Abstract
Osteosarcoma (OS) is the most common form of bone malignancy in children and adolescents. MicroRNAs (miRNAs) have been associated with the development and progression of OS. In the present study, reverse transcription-quantitative PCR, western blotting, Cell Counting Kit-8, luciferase and Transwell assays were performed to investigate the biological function of microRNA-150 (miR-150) in OS. The results revealed that miR-150 was significantly downregulated in OS cell lines (HOS, SAOS2, MG-63 and U2OS) in comparison with the normal osteoblast cells (hFOB1.19). Overexpression of miR-150 significantly inhibited cell proliferation in OS cells. miR-150 could sensitize OS cells to chemotherapy treatment of doxorubicin. Runt-related transcription factor 2 (RUNX2) was identified as a target gene of miR-150. RUNX2 knockdown exhibited similar inhibitory effects on both OS cell proliferation and chemotherapy sensitivity. Restoration of RUNX2 reversed the biological function of miR-150. Finally, miR-150 overexpression and RUNX2 knockdown enhanced caspase-3 cleavage. Taken together, the present study established a novel molecular mechanism, in that miR-150 plays tumor suppressor and chemoprotective roles by targeting RUNX2 in OS, indicating that miR-150 may be a potential therapeutic target for OS therapy in the future.
Keywords: osteosarcoma, microRNA-150, runt-related transcription factor 2, proliferation, chemotherapy
Introduction
Osteosarcoma (OS) is a bone tumor that is primarily observed in children and adolescents (1), making up 2.4% of all pediatric patient malignancies and ~20% of all types of primary bone cancer (2–4). OS is the most common type of primary bone malignancy, and normally affects the long bones of legs and arms (4). Although the therapy of OS has improved over the past decade, the prognosis of OS remains poor. The main cause of the poor prognosis is the occurrence of metastases following surgical resection and intensive chemotherapy (5,6). OS pathogenesis, progression and prognosis have been revealed to be regulated by a number of tumor-associated signaling pathways. However, the detailed molecular mechanisms underlying OS formation and development remain poorly understood (7,8). In order to improve the therapy and develop better prognoses for patients with OS, the potential underlying molecular mechanisms must be elucidated and new therapeutic targets must be identified for OS treatment.
MicroRNAs (miRNAs) have been demonstrated to regulate gene expression via binding to the 3′-untranslated region (3′-UTR) of their target mRNAs (9). Previous studies have demonstrated that miRNAs are important cancer biomarkers and play a key role in cancer cell growth and metastasis (10,11). Tumor-associated miRNAs can function as both or either tumor suppressors and oncogenes, depending on the biological function of the target genes (12,13), meaning the function of miRNAs in tumors can be two-sided (14). miRNA-150 (miR-150) is a tumor-associated miRNA, which has been reported as a biomarker in several different types of cancer, including osteosarcoma (15), gastric cancer (16), non-small cell lung cancer (17). Colorectal cancer (18) and leiomyoma (19). However, the biological function and the underlying mechanisms of miR-150 in OS have not yet been investigated.
The runt-related transcription factor 2 (RUNX2) gene has been suggested to be a cancer-associated gene that is implicated in chemotherapeutic resistance. RUNX2 is often amplified and aberrantly expressed in OS, and is the master regulator of skeletal development, directly regulating cell proliferation, apoptosis and differentiation in osteoblasts (20). However, in order to evaluate whether RUNX2 is a viable biomarker and therapeutic target for OS treatment, it is necessary to investigate the specific biological role of RUNX2 in OS. Therefore, the aim of the present study was to investigate the biological function and associated mechanism of miR-150 in OS doxorubicin (DOX) sensitivity of OS.
Materials and methods
Tissue samples and cell lines
A total of 26 paired OS and adjacent non-tumor tissues were collected from patients (sex, 15 males and 11 females; age, 60±5 years) with OS who underwent curative tumor resection between September 2010 and August 2014 at the Jinling Hospital (Nanjing, China). The tissues samples were obtained once the patients had provided written informed consent, and the process was approved by the Ethics Committee of Jinling Hospital (Nanjing, China). SAOS2, MG-63, HOS and U2OS human OS cell lines and hFOB1.19 normal osteoblast cells were purchased from the American Type Culture Collection.
Cell proliferation and cell viability assay
All cells were cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Sigma-Aldrich; Merck KGaA) at 37°C with 5% CO2. For plasmid transfection, cells were first seeded in six-well plates, and transfection was performed when 80% confluence was achieved. A total of two 8 µl (500 ng/µl) plasmids (pcDNA3.1-RUNX2, RUNX2-shRNA (5′-AAAAGCGCATTCCTCATCCCAGTATTTCGATACTGGGATGAGGAATGCGC-3′, Shanghai GenePharma Co., Ltd.) or miR-150 mimics and NC (hsa-miR-150 mimics, 5′-UCUCCCAACCCUUGUACCAGUG-3′; hsa-miR-150 mimics NC, 5′-UUCUCCGAACGUGUCACGUTT-3′; Shanghai GenePharma Co., Ltd.) and 8 µl Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) were suspended in 100 µl Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) and the final concentration of RUNX2 plasmids or mimics used was 1 mg/ml. Following incubation at room temperature for 30 min, the mixture was added into cell culture. After 6 h of culture, the cell medium was replaced by fresh medium. After another 24 h of culture, the cells were used to detect the rates of proliferation. The proliferation rates in the different groups of HOS and U2OS cells (blank, vector and RUNX2 overexpression groups) were detected via MTT assay. Cells were suspended in 200 µl culture medium in a 96-well plate at 1×104 cells/well. At every 24 h after cell seeding for 72 h, the culture medium of the 96-well plate was replaced with DMEM plus 0.5 mg/ml MTT and incubated at 37°C for 4 h. DMEM medium was then replaced with isopycnic DMSO. Following mixing by extensive shaking, absorbance at 490 nm was detected for each well using a SpectraMax 190 (Molecular Devices in Sunnyvale) to measure the optical density value at 0, 24, 48 and 72 h.
Cell Counting Kit-8 (CCK-8) assay
DOX (Selleck Chemicals) was used to construct an OS cell chemotherapy resistance model, where cell viability was measured using CCK8 assay. A dose-dependent assay was performed using 0, 25, 50, 75 and 100 µM DOX treatment at 37°C for 4 h. Since cell viability did not reduce further on increasing the concentration of DOX beyond 50 µM, this concentration was chosen for subsequent experiments. OS cell lines HOS and U2OS were seeded in 96-well plates with 5,000 cells/well. Following DOX treatment, cell proliferation was assessed using a CCK-8 assay (Dojindo Molecular Technologies). Every 24 h for 72 h after seeding, 10 µl CCK-8 solution was added to the culture medium, and the cultures were incubated for 30 min at 37°C in 5% CO2. The absorbance was measured at 450 nm using a Microplate Reader (Bio-Rad Laboratories, Inc.).
Reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted from the HOS and U2OS cell lines and tissues using TRIzol® reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Reverse transcription was performed using a Takara PrimeScript™ RT reagent kit (Takara Bio Inc.) at 37°C for 1 h and 85°C for 15 sec. qPCR was performed using SYBR Green (Takara Bio Inc.). The thermocycling conditions were as follows: Initial denaturation at 90°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec) with the ABI Prism 7700 Sequence Detection system (Applied Biosystems; Thermo Fisher Scientifc, Inc.). RUNX2 or miR-150 expression was quantified using the 2−∆∆Cq method (21). GAPDH and U6 were used as internal controls of the mRNA or miRNA, respectively. Sequences of the primers used in the present study were as follows: miR-150 forward, 5′-TGTCGTGGAGTCGGCAATTCAGTTGAGCACTGG-3′ and reverse, 5′-ACACTCCAGCTGGGTCTCCCAACCCTTGTA-3; GAPDH forward, 5′-TGCACCACCAACTGCTTAGC-3′ and reverse, 5′-GGCATGGACTGTGGTCATGAG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Luciferase assay
TargetScan bioinformatics algorithm (version 7.2; http://www.targetscan.org/vert_72/) was used to identify the potential targets of miR-150. The 293T cells (Type Culture Collection of the Chinese Academy of Sciences) were seeded in 24-well plates and cultured until they reached 60% confluence. After incubation overnight at 37°C, the cells were co-transfected pmirGLO plasmids encoding RUNX2 wild-type (WT) 3′-UTR or mutant (Mut) 3′-UTR (Shanghai GenePharma Co., Ltd.) and miR-150 mimics (Shanghai GenePharma Co., Ltd.) or the control mimics (Shanghai GenePharma Co., Ltd.) using Lipofectamine 2000. Firefly and Renilla luciferase activities were detected using Dual-Luciferase Reporter assay system (Promega Corporation) 48 h after transfection, according to the manufacturer's protocol. All transfection assays were performed in triplicate.
Western blot analysis
RIPA lysis buffer (Beyotime Institute of Biotechnology) with protease inhibitor cocktail (Beyotime Institute of Biotechnology) was used for cell lysis. A BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.) was used to detect the protein concentration. In total, 4 µg all proteins were resolved by 10% SDS-PAGE and transferred onto PVDF membranes. The membranes were subsequently incubated with blocking buffers, consisting of 10% skimmed milk powder (dissolved in TBS supplemented with 0.1% Tween-20) for 2 h at room temperature, prior to incubation with the respective primary antibodies. The primary antibodies used in the present study were as follows: Anti-RUNX2 (1:1,000; cat. no. D1H7; Cell Signaling Technology, Inc.), anti-β-actin (1:2,000; cat. no. 20536-1-AP; Proteintech Group, Inc.), anti-caspase-8 (1:1,000; cat. no. 9746; Cell Signaling Technology, Inc.), anti-caspase-3 (1:1,000; cat. no. 9662; Cell Signaling Technology, Inc.). The membranes were incubated with the primary antibodies at 4°C overnight. Subsequently, the membranes were incubated with horseradish peroxidase conjugated-goat anti-mouse IgG anti-rabbit secondary antibodies (1:4,000; cat. no. sc-2030; Santa Cruz Biotechnology, Inc.) for 2 h at room temperature. The membranes were exposed using an ECL kit (EMD Millipore) and assessed using Image Lab™ Software 2.0 (Bio-Rad Laboratories, Inc.).
Statistical analysis
Data are presented as the mean ± standard error of the mean, and experiments were repeated at least three times. Significance between groups was analyzed by one-way analysis of variance followed by Student-Newman-Keuls test. Paired Student's t-test was used to analyze differences between paired samples. SPSS software (version 17.0; SPSS, Inc.) was used for the data analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-150 is decreased in OS tissues and cell lines
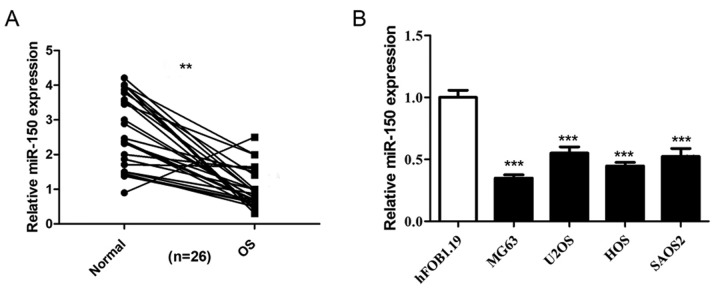
The expression levels of miR-150 in 26 OS tissues and non-tumor tissues were detected by RT-qPCR, and the results revealed that miR-150 was significantly decreased in OS tissues compared with non-tumor tissues (Fig. 1A). The expression levels of miR-150 were also significantly decreased in the four human OS cell lines (HOS, SAOS2, MG-63 and U2OS) compared with the normal osteoblast cells (hFOB1.19) (Fig. 1B), indicating that miR-150 exhibits low expression levels in OS.
Figure 1.
miR-150 is downregulated in the OS tissues and cell lines. (A) RT-qPCR analysis to detect the expression levels of miR-150 in 26 OS patient tissue samples and adjacent normal tissues. (B) Relative expression of miR-150 in HOS, SAOS2, MG63 and U2OS cell lines and the normal hFOB1.19 cell line were determined using RT-qPCR analysis. Data are presented as the mean ± SEM. All assays were performed in triplicate. ***P<0.001 vs. hFOB1.19. **P<0.01. miR, microRNA; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative PCR.
miR-150 inhibits OS cell proliferation and sensitizes OS cells to DOX
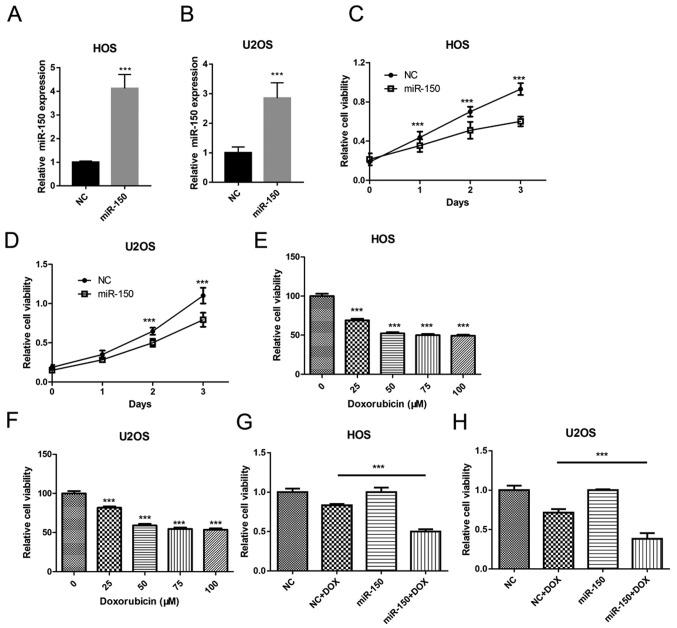
In order to investigate the biological role of miR-150 in OS tumorigenesis, miR-150 was overexpressed in OS cell lines HOS and U2OS in the present study (Fig. 2A and B). Cell proliferation was assessed in order to determine the effects of miR-150 on OS cell growth using CCK-8. The data revealed that overexpression of miR-150 significantly inhibited the proliferation of HOS and U2OS cells (Fig. 2C and D). In order to investigate the role of miR-150 in OS cell chemotherapy resistance, an OS cell chemotherapy model was constructed using DOX. Cell viability was assessed in order to determine the effect of DOX treatment on OS cell viability. A dose-dependency assay was performed using 0, 25, 50, 75 and 100 µM DOX treatment. The results revealed that the cell viability of both HOS (Fig. 2E) and U2OS (Fig. 2F) was significantly decreased with DOX treatment in a dose-dependent manner. No further reductions in cell viability was observed when the concentration of DOX exceeded 50 µM. Furthermore, miR-150 overexpression significantly decreased the cell viability of HOS and U2OS in the DOX treatment group, compared with the NC mimic + DOX group (Fig. 2G and H). Thus, these data suggested that miR-150 inhibited OS cell proliferation and sensitized OS cells to DOX.
Figure 2.
miR-150 inhibits OS cell growth, as demonstrated using a Cell Counting Kit-8 assay. Relative miR-150 expression in (A) HOS and (B) U2OS cells. ***P<0.001 vs. miR-NC. Cell proliferation in (C) HOS and (D) U2OS cells. (E) HOS cell viability at different concentrations of DOX. (F) U2OS cell viability at different concentrations of DOX. (G) HOS cell viability at 25 µM DOX. (H) U2OS cell viability at 25 µM DOX. All assays were performed in triplicate. ***P<0.001. miR, microRNA; OS, osteosarcoma; DOX, doxorubicin; miR-NC, negative control miR mimic.
RUNX2 is a target gene of miR-150
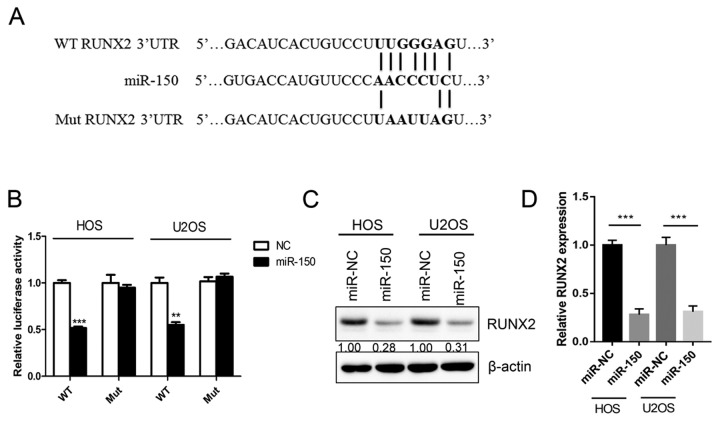
In order to assess the downstream target and underlying molecular mechanism of miR-150 in OS, a TargetScan bioinformatics algorithm was used to identify the potential target gene of miR-150. The results revealed that RUNX2 was a potential target of miR-150 based on a putative 3′-UTR sequence (Fig. 3A). A luciferase assay was performed in order to investigate the association between miR-150 and RUNX2. Luciferase reporter constructs containing the WT or Mut 3′-UTR of the RUNX2 were constructed. The results revealed that miR-150 significantly decreased the luciferase activity of the WT RUNX2 3′-UTR but not the Mut RUNX2 3′-UTR (Fig. 3B). Overexpression of miR-150 significantly downregulated the expression of RUNX2 both in HOS and U2OS cells (Fig. 3C and D). The results indicated that RUNX2 was a target gene of miR-150 in OS.
Figure 3.
RUNX2 is a direct target of miR-150. (A) Diagram of the WT and Mut RUNX2 3′-UTR constructs. (B) Luciferase reporter assay of the inhibitory effect of miR-150 on RUNX2-3′-UTR luciferase activity in HOS and U2OS cells. (C) RUNX2 protein expression was analyzed in HOS and U2OS cells following transfection with miR-150. (D) Statistical analysis of RUNX2 expression. All assays were performed in triplicate. ***P<0.001 vs. control; **P<0.01 vs. control. RUNX2, runt-related transcription factor 2; 3′-UTR, 3′-untranslated region; miR, microRNA; WT, wild-type RUNX2 3′-UTR; Mut, RUNX2 3′-UTR mutation.
RUNX2 is involved in miR-150-induced OS cell proliferation suppression and chemotherapy sensitization
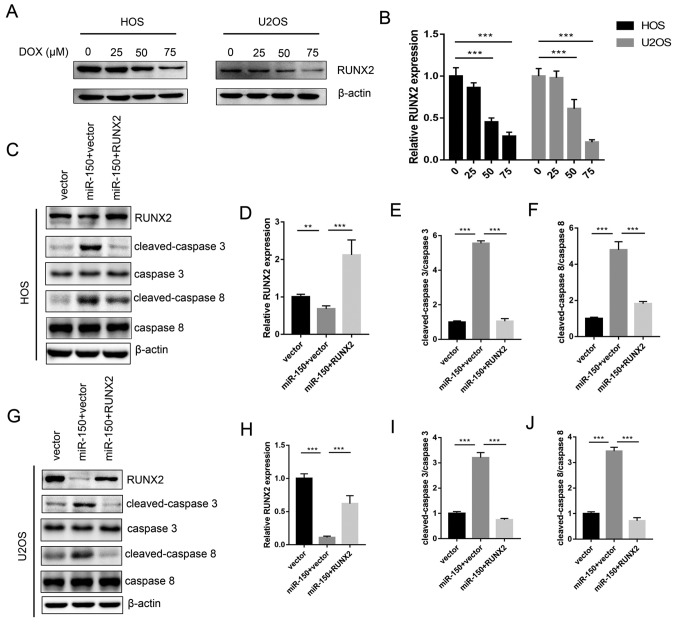
The aforementioned results indicated that miR-150 inhibited OS cell growth by targeting RUNX2. Therefore, the roles of RUNX2 in OS cell proliferation and chemotherapy resistance were further detected in the present study. The data revealed that RUNX2 expression was significantly decreased by transfection with miR-150 mimic in both HOS (Fig. 4A and B) and U2OS (Fig. 4E and F) cells, compared with the NC mimic group. Treatment of the miR-NC group with DOX also significantly reduced the expression of RUNX2 (Fig. 4B and F), suggesting that sensitization of OS to DOX chemotherapy is associated with RUNX2 expression. RUNX2 was then overexpressed using the pcDNA3.1 vector to rescue RUNX expression. Proliferation assays revealed that the suppressive effect of miR-150 on HOS (Fig. 4C) and U2OS (Fig. 4G) cells was comparable to that of shRUNX2. As presented in Fig. 4D and H, overexpression of RUNX2 reversed the effects of miR-150 on the DOX sensitization of HOS and U2OS cells, respectively. Furthermore, following RUNX2 knockdown (Fig. 4I and J), sensitization of HOS and U2OS cells to DOX was also significantly increased (Fig. 4K and L). Taken together, the results indicated that RUNX2 was an important functional target of miR-150 involved in the proliferation and chemotherapy sensitization of OS cells.
Figure 4.
RUNX2 is involved in miR-150-induced OS cell proliferation suppression and chemotherapy sensitization. (A) Western blot analysis to detect RUNX2 protein expression in HOS cells. (B) Statistical analysis of RUNX2 expression in HOS cells. (C) HOS cell proliferation was detected using a CCK-8 assay. (D) HOS cell viability was measured using a CCK-8 assay following treatment with or without DOX. (E) Western blot analysis to detect RUNX2 protein expression in U2OS cells. (F) Statistical analysis of RUNX2 expression in U2OS cells. (G) U2OS cell proliferation was detected using a CCK-8 assay. (H) U2OS cell viability was measured using a CCK-8 assay following treatment with or without DOX. (I) Western blot analysis to detect RUNX2 protein expression in RUNX2 shRNA-transfected HOS and U2OS cells. (J) Statistical analysis of RUNX2 expression in HOS cell and U2OS cells. (K) RUNX2 shRNA-transfected HOS cell viability was measured using a CCK-8 assay following treatment with or without DOX. (L) RUNX2 shRNA-transfected U2OS cell viability was measured using a CCK-8 assay following treatment with or without DOX. All assays were performed in triplicate. ***P<0.001 and **P<0.01. RUNX2, runt-related transcription factor 2; miR, microRNA; OS, osteosarcoma; CCK-8, Cell Counting Kit-8; DOX, doxorubicin; shRNA, short hairpin RNA; miR-NC, negative control miR mimic.
miR-150 and RUNX2 affect OS cell chemosensitivity by regulating apoptosis proteins
Expression levels of RUNX2 were decreased with increased concentrations of DOX treatment in HOS and U2OS cells in a dose-dependent manner (Fig. 5A and B). The levels of apoptosis proteins, including cleaved caspase-3 and cleaved caspase-8 were tested. The results revealed that miR-150 significantly increased the expression of cleaved caspase-3 and cleaved caspase-8 in both HOS and U2OS cells, while RUNX2 significantly decreased the expression levels of cleaved caspase-3 and cleaved caspase-8 (Fig. 5C-J). Therefore, the data from the present study demonstrated the molecular mechanism underlying the miR-150-RUNX2 axis in OS resistance to DOX.
Figure 5.
Effects of miR-150 and RUNX2 on the expression of apoptosis proteins. (A) Protein expression of RUNX2 at different concentrations of DOX in HOS and U2OS cells. (B) Statistical analysis of RUNX2 expression. (C) The expression of cleaved-caspase-3 and cleaved-caspase-8 in HOS cells was detected via western blotting. (D) Statistical analysis of RUNX2 expression in HOS cells. (E) Statistical analysis of cleaved-caspase-3 expression in HOS cells. (F) Statistical analysis of cleaved-caspase-8 expression in HOS cells. (G) The expression of cleaved-caspase-3 and cleaved-caspase-8 in U2OS cells was detected via western blotting. (H) Statistical analysis of RUNX2 expression in U2OS cells. (I) Statistical analysis of cleaved-caspase-3 expression in U2OS cells. (J) Statistical analysis of cleaved-caspase-8 expression in U2OS cells. **P<0.01 and ***P<0.001. miR, microRNA; RUNX2, runt-related transcription factor 2; DOX, doxorubicin.
Discussion
In the present study, it was revealed that RUNX2 knockdown sensitized OS to the treatment of DOX. In addition, a novel mechanism for the miR-150-RUNX2 axis was indicated, which demonstrated that RUNX2 is a target gene of miR-150. miR-150 significantly suppressed cell proliferation and invasion in OS cells by directly targeting RUNX2. Restoration of RUNX2 in OS cells reversed the tumor suppressor and chemoprotective roles of miR-150. In summary, miR-150 functions as a tumor suppressor and sensitizes OS to chemotherapy-induced apoptosis by targeting RUNX2.
In recent years, miRNAs have been demonstrated as important cancer biomarkers in a wide range of different types of cancer (12–17). During cancer progression, miRNAs regulate the transcription and expression of a large number of genes, a number of which are vital for the genesis and development of cancer cells (22,23). There are abundant reports of the aberrant expression and the function of miRNAs in OS; certain miRNAs act as oncogenes, while others serve as tumor suppressors (24,25). miR-150 is also a tumor-associated miRNA, but the biological function and the underlying molecular mechanisms of miR-150 in OS have not yet been fully investigated.
miR-150 has been reported as downregulated in OS tissues and cells (15). miR-150 has also been reported as a tumor suppressor by suppressing the PI3K-AKT pathway (26). miR-150, insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) and combined miR-150/IGF2BP1 expressions were all identified as independent prognostic factors for overall and disease-free survival of patients with osteosarcoma (27). In a previous study, exogenous miR-150 expression stimulated cell apoptosis and inhibited proliferation, invasion and migration (28). However, to the best of our knowledge, the downstream targets and details regarding the molecular mechanisms underlying tumor suppression by miR-150 have not yet been elucidated.
RUNX2 is an important transcription factor, which regulates the cell fate during normal osteoblast differentiation, such as cell apoptosis in response to tumor necrosis factor-α (29,31). Furthermore, RUNX2 has been implicated as a putative oncogene and a cancer biomarker in OS (32,33). However, until now, the biological consequences of RUNX2 overexpression and its molecular role in the chemoprotection of OS had not been clearly defined.
In the present study, a TargetScan bioinformatics algorithm was used to investigate whether RUNX2 is a target of miR-150, and it was revealed that miR-150 may bind to RUNX2 3′-UTR. The subsequent luciferase reporter assay demonstrated that miR-150 could specifically decrease the luciferase activity of the RUNX2 3′-UTR, but not that of the Mut, and that miR-150 could suppress RUNX2 in OS cell lines, indicating that RUNX2 was a direct target of miR-150. Proliferation assays revealed that miR-150 overexpression and RUNX2 knockdown exhibited similar inhibiting effects on OS cell proliferation and chemotherapy resistance. Furthermore, restoration of RUNX2 reversed the suppressive effects of miR-150 on OS cell proliferation and chemotherapy treatment.
In the present study, although it was demonstrated that the miR-150-RUNX2 axis played an important role in OS cell chemotherapeutic agent resistance, several issues remain to be investigated. The regulators of the miR-150-RUNX2 axis require further investigation, and this theory must be verified by additional clinical experiments. A biological function study of the network of miR-150-RUNX2 axis would facilitate the pathogenesis research of human OS. New molecular targeting drugs may be developed based on this miR-150-RUNX2 axis network.
Taken together, the present study identified miR-150 as a tumor suppressor in OS, and that it inhibits OS growth by targeting RUNX2. miR-150 can be used as a potential therapy in future OS treatment.
Acknowledgements
Not applicable.
Funding
This article is supported by the national natural science foundation of China (grant no. 81802693).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YC conceived and designed the present study. YC, JF and GZ developed the methodology. ZL and GF performed the majority of the experiments. GF, DY, JZ and YZ performed the data analysis, ZL wrote the manuscript. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Jinling Hospital (Nanjing, China). All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mousa SA, Gallati C, Simone T, Dier E, Yalcin M, Dyskin E, Thangirala S, Hanko C, Rebbaa A. Dual targeting of the antagonistic pathways mediated by Sirt1 and TXNIP as a putative approach to enhance the efficacy of anti-aging interventions. Aging (Albany NY) 2009;1:412–424. doi: 10.18632/aging.100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielack SS, Marina N, Ferrari S, Helman LJ, Smeland S, Whelan JS, Reaman GH. Osteosarcoma: The same old drugs or more? J Clin Oncol. 2008;26:3102–3105. doi: 10.1200/JCO.2008.17.1108. [DOI] [PubMed] [Google Scholar]
- 3.Longhi A, Errani C, De Paolis M, Mercuri M, Bacci G. Primary bone osteosarcoma in the pediatric age: State of the art. Cancer Treat Rev. 2006;32:423–436. doi: 10.1016/j.ctrv.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Gorlick R, Khanna C. Osteosarcoma. J Bone Mineral Res. 2010;25:683–691. doi: 10.1002/jbmr.77. [DOI] [PubMed] [Google Scholar]
- 5.Bacci G, Briccoli A, Rocca M, Ferrari S, Donati D, Longhi A, Bertoni F, Bacchini P, Giacomini S, Forni C, et al. Neoadjuvant chemotherapy for osteosarcoma of the extremities with metastases at presentation: Recent experience at the Rizzoli Institute in 57 patients treated with cisplatin, doxorubicin, and a high dose of methotrexate and ifosfamide. Ann Oncol. 2003;14:1126–1134. doi: 10.1093/annonc/mdg286. [DOI] [PubMed] [Google Scholar]
- 6.Rainusso N, Wang LL, Yustein JT. The adolescent and young adult with cancer: State of the art e bone tumors. Curr Oncol Rep. 2013;15:296–307. doi: 10.1007/s11912-013-0321-9. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25:398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 8.Wang T, Ji F, Dai Z, Xie Y, Yuan D. Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer Biomark. 2015;15:543–550. doi: 10.3233/CBM-150493. [DOI] [PubMed] [Google Scholar]
- 9.Muoio DM. TXNIP links redox circuitry to glucose control. Cell Metab. 2007;5:412–414. doi: 10.1016/j.cmet.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Forrester MT, Seth D, Hausladen A, Eyler CE, Foster MW, Matsumoto A, Benhar M, Marshall HE, Stamler JS. Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. J Biol Chem. 2009;284:36160–36166. doi: 10.1074/jbc.M109.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreyer F, Baur A. Biogenesis and functions of exosomes and extracellular vesicles. Methods Mol Biol. 2016;1448:201–216. doi: 10.1007/978-1-4939-3753-0_15. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: From functions to targets. PLoS One. 2010;5(pii):e13067. doi: 10.1371/journal.pone.0013067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li CH, Yu TB, Qiu HW, Zhao X, Zhou CL, Qi C. miR-150 is downregulated in osteosarcoma and suppresses cell proliferation, migration and invasion by targeting ROCK1. Oncol Lett. 2017;13:2191–2197. doi: 10.3892/ol.2017.5709. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Quan X, Chen D, Li M, Chen X, Huang M. MicroRNA-150-5p and SRC kinase signaling inhibitor 1 involvement in the pathological development of gastric cancer. Exp Ther Med. 2019;18:2667–2674. doi: 10.3892/etm.2019.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin M, Shi C, Yang C, Liu J, Huang G. Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in NSCLC through regulation of the miR-150-5p/GLUT-1 axis. Mol Ther Nucleic Acids. 2019;18:219–231. doi: 10.1016/j.omtn.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Chen X, Xu X, Pan B, Zeng K, Xu M, Liu X, He B, Pan Y, Sun H, Wang S. miR-150-5p suppresses tumor progression by targeting VEGFA in colorectal cancer. Aging (Albany NY) 2018;10:3421–3437. doi: 10.18632/aging.101656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Choi YS, Park JH, Kim H, Lee I, Won YB, Yun BH, Park JH, Seo SK, Lee BS, Cho S. miR-150-5p may contribute to pathogenesis of human leiomyoma via regulation of the Akt/p27Kip1 pathway in vitro. Int J Mol Sci. 2019;20(pii):E2684. doi: 10.3390/ijms20112684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadikovic B, Thorner P, Chilton-Macneill S, Martin JW, Cervigne NK, Squire J, Zielenska M. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer. 2010;10:202. doi: 10.1186/1471-2407-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Villadsen SB, Bramsen JB, Ostenfeld MS, Wiklund ED, Fristrup N, Gao S, Hansen TB, Jensen TI, Borre M, Ørntoft TF, et al. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br J Cancer. 2012;106:366–374. doi: 10.1038/bjc.2011.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiyomaru T, Tatarano S, Kawakami K, Enokida H, Yoshino H, Nohata N, Fuse M, Seki N, Nakagawa M. SWAP70, actin-binding protein, function as an oncogene targeting tumor-suppressive miR-145 in prostate cancer. Prostate. 2011;71:1559–1567. doi: 10.1002/pros.21372. [DOI] [PubMed] [Google Scholar]
- 24.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and −145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 25.Tang M, Lin L, Cai H, Tang J, Zhou Z. MicroRNA-145 downregulation associates with advanced tumor progression and poor prognosis in patients suffering osteosarcoma. Onco Targets Ther. 2013;6:833–838. doi: 10.2147/OTT.S40080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Aireti A, Aihaiti A, Li K. Expression of microRNA-150 and its Target Gene IGF2BP1 in human osteosarcoma and their clinical implications. Pathol Oncol Res. 2019;25:527–533. doi: 10.1007/s12253-018-0454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Chen L, Wang W, Meng FB, Zhao RT, Chen Y. MicroRNA-150 inhibits cell invasion and migration and is downregulated in human osteosarcoma. Cytogenet Genome Res. 2015;146:124–135. doi: 10.1159/000437379. [DOI] [PubMed] [Google Scholar]
- 29.Ghali O, Chauveau C, Hardouin P, Broux O, Devedjian JC. TNF-alpha's effects on proliferation and apoptosis in human mesenchymal stem cells depend on RUNX2 expression. J Bone Miner Res. 2010;25:1616–1626. doi: 10.1002/jbmr.52. [DOI] [PubMed] [Google Scholar]
- 30.Olfa G, Christophe C, Philippe L, Romain S, Khaled H, Pierre H, Odile B, Jean-Christophe D. RUNX2 regulates the effects of TNFalpha on proliferation and apoptosis in SaOs-2 cells. Bone. 2010;46:901–910. doi: 10.1016/j.bone.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 31.Nathan SS, Pereira BP, Zhou YF, Gupta A, Dombrowski C, Soong R, Pho RW, Stein GS, Salto-Tellez M, Cool SM, van Wijnen AJ. Elevated expression of Runx2 as a key parameter in the etiology of osteosarcoma. Mol Biol Rep. 2009;36:153–158. doi: 10.1007/s11033-008-9378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira BP, Zhou Y, Gupta A, Leong DT, Aung KZ, Ling L, Pho RW, Galindo M, Salto-Tellez M, Stein GS, et al. Runx2, p53, and pRB status as diagnostic parameters for deregulation of osteoblast growth and differentiation in a new pre-chemotherapeutic osteosarcoma cell line (OS1) J Cell Physiol. 2009;221:778–788. doi: 10.1002/jcp.21921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee DH, Qi J, Bradner JE, Said JW, Doan NB, Forscher C, Yang H, Koeffler HP. Synergistic effect of JQ1 and rapamycin for treatment of human osteosarcoma. Int J Cancer. 2015;136:2055–2064. doi: 10.1002/ijc.29269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.