively high levels of KRAS4A, but only the latter harbors oncogenic KRAS. Endogenous HK1 was co-immunoprecipitated with endogenous RAS in GP5d but not HT55 cells (Fig. 1c).
Fig. 1. |. KRAS4A binds HK1 and HK2 in a GTP- and prenylation-dependent fashion.
a, The indicated FLAG-tagged RAS constructs (±G12V mutations) were expressed in HeLa cells and immunoprecipitated with anti-FLAG beads. Blots were probed for endogenous HK1. FLAG-RAC61L served as the negative control. Data shown are representative of 4 independent experiments (n=4). b, GFP-tagged HK1 or HK2 were co-expressed with the indicated FLAG-tagged RAS constructs, immunoprecipitated with anti-GFP beads, and blots probed with anti-FLAG antibody. KRAS4AG12V,C186S is not prenylated and therefore cannot associate with membranes. n=3. c, Co-immunoprecipitation of endogenous HK1 with endogenous KRAS4A (captured by MAb Y13–259) in colorectal tumor cells harboring oncogenic (GP5d) but not wild-type (HT55) KRAS. n=5.
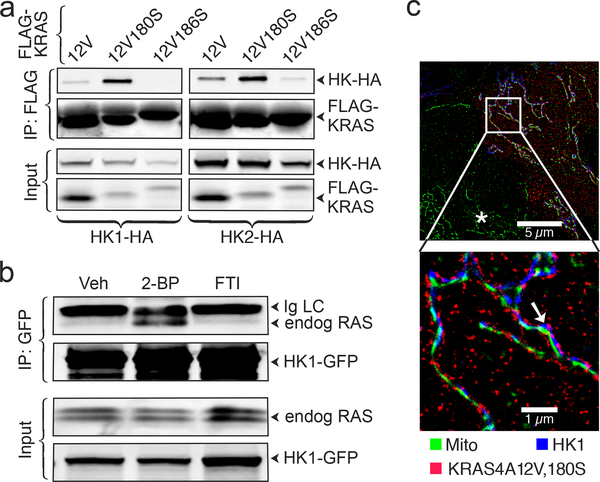
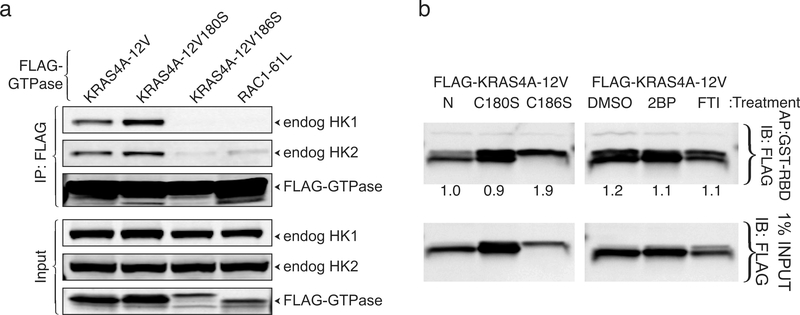
Differential trafficking of RAS proteins is driven by post-translational modifications that include palmitoylation, which, among the KRAS splice variants, is unique to KRAS4A. Palmitoylation of KRAS4A on cysteine 180 in the C-terminal membrane-targeting region is required for efficient plasma membrane association 1. Because palmitoylation is reversible and short-lived 9, palmitoylated RAS proteins continuously cycle between membrane compartments 10. Whereas a C186S mutation of FLAG-KRAS4A, which blocks prenylation and therefore all membrane association, completely blocked associations with HK1 and HK2, a C180S mutation that blocks palmitoylation enhanced the associations (Fig. 2a). A similar result was obtained for endogenous HK1 and HK2 (Extended Data Fig. 2). Inhibition of palmitoylation with 2-bromopalmitate (2-BP) 11 enhanced the association of endogenous RAS with HK1 (Fig. 2b). Thus, whereas prenylation was required for the interaction of KRAS4A with HKs, palmitoylation, which drives efficient association with the plasma membrane, negatively regulated the interaction.
Fig. 2. |. Depalmitoylated KRAS4A interacts with HK1 on the outer mitochondrial membrane (OMM).
a, HA-tagged HK1 or HK2 were co-expressed in HEK293 cells with the indicated FLAG-tagged KRAS4A constructs. KRAS was immunoprecipitated with anti-FLAG beads and analyzed by immunoblot probed with anti-HA and anti-FLAG antibodies. n=3. b, GFP-tagged HK1 was expressed in HCT-116 cells treated with vehicle (Veh), 25 μM 2-bromopalmitate (2-BP) or 20 μM farnesyl transferase inhibitor (FTI). HK1-GFP was immunoprecipitated and blots were probed with an anti-pan-RAS antibody. n=2. c, U2OS cells expressing FLAG-KRAS4A12V,180S and HK1-GFP were treated with MitoTracker, fixed, stained for FLAG and GFP, and imaged by STORM super-resolution microscopy. Asterisk indicates an untransfected cell. Arrow indicates colocalization of KRAS4A and HK1 on OMM. Representative image of n=5.
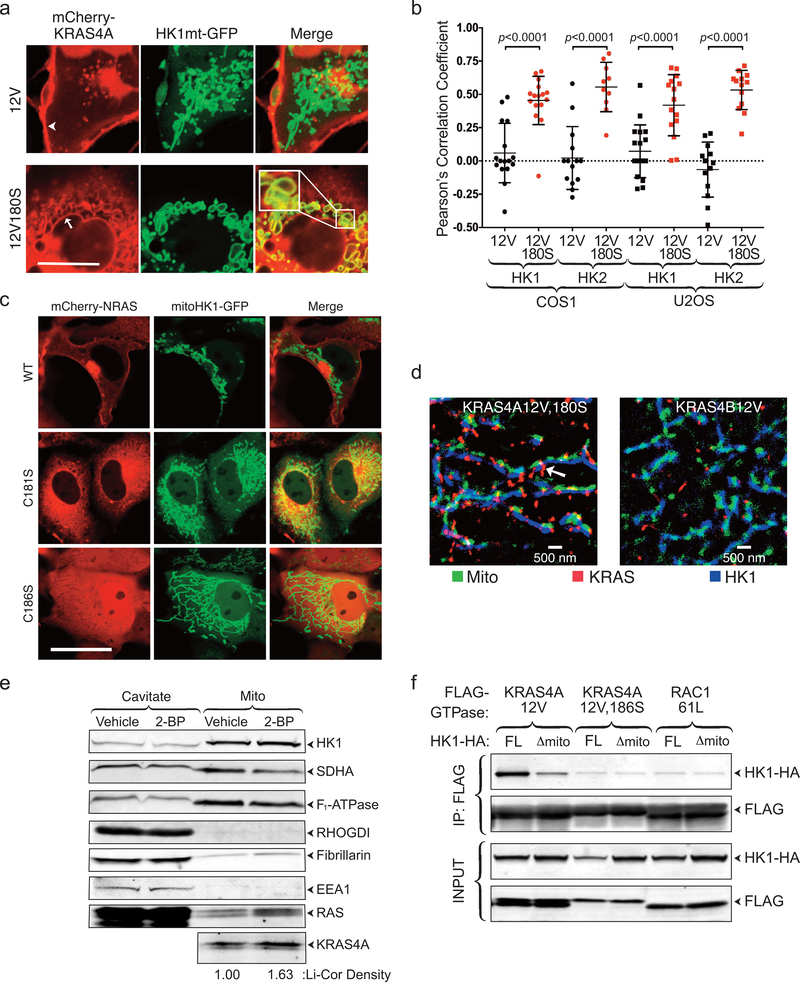
KRAS4A and HK1 interact on mitochondria
Because HK1 and HK2 are targeted to the OMM 12, our data suggest that depalmitoylated KRAS4A may have affinity for the OMM, thereby supporting association with HKs. To test this hypothesis, we co-expressed mCherry-KRAS4A12V ± palmitoylation (±C180S), with GFP targeted to the OMM via the HK1 mitochondrial targeting sequence (aa 1–16). Whereas mCherry-KRAS4A12V was predominantly observed on the plasma membrane, and to a lesser degree on intracellular vesicles, mCherry-KRAS12V180S colocalized with HK1mt-GFP on the OMM (Extended Data Fig. 3a,b). In contrast, although depalmitoylated NRAS also accumulated on endomembranes 1, the OMM was not among them (Extended Data Fig. 3c). Super-resolution immunofluorescent microscopy confirmed colocalization of palmitoylation-deficient KRAS4A and HK1 on the OMM (Fig. 2c). KRAS4B did not decorate the OMM (Extended Data Fig. 3d). HCT-15 cells are KRAS-mutant colorectal cancer cells with relatively high KRAS4A expression. We detected endogenous KRAS4A on mitochondria rapidly isolated from these cells (Extended Data Fig. 3e) and the mitochondria-associated pool was augmented 63% by pre-treatment with 2-BP. Thus, endogenous de-palmitoylated KRAS4A colocalizes with HK1 on the OMM.
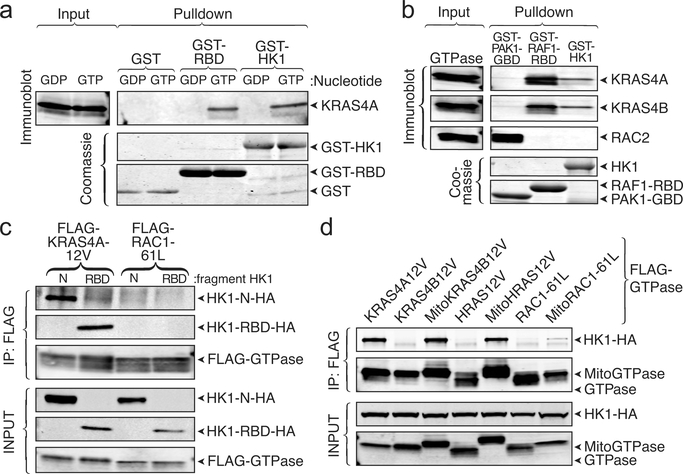
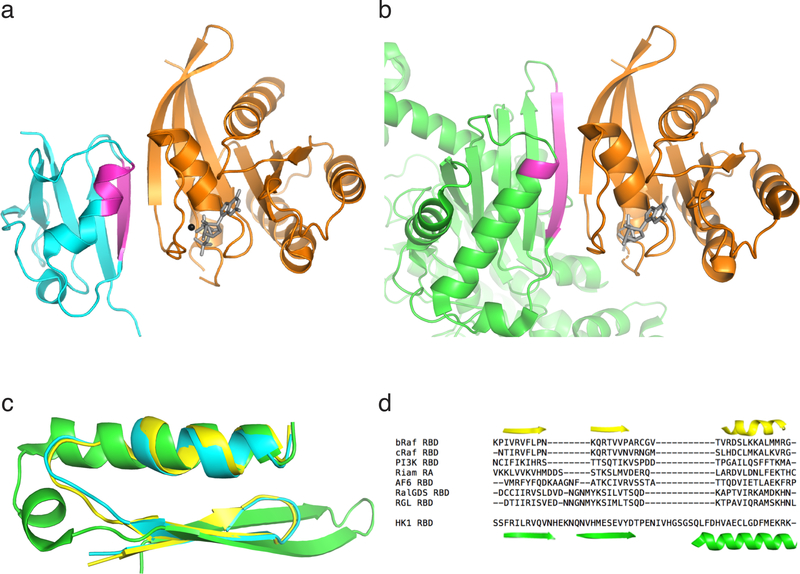
To determine if the interaction between KRAS4A and HK1 is direct, we utilized a fully recombinant system. Bacterially expressed KRAS4A was affinity captured by GST fused with the RAS-binding domain (RBD) of RAF1 or HK1, but not GST alone (Fig. 3a). Importantly, the interaction was observed only when KRAS4A was loaded with GTP. The GTP-dependence suggests that the KRAS4A G-domain interacts with HK1. Given that the G-domains of all four RAS proteins are nearly identical, we hypothesized that without membrane targeting, there should be no isoform specificity. Indeed, in the in vitro assay driven by mass action, recombinant, non-prenylated KRAS4B but not RAC2 behaved like KRAS4A and interacted with GST-HK1 in a GTP-dependent fashion (Fig. 3b). The GTP-dependent, direct protein-protein interaction suggests that HK1 may contain a structural analog of an RBD. Examination of published HK1 structures revealed a surface-exposed strand-helix motif characteristic of RBDs (Extended Data Fig. 4) 13. We expressed the isolated, putative RBD region (HK1 aa 76–206) tagged with HA and found that it was as efficient in pulling down KRAS4A12V as was the N-terminal kinase domain of HK1 (Fig. 3c). These data suggest that the region of HK1 comprised of aa 76–206 functions as an RBD.
Fig. 3. |. The interaction between KRAS4A and HK1 is direct, GTP-dependent, mediated by an RBD-like region of HK1, and requires OMM localization.
a, Recombinant KRAS4A was loaded with GDP or GTPɣS, and then incubated with GST alone, GST-RAF1-RBD, or GST-HK1. Affinity capture was assessed by immunoblot for RAS. GST loading is shown by a Coomassie-stained gel. n=3. b, Recombinant KRAS4A, KRAS4B and RAC2 were loaded with GTPɣS, and then incubated with GST-PAK1-RBD, GST-RAF1-RBD, or GST-HK1. Affinity capture was assessed by immunoblots for RAS or RAC2. n=2. c, HA-tagged N-terminal domain of HK1 or the isolated, putative RBD (aa 76–206) were expressed with FLAG-tagged KRAS4A12V or RAC161L and FLAG immunoprecipitates were blotted as indicated. n=3. d, HA-tagged HK1 was co-expressed in HEK293 cells with FLAG-tagged KRAS4A12V or KRAS4B12V, HRAS12V, or RAC161L with or without N-terminal extension with the mitochondrial targeting sequence of HK1 (Mito; aa 1–16). The FLAG-tagged GTPases were immunoprecipitated and binding to HK1 was assessed with anti-HA antibody. n=4.
The binding affinities of RBDs for RAS vary widely 13. The two-dimensional surface of the mitochondrion might promote the interaction of KRAS4A with HK1 even if the affinity is relatively weak. To confirm this model, we artificially targeted constitutively GTP-bound KRAS4B and HRAS to the OMM by extending them at their N-termini with the mitochondrial targeting sequence of HK1 (Mito). Whereas HK1 was co-immunoprecipitated neither with FLAG-KRAS4B12V nor FLAG-HRAS12V, a robust association was detected between HK1 and both MitoKRAS4B12V-FLAG and MitoHRAS12V-FLAG (Fig. 3d). In contrast, GTP-loaded RAC1–61L targeted to the OMM in the same way failed to engage HK1. Conversely, removing the mitochondrial targeting sequence from HK1 diminished its ability to interact with KRAS4A (Extended Data Fig. 3d). Thus, the isoform specificity of the interaction with regard to KRAS4A is driven by its membrane targeting sequence that permits colocalization with HK1 on the OMM.
KRAS4A blocks allosteric inhibition of HK1
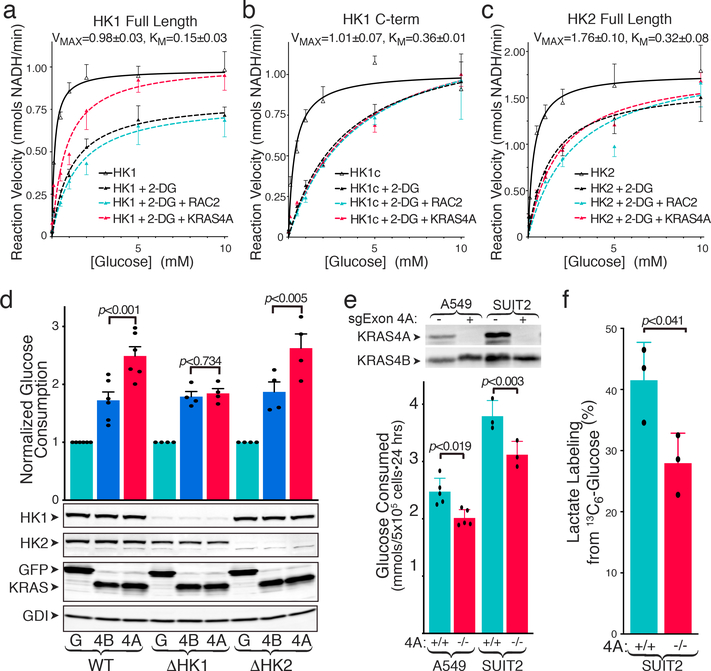
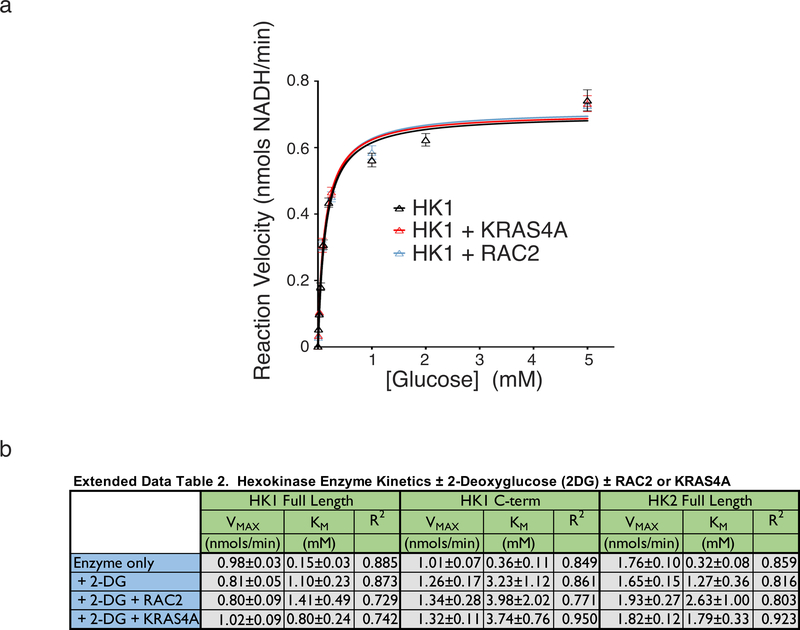
To determine if KRAS4A directly affects HK1 enzymatic activity we studied recombinant, GST-tagged enzymes associated with glutathione-agarose beads (to mimic OMM and allow multimerization) 14. Neither recombinant, GTP-loaded KRAS4A nor RAC2 affected the kinetics of full-length HK1 (Extended Data Fig. 5a). Both HK1 and HK2 consist of two tandem kinase domains. Whereas both domains are catalytic in HK2 15, only the C-terminal domain of HK1 can phosphorylate glucose. The HK1 N-terminal domain provides allosteric product feedback inhibition 16. As expected, 2-deoxyglucose (2-DG) slowed the reaction (Fig. 4a–c). 2-DG is both a competitive and non-competitive inhibitor of HK1: aside from directly competing with glucose it is also converted by HKs to 2-deoxyglucose-6-phosphate, which cannot be further metabolized and therefore accumulates and acts as an analog of G6P to mediate allosteric feedback inhibition 16. Whereas recombinant RAC2 had no effect on the 2-DG-inhibited reaction, recombinant KRAS4A partially reversed 2-DG inhibition (Fig. 4a). In contrast, KRAS4A reversed 2-DG inhibition of neither the isolated C-terminal kinase domain of HK1 (Fig. 4b) nor full-length HK2 (Fig. 4c), suggesting its action on the full-length HK1 is through the allosteric site. Consistent with this interpretation, KRAS4A restored the VMAX diminished by 2-DG but had relatively little effect on the KM (Extended Data Fig. 5b). The mitigating effect of KRAS4A on the allosteric inhibition induced by 2-DG demonstrates that the direct protein-protein interaction between KRAS4A and HK1 has a functional consequence.
Fig. 4. |. KRAS4A increases HK1 activity in vitro and in vivo.
a-c, Activity of recombinant full-length HK1 (a), C-terminal kinase domain of HK1 (b) or full-length HK2 (c). Reaction velocity is plotted (mean±SE) as a function of glucose concentration. Velocities ±2-DG (20 mM) are shown ±addition of recombinant, GTP-loaded RAC2 or KRAS4A. Plots combine independent assays, n=4 (a) or n=3 (b,c). d, WT (n=6) or HK1- or HK2-deficient (n=4) HEK293 cells were transfected with FLAG-tagged GFP, KRAS4B12V, or KRAS4A12V, and 24-h glucose consumption (mean±SE) was determined. Immunoblot shows relative expression. e, Glucose consumption (mean±SE) of parental and KRAS4A-targeted A549 (n=5) and SUIT2 (n=3) human cancer cells. f, Incorporation of 13C from glucose into lactate (mean±SE) over 15 min in SUIT2 cells ±KRAS exon 4A (n=3). Immunoblot shows absence of KRAS4A. d-f, Significance determined by two-sided student’s t-test (d-e paired; f unpaired). Growth rates of the two genotypes were equal over 24 hr assessment.
KRAS4A enhances glycolytic flux
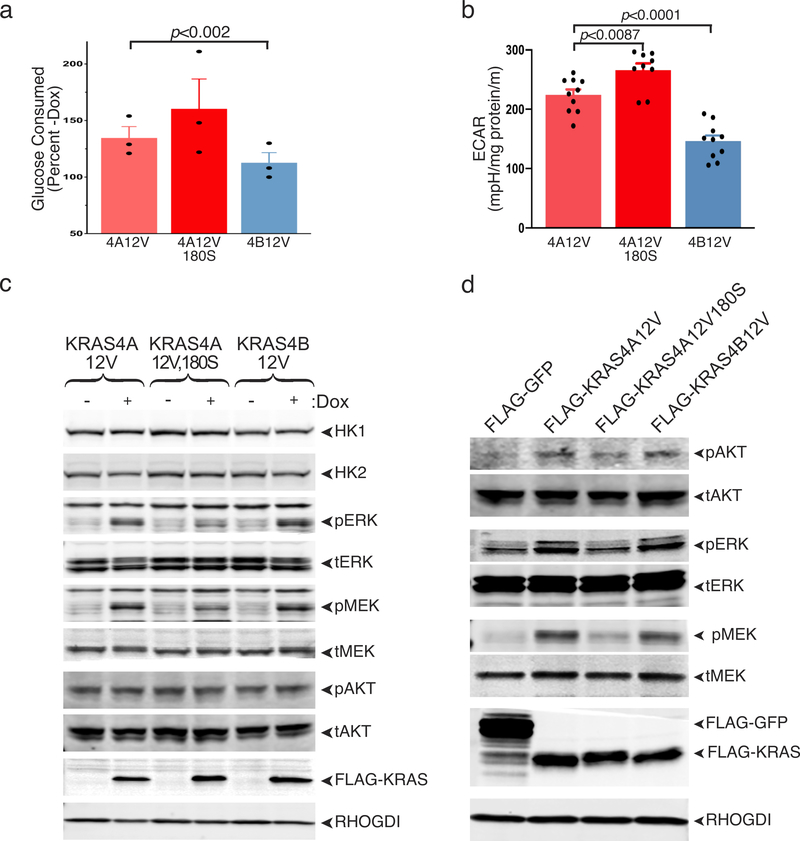
To correlate these in vitro results with cellular glucose metabolism, we expressed oncogenic forms of the KRAS splice variants in HEK293 cells. As expected, both oncogenic KRAS4A and KRAS4B enhanced glucose consumption; however, when expressed at similar levels, KRAS4A had twice the impact of KRAS4B (Fig. 4d). When HK1, but not HK2, was silenced, this differential was lost, suggesting that the mechanism whereby KRAS4A exceeds KRAS4B in promoting glucose consumption requires HK1. This result was confirmed by metabolic analysis of Flp-In T-REx293™ cells induced with doxycycline to express equivalent levels of KRAS4A12V, KRAS4A12V,180S or KRAS4B12V. Both glucose consumption (Extended Data Fig. 6a) and basal extracellular acidification rate (ECAR) (Extended Data Fig. 6b) were increased in KRAS4A12V expressing cells relative to those expressing KRAS4B12V. Importantly, palmitoylation-deficient KRAS4A12V,180S was more potent than KRAS4A12V, demonstrating an association between OMM localization and enhanced glucose consumption. Whereas KRAS4A and KRAS4B were equivalent in inducing phosphorylation of MEK, ERK and AKT, KRAS4A12V,180S was less effective (Extended Data Fig. 6c,d). Moreover, the levels of HK1 and HK2 were increased by none of the KRAS proteins. Thus, increased glucose metabolism was dissociated in this system from KRAS signaling down the MAPK and PI3K pathways and from transcriptional activation of HKs.
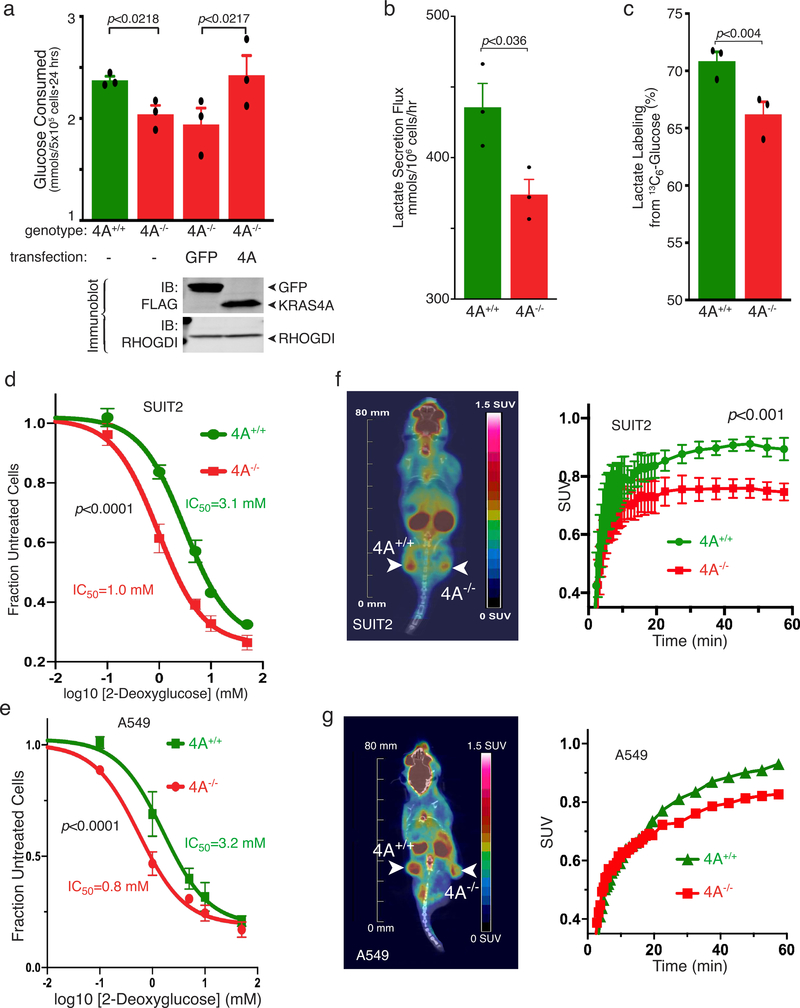
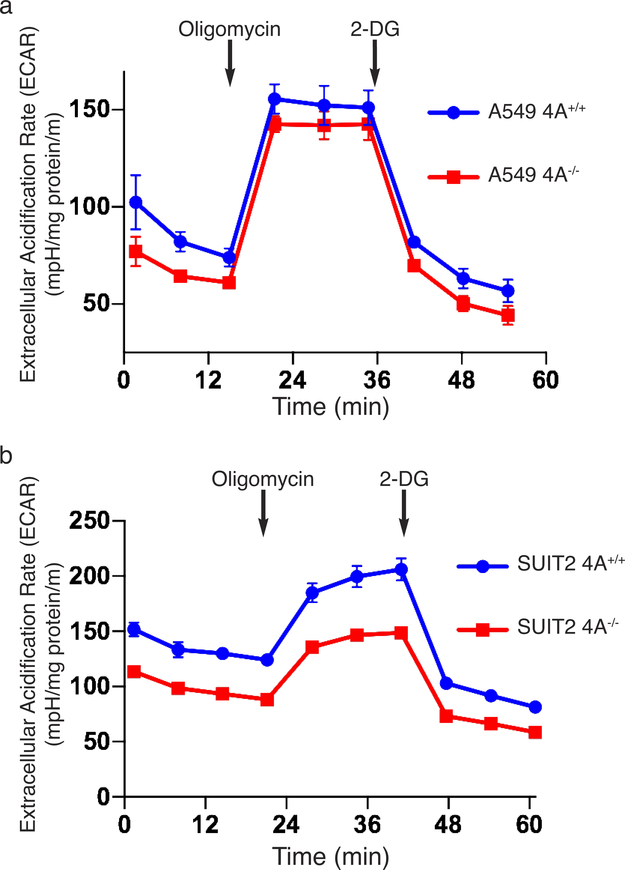
We performed the converse experiments by targeting the 4A exon with CRISPR/Cas9 in two human tumor cell lines that harbor oncogenic KRAS mutations, A549 (lung) and SUIT2 (pancreas). In both cases, disrupting exon 4A diminished glucose utilization (Fig. 4e and Extended Data Fig. 7a), lactate secretion (Extended Data Fig. 7b), and ECAR (Extended Data Fig. 8). Glucose consumption was restored by forced expression of KRAS4A (Extended Data Fig. 7a). To determine if these results reflect alterations in glycolytic flux we employed stable isotope (13C) labeled glucose and found diminished incorporation into lactate upon disruption of exon 4A (Fig. 4f and Extended Data Fig. 7c). In addition, both KRAS4A−/− cells were more sensitive to growth inhibition by 2-DG than KRAS4A+/+ cells (Extended Data Fig. 7d,e), consistent with diminished glycolytic capacity. We measured glucose uptake in vivo by 18F-deoxyglucose positron emission tomography (18FDG-PET) in xenograft tumors generated with SUIT2 (Extended Data Fig. 7f) or A549 (Extended Data Fig. 7g) cells with the genotypes KRAS4A+/+ or KRAS4A−/−. KRAS4A-expressing tumors took up glucose at a faster rate per unit volume than KRAS4A−/− tumors.
Because the regulation of HK1 by KRAS4A appears to be stoichiometric, as is the case for all RAS effectors 17, we sought to determine the relative abundance of KRAS4A and HK1. Using quantitative immunoblotting we determined that A549 and HCT-15 cells contain on average 7×105 and 2×105 molecules of KRAS, and 2×105 and 1.5×105 molecules of HK1, respectively (Extended Data Fig. 9). Because KRAS4A ranges from 15–50% of total KRAS expression 1 and HK1 functions as a homodimer, we conclude that there is sufficient KRAS4A in these cells to have a significant impact on catalytic activity.
Discussion
An effector of a small GTPase such as RAS must meet three requirements: binding must 1) be direct; 2) occur only with the GTP-bound form; and 3) alter activity of the effector. Our data demonstrate that HK1 meets these requirements and is therefore a KRAS4A effector. HK1 is unique in two ways: it is the first metabolic enzyme shown to be an effector of a GTPase and it is the first effector that discriminates between the two KRAS splice variants. Although, like all effector interactions, binding takes place through the G-domain, it is the distinct subcellular trafficking of KRAS4A and KRAS4B that establishes the differential engagement. We and others have previously reported compartmentalized RAS signaling based on subcellular trafficking 18–20. KRAS4A signaling to HK1 on the OMM is another clear example of this phenomenon.
The metabolic rewiring of human tumors by oncogenic KRAS is likely the consequence of many factors, including increased expression of transporters and enzymes 8,21. Our data suggest that direct regulation of HK1 by KRAS4A represents an additional mechanism. In support of this model (Extended Data Fig. 10), whereas maximal MAPK signaling does not correlate with KRAS expression, the ability of oncogenic KRAS to upregulate glycolysis depends on mutant KRAS copy number 5.
Distinct roles in metabolism may be among the reasons for the persistence throughout vertebrate evolution of alternative KRAS splicing. Preferential KRAS4A expression in the gastrointestinal track 22 may be due to the specific metabolic requirements of these tissues. Recently, HK1 and phosphofructokinase were found to be the glycolytic enzymes that control flux through the pathway 23, suggesting that direct regulation of HK1 by KRAS4A would have a significant impact. Moreover, because one of the desired outcomes of metabolic rewiring from the standpoint of sustaining rapid tumor growth is diverting glucose into the pentose phosphate pathway 24, HK1 is an ideal regulatory node. KRAS-mediated metabolic reprogramming has long been seen as a potential vulnerability in cancer, and both HK1 and HK2 inhibitors have been examined in this regard 25. Our findings suggest that efforts along these lines may prove most effective in tumors that express relatively high levels of KRAS4A and that a better understanding of the regulation of KRAS splicing and KRAS4A palmitoylation may reveal new modes of therapy.
Methods
Cell lines, culture and transfection
The cell lines A549, COS1, HCT-15, HCT-116, HEK293, HeLa, SUIT2 and U2OS were from ATCC (Manassas, VA, USA) where each line was validated by short tandem repeat profiling. Flp-In T-REx293™ cells were from Thermo Fisher Scientific (Waltham, MA, USA) and validated by successful insertion of a gene of interest upon ectopic expression of Flp recombinase. KRAS4A null A549 and SUIT2 cells were generated by introducing an indel into exon 4A with CRISPR/Cas9, and HK1 and HK2 deficient HEK293 cells were generated by targeting the second and third exons of HK1 and HK2, respectively, with the same strategy. All cell lines were maintained in DMEM, except HCT-15 cells, which were grown in RPMI medium. All media were supplemented with 10% FBS and 1× penicillin/streptomycin. Transgene expression was induced in Flp-In T-REx293™ cells with 0.75 mg/mL doxycycline treatment for 24 h. Serum starving was performed with media containing 0.1%–1.0% FBS. All cell lines were transfected with Lipofectamine 3000 reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Recombinant proteins
All GST-tagged proteins (HK1-FL, HK1-Cterm, HK2-FL, PAK1-RBD, RAF1-RBD, and free GST) and 6xHis-RAC2 were produced in the BL21 strain of E.coli and purified in HEPES buffer (20 mM HEPES, pH 7.3, 150 mM NaCl, 5 mM MgCl2, and 0.2% TritonX-100). HK1 proteins were induced with 0.75 mM IPTG at 18°C for 24 h, while all other constructs were induced at 37°C for 3 h. E.coli were lysed in HEPES buffer containing 2% TritonX-100 via sonication, and GST-tagged proteins were isolated from clarified lysate using glutathione-agarose beads (Sigma-Aldrich, St. Louis, MO, USA). Recombinant RAS proteins were generously provided by William Gillette at the NCI RAS Initiative (Fredrick, MD, USA). For GDP/GTP-loading experiments RAS proteins were incubated at 4°C in HEPES buffer supplemented with 10 mM EDTA and a 10-molar excess (with respect to RAS) of GDP or GTPɣS (Sigma-Aldrich) for 15 min to dissociate bound nucleotide. Then, 1M MgCl2 was added to the solution and incubated for 30 min at 4°C to allow nucleotide re-binding.
Immunoprecipitation, in vitro binding and western blotting
Cells were lysed in co-IP Buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM NaF, 1 mM EDTA and 1% NP40) supplemented with 1× protease inhibitor tablet (Roche, Basel, Switzerland) and 1× PhosStop tablet (Sigma-Aldrich). Immunoprecipitation was performed in cleared cellular lysate using anti-FLAG- (Sigma-Aldrich), anti-GFP- (MBL Life Science, Woburn, MA, USA) or Y13–259 anti-RAS (Sigma-Aldrich) conjugated agarose beads for 1–3 hr at 4°C. For in vitro binding assays, 0.5 μg of GST-tagged bait proteins bound to glutathione-agarose beads were incubated with 1.0 μg of the indicated recombinant GTPases for 1 hr at 4°C.
After washing beads with co-IP or HEPES buffer, bound proteins were eluted with SDS sample buffer and assayed by western blotting using 4%–20% SDS-PAGE gels (Invitrogen, Carlsbad, CA, USA) and PVDF membranes (Millipore, Burlington, MA, USA). The following antibodies were used for western blotting: anti-FLAG (Sigma-Aldrich, F7425, Lot:085M4774V 1:2000), anti-GFP (ThermoFisher, A-6455, Lot:1826342, 1:2000), anti-HK1 (CST, #2024, Clone:C35C4, Lot:3 1:2000), anti-HK2 (CST, #2867, Clone:C64G5, Lot:3, 1:2000), anti-SDHA (CST, #11998, Clone:D6J9M, Lot:2, 1:1000), anti-pMEK (CST, #9121, Lot:47, 1:1000), anti-tMEK (CST, #4694, Clone:L38C12, 1:1000), anti-pERK (CST, #9106, Clone:E10, 1:1000), anti-pAKT (CST, #9271, Clone:S473, Lot:14, 1:1000), anti-tAKT (CST, #9272, Lot:27, 1:1000), anti-pan-RAS (CalBiochem, OP40, Clone:Ab-3, Lot: D00119097, 1:2000), anti-Fibrillarin (Santa Cruz Biotechnology, sc-374022, Clone:G-8 Lot:GO116, 1:1000), anti-EEA1 (Santa Cruz Biotechnology, sc-137130, Clone:G-4, Lot:F2716, 1:1000), anti-F1-ATPase (Santa Cruz Biotechnology, sc-514419, Clone:C-12, Lot:E1016, 1:1000), anti-tERK (Santa Cruz Biotechnology, sc-94, K-23, 1:1000), anti-Rho GDI (Santa Cruz Biotechnology, sc-360, Clone:A-20, Lot:J0313, 1:5000), anti-HA (Santa Cruz Biotechnology, sc-7392, Clone: F-7, Lot:K3012 1:1000), anti-RAC2 (Abcam, 130415, Lot:GR310183–1, 1:1000), and anti-KRAS4A, polyclonal rabbit antibody developed by our laboratory 1 and licensed for commercial distribution (Sigma-Aldrich ABC1442, 1:500).
Mass spectrometry
Affinity purified proteins were reduced, alkylated, and loaded onto an SDS-PAGE gel to remove any detergents and LCMS incompatible reagents. The gel plugs were excised, de-stained, and subjected to proteolytic digestion with trypsin. The resulting peptides were extracted, desalted, and an aliquot was analyzed with LCMS coupled to a Thermo Fisher Scientific Orbitrap QExactive Mass Spectrometer operated in data-dependent mode as previously described 26. The data was searched against a UniProt human database, using Sequest within Proteome Discoverer.
Cellular and biochemical assays
Hexokinase activity assays were performed using the Hexokinase Assay Kit (BioVision, Milpitas, CA, USA); assays that included 2-DG as an HK1 inhibitor were performed with 20 mM 2-DG (Sigma-Aldrich). In vitro glucose uptake assays were performed by culturing 5×105 cells in 2 mL of low-serum, 5 mM glucose DMEM for 24 h. After culture, the media was removed, cells and debris were cleared by centrifugation, and remaining glucose in the media was measured using Infinity™ Glucose Oxidase Liquid Stable Reagent. Mitochondria were isolated from the post-nuclear supernatants of homogenates of cells disrupted by nitrogen cavitation as we have described 27 using a Mitochondria Isolation Kit, that relies on immunoaffinity purification with magnetic beads conjugated with anti-TOM70 antibodies (Miltenyi Biotec, Bergisch Gladbach, Germany). The inhibition of cell growth by 2-DG was tested by growing cells in different concentrations of 2-DG and counting cells after 24-h treatment with a Beckman Coulter cell counter (Indianapolis, IN, USA).
Microscopy
Live cell imaging was performed on cells transiently transfected in 35 mm plates that incorporate a No. 1.5 cover-slip-covered well (MatTek Copr., Ashland, MA) with an inverted Zeiss 800 laser scanning confocal microscope (Oberkochen, Germany). Zeiss Zen Blue 2 version 2.1 software was used for data acquisition and analysis.
Super-resolution imaging by stochastic optical reconstruction microscopy (STORM)
Cells were stained with MitoTracker, then fixed (3% PFA, 0.2% Glutaraldehyde in PBS) and permeabilized (0.1% Triton X100 in PBS). 0.4 M glycine was added for 20 min to quench the glutaraldehyde. Cells were incubated in blocking buffer (2% glycine, 2% BSA, 0.2% gelatin, 50 mM NH4Cl in PBS) overnight at 4 °C. FLAG-KRAS4A and FLAG-KRAS4A12V were labelled with Rabbit anti-FLAG primary antibody (Thermo Fisher) then incubated with secondary antibody (Goat anti-rabbit conjugated to Alexa 647; Abcam). HK1-GFP was stained with Rabbit anti-GFP antibody conjugated to Alexa 488 (Abcam). All antibodies used were prepared and tested commercial and used at low concentration to ensure specificity. Coverslips were mounted on to a slide to make a flow chamber, and freshly mixed imaging buffer (1 mg/mL glucose oxidase, 0.02 mg/mL catalase, 10% glucose, and 100 mM mercaptoethylamine) was added to the sample chamber.
Imaging was performed on a custom-built setup based on a Leica DMI 300 inverse microscope. A 639 nm (UltraLaser, MRL-FN-639–800) and 488 nm (OBIS) laser was collimated into the microscope objective (Zeiss, HCX PL APO 63X NA = 1.47 OIL CORR TIRF), with the illumination at the objective focus adjusted to ~1.5 and 0.8 kW cm−2 for 639 and 488, respectively. Emitted photons were sequentially collected by a sCMOS camera (Prime 95B, Photometrics) at 33 Hz (30 ms per frame) for 2000 frames. STROM reconstruction was performed using an in-house written MATLAB script 28. Each point-spread-function (PSF) was fitted via the Maximum Likelihood Estimation (MLE). Note that the pixel-specific noise of each pixel was calibrated, characterized as a Gaussian distribution, and convolved with the Poisson short-noise distribution for MLE fitting 29. Broad-spectrum fluorescent beads (Diameter ~ 100 nm, TetraSpec, Thermofisher) were imaged in both the AlexaFluor 488 and 647 channels and the positions of each bead in the two different channels were mapped via a 2nd polynomial mapping algorithm were used to align images from different illuminations 28,29. Code is available at GitHub: https://github.com/yiny02/direct-Triple-Correlation-Algorithm and in the Supplementary materials to Ref. 29.
Stable-isotope tracing, lactate secretion, metabolite extraction, and GC-MS analysis
For stable-isotope labeling experiments, DMEM (D5030, Sigma; St. Louis, MO, USA) containing uniformly 13C-labeled glucose (13C6-glucose; Cambridge Isotope Laboratories, Tewksbury, MA, USA) and 10% dialyzed FBS (ThermoFisher, Waltham, MA, USA) was added to cells prior to metabolite extraction and GC-MS analysis. Cellular metabolites were extracted after a rinse with cold 0.9% saline solution using a methanol/water/chloroform extraction 30. For lactate secretion analysis, cells were cultured in DMEM with 10% dialyzed FBS for 24 hours, and 5μl of initial and conditioned media was extracted in 250 μl of 80% methanol:water containing 5 nmol of 13C3-lactate (Cambridge Isotope Laboratories, Tewksbury, MA, USA). Conditioned media was subjected to 1,000 x g centrifugation for 15 minutes at 4oC to remove cellular debris prior to extraction. Lactate secretion flux (nmol/cell*hr) was calculated by measuring the molar accumulation of lactate in conditioned media divided by the viable cell density over the 24 hours incubation 31. Post extraction, the aqueous phase was evaporated to dryness under vacuum by SpeedVac (Thermo Fisher, Waltham, MA, USA) followed by methoxyamine (MOX)-tBDMS derivatization as previously described 32. Derivatized samples were analyzed by GC-MS using a DB-35MS column (30 m x 0.25 mm i.d. x 0.25 μm) installed in an Agilent 7890B gas chromatograph (GC) interfaced with an Agilent 5977B mass spectrometer (MS) 31 and corrected for natural isotope abundance using in-house algorithms adapted from 33.
Extracellular acidification rate (ECAR)
ECAR was measured using a Seahorse XFe96 analyzer (Agilent, Santa Clara, CA, USA). Cells were seeded into Seahorse XFe96 plates at either 1.0, 1.5, or 2.0×104 cells/well 24 h prior to measurements. The following day, media was exchanged with DMEM (Sigma-Aldrich) supplemented with 25 mM glucose and 2 mM glutamine and incubated for 30 min at 37°C in a non-CO2-containing incubator. Respiratory and glycolytic rates were measured in response to sequential injections of oligomycin (2 μM) and 2-D-deoxyglucose (50 mM). Cells were immediately lysed using 30 μL/well Reagent A (Bio-Rad, Hercules, CA, USA) following each experiment, and protein was quantified using the DC protein assay kit (Bio-Rad). Individual well glycolytic rates were normalized to milligrams total protein, as quantified by a standard curve.
Xenografts
SUIT2 and A549 xenografts were established in 4–8-week-old male NCG mice (Charles River, Wilmington, MA, USA). Mice were injected with 0.5×106 KRAS4A−/− or KRAS4A+/+ cells into their contralateral flanks, and xenografts were grown for at most 6 weeks. Tumors were measured every 3–4 d with calipers. When tumors reached approximated 1 cm along their longest axis the mice were analyzed for glucose uptake by 18FDG PET/CT imaging. After imaging, the mice were sacrificed and tumors were excised, weighed and fixed in formalin. All animal protocols were approved by the NYU School of Medicine Institutional Animal Care and Use Committee (IACUC). Tumors were generated in mice under IACUC protocol IA16–00051, which allows tumors to grow to 2.5 cm3 or 2 cm in diameter. In no mouse did the tumor size exceed that allowable by the protocol.
microPET imaging
All mice were subjected to 18FDG-PET scans under IACUC protocol IA17–00566 using a microPET/microX-ray computed tomography (μPET/μCT) scanner (Inveon MM, Siemens Medical Solutions, Erlangen, Germany), equipped with a PET detector ring comprising of 16 detector blocks each containing a row of four lutetium oxyorthosilicate detectors for a total of 64 detectors capable of 1.4-mm isotropic spatial resolution. The scan consisted of a 60-min whole body μPET acquisition after the injection of 18FDG. This was followed by a 100-μm μCT scan after each μPET acquisition to assess the attenuation correction for the 18FDG datasets. The mice were fasted for at least 8 h before an intravenous injection of 18FDG (300–400 μCi dose diluted in physiological saline to a volume of 200 μL). The injection of 18FDG was administered via an infusion pump (PHD2000, Harvard Apparatus, Cambridge, MA, USA) at a rate of 60 μL/min. Each subject was administered the radiotracer right after the start of the PET acquisition, allowing the capture of the pharmacokinetics of 18FDG uptake in the whole body of the mouse. The hybrid scanner was equipped with a M2M BIOVET module used to continuously monitor vital signs of the examined subject. The imaging scan consisted of initially placing each mouse in an induction chamber using 3%–5% isoflurane exposure during 2–3 min until the onset of anesthesia. The animal was subsequently positioned prone on the bed palate over a recirculating warm water blanket in which 1.0%–1.5% isoflurane was administered by a nose cone throughout the scan. Mice were monitored continuously throughout the scanning session via a respiration pad and a rectal temperature probe as a feedback for temperature regulation of the whole stage. The body temperatures of the animals were maintained between 36°C–37°C using a circulating water-heating pad.
PET data analysis
FDG-PET data were analyzed using the Inveon Research Workplace software (Siemens Medical Solutions). Each PET dataset was reconstructed as a 3D dataset co-registered to a 100-μm isotopic μCT dataset for attenuation correction and overlaid to provide anatomical context. Tumor uptake was calculated as the standard uptake value (SUV), a common metric in PET analysis to account for the variations introduced by the injected dose, the time of injection, and the animal weight. SUV was measured over a region of interest (ROI) circumventing the tumor mass by thresholding each individual volume from the μCT dataset using Hounsfield units. SUV time activity curves of each ROI were interpreted as a surrogate marker of glucose utilization.
Statistical analysis
All statistical analyses were performed with GraphPad Prism version 8.1.1 (San Diego, CA, USA). Differences between groups of two and more than two were assessed with Student’s t-tests and two-way ANOVA with the Tukey post hoc analysis for multiple comparisons, respectively. A p-value <0.05 was the cutoff used for statistical significance.
Extended Data
Extended Data Figure 1. |. KRAS4A binds HK2 in an isoform-specific and GTP-dependent fashion.
The indicated FLAG-tagged RAS constructs (±G12V mutations) were expressed in HeLa cells and immunoprecipitated with anti-FLAG beads. Blots were probed for FLAG-tagged proteins and endogenous HK2. FLAG-RAC161L served as the negative control. The immunoblot shown is representative of 4 independent experiments.
Extended Data Figure 2. |. Association of KRAS4A with HK1 and HK2 requires prenylation but is diminished by palmitoylation.
a, HeLa cells expressing the indicated, FLAG-tagged GTPase were lysed, KRAS4A or RAC1 were immunoprecipitated, and the precipitates blotted for FLAG-tagged GTPases or endogenous HK1 or HK2. b, To confirm that the results in (a) reflect membrane targeting rather than GTP loading, the relative GTP loading of the KRAS4A proteins was determined by GST-RBD affinity capture. FLAG-KRAS4A with an activating 12V mutation and either a native (N) membrane-targeting sequence or one with the indicated substitution were expressed in HEK293 cells (left). In addition, FLAG-KRAS4A12V expressing cells were treated with 2BP to inhibit palmitoylation or FTI to inhibit farnesylation (right). Total FLAG-KRAS12V was measured by anti-FLAG immunoblot of 1% of the lysate (bottom) and GTP-bound FLAG-KRAS4A was measured by affinity purification of the remaining lysate with GST-RBD (top). The number under each lane is the amount of GTP-bound KRAS4A relative to lane 1 after normalization for expression (bottom). a-b, Immunoblots shown are representative of 2 independent experiments.
Extended Data Figure 3. |. Colocalization of palmitoylation-deficient KRAS4A but not NRAS with HK1 on mitochondria.
a, Representative live-cell images of COS1 cells co-transfected with the indicated mCherry-KRAS4A constructs and GFP extended with the mitochondrial-targeting sequence of HK1 (HK1mt-GFP). Arrowhead and arrow indicate plasma membrane and nuclear envelope, respectively; scale bar represents 10 μm. The cell shown is representative of hundreds on each plate of 5 independent transfections. b, Colocalization of KRAS4A ±palmitoylation with HK1 and HK2. COS1 or U2OS cells were co-transfected with GFP-tagged HK1 or HK2 and mCherry-tagged, constitutively active KRAS4A12V, with or without mutation of cysteine 180 to serine (180S) to block palmitoylation. The cells were imaged alive with a Zeiss LSM 800 and the Pearson’s correlation coefficient between the red and green channels was measured. Data plotted as mean±SD of the values measured in 15 cells examined (n=15). Significance was determined by unpaired, two-tailed student’s t-test. c, Neither WT nor palmitoylation-deficient NRAS colocalizes with HK1-targeted GFP on mitochondria. mCherry tagged WT, palmitoylation-deficient (C181S), or prenylation-deficient (C186S) NRAS were coexpressed in COS1 cells with GFP extended with the mitochondrial targeting region of HK1 (mitoHK1-GFP) and imaged alive with an inverted Zeiss 800 laser scanning confocal microscope. Bar indicates 10 μm. Images shown are representative of hundreds of transfected cells on each plate in 2 independent experiments. d, Super-resolution (STORM) image of U2OS cells transfected with FLAG-KRAS4A12V,180S or FLAG-KRAS4B12V showing colocalization with HK1 on the OMM (arrow) of KRAS4A12V,180S but not KRAS4B12V. n=3. e, Mitochondria were purified from HCT-15 cells pre-treated with vehicle or 2-BP and analyzed by immunoblot with the indicated antibodies: succinate dehydrogenase (SDHA, mitochondrial matrix), F1-ATPase (mitochondrial inner membrane), RHOGDI (cytosol), fibrillarin (nucleolus), EEA1 (endosomes). RAS indicates total RAS detected by a pan-RAS antibody. KRAS4A immunoblot was quantified by Li-Cor Odyssey infrared scanner. Immunoblot shown is representative of 2 independent experiments. f, The interaction of KRAS4A with HK1 requires the HK1 OMM targeting sequence. The indicated FLAG-tagged KRAS4A constructs were co-expressed in HEK293 cells with HA-tagged full-length HK1 (FL) or HK1 missing its OMM-targeting region (Δ1–21; Δmito). FLAG-KRAS4A was immunoprecipitated, and binding to HK1 was assessed with an anti-HA immunoblot. The immunoblot shown is representative of 4 independent experiments.
Extended Data Figure 4. |. A putative RAS binding region in HK1.
a, Crystal structure (PDB 4G0N) of the CRAF RAS binding domain (RBD, cyan and magenta) in complex with the G-domain of HRAS (orange; nucleotide (gray) and magnesium (black)). The regions of the CRAF RBD that mediate the interaction with HRAS are colored magenta. b, Superposition of the N-terminal lobe of HK1 (green and magenta; PDB 4F9O) alongside HRAS (orange). The putative region of HK1 that interacts with RAS, corresponding to that of CRAF RBD, are colored magenta. c, A section of the helix-loop-sheet structure common to RBDs are superimposed on the region of HK1 highlighted in (b). The RBD of CRAF is shown in cyan, BRAF (PDB 3NY5) in yellow and the putative RBD of HK1 in green. d, Sequence alignments of validated RBDs and the HK1 putative RBD shown with structural motifs of BRAF (yellow) and HK1 (green). Despite the highly conserved structural features shown in (c), RBDs have little sequence homology.
Extended Data Figure 5. |. Hexokinase enzyme kinetics with and without recombinant KRAS4A and with and without 2-deoxyglucose (2-DG).
a, Activity of recombinant full-length hexokinase 1 is unaffected by recombinant KRAS4A. Reaction velocity is plotted (mean±SE) as a function of glucose concentration. Velocities are plotted ±addition of recombinant, GTP-loaded KRAS4A or RAC2. Plots combine independent assays, n=4. b, Hexokinase enzyme kinetics. Full-length hexokinase 1 and 2 and the catalytic C-terminal domain of HK1 were expressed in E. coli as GST fusion proteins and affinity purified with glutathione-agarose beads. Hexokinase activities on the beads was measured with a linked assay kit (BioVision, Milpitas, CA, USA) in which the glucose-6-phophate produced is oxidized by glucose-6-phosphate dehydrogenase to form NADH, which reduces a colorless probe to a colored product with strong absorbance at 450 nm. VMAX and KM were calculated by nonlinear regression using GraphPad Prism software (v 8.1.1) and goodness of fit is given as R2.
Extended Data Figure 6. |. Dissociation between MAPK signaling and differential stimulation of glucose consumption and basal extracellular acidification rate (ECAR) by KRAS4A versus palmitoylation-deficient KRAS4A and KRAS4B.
a-c, Flp-In™ T-REx™ 293 cells were generated that express the indicated KRAS proteins upon doxycycline induction. a,b, Glucose consumption, mean±SE, n=5 (a), and basal ECAR, mean±SE, n=10 (b), were measured in doxycycline induced cells revealing the order of potency as KRAS4A12V,180S> KRAS4A12V> KRAS4B12V. Significance was determined by student’s t-test, paired (a) and unpaired (b). c, Immunoblot reveals equivalent expression of the three KRAS proteins in the cells used in a and b. Whereas KRAS4A12V and KRAS4B12V induced equivalent levels of phospho-ERK and phospho-MEK, KRAS4A12V,180S (palmitoylation deficient) was less potent. These cells have constitutively high levels of AKT phosphorylation that were not altered by expression of any form of KRAS. Note also that, despite MAPK stimulation, protein levels of HK1 and HK2 were not altered. Immunoblots shown are representative of two independent experiments (n=2). d, Parental HEK293 cells with lower basal pAKT were transfected with the indicated constructs, transferred to 0.1% serum 18 hrs after transfection and lysed 24 hrs later. Lysates were analyzed for the indicated proteins by immunoblot. n=2.
Extended Data Figure 7 |. Glucose utilization and 2DG sensitivity in KRAS mutant human tumor cells with or without exon 4A.
a, Rate of glucose consumption (mean±SE, n=3) in parental (4A+/+), exon 4A targeted (4A−/−) A549 and 4A−/− cells transfected with FLAG-GFP or FLAG-KRAS4A12V. Representative expression is shown by immunoblot. b, Lactate secretion flux (mean±SE, n=3) measured over 24 hrs in A549 cells with the indicated genotype. c, Incorporation of 13C from glucose into lactate (mean±SE, n=3) in A549 cells with the indicated genotype. a-c, Mean±SE is plotted, n=3, significance determined by paired student’s t-test. d-e, Growth inhibition by 2DG of SUIT2 (d, 48 hrs) and A549 (e, 24 hrs) cells with or without KRAS4A (mean±SE, n=3, significance by two-way ANOVA). f-g, SUIT2 (f) or A549 (g) cells were used to establish xenograft tumors on the contralateral flanks (4A+/+ versus 4A−/−) of NCG mice. Six weeks later, when the tumors were established and of equivalent size, glucose uptake was measured by 18FDG PET-CT scan. f, Representative coronal scan or mouse with SUIT2 xenografts (left). Glucose uptake represented by a color look up table. Standardize uptake values (SUVs) of the entire tumor are plotted (mean±SD, n=5 mice, significance by two-way ANOVA) as a function of time after 18FDG injection (right). g, Coronal scan (left) of representative mouse (n=5) with A549 xenografts. SUV versus time after injection plotted for that mouse (right).
Extended Data Figure 8. |. Diminished basal extracellular acidification rate (ECAR) in KRAS mutant tumor cells null for KRAS4A.
a-b, ECAR measured by Seahorse XFe96 of (a) A549 and (b) SUIT2 cells with or without disruption of KRAS exon 4A by CRISPR/Cas9. Oligomycin inhibits oxidative phosphorylation and allows measurement of glycolytic reserve. 2-DG inhibits glycolysis. Data plotted are mean±SE, n=10 technical replicates.
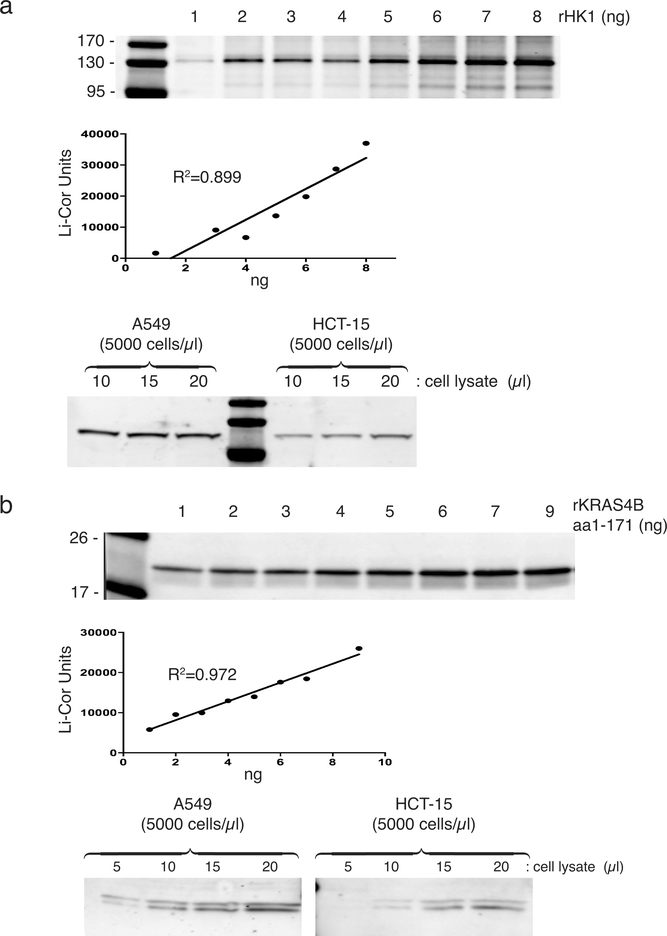
Extended Data Figure 9. |. Quantification of HK1 and total KRAS in tumor cell lines.
a, (top) A standard curve for the quantification of HK1 by immunoblot was generated by titrating recombinant HK1 and probing with a rabbit monoclonal Ab (Cell Signaling C3534). (bottom) Immunoblots of the indicated amounts of lysate (5000 cells per μl). Calculations based on these results and MW=100 kDa indicate 200,000 and 150,000 molecules per cell for A549 and HCT-15 cells, respectively. b, (top) A standard curve for the quantification of total KRAS by immunoblot was generated by titrating recombinant KRAS4B truncated at aa171 and probing with a mouse monoclonal Ab (Sigma WH0003845M1). (bottom) Immunoblots of the indicated amounts of lysate (5000 cells per μl). Calculations based on these results and MW=21 kDa indicate 700,000 and 200,000 molecules per cell for A549 and HCT-15 cells, respectively. a,b, Standard curves were plotted by linear regression using GraphPad Prism software v. 8.1.1, with goodness of fit given as R2.
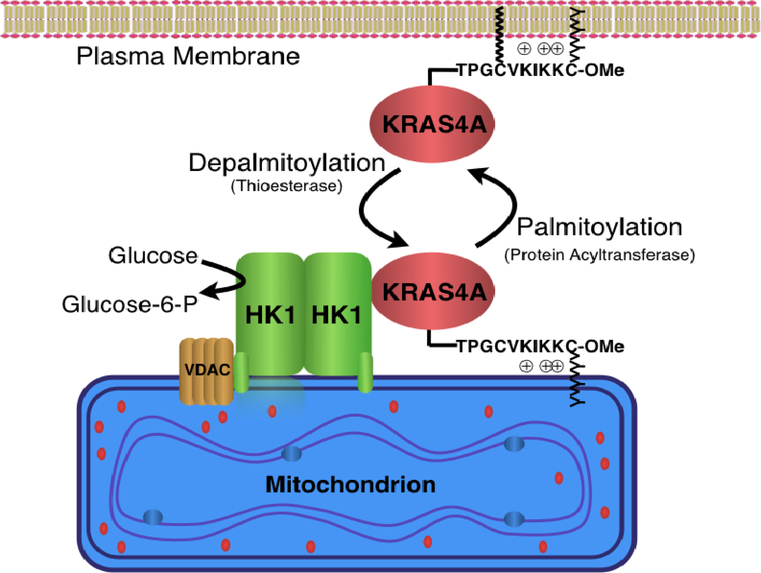
Extended Data Figure 10. |. Model of KRAS4A regulation of hexokinase 1 (HK1).
Like all palmitoylated GTPases, KRAS4A cycles between a palmitoylated and depalmitoylated state. When palmitoylated, the protein has relatively high affinity for the plasma membrane by virtue of farnesylation of the C-terminal CAAX sequence and an adjacent polybasic region that operate in conjunction with palmitoylation. Upon depalmitoylation, KRAS4A loses affinity for the plasma membrane and gains affinity for endomembranes, including the outer mitochondrial membrane (OMM). Tethering of KRAS4A to the OMM allows it to interact with HK1, resident on this compartment by virtue of an N-terminal OMM targeting sequence and protein-protein interaction with VDAC. Interaction of KRAS4A with HK1 on the OMM decreases allosteric inhibition by glucose-6-phosphate and thereby enhances HK1 activity and glycolytic flux.
Supplementary Material
Acknowledgments
We thank Dominic Esposito and William Gillette of NCI Frederick for supplying highly purified recombinant KRAS proteins. This work was funded by the National Institutes of Health (R01CA163489 and R01CA116034 to M.R.P.; R01CA157490, R01CA188048, P01CA117969, R35CA232124, R01GM095567 to A.C.K.; R35CA210018 and U01CA217864 to A.B.; R01CA214948 to R.P.; T32CA009161 to J.P.M.; T32GM088118 to C.R.A.), the AACR Basic Cancer Research Fellowship (grant number 15-40-01-MAHA, to J.P.M.), the Charles H. Revson Senior Fellowship in Biomedical Science (to J.P.M.), UCSF Pancreas Center and the Schwartz Family Foundation (to W-C.C.) and the Lustgarten Foundation and SU2C (to A.C.K.). Proteomic analysis and in vivo imaging were performed in the Proteomics and DART Preclinical Imaging Cores, respectively, each partially funded by the NYU Laura and Isaac Perlmutter Cancer Center Support Grant, NIH/NCI P30CA016087. The Center for Advanced Imaging Innovation and Research (CAI2R, www.cai2r.net) at New York University School of Medicine is supported by NIH/NIBIB P41 EB017183.
Footnotes
Competing Interests: A.C.K. has financial interests in Vescor Therapeutics, LLC. A.C.K. is an inventor on patents pertaining to KRAS regulated metabolic pathways, redox control pathways in pancreatic cancer, targeting GOT1 as therapeutic approach, and the autophagic control of iron metabolism. A.C.K. is on the SAB of Rafael/Cornerstone Pharmaceuticals.
Data Availability: The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.
Supplemental Information: is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Tsai FD et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci U S A 112, 779–784, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voice JK, Klemke RL, Le A & Jackson JH Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J Biol Chem 274, 17164–17170. (1999). [DOI] [PubMed] [Google Scholar]
- 3.Kimmelman AC Metabolic Dependencies in RAS-Driven Cancers. Clin Cancer Res 21, 1828–1834, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ying H et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerr EM, Gaude E, Turrell FK, Frezza C & Martins CP Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 531, 110–113, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottlob K et al. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev 15, 1406–1418, (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vyssokikh MY & Brdiczka D The function of complexes between the outer mitochondrial membrane pore (VDAC) and the adenine nucleotide translocase in regulation of energy metabolism and apoptosis. Acta Biochim Pol 50, 389–404, (2003). [PubMed] [Google Scholar]
- 8.Patra KC et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 24, 213–228, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahearn I, Zhou M & Philips MR Posttranslational Modifications of RAS Proteins. Cold Spring Harb Perspect Med, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rocks O et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141, 458–471, (2010). [DOI] [PubMed] [Google Scholar]
- 11.Jennings BC et al. 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J Lipid Res 50, 233–242, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.John S, Weiss JN & Ribalet B Subcellular localization of hexokinases I and II directs the metabolic fate of glucose. PLoS One 6, e17674, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wohlgemuth S et al. Recognizing and defining true Ras binding domains I: biochemical analysis. J Mol Biol 348, 741–758, (2005). [DOI] [PubMed] [Google Scholar]
- 14.Zhu A, Romero R & Petty HR An enzymatic colorimetric assay for glucose-6-phosphate. Anal Biochem 419, 266–270, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai HJ & Wilson JE Functional organization of mammalian hexokinases: both N- and C-terminal halves of the rat type II isozyme possess catalytic sites. Arch Biochem Biophys 329, 17–23, (1996). [DOI] [PubMed] [Google Scholar]
- 16.Sebastian S, Wilson JE, Mulichak A & Garavito RM Allosteric regulation of type I hexokinase: A site-directed mutational study indicating location of the functional glucose 6-phosphate binding site in the N-terminal half of the enzyme. Arch Biochem Biophys 362, 203–210, (1999). [DOI] [PubMed] [Google Scholar]
- 17.Rajalingam K, Schreck R, Rapp UR & Albert S Ras oncogenes and their downstream targets. Biochim Biophys Acta 1773, 1177–1195, (2007). [DOI] [PubMed] [Google Scholar]
- 18.Chiu VK et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol 4, 343–350. (2002). [DOI] [PubMed] [Google Scholar]
- 19.Mor A & Philips MR Compartmentalized Ras/MAPK Signaling. Annual review of immunology 24, 771–800 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Aran V & Prior IA Compartmentalized Ras signaling differentially contributes to phenotypic outputs. Cell Signal 25, 1748–1753, doi: 10.1016/j.cellsig.2013.05.004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yun J et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 325, 1555–1559, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newlaczyl AU, Coulson JM & Prior IA Quantification of spatiotemporal patterns of Ras isoform expression during development. Sci Rep 7, 41297, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanner LB et al. Four Key Steps Control Glycolytic Flux in Mammalian Cells. Cell Syst 7, 49–62 e48, doi: 10.1016/j.cels.2018.06.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vander Heiden MG, Cantley LC & Thompson CB Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hay N Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer 16, 635–649, doi: 10.1038/nrc.2016.77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peled M et al. Affinity purification mass spectrometry analysis of PD-1 uncovers SAP as a new checkpoint inhibitor. Proc Natl Acad Sci U S A 115, E468–E477, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M & Philips MR Nitrogen Cavitation and Differential Centrifugation Allows for Monitoring the Distribution of Peripheral Membrane Proteins in Cultured Cells. JoVE (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y, Lee WTC & Rothenberg E Ultrafast data mining of molecular assemblies in multiplexed high-density super-resolution images. Nature communications 10, 119, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang F et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat Methods 10, 653–658, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metallo CM et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grassian AR et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res 74, 3317–3331, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis CA et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell 55, 253–263, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez CA, Des Rosiers C, Previs SF, David F & Brunengraber H Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom 31, 255–262, (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.