Abstract
Triple-negative breast cancers (TNBCs) represent 15–20% of all breast cancers and are often associated with poor prognosis. The lack of targeted therapies for TNBCs contributes to higher mortality rates. Aberrations in the Phosphoinositide-3-kinase (PI3K) and Mitogen Activated Protein Kinase (MAPK) pathways have been linked to increased breast cancer proliferation and survival. It has been proposed that these survival characteristics are enhanced through compensatory signaling and crosstalk mechanisms. While the crosstalk between PI3K and ERK1/2 pathways has been characterized in several systems, new evidence suggests that MEK5/ERK5 signaling is a key component in the proliferation and survival of several aggressive cancers. In this study, we examined the effects of dual inhibition of PI3K/Akt and MEK5/ERK5 in the MDA-MB-231, BT-549, and MDA-MB-468 TNBC cell lines. We used the Akt inhibitor Ipatasertib, ERK5 inhibitors XMD8-92 and AX15836, and the novel MEK5 inhibitor SC-1-181 to investigate the effects of dual inhibition. Our results indicate that dual inhibition of PI3K/Akt and MEK5/ERK5 signaling was more effective at reducing proliferation and survival of TNBCs than single inhibition of either pathway alone. In particular, a loss of Bad phosphorylation at two distinct sites was observed with dual inhibition. Furthermore, the inhibition of both pathways led to p21 restoration, decreased cell proliferation, and induced apoptosis. Additionally, the dual inhibition strategy was determined to be synergistic in MDA-MB-231 and BT-549 cells and was relatively nontoxic in the non-neoplastic MCF-10 cell line. In summary, the results from this study provide a unique prospective into the utility of a novel dual inhibition strategy for targeting TNBCs.
Keywords: Triple-negative breast cancer (TNBC), PI3K/Akt, MEK5/ERK5, Synergy
1. Introduction
Triple-negative breast cancers (TNBCs) represent 15–20% of breast cancers and are often associated with a poor prognosis and increased mortality [Voduc et al., 2010; Schnitt, 2010]. Treatment options for TNBCs are primarily limited to chemotherapy, radiation, and surgery [Anders and Carey, 2009]. Identification of driver mutations in the phosphatidylinositol-3-kinase (PI3K) and mitogen activated protein kinase (MAPK) pathways have led to FDA approvals of drugs targeting MEK1/2 (Trametinib) and BRAF (Dabrafenib) in melanoma and PI3K delta (Idelalisib) in leukemia [Khalili et al., 2012; Rheault et al., 2013; Lannutti et al., 2011]. Genomic alterations leading to constitutive activation of BRAF, a component of the MAPK pathway, or a loss of the tumor suppressor PTEN, make the PI3K and MAPK pathways promising leads for applying targeted therapy to TNBCs [Cancer Genome Atlas, 2012].
It has been proposed that anti-apoptotic, pro-proliferative, and migratory signaling is enhanced through compensatory signaling and crosstalk between intracellular pathways. The crosstalk between the PI3K/Akt and MEK1/2/ERK1/2 pathways has been characterized in several systems. For example, MEK1/2 inhibition results in an increase in active Akt by decreasing the regulatory capacity of the phosphatase PTEN in HEK-293 (Kidney), HeLa (Lung), and BT474 (Breast) cells [Aksamitiene et al., 2012]. The discovery of these interactions, between the PI3K/Akt and MEK1/2/ERK1/2 pathways, has led to cancer treatments that combine inhibitors of these signaling cascades to enhance the therapeutic effect vs. each inhibitor alone. However, targeting both pathways in parallel has yielded mixed results with observed synergy in Non-small cell lung cancer (NSCLC) while parallel combinations of either MEK1/2 and Akt or MEK1/2 and PI3K inhibition were less efficacious than expected and exhibited high toxicity in breast cancer patients [Meng et al., 2010; Saini et al., 2013]. Therefore, there is a need to identify both efficacious and less-toxic kinase inhibition combinations for breast cancer treatment.
The MEK5/ERK5 pathway is a distinct member of the MAPK family consisting of a tiered kinase cascade that is activated in response to stress, growth factors, and cytokines [Hoang et al., 2017]. MEK5/ERK5 signaling has been shown to regulate the proliferation and survival of aggressive prostate cancers [McCracken et al., 2008]. Relevant to our current study, ERK5 expression is higher in TNBCs when compared to healthy breast tissue and correlates with a poorer prognosis [Al-Ejeh et al., 2014]. Targeting MEK/5ERK5 has been limited to shRNA and molecular tools such as BIX02189 and XMD8-92, which target the ATP site of MEK5 and ERK5, respectively [Hoang et al., 2017]. ERK5 shRNA induced apoptosis in TNBCs via a BCL-2/caspase dependent mechanism [Ortiz-Ruiz et al., 2014]. Pharmacological inhibition of ERK5 with XMD8-92 decreased proliferation in HeLa cells by inducing p21 expression [Yang et al., 2010]. In addition to its promise as a single target, MEK5/ERK5 has been linked to Akt signaling with the latter phosphorylating MEKK3, the upstream kinase of MEK5, in neuroblastoma cells [Umapathy et al., 2014].
In this study, we used a variety of PI3K/Akt and MEK5/ERK5 inhibitors to determine the effect of the inhibitors alone or in combination on the viability of TNBC cells. In particular, we examined the effect of the Akt inhibitor Ipatasertib, which is currently in Phase III trials for TNBC []. We also examined the downstream effects on Bad at two distinct phosphorylation distinct sites: serine 112 and serine 136. Overexpression of constitutively active MEK5 in endothelial cells suppressed caspase 3 activity via hyperphosphorylation of the proapoptotic protein Bad at serine 112, in a p90RSK independent manner [Pi et al., 2004]. Conversely, ERK5 knockdown in endothelial cells reduced the phosphorylation of Bad at serine 112, leading to cleaved caspase-3 dependent cell death [Hayshi et al., 2004]. A second phosphorylation site on Bad, serine 136, is phosphorylated by Akt, leads to a decrease in caspase-mediated apoptosis [Datta et al., 1997]. Therefore, we propose that serine 112 and serine 136 are phosphorylated by ERK5 and Akt, respectively, and targeting both ERK5 and Akt will decrease TNBC cell survival more than targeting either pathway alone. Lastly, we evaluated the efficacy of PI3K/Akt and MEK5/ERK5 inhibition in combination with conventional TNBC therapy, paclitaxel, and measured collateral toxicity in a non-neoplastic cell line.
2. Materials and Methods
2.1. Cell culture
The human triple-negative breast cancer cell lines (TNBC): MDA-MB-231, BT-549, and MDA-MB-468 were obtained from ATCC. MDA-MB-231 cells were maintained in DMEM:F-12 (1:1) (Life Technologies) while BT-549 and MDA-MB-468 were maintained in 1640 RPMI (Life Technologies) and L-15 (ATCC) media, respectively.. Cultures were supplemented with 10% Fetal Bovine Serum (Atlanta Biologicals), and 1% Penicillin/Streptomycin (Sigma). The MCF-10A cell line was also obtained from ATCC and was used as a normal tissue control. Cells were incubated at 5% CO2 and 37°C, except the MDA-MB-468 cell line: 0% CO2. Each cell line was utilized for fewer than 15 passages after thawing.
2.2. Inhibitors
The following inhibitors were obtained and used in the described experiments. Abbreviations in bold: Pan-Akt ATP-site inhibitor Ipatasertib (Ipat; Selleckchem), BRD4 inhibitor CPI203 (CPI; Sellekchem), ATP-site ERK-5 inhibitors XMD8-92 (XMD; Tocris) and AX15836 (AX; Tocris), MEK1/2 inhibitor Trametinib (Selleckchem), and novel MEK5 inhibitor SC-1-181 [Chakrabarty et al., 2018]. Additionally, the microtubule inhibitor paclitaxel (Sigma) was used for select experiments. All inhibitors were dissolved in DMSO (Fisher) and used in the concentrations noted in the text and figures. Control groups were treated with a percentage of DMSO (0.2% maximum in combination groups) equal to the treatment groups.
2.3. Cell viability
TNBC and MCF-10A cells were plated at a density of 5×103 cells per well. Cells were allowed to attach overnight and were treated with inhibitors alone or in combination with 5% FBS stimulation for 72 hours. DMSO was used as a vehicle control. After treatment, 10µL MTT (Sigma) was added to each well (0.5 mg/mL final concentration) and the plates were incubated for 3 hours (5% CO2 and 37°C). The medium was removed and the MTT-formazan crystals were dissolved with 100µL DMSO per well. The absorbance was measured at 570 nm with a VICTOR3 1420 multilabel counter (Perkin Elmer). Three wells were analyzed for each condition, and wells containing medium-MTT only (no cells) and vehicle were used as controls. IC50 values from the MTT experiments were calculated with Microsoft Excel.
2.4. Immunofluorescence microscopy
Cells were plated and treated as described in the MTT assay. After treatment, cells were fixed in 4% Paraformaldehyde for 15 minutes. Cells were then permeabilized with 0.3% Triton-X followed by the addition of rabbit Ki67 and mouse ∝-Tubulin primary antibodies (1:1000, Cell Signaling). Goat anti-mouse Alexa Flour 488nm and goat anti-Rabbit Alexa Flour 555nm (1:1000, Invitrogen) were used as secondary antibodies. A Hoechst (Fisher) stain was used to visualize the nucleus. Images were obtained with the EVOS fl inverted microscope (Life Technologies) under 10x objective. A blinded observer counted Hoechst+ and Ki67+ cells using Image J software.
2.5. Detection of Cleaved Caspase 3
MDA-MB-231 cells were treated with inhibitors for 48 hours then lysed in accordance with the PathScan Sandwich ELISA (Asp 175) protocol (Cell Signaling; 7190C). The absorbance was measured at 450 nm with a VICTOR3 1420 multilabel counter (Perkin Elmer). Three wells were analyzed for each condition and the results were normalized to lysate protein concentration.
2.6. Migration assay
Cells were seeded at a density of 200,000 cells/well in a 12 well plate. Cells were allowed to attach and grow to 70% confluence. Prior to treatment, the medium was removed and a “scratch” was made with a 200 μL pipet tip. The underside of the plate was marked to denote the location of the initial wound. Detached cells and debris were washed off and removed with 1x PBS. After the wash, treatments were added to each well and initial images were obtained with an EVOS-fl inverted microscope under 10x magnification. The assay was ended with images were obtained after 24 hours. Wound closure was calculated by the formula: (border at 24 hr – border at 0 hr)/(border at 0 hr) x 100.
2.7. Western blot analysis
Cells were plated at a density of 5×105 per well. Cells were allowed to attach overnight and were serum starved for 24 hours before treatment. Cells were treated with vehicle or inhibitors one hour prior to 5% FBS stimulation for 24 hours. For Bad signaling, 50ng/mL EGF for 4 hours was used as a stimulus. Following treatment, cells were lysed and protein content was determined by a Bradford assay (Bio-Rad). 30µg of protein was loaded on a 4–15% SDS-PAGE gradient gel (Bio-Rad). The contents of the gel were transferred to a membrane and then probed with various antibodies: anti-phospho-ERK1/2, anti-total ERK1/2, anti-total ERK5, anti-phospho-ERK5, anti-phospho-Akt (Ser 473), anti-Akt, anti-pS6 (Ser 240/244), anti-p21, anti-cMYC, anti-Bad, and anti-phospho-Bad (Ser136) (1:1000, Cell Signaling). Anti-GAPDH (Millipore) was used as a loading control. The binding of antibody to antigen was detected by incubating membranes with secondary antibodies and scanning on an Odyssey Infrared Imager (LICOR biosciences). Blots were analyzed using ImageStudioLite by a blinded observer (LICOR biosciences). Phospho-Bad Ser112 was quantified with PathScan ELISA (Cell Signaling; 7182C).
2.8. Synergy calculations
Synergy data were analyzed with CompuSyn (v 1.4) software using the Chou Talalay method (1984). Combination indexes (CI) were obtained. Combination Indexes (CI) below 1 were considered to be synergistic.
2.9. Statistical inference
The data are presented as the mean +/− SEM of at least three independent experiments run in triplicate (n=3). Results were analyzed using Graph Pad Prism software (La Jolla, CA). Data were compared to respective controls with a One-Way ANOVA (Boferroni post hoc analysis), Two-Way ANOVA, or t-test where appropriate. A p value <0.05 was considered significant.
3. Results
3.1. Akt inhibitor Ipatasetib plus ERK5 inhibitor XMD8-92 synergistically decreased TNBC cell viability while sparing MCF-10 cells.
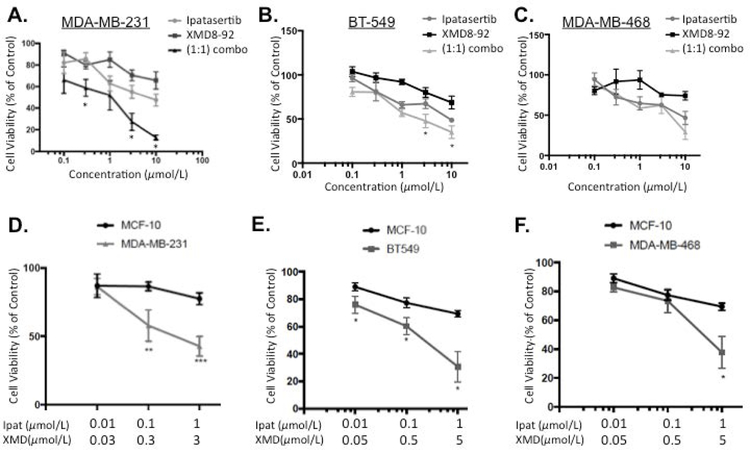
We preliminarily investigated the Akt inhibitor Ipatasertib and the ERK5 inhibitor XMD8-92 alone or in combination in TNBC cell lines (Figure 1A: MDA-MB-231; Figure 1B: BT-549; and Figure 1C: MDA-MB-468). Ipatasertib decreased viability in a concentration dependent manner in all three TNBC with IC50s of 10.4 µM, 8.1 µM, and 5.4 µM, respectively. The ERK5 inhibitor XMD8-92 decreased viability in the basal B cell lines MDA-MB-231 and BT-549 with IC50s of 31.3 µM and 44 µM, respectively. Equal molar ratios (1:1) of Ipat:XMD was most effective in the MDA-MB-231 and BT-549 cell lines. In particular, 0.3,0.3 µM, 3,3 µM, and 10,10 µM combinations were synergistic in MDA-MB-231 cells with CIs of 0.69, 0.54, 0.14, respectively (Table 1). Additionally, 3,3 µM and 10,10 µM combinations were synergistic in BT-549 cells with CIs of 0.44 and 0.28, respectively (Table 1). However, the MDA-MB-468 cell line did not exhibit synergy in equal molar combinations (1:1) of Ipat:XMD. This was in part due to the fact an IC50 XMD8-92 was not obtained. Additionally, Ipatasertib dominated the combination effect in the MDA-MB-468 cell line (Figure 1C). Therefore, we examined 1:3 and 1:5 ratios in the TNBC cell lines to account for the respective potencies of Ipatasertib and XMD8-92. In the MDA-MB-231 cell line the 1:3 combinations exhibited the strongest synergy with CIs of 0.11 and 0.25, respectively (Figure 1 D and Table 1). This synergy was noted with both inhibitors being administered two log units below their IC50s. Next, we established the preferential targeting of TNBC cells compared to normal MCF-10 cells with the Ipat+XMD combination in Figure 1D. The Ipat+XMD combination (0.1 µM and 0.3 µM), the viability was 56% in the MDA-MB-231 cell line and 86% in the MCF-10 cell line (30% difference). At the highest concentrations of the Ipat+XMD combination (1 µM and 3 µM), the viability was 42% in the MDA-MB-231 cell line and 77% in the MCF-10 cell line (35% difference). Similar results were achieved in the BT-549 and MDA-MB-468 cell lines in 1:5 combinations (Figure 1E and 1F). In particular, the 1,5 μM combination decreased viability by 70% in BT-549 cells vs 30% in the MCF-10A cells (40% difference). Additionally, the 1,5μM combination was synergistic with a CI of 0.23. Although synergy was not achieved in the MDA-MB-468 cell line, significant reductions in viability were observed with the 1,5μM combination of Ipat+XMD (Figure 1F).
Figure 1. Akt inhibitor Ipatasetib plus ERK5 inhibitor XMD8-92 synergistically decreased TNBC cell viability while sparing MCF-10 cells.
A-C. Ipatasertib and XMD8-92 were added alone or in 1:1 combinations to TNBC cell lines: MDA-MB-231 (A), BT-549 (B), and MDA-MB-468 (C). Cells were treated for 72 hours under 5% FBS stimulation and viability was analyzed using the MTT assay. Data represent the mean value +/− SEM of three different experiments (n=3). *P<0.05 vs drugs alone. 1:3 and 1:5 combinations of Ipatasertib and XMD8-92 were added to MCF-10 and TNBC cell lines: MDA-MB-231 (D), BT-549 (E), and MDA-MB-468 (F). Cell viability was analyzed after 72 hours using the MTT assay. *P<0.05, **P<0.01, and ***P<0.001 vs MCF-10 cells.
Table 1. Akt and ERK5 combination inhibition was synergistic in MDA-MB-231 and BT-549 TNBC cells.
Combinational analysis was performed using the Chou and Talalay method. Combination Indexes (CI) below 1 were considered to be synergistic. Values calculated using CompuSyn (v 1.4).
| MDA-MB-231 | BT-549 | ||||
|---|---|---|---|---|---|
| Ratio | Ipat+XMD (µmol/L) | CI (Combination index) | Ratio | Ipat+XMD (µmol/L) | CI (Combination index) |
| 1:1 | 0.3+0.3 | 0.69 | 1:1 | 3+3 | 0.44 |
| 3+3 | 0.54 | 10+10 | 0.28 | ||
| 10+10 | 0.14 | ||||
| 1:3 | 0.1+0.3 | 0.11 | 1:5 | 1+5 | 0.23 |
| 1+3 | 0.25 | ||||
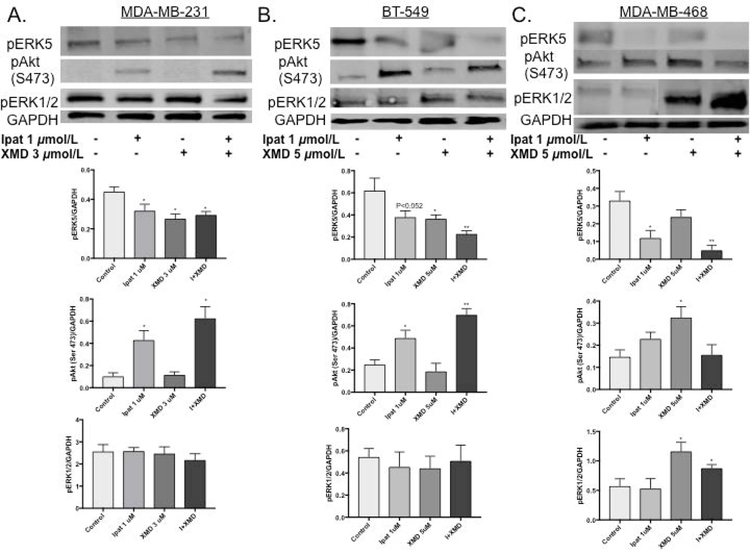
3.2. Effects of treatment with Ipatasertib and XMD8-92 on activation of signaling kinases in TNBC cell lines.
To assess the mechanism of synergy in the TNBC cell lines, we examined the phosphorylation sate of ERK5, ERK1/2 and Akt (Figure 2). 1:3 and 1:5 ratios of Ipat:XMD were used and, the kinase activity was examined after 24 hours. The ERK5 inhibitor worked as expected and reduced phosphorylation levels alone or in combination in all three cell lines. Interestingly, Ipatasertib decreased ERK5 phosphorylation in the MDA-MB-468 cell line. Ipatasertib preferentially binds to Akt in the active conformation [Kui et al., 2012]. This conformation protects the Serine 473 (Ser 473) and Threonine 308 (Thr 308) phosphorylation sites from becoming dephosphorylated by the phosphatase PP2A. Therefore, pAkt (Ser 473) is observed in the presence of Ipatasertib [Kui et al., 2012; Lin et al. 2013]. Ipatasertib also worked as expected and increased the phosphorylated Akt levels in the TNBC cell lines. Across all treatment groups, pERK1/2 signaling was not affected in the MDA-MB-231 and BT-549 cell lines. However, pERK1/2 signaling was increased in XMD alone or Ipat+XMD in the MDA-MB-468 cell line.
Figure 2. Analysis of ERK5, Akt and ERK1/2 activation in TNBC cells.
MDA-MB-231 (A), BT-549 (B), and MDA-MB-468 (C) cells were treated with Akt inhibitor, Ipatasertib, and ERK5 inhibitor, XMD8-92, for 24 hours under 5% FBS stimulation. Western blot analysis was performed on cellular lysates. GAPDH was used as a loading control. *P<0.05 and **P<0.01 vs control.
3.3. Effects of Ipatasertib alone or in combination with XMD8-92 on the proliferation of TNBC cell lines.
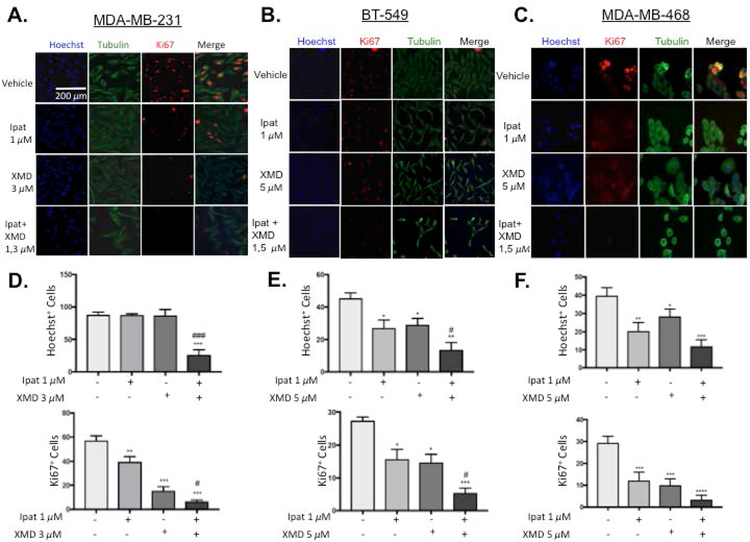
To examine the effects of the Ipatasertib and XMD8-92 combination on proliferation, immunofluorescent images of the proliferative marker Ki67 and cell counts were obtained (Figure 3). In MDA-MB-231 cells, neither Ipatasertib nor XMD8-92 alone decreased Hoechst+ cell counts. However, the Ipat+XMD combination significantly decreased Hoechst+ cells with respect to the control and both drugs alone (Figure 3D). These were results were consistent with the 1,3 uM viability data from Figure 1 D. XMD8-92 significantly decreased Ki67+ cells both alone and in combination (Figure 3D). In BT-549 cells, Ipatasertib and XMD8-92 alone or in combination significantly decreased Hoechst+ cell counts and Ki67+ cells. The combination also significantly decreased Hoechst+ cell counts and Ki67+ cells more than each drug alone (Figure 3E). In MDA-MB-468 cells, Ipatasertib and XMD8-92 alone or in combination significantly decreased cell counts and Ki67 cells (Figure 3 F).
Figure 3. The combination of Ipatasertib and XMD8-92 decreased proliferation in TNBC cells.
1:3 and 1:5 combinations of Ipatasertib and XMD8-92 were added to TNBC cell lines MDA-MB-231 (A,D), BT-549 (B,E), and MDA-MB-468 (C,F). After 72 hours of treatment, cells were fixed and Ki67 and alpha-tubulin were determined by immunofluorescence. The nuclei were stained with Hoechst. Images were obtained at 10x magnification with an EVOS inverted microscope. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 vs control. #P<0.05 and ###P<0.001 vs drugs alone.
One limitation of XMD8-92 is that it has also been shown to inhibit BRD4, an epigenetic regulator of many proliferative genes [Wong et al., 2014]. We addressed this by using the inhibitors AX15836 and CPI203 that selectively target ERK5 and BRD4, respectively (Lin et al. 2016). In MDA-MB-231 cells, the BRD4 inhibitor CPI-203 increased p21 expression alone (4.75 fold over control) and in combination (7.25 fold over control) with ERK5 inhibitor AX15836 after 24 hours of treatment (Supplemental Figure 1 A). Neither CPI nor AX was able to significantly reduce cMYC expression alone. However, both XMD8-92 (66% of control) and CPI+AX (62% of control) were able to significantly reduce cMYC expression (Supplemental Figure 1A). The BRD4 inhibitor, CPI-203, decreased viability starting at 1uM (77% viable) but was only able to modestly reduce viability even at 100 µM (65% viable, Supplemental Figure 1 B). Similarly, ERK5 inhibitor AX15836 was unable to reduce viability below 50% event at 100 µM. Interestingly, the combination of Ipat+CPI+AX, 100nM each, (61% viable) approximately recapitulated the effect of Ipat+XMD in MDA-MB-231 cells (66% viable; Supplemental Figure 1 C and D).
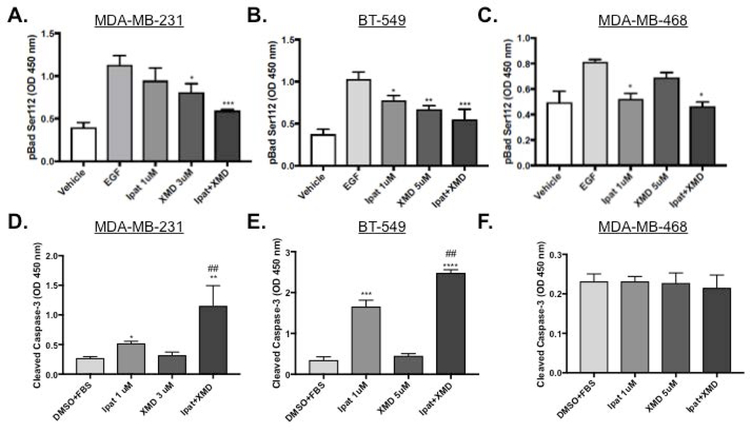
3.4. Evaluation of apoptotic markers pBad S112 and cleaved caspase-3 in TNBC cells.
Next, we examined the mechanism Ipat+XMD decreased viability by examining apoptotic markers. Ipatasertib significantly decreased the phosphorylation of Bad at serine 136 by 63% alone and 68% in combination with XMD8-92 (Supplemental Figure 2 C). XMD8-92 significantly decreased phosphorylation of Bad at serine 112 by 28% alone and 47% in combination with Ipatasertib (Figure 4 A). Additionally, the decrease in phosphorylation of Bad at serine 112 occurred in a p90RSK and ERK1/2 independent manner (Supplemental Figure 2 A and B). Ipatasertib increased cleaved caspase-3 levels 1.9 fold alone and 4.25 fold in combination with XMD8-92 (Figure 4 D). In BT-549 cells, both Ipatasertib and XMD8-92 contributed to the decrease in pBad S112. (Figure 4 B). Additionally, the decrease in pBad S112 corresponded to an increase in cleaved caspase-3: 4.9 fold vs control for Ipatasertib alone and 7.3 fold vs control for the combination (Figure 4 E). These results were similar to what was previously observed in the MDA-MB-231 cell line (also a Basal B TNBC cell type). In MDA-MB-468 cells, Ipatasertib significantly decreased pBad S112 both alone by 37% and in combination with XMD8-92 by 41% vs control (Figure 4 C). XMD8-92 only modestly decreased pBad S112 by 16% however, this was not significant vs control. Surprisingly, Cleaved caspase-3 was not elevated above control in any of the treatment groups at 48 hours (Figure 4 F).
Figure 4. Evaluation of apoptotic markers pBad S112 and cleaved caspase-3 in TNBC cells.
1:3 and 1:5 combinations of Ipatasertib and XMD8-92 were added to TNBC cell lines MDA-MB-231 (A,D), BT-549 (B,E), and MDA-MB-468 (C,F). After 4 hours of treatment with EGF cells were lysed and analyzed for pBad S112 expression using an ELISA assay (A-C). DMSO was used as a vehicle control. *P<0.05, **P<0.01, and ***P<0.001 vs EGF control. One-way ANOVA with Bonferroni post hoc analysis. For caspase activation, cells were treated for 48 hours with 5% FBS and analyzed with and ELISA for cleaved caspsae-3 (D-F). *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 vs DMSO+FBS control. ##P<0.01 vs drugs alone.
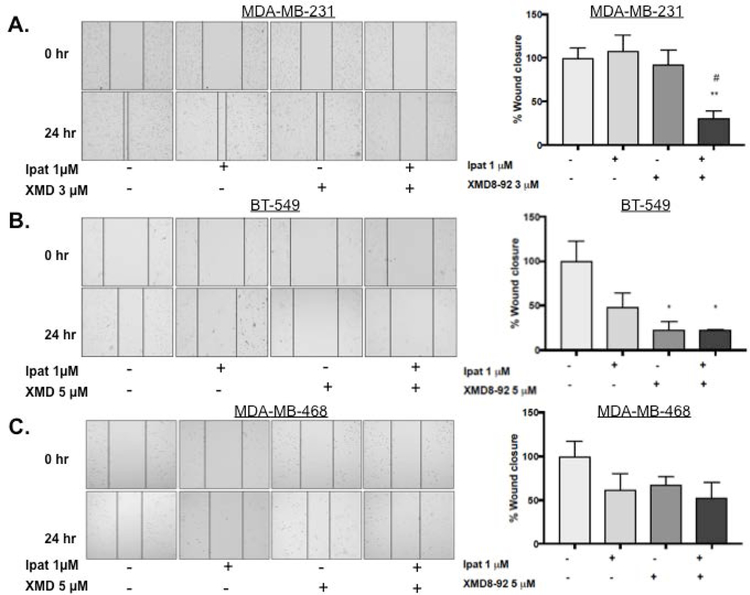
3.5. Effect of Ipatasertib and XMD8-92 treatment on TNBC migration.
Due to the metastatic potential of TNBCs, we sought to investigate the effect of Ipatasertib plus XMD8-92 on TNBC cell migration. In the MDA-MB-231 cell line neither drug alone significantly decreased invasion. However, the combination of Ipatasertib and XMD8-92 significantly decreased migration compared to control and each drug alone (Figure 5 A). In the BT-549 cell line, XMD8-92 decreased migration alone and in combination with Ipatasertib. The latter was also significantly different than each drug alone (Figure 5 B). Lastly, in the MDA-MB-468 cell line, migration was reduced by 40% in the combination group although this result was not statistically significant (Figure 5 C).
Figure 5. Effect of Ipatasertib and XMD8-92 treatment on TNBC migration.
MDA-MB-231 (A), BT-549 (B), and MDA-MB-468 (C) cells were treated with Ipatasertib, and XMD8-92 alone or in combination for 24 hours under 5% FBS stimulation. DMSO was used as a vehicle control. Migration was measured using the scratch/wound healing assay. Images at 0 and 24 hours were obtained at 10x magnification with an EVOS inverted microscope. *P<0.05 and **P<0.01 vs control. #P<0.05 vs drugs alone.
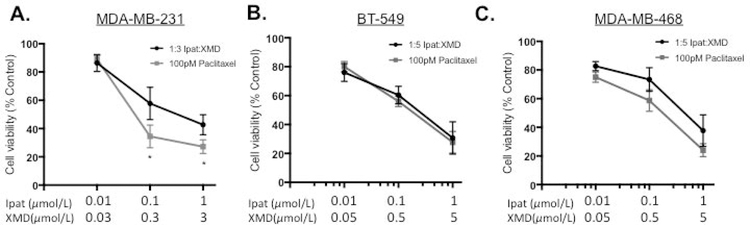
3.6. The combination of Ipatasertib and XMD8-92 enhanced the effect of the conventional chemotherapeutic agent Paclitaxel in MDA-MB-231 TNBC cells.
We established the preferential targeting of TNBC cells vs MCF-10 with the Ipat+XMD combination in Figure 1 D–F. Therefore, we sought to improve the efficacy of the Ipat+XMD combination by adding a conventional TNBC chemotherapy, paclitaxel. Paclitaxel was added for 24 hours followed by 1:3 or 1:5 combinations of Ipat:XMD for 48 hours (Figure 6 A–C). In the MDA-MB-231 cell line, 100 pM paclitaxel significantly enhanced the response of 1:3 Ipat+XMD (Figure 6A). We observed a decrease in viability following treatment with Paclitaxel and Ipat+XMD (34% viability; Figure 6A; Three drug synergy CI=0.08) as compared to the Ipat+XMD combination alone (56% viability). In MCF-10 cells, the adjuvant Ipat+XMD with Paclitaxel resulted in 81% viability (Supplemental Figure 3) thus widening the therapeutic index (47% difference). However, in the BT549 and MDA-MB-468 cell lines the effect of paclitaxel was not significant compared to the 1:5 combinations (Figure 6 B and C).
Figure 6. The combination of Ipatasertib and XMD8-92 enhanced the effect of the conventional chemotherapeutic agent Paclitaxel in MDA-MB-231 TNBC cells.
MDA-MB-231 (A), BT-549 (B), and MDA-MB-468 (C). Cells were pretreated with 100 pM Paclitaxel for 24 hours followed by Ipatasertib and XMD8-92 combinations for 48 hours. Cell viability was determined by the MTT assay. *P<0.05 vs MDA-MB-231 Ipat+XMD only.
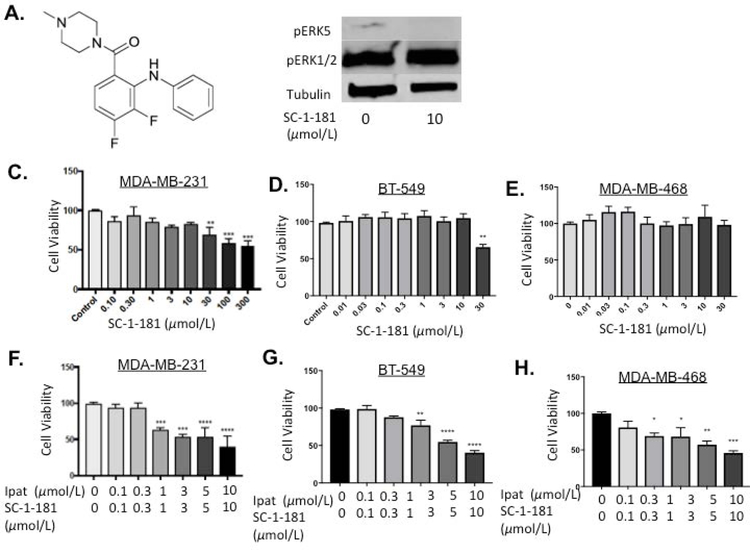
3.7. Evaluation of novel MEK5 inhibitor, SC-1-181, in combination with Ipatasertib in TNBC cells.
Our previous work established that SC-1-181 was the most selective MEK5 inhibitor, in a series of, diphenylamines (Figure 7 A, SC-1-181 structure). In MDA-MB-231 cells treated with SC-1-181, pERK5 was inhibited 82.4% vs EGF stimulated control [Chakrabarty et al., 2018]. Inhibition of ERK5 phosphorylation was achieved while sparing ERK1/2 activity (Figure 7B). SC-1-181 significantly decreased the viability of TNBC cells at concentrations equal to and greater than 30 µM in MDA-MB-231 and BT-549 cells (Figure 7 C and D). Similar to the Ipat+XMD combination, significant decreases in viability were observed at the 1:1 ratio of Ipat+SC-1-181 in all three TNBC cell lines (Figure 7 F–H). In particular, the 3,3 µM combination in the BT-549 cell line was significantly better than either drug alone (Figure 7 G).
Figure 7. Evaluation of novel MEK5 inhibitor, SC-1-181, in combination with Ipatasertib in TNBC cells.
Molecular structure of novel MEK5 inhibitor SC-1-181 (A). SC-1-181 inhibits pERK5 while sparing pERK1/2 (B). MDA-MB-231 (C), BT-549 (D), and MDA-MB-468 (E) TNBC cells were treated with increasing concentrations of SC-1-181 for 72 hours. Cell viability determined by the MTT assay. **P<0.01 and ***P<0.001 vs control. DMSO was used as a vehicle control. MDA-MB-231 (F), BT-549 (G), and MDA-MB-468 (H) TNBC cells were treated with SC-1-181 and Ipatasertib combinations for 72 hours. Cell viability determined by the MTT assay. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs control. DMSO was used as a vehicle control.
4. Discussion
In this study we observed the synergistic activity of PI3K/Akt and MEK5/ERK5 pathway inhibitor combinations in MDA-MB-231 and BT-549 TNBC cells. Specifically, the combination of the Akt inhibitor Ipatasertib and the ERK5 inhibitor XMD8-92 more effectively decreased TNBC cell viability, proliferation, and migration than either inhibitor alone. The 1:1 ratios of Ipat:XMD displayed CIs less than one over a wide concentration range (Figure 1 and Table 1). Interestingly, the 1:3 and 1:5 ratios exhibited the strongest synergy (Figure 1) in MDA-MB-231 and BT-549 cells, respectively. These results were similar to previous studies, which examined combinations of Akt and MEK1/2 inhibitors and observed synergy at 1:2, 1:4, and 1:8 ratios and antagonism with 4:1 and 8:1 ratios in lung cancer cell lines [Meng et al., 2010]. Additionally, we hypothesize that the Akt and ERK5 inhibition produces synergy in a serial rather than parallel manner. Additional work has revealed that the synergy from MEK1/2 and Akt inhibition occurs in a parallel manner. Whereas, synergy observed by targeting BRAF and MEK1/2 simultaneously occurs in a serial manner [Yin et al. 2014]. Serial synergy is less exploited than parallel but it has been successfully employed clinically in BRAF driven melanomas [Eroglu and Ribas 2016]. One key distinction is that in our study, we were able to spare ERK1/2 signaling with the Ipat+XMD combination in the basal B TNBC cell lines MDA-MB-231 and BT-549 (Figure 2). However, this was not the case in basal A MDA-MB-468 cells where ERK1/2 was activated by XMD8-92 alone or in combination with Ipatasertib (Figure 2).
In addition to the viability results (Figure 1) both the proliferation and migration (Figures 3 and 4) data suggest synergy with the 1:3 and 1:5 combinations of Ipatasertib and XMD8-92. In MDA-MB-231 cells, the Ki67+ and Hoechst+ cell counts were lowest in the combination group and the protein levels of p21 were elevated, thus indicating a decrease in proliferation (Figure 3 and Supplemental Figure 1). XMD8-92 has previously been shown to decrease proliferation in HeLa cells via a p21 mechanism [Yang et al., 2010]. We show in Supplemental Figure 1 that the induction of p21 is primarily mediated through BRD4 inhibition, a known off-target effect of XMD8-92. In addition to p21 induction, XMD8-92 has also been shown to suppress cMYC in neuroblastoma cells [Umapathy et al., 2014]. In our studies, we observed that both ERK5 and BRD4 inhibition appear to be required for cMYC suppression in MDA-MB-231 cells (Supplemental Figure 1). We achieved this using XMD8-92 alone and by combining the BRD4 inhibitor CPI-203 with the ERK5 inhibitor AX15836. Due to the limited tools used to study MEK5/ERK5, we examined the effect of combining novel MEK5 inhibitor SC-1-181 and Ipatasetib in TNBC cell lines. SC-1-181 exhibited similar viability data as the ERK5 inhibitor AX15836 with both compounds reducing viability by 30% at 30 µM in MDA-MB-231 and BT549 cells (Figure 7). Additionally, SC-1-181 combined with Ipatasertib decreased viability 35% starting at 1 µM each (Figure 7).
We also observed a convergence of the PI3K/Akt and MEK5/ERK5 pathways on the pro-apoptotic protein Bad. Each kinase inhibited the phosphorylation Bad at two separate sites, serine 112 and 136. Increased levels of cleaved caspase-3 accompanied the inhibition of Bad phosphorylation by Ipat+XMD (Figure 4 and Supplemental Figure 2). Akt has been shown to phosphorylate Bad at serine 136 and ERK5 has been shown to phosphorylate Bad at serine 112 independently of PKA, Akt, and p90RSK in endothelial cells [Hayashi et al., 2004; Pi et al., 2004]. In MDA-MB-468 TNBC cells synergistic MEK and Akt combinations was likely due to the blockade of phosphorylation of Bad at two separate sites [She et al., 2005]. Previous studies have shown the phosphorylation of Bad at serine 112 was mediated by p90RSK, which is a known target of ERK1/2; however the time-point of activation may be of importance [Manhas et al., 2010]. Therefore, there could be a temporal component to the ERK5 regulation of Bad phosphorylation (Supplemental Figure 2). Furthermore, we propose that combinations of Akt and MEK5/ERK5 inhibition reduce phosphorylation of Bad and increase apoptosis in a synergistic fashion. This is supported by the fact that we observed a 4.25 fold increase in cleaved caspase-3 levels in the Ipat+XMD combination group (Figure 4). Increased caspase-3 activity is likely due to cell cycle arrest followed by inhibition of pro-survival anti-apoptotic proteins such as Bad [Coulombe et al., 2007]. The kinase activity, pBad signaling, and caspase activation in BT-549 cells were similar to the MDA-MB-231 cells (Figures 2 and 4). Conversely, MDA-MB-468 cells treated with Ipat+XMD did not exhibit increased cleaved caspase activity. Perhaps this is due to the unique receptor expression profile of MDA-MB-468 cells. MDA-MB-468 cells overexpress cMET. cMET is the receptor for HGF and is observed in many cancers. In addition to activating growth factor signaling, cMET contains a domain that inhibits caspases [Jung et al., 2012]. The Ipat+XMD combination in MDA-MB-468 cells also activated pERK1/2 signaling (Figure 2). This was in contrast with MDA-MB-231 and BT-549 cell both of which pERK1/2 were spared (Figure 2). Another unique feature of MDA-MB-468 cells is that they have an EGFR amplification [Chavez et al. 2010]. This may be what was responsible for the increased pERK1/2 activity in the XMD8-92 treated groups. Therefore, inhibiting EGFR may be needed to achieve full efficacy since ERK1/2 appears to be essential to survival in this cell line (She et al., 2005). Sohn et al. 2014, achieved synergy with EGFR and cMET inhibitor combinations in MDA-MB-468 cells. In their study, MDA-MB-468 cells had hyperactive pMET and pEGFR vs MDA-MB-231 cells. This led to increased downstream signaling in the presence of EGF in the MDA-MB-468 cells. Therefore, EGFR and cMET may need to be inhibited for Ipat+XMD to achieve an apoptotic effect. There could also be other mechanisms contributing to apoptosis by blocking multiple phosphorylation sites of FOXO3a [Brunet et al., 2001; Hayashi et al., 2001; Leong et al., 2003]. Further studies with genetic ablation of ERK5 in TNBCs are needed to precisely determine its role in the apoptotic cascade of TNBCs.
The last focus of this study was to examine the efficacy of Ipat+XMD in combination with conventional therapy and to measure collateral toxicity. In our study, synergy was observed with Ipat+XMD and paclitaxel (Figure 6; CI=0.08) in TNBC cells at concentrations well below paclitaxel’s IC50 of 5 nM (Supplemental Figure 3). Furthermore, the combination of Ipat+XMD enhanced the effect of the paclitaxel such that a concentration 50-fold lower than the IC50 of paclitaxel significantly decreased cell viability in combination with Ipat+XMD. Taken together, these results suggest the inhibition of both the PI3K/Akt and MEK/ERK5 pathways is a promising strategy for targeting TNBC. Additionally, Paclitaxel has been shown to synergize with Ipatasertib in TNBC cell lines and is currently in clinical trials as a combination [Morgillo et al., 2017] We hypothesize that this combination of Paclitaxel with Ipat + XMD will cause less collateral toxicity because of the low concentration of Paclitaxel and the low toxicity of Ipat + XMD in MCF10 cells. Perhaps the preference for TNBC vs MCF-10 cells is due to having spared ERK1/2 activity and the differential expression of pBad serine 112 and 136. Both pBad serine 112 and 136 expression has been shown to be greater in MDA-MB-231 cells vs MCF10 cells, [Stickles et al., 2015]. Moreover, while Ipat + XMD inhibit Akt and ERK5 activity, ERK1/2 activity is spared and the sparing of ERK1/2 activity may decrease the collateral toxicity of this combination.
In summary, these results provide a biological rationale for targeting the PI3K/Akt and MEK5/ERK5 pathways in TNBC cells with the potential for rapid translational impact.
Supplementary Material
Supplemental Figure 1. BRD4 inhibition increased p-21 expression while simultaneous BRD4 and ERK5 inhibition decreased cMYC expression in MDA-MB-231 cells. BRD4 inhibitor CPI-203 increased p-21 expression in MDA-MB-231 cells (A). Combination BRD4 and ERK5 inhibition with CPI and AX15836 respectively decreased cMYC expression. Cells were treated with inhibitors alone or in combination for 24 hours under 5% FBS stimulation prior to lysis and analysis via western blot. *P<0.05, **P<0.01 vs 5% FBS control. MDA-MB-231 cells were treated with the Akt inhibitor, Ipat, ERK5 inhibitor, AX15836, and BRD4 inhibitor, CPI-203, alone or in combination for 72 hours under 5% FBS stimulation (B-D). Cell viability was analyzed using the MTT assay. **P<0.01; ***P<0.001; ****P<0.0001 vs control group, #P<0.05 vs drugs alone.
Supplemental Figure 2. Bad phosphorylation at serine 112 was independent of ERK1/2 and p90RSK at 4 hours in MDA-MB-231 cells. MDA-MB-231 cells were treated with MEK1/2 inhibitor Trametinib 100nM to 10uM for 1 hour followed by 50ng/mL EGF stimulation for 0.25, 1, and 4 hours (A). Protein levels of pBad (S112) were determined by western blot. GAPDH was used as a loading control. Normalized values shown for each time point are shown (A). *P<0.05 vs 15 min and 1 hr EGF controls. Western blot of p90RSK, pERK1/2, pBad (S112) expression at 4 hours (B). Protein levels of pBad (S136) were determined by western blot (C). GAPDH was used as a loading control. *P<0.05 vs EGF control. Proposed mechanism of synergy in MDA-MB-231 cells (D).
Supplemental Figure 3. Ipatasertib plus XMD8-92 enhanced the effect of the, Paclitaxel and exhibited low collateral toxicity. MDA-MB-231 cells were treated with Paclitaxel for 72 hours under 5% FBS stimulation (A). Viability determined using the MTT assay. *P<0.05; **P<0.01; ***P<0.001 vs 5% FBS control. MCF-10 and 231 cells were treated with drugs alone or in combination: pretreated with 0.1 nM Paclitaxel for 24 hours followed by 100,300 nM Ipat+XMD for 48 hours (B). Viability determined using the MTT assay. *P<0.05; **P<0.01; ***P<0.001 vs control; #P<0.05 vs 100,300 nM Ipat+XMD and 0.1 nM Paclitaxel.
Acknowledgements:
Supported by the Department of Pharmacology and Toxicology at Duquesne University and NIH grant R15CA176496 (JEC).
Footnotes
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of Interest statement
The authors have no conflict of interest to report.
References
- Anders CK and Carey LA. 2009. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer 9:S73–S81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksamitiene E, Kiyatkin A, and Kholodenko BN. 2012. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: a fine balance. Biochem Soc Trans 40(1):139–46. DOI: 10.1042/BST20110609. [DOI] [PubMed] [Google Scholar]
- Al-Ejeh F, Miranda M, Shi W, Simpson PT, Song S, Vargas AC, Saunus JM, Smart CE, Mariasegaram M, Wiegmans AP, Chenevix-Trench G, Lakhani SR, Khanna KK. 2014. Kinome profiling reveals breast cancer heterogeneity and identifies targeted therapeutic opportunities for triple negative breast cancer. Oncotarget 5(10):3145–58. DOI: 10.18632/oncotarget.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. 2001. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol Cell Biol 21(3): 952–965. DOI: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas, N. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490(7418): 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty S, Monlish DA, Gupta M, Wright TD, Hoang VT, Fedak M, Chopra I, Flaherty PT, Madura J, Mannepelli S, Burow ME, Cavanaugh JE. 2018. Structure activity relationships of anthranilic acid-based compounds on cellular and in vivo mitogen activated protein kinase-5 signaling pathways. Bioorganic & Medicinal Chemistry Letters 28; 2294–2301. DOI: 10.1016/j.bmcl.2018.05.029 [DOI] [PubMed] [Google Scholar]
- Chavez KJ, G. S, and Lipkowitz S (2010). Triple Negative Breast Cancer Cell Lines: One Tool in the Search for Better Treatment of Triple Negative Breast Cancer. Breast Dis, 32(1–2), 35–48. DOI: 10.3233/BD-2010-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC and Talalay P. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22:27–55. [DOI] [PubMed] [Google Scholar]
- ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US) 2017. November 09 -. Identifier “Study of Ipatasertib in Combination With Paclitaxel” Available from: https://clinicaltrials.gov/ct2/show/NCT03337724?term=Ipatasertib&rank=8 [Google Scholar]
- Coulombe P and Meloche S. 2007. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta 1773(8): 1376–1387. DOI: 10.1016/j.bbamcr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. 1997. Akt phosphorylation of BAD couples survival signals to the cell: intrinsic death machinery. Cell 91:231–241. [DOI] [PubMed] [Google Scholar]
- Eroglu Z and Ribas A. 2016. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Therapeutic Advances in Medical Oncology 8(1) 48–56. DOI: 10.1177/1758834015616934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Iwashita T, Murakamai H, Kato Y, Kawai K, Kurokawa K, Tohnai I, Ueda M, Takahashi M. 2001. Activation of BMK1 via tyrosine 1062 in RET by GDNF and MEN2A mutation. Biochem Biophys Res Commun 281(3): 682–689. DOI: 10.1006/bbrc.2001.4338. [DOI] [PubMed] [Google Scholar]
- Hayashi M and Lee JD. 2004. Role of the BMK1/ERK5 signaling pathway: lessons from knockout mice. J. Mol. Med 82 (12): 800–808. DOI: 10.1007/s00109-004-0602-8. [DOI] [PubMed] [Google Scholar]
- Hoang VT, Yan TJ, Cavanaugh JE, Flaherty PT, Beckman BS, Burow ME. 2017Oncogenic signaling of MEK5-ERK5 (392): 51–59. DOI: 10.1016/j.canlet.2017.01.034. [DOI] [PMC free article] [PubMed]
- Jung H-Y, J H-J, Park JK, and Kim YH (2012). The Blocking of c-Met Signaling Induces Apoptosis through the Increase of p53 Protein in Lung Cancer. Cancer Res Treat, 44(4), 251–261. DOI: 10.4143/crt.2012.44.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili JS, Yu X, Wang J, Hayes BC, Davies MA, Lizee G, Esmaeli B, Woodman SE. 2012. Combination small molecule MEK and PI3K inhibition enhances uveal melanoma call death in a mutant GNAQ and GNA11-dependent manner. Clinical Cancer Research 18(16):4345–55. DOI: 10.1158/1078-0432.CCR-11-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kui L, Lin J, Wu WI, Ballard J, Lee BB, Gloor SL, Vigers GP, Morales TH, Friedman LS, Skelton N, Brandhuber BJ. 2012. An ATP site on-Off Switch That Restricts Phosphatase Accessibility of Akt. Sci. Signal 5(223). DOI: 10.1126/scisignal.2002618. [DOI] [PubMed] [Google Scholar]
- Lannutti BJ, Meadows SA, Herman SE, Kashishian A, Steiner B, Johnson AJ, Byrd JC, Tyner JW, Loriaux MM, Deininger M, Druker BJ, Puri KD, Ulrich RG, Giese NA. 2011. CAL-101, a p100delta selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood 117(2):591–4. DOI: 10.1182/blood-2010-03-275305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong ML, Maiyar AC, Kim B, O’Keeffe BA, Firestone GL. 2003. Expression of the serum- and glucocorticoid-inducible protein kinase, Sgk, is a cell survival response to multiple types of environmental stress stimuli in mammary epithelial cells. J Biol Chem 278(8): 5871–5882. DOI: 10.1074/jbc.M211649200. [DOI] [PubMed] [Google Scholar]
- Li Q, Chow AB, Mattingly RR. 2010. Three-dimensional overlay culture models of human breast cancer reveal a critical sensitivity to mitogen-activated protein kinase kinase inhibitors. J Pharmacol Exp Ther 332:821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EC, Amantea CM, Nomanbhoy TK, Weissig H, Ishiyama J, Hu Y, Sidique S, Li B, Kozarich JW, Rosenblum JS. 2016. ERK5 kinase activity is dispensable for cellular immune response and proliferation. PNAS 113 (42): 11865–11870. DOI: 10.1073/pnas.1609019113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Sampath D, Nannini MA, Lee BB, Degtyarev M, Oeh J, Savage H, Guan Z, Hong R, Kassees R, Lee LB, Risom T, Gross S, Liederer BM, Koeppen H, Skelton NJ, Wallin JJ, Belvin M, Punnoose E, Friedman LS, Lin K. 2013. Targeting activated Akt with GDC-0068, a novel selective Akt inhibitor that is efficacious in multiple tumor models. Clin Cancer Res 19(7): 1760–1772. DOI: 10.1158/1078-0432.CCR-12-3072. [DOI] [PubMed] [Google Scholar]
- Manhas N, Kintner DB, Shull GE, Sun D. 2010. ERK1/2-p90RSK activation contributes to cerebral ischemic damage via phosphorylation of Na+/H+ exchanger isoform 1. J Neurochem 114(5): 1476–1486. DOI: 10.1074/jbc.M702373200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken SR, Ramsay A, Heer R, Mathers ME, Jenkins BL, Edwards J, Robson CN, Marquez R, Cohen P, Leung HY. 2008. Aberrant expression of extracellular signal-regulated kinase 5 in human prostate cancer. Oncogene 27(21): 2978–2988. DOI: 10.1038/sj.onc.1210963. [DOI] [PubMed] [Google Scholar]
- Meng J, Dai B, Fang B, Bekele BN, Bornmann WG, Sun D, Peng Z, Herbst RS, Papadimitrakopoulou V, Minna JD, Peyton M, Roth JA. 2010. Combination treatment with MEK and AKT inhibitors is more effective than each drug alone in human non-small cell lung cancer in vitro and in vivo. PLoS One 5(11): e14124 DOI: 10.1371/journal.pone.0014124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgillo F, Della Corte CM, Diana A, Mauro CD, Ciaramella V, Barra G, Belli V, Franzese E, Bianco R, Maiello E, de Vita F, Ciardiello F, Orditura M. 2017. PI3Ka/Akt axis blockade with taselisib or ipatasertib enhances the efficacy of anti-microtubule drugs in human breast cancer cells. Oncotarget 8 (44):76479–76491. DOI: 10.18632/oncotarget.20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ruiz MJ, Álvarez-Fernández S, Parrott T, Zaknoen S, Burrows FJ, Ocaña A, Pandiella A, Esparís-Ogando A. 2014. Therapeutic potential of ERK5 targeting in triple negative breast cancer. Oncotarget 5(22):11308–18. DOI: 10.18632/oncotarget.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi X, Yan C, Berk BC. 2004. Big mitogen-activated protein kinase (BMK1)/ERK5 protects endothelial cells from apoptosis. Circ Res 94(3): 362–369. DOI: 10.1161/01.RES.0000112406.27800.6F. [DOI] [PubMed] [Google Scholar]
- Rheault TR, Stellwagen JC, Adjabeng GM, Hornberger KR, Petrov KG, Waterson AG, Dickerson SH, Mook RA Jr, Laquerre SG, King AJ, Rossanese OW, Arnone MR, Smitheman KN, Kane-Carson LS, Han C, Moorthy GS, Moss KG, Uehling DE. 2013. Discovery of Dabrafenib; A Selective Inhibitor of Raf Kinases with Antitumor Activity against B-Raf-Driven Tumors. ACS Medicinal Chemistry Letters 4(3):358–62. doi: 10.1021/ml4000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini KS, Loi S, de Azambuja E, Metzger-Filho O, Saini ML, Ignatiadis M, Dancey JE, Piccart-Gebhart MJ. 2013. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev 39(8): 935–946. DOI: 10.1016/j.ctrv.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Schnitt SJ. 2010. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol 23 Suppl 2: S60–64. DOI: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- She QB, Solit DB, Ye Q, O’Reilly KE, Lobo J, Rosen N. 2005. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell 8(4): 287–297. DOI: 10.1016/j.ccr.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, S. L, Napa Parinyanitikul, Jangsoon Lee, Hortobagyi Gabriel N., Gordon B, & Mills NTU, Gonzalez-Angulo Ana M. (2014). cMET Activation and EGFR-Directed Therapy Resistance in Triple-Negative Breast Cancer. Journal of Cancer, 5(9), 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickles XB, Marchion DC, Bicaku E, Al Sawah E, Abbasi F, Xiong Y, Bou Zgheib N, Boac BM, Orr BC, Judson PL, Berry A, Hakam A, Wenham RM, Apte SM, Berglund AE, Lancaster JM. 2015. BAD-mediated apoptotic pathway is associated with human cancer development. International Journal of Molecular Medicine 35: 1081–1087. DOI: 10.3892/ijmm.2015.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umapathy G, El Wakil A, Witek B, Chesler L, Danielson L, Deng X, Gray NS, Johansson M, Kvarnbrink S, Ruuth K, Schönherr C, Palmer RH, Hallberg B. 2014. The kinase ALK stimulates the kinase ERK5 to promote the expression of the oncogene MYCN in neuroblastoma. Science Signal 7(349). DOI: 10.1126/scisignal.2005470. [DOI] [PubMed] [Google Scholar]
- Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. 2010. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol 28(10): 1684–1691. DOI: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- Wong C, Laddha SV, Tang L, Vosburgh E, Levine AJ, Normant E, Sandy P, Harris CR, Chan CS, Xu EY. 2014. The bromodomain and extra-terminal inhibitor CPI203 enhances the antiproliferative effects of rapamycin on human neuroendrocrine tumors. Cell Death and Disease 5, e1450 DOI: 10.1038/cddis.2014.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Deng X, Lu B, Cameron M, Fearns C, Patricelli MP, Yates JR 3rd, Gray NS, Lee JD. 2010. Pharmacological inhibition of BMK1 suppresses tumor growth through promyelocytic leukemia protein. Cancer Cell 18(3):258–67. DOI: 10.1016/j.ccr.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin N, Ma W, Pei J, Ouyang Q, Tang C, Lai L. 2014. Synergistic and Antagonistic Drug Combinations Depend on Network Topology. PLoS ONE 9(4): e93960 DOI: 10.1371/journal.pone.0093960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. BRD4 inhibition increased p-21 expression while simultaneous BRD4 and ERK5 inhibition decreased cMYC expression in MDA-MB-231 cells. BRD4 inhibitor CPI-203 increased p-21 expression in MDA-MB-231 cells (A). Combination BRD4 and ERK5 inhibition with CPI and AX15836 respectively decreased cMYC expression. Cells were treated with inhibitors alone or in combination for 24 hours under 5% FBS stimulation prior to lysis and analysis via western blot. *P<0.05, **P<0.01 vs 5% FBS control. MDA-MB-231 cells were treated with the Akt inhibitor, Ipat, ERK5 inhibitor, AX15836, and BRD4 inhibitor, CPI-203, alone or in combination for 72 hours under 5% FBS stimulation (B-D). Cell viability was analyzed using the MTT assay. **P<0.01; ***P<0.001; ****P<0.0001 vs control group, #P<0.05 vs drugs alone.
Supplemental Figure 2. Bad phosphorylation at serine 112 was independent of ERK1/2 and p90RSK at 4 hours in MDA-MB-231 cells. MDA-MB-231 cells were treated with MEK1/2 inhibitor Trametinib 100nM to 10uM for 1 hour followed by 50ng/mL EGF stimulation for 0.25, 1, and 4 hours (A). Protein levels of pBad (S112) were determined by western blot. GAPDH was used as a loading control. Normalized values shown for each time point are shown (A). *P<0.05 vs 15 min and 1 hr EGF controls. Western blot of p90RSK, pERK1/2, pBad (S112) expression at 4 hours (B). Protein levels of pBad (S136) were determined by western blot (C). GAPDH was used as a loading control. *P<0.05 vs EGF control. Proposed mechanism of synergy in MDA-MB-231 cells (D).
Supplemental Figure 3. Ipatasertib plus XMD8-92 enhanced the effect of the, Paclitaxel and exhibited low collateral toxicity. MDA-MB-231 cells were treated with Paclitaxel for 72 hours under 5% FBS stimulation (A). Viability determined using the MTT assay. *P<0.05; **P<0.01; ***P<0.001 vs 5% FBS control. MCF-10 and 231 cells were treated with drugs alone or in combination: pretreated with 0.1 nM Paclitaxel for 24 hours followed by 100,300 nM Ipat+XMD for 48 hours (B). Viability determined using the MTT assay. *P<0.05; **P<0.01; ***P<0.001 vs control; #P<0.05 vs 100,300 nM Ipat+XMD and 0.1 nM Paclitaxel.