Abstract

Various β-galactosidase enzymes catalyze the trans-glycosylation reaction with lactose. The resulting galactooligosaccharide (GOS) mixtures are widely used in infant nutrition to stimulate growth of beneficial gut bacteria. GOS consists mainly of compounds with a degree of polymerization (DP) varying from 2–8 and with diverse glycosidic linkages. In recent years, we have elucidated in detail the composition of several commercial GOS mixtures in terms of DP and the structural identity of the individual compounds. In this work, 13 (single) probiotic strains of gut bacteria, belonging to 11 different species, were grown to stationary phase with a Vivinal GOS-derived sample purified to remove lactose and monosaccharides (pGOS). Growth among the probiotic strains varied strongly between 30 and 100% of OD600nm relative to positive controls with glucose. By identifying the components of the pGOS mixture that remain after growth, we showed that strains varied in their consumption of specific GOS compounds. All strains commonly used most of the GOS DP2 pool. Lactobacillus salivarius W57 also utilized the DP3 branched compound β-d-Galp-(1 → 4)-[β-d-Galp-(1 → 2)]-d-Glc. Bifidobacterial strains tended to use GOS with higher DP and branching than lactobacilli; Bifidobacterium breve DSM 20091, Lactobacillus acidophilus W37, and Bifidobacterium infantis DSM 20088 were exceptional in using 38, 36, and 35 compounds, respectively, out of the 40 different structures identified in pGOS. We correlated these bacterial GOS consumption profiles with their genomic information and were able to relate metabolic activity with the presence of genome-encoded transporters and carbohydrate-active enzymes. These detailed insights may support the design of synbiotic combinations pairing probiotic bacterial strains with GOS compounds that specifically stimulate their growth. Such synbiotic combinations may be of interest in food/feed and/or pharmacy/medicine applications.
Keywords: galactooligosaccharides, glycosidic linkages, bifidobacteria, lactic acid bacteria, synbiotics, catabolic pathways
Introduction
Probiotic bacteria influence human health in various ways, that is, by modulating the immune system, assisting in fermentation of dietary nondigestible elements into short-chain fatty acids, and by inhibiting growth of pathogens. Imbalanced microbiome composition (dysbiosis) has been related to many diseases, including coronary heart disease, fatty liver diseases, rheumatoid arthritis, irritable bowel syndrome, and inflammatory bowel disease.1−4 Changes in the microbiota composition may occur naturally during a lifetime but can also be caused by diseases and through the use of antibiotics. These changes require readjustment toward a healthy bacterial composition in the recovery phase. Stimulating growth and activity of beneficial bacteria in the human gut has become a valuable approach to sustain and restore human health.5 A growing number of prebiotic molecules, mostly carbohydrates, have been shown to affect the presence and to increase the numbers of distinct bacterial groups in the human colon that may ultimately drive the health effect.6
At the beginning of life the human colon is a nearly sterile environment that is rapidly colonized primarily by bifidobacteria forming the microbiota of the healthy infant gut.7 Human milk oligosaccharides were found to be the main drivers in infant gut microbiota development.8 Infant nutritional supplements, as replacement for human milk, aim to stimulate the development of an infant gut microbiota with a composition as close to the natural situation as possible. For example, current infant formulas often contain mixtures of galactooligosaccharide (GOS) and fructooligosaccharides which have been shown to support the formation of a gut microbial environment that closely resembles that of breastfed infants.9,10 GOS are primarily produced by incubating β-galactosidase enzymes at high lactose concentrations. These enzymes use lactose as the donor substrate for transfer of galactose residues onto lactose or any prior formed GOS. These GOS molecules exhibit often a terminal reducing glucose residue elongated with galactose residues toward the nonreducing end. The GOS yield and product composition are strongly influenced by the microbial origin of the (mutant) enzyme, reaction conditions, and substrate concentrations used, resulting in a growing number of industrially produced GOS mixtures of different compositions.11,12 These GOS mixtures vary mostly in their degree of polymerization (DP) ranging from DP2-8 and the presence of different ratios of glycosidic linkages of β-(1 → 2), β-(1 → 3), β-(1 → 4), and/or β-(1 → 6). It is unknown whether and how these individual GOS structures differ functionally.
To study the selective stimulatory effects of GOS on the strains of bacteria that are associated with the human gut, we followed the bacterial growth in vitro using a GOS mixture as the sole carbon source. Previously, we have analyzed the composition of 7 commercial GOS mixtures.20 Vivinal GOS was the most diverse in GOS composition (with over 40 different molecules), and hence, it was selected for this study. This allowed us to identify the specific GOS compounds, differing in DP and glycosidic linkages present, that these bacteria use for growth. Furthermore, we correlated bacterial growth to the distinct set of genome-encoded enzymes and transporters that the bacteria may produce in the presence of GOS substrates.13 In previous work, Gopal et al. studied two strains, Bifidobacterium lactis DR10 and Lactobacillus rhamnosus DR20 which utilized different GOS components within GOS mixtures: B. lactis DR10 consumed GOS with a higher DP while L. rhamnosus preferred the use of galactose and GOS disaccharides.14 Another study tested the growth of 68 human derived strains from the genera Lactobacillus and Bifidobacterium on a broad range of prebiotics including GOS. While many strains grew well on GOS compared to other substrates, no clear selectivity of the strains toward certain components of GOS was found.15 Recently, a study revealed three subsets of strains of lactobacilli and bifidobacteria regarding GOS utilization: one group utilizing only GOS DP2, a second utilizing GOS ≤ DP3, and a third utilizing all GOS oligomers.16 Another study has observed that different strains of bifidobacteria have a different preference for GOS molecules but was unable to identify the specific GOS structures.17 Kittibunchakul et al. (2018) tested fermentability of 3 GOS mixtures (including Vivinal GOS) using 8 Lactobacillus spp. strains and 3 Bifidobacterium spp. strains. The highest growth scores were obtained with a novel GOS mixture that contained mostly oligosaccharides with β-(1 → 3) and β-(1 → 6) glycosidic linkages. The precise identity of the GOS molecules degraded was not studied however.18 It, thus, is clear that there is a strain-based preference for specific GOS DPs, but detailed information about the structural identity of GOS molecules consumed in terms of glycosidic linkages is still missing.
We recently introduced methodologies for rapid identification of the complete structural composition of GOS mixtures.19,20 Here, we applied this comparative high performance anion-exchange chromatography with pulsed amperometric detection (HPAEC–PAD) analysis to identify the GOS structures that were specifically utilized by probiotic bifidobacteria and lactic acid bacteria (LAB). The results show that individual strains differ strongly in utilization of pGOS in terms of the polymerization degree and the type of glycosidic linkages. These results provide a more detailed understanding of how GOS structures found in (commercial) prebiotic samples stimulate growth of (commercial) probiotic bacteria at the level of individual strains and may be of interest in designing novel synbiotic mixtures.
Materials and Methods
Materials
The commercial probiotic strains Lactobacillus paracasei subsp. paracasei W20, Lactobacillus acidophilus W37, Lactobacillus salivarius W57, Lactobacillus casei W56, Enterococcus faecium W54, and Pediococcus acidilactici W143, Bifidobacterium animalis subsp. lactis W51, B. animalis subsp. lactis, and B. animalis subsp. lactis W53 were supplied by Winclove Probiotics (Amsterdam, The Netherlands). Bifidobacterium longum subsp. infantis DSM 20088, Bifidobacterium breve DSM 20091, Bifidobacterium adolescentis DSM 20083, and Bifidobacterium bifidum DSM 20456 were purchased from DMSZ (Braunschweig, Germany). Purified GOS (pGOS), derived from Vivinal GOS with galactose, glucose and lactose largely removed, was kindly provided by FrieslandCampina Domo (Amersfoort, The Netherlands). The modified de Man–Rogosa–Sharpe-medium (mMRS-medium) was prepared as described previously.21 In brief, 1 L of mMRS-medium contained: peptone 10 g; granulated yeast extract 2.5 g; tryptose 3 g; Tween80 1 g; K2HPO4 3 g; KH2PO4 3 g; ammonium citrate 2 g; pyruvic acid sodium salt 0.2 g; MgSO4·7H2O 0.575 g; MnSO4·H2O 0.12 g; and FeSO4·7H2O 0.034 g. All the components were dissolved in aqua bidest. H2O and medium heated to 60 °C to dissolve all the components. After adjusting the pH to 6.8, the medium was sterilized by autoclaving (15 min, 121 °C) and supplemented with filter-sterilized 0.5 g/L Cys-HCl (final concentration). The carbon-source free Bifidobacterium medium (cfBM) contained (g/L):22 trypticase peptone 10 g; yeast extract 2.5 g; tryptose 3 g; K2HPO4 3 g; KH2PO4 3 g; triammonium citrate 2 g; pyruvic acid 0.3 g; Tween 80 1 g; MgSO4·7H2O 0.574 g; MnSO4·H2O 0.12 g; and NaCl 5 g. All the components were dissolved in aqua bidest. H2O, and after boiling the medium, the pH was adjusted to 6.8. The medium was sterilized by autoclaving as stated for the mMRS-medium and afterward supplemented with sterile 0.5 g/L Cys-HCl.
Growth of Probiotic Bacterial Strains
LAB were cultured in MRS-medium (Oxoid, Basingstoke, UK) in anaerobic culture tubes flushed with nitrogen using the Hungate technique.23 Under these conditions, strains were precultured twice overnight at 37 °C before the growth experiments with GOS; purity of the cultures was frequently checked under a microscope. For growth experiments with pGOS, cultures of LAB strains were harvested by centrifugation (2300g, 2 min) and bacterial pellets were diluted 25-fold in 2× sterilized mMRS. Diluted cultures were mixed 1:1 with sterile pGOS (dissolved in MilliQ water at 10 mg/mL) in microtiter plates (96 well, flat-bottom) (Greiner Bio-One, Frickenhausen, Germany) yielding final volumes of 160–200 μL per well.24 Glucose (5 mg/mL) and the medium without any carbon source added served as the positive and negative control, respectively. Inoculation of microtiter plates was carried out inside a glovebox (Bohlender, Grünsfeld, Germany) constantly flushed with N2 in order to ensure anaerobic conditions. Afterwards, plates were sealed airtight (Simport, Beloeil, Canada) and transferred into a microtiter plate reader (BioTek, Winooski, VT) incubating plates at 37 °C. Plates were shaken continuously at medium speed and OD600nm measured every 5 min. Bifidobacteria were cultured in Medium 58 following the recipe of the supplier (DSMZ, Braunschweig, Germany), using anaerobic culture tubes flushed with CO2. Every strain was precultured twice overnight at 37 °C prior to growth experiments with pGOS. Bacterial cultures were harvested as described above for LAB strains and bacterial pellets diluted 25-fold in 2× carbon source-free Bifidobacterium medium. For the actual growth experiments, the diluted bacterial cultures were mixed 1:1 with 10 mg/mL of sterilized pGOS in anaerobic glass tubes and cultures flushed with 100% CO2. During the growth experiments, the cultures were maintained at 37 °C and a cell density meter WPA CO 8000 (Biochrom, Cambridge, UK) was used for direct measurement of OD600nm within anaerobic glass tubes. All the cultures were inoculated as independent n = 3 triplicates.
pH measurements of cultures were carried out at the time of inoculation (0 h) and stationary growth phase (18 h for LAB strains, 25–32 h Bifidobacterium strains) using a pH electrode VWR pH100 (VWR International, Leuven, Belgium).
Carbohydrate Structural Analysis
GOS composition in commercial mixtures and bacterial cultures was profiled using HPAEC–PAD as described.20 Bacterial cultures were harvested at the stationary phase (see Figure 1 for time points) by centrifugation (2 min, 16 000g). Supernatants were transferred immediately into HPAEC vials and diluted fivefold in 80% DMSO. GOS molecules were separated on a CarboPac PA-1 column (250 by 2 mm; Dionex, Amsterdam, The Netherlands) with a complex gradient of buffer A = 0.1 M NaOH, buffer B = 0.6 M NaOAc in 0.1 M NaOH, buffer C = deionized water, and buffer D = 50 mM NaOAc as described.20 The remaining pGOS composition after growth was analyzed per strain in n = 3 independent samples, each derived from a different biological replicate. The samples containing pure pGOS and the medium without a carbon source added served as positive and negative controls, respectively.
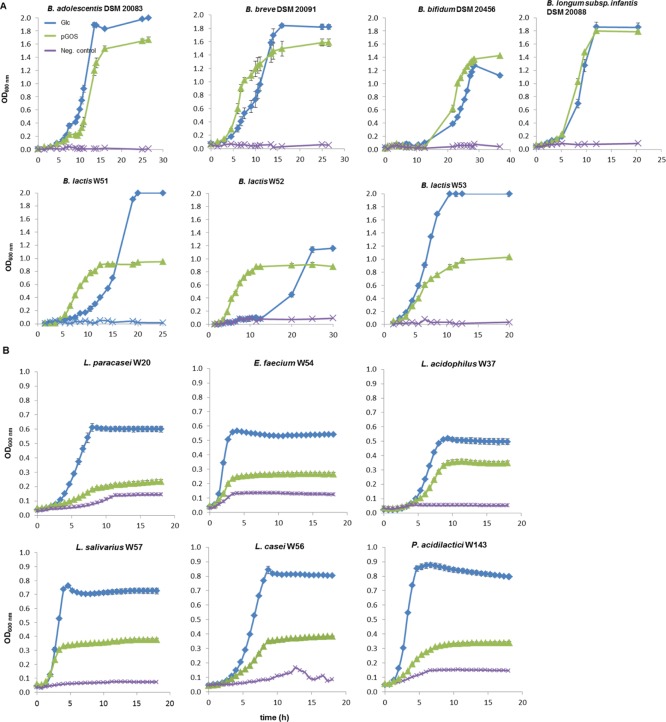
Figure 1.
(A) Bifidobacterial strains grown with 5 mg/mL pGOS; glucose (5 mg/mL) and modified MRS-medium served as positive and negative controls (Neg. control), respectively. Growth was followed by measuring ΔOD manually in anaerobic glass tubes (s. Material and Methods). (B) Strains from LAB grown with 5 mg/mL pGOS; glucose (5 mg/mL) and modified MRS-medium served as positive and negative controls (Neg. control), respectively. OD600nm values were acquired by growing strains in microtiter plates and following ΔOD using a microtiter plate reader. All the values shown are means from 3 biological replicates. Most standard deviations are smaller than the size of the symbols and therefore not apparent.
Genome Analysis
Genbank files of bacterial genomes including plasmids for the DSMZ strains were downloaded (https://www.ncbi.nlm.nih.gov) at 07-11-2018 or supplied by Winclove. A comparative analysis for different GOS catabolic pathways identified in the reference strains was performed. Genomes were submitted to RAST (Rapid Annotation using Subsystem Technology; http://rast.nmpdr.org/rast.cgi) using RASTtk as the default method.25 Annotated bacterial genomes were compared for lactose and galactose utilization to identify candidate genes encoding the LacS/LacZ, LacEF/LacG, and LacY pathways. Whole CAZomes were established using dbCAN2 (automated CAZyme annotation; http://cys.bios.niu.edu/dbCAN2/blast.php) to identify candidate genes encoding galactose-specific glycoside hydrolases (GH) of families 1, 2, 35, 42, and 53.26 Searches for signal sequences for secretion in GH proteins were performed with dbCAN2 and PSORTb version 3.0.2 (bacterial localization prediction tool; http://www.psort.org/psortb/).27 As the RAST subsystem for lactose and galactose utilization does not contain genes of the GosDEC and GalCDE pathways, the presence of these genes in bacterial genomes was checked by cross BLAST searches using the GosDEC gene from B. lactis 04 (Balac_0485, Acc. nr. WP_004268783.1; Balac_0486, Acc. nr. WP_004269047.1) and GalCDE gene from B. breve UCC2003 (Bbr_0417_galc, Acc. nr. WP_015438440.1; Bbr_0418_galD, Acc. nr. WP_025262769.1; Bbr_0419_galE, Acc. nr. WP_003828491.1) as a reference. Total number of candidate genes for each enzyme in these pathways was retrieved and on this basis a heat map was created using the program GraphPrism (version 7.0).
Statistical Analysis
All growth experiments are biological triplicates; the OD600 values are expressed as averages. The medium without bacterial inoculation was used to obtain blank values during OD600 measurements.
Results and Discussion
Growth of Probiotic Bacterial Strains on pGOS
Utilization of GOS compounds was studied by growing the strains under appropriate anaerobic conditions with 5 mg/mL pGOS or appropriate controls. Most bifidobacterial strains grew well on pGOS with lag phases and growth rates close to positive controls (glucose 5 mg/mL) (Figure 1A). B. breve DSM 20091 and B. lactis W51-52 preferred pGOS as growth substrates over glucose clearly indicating saccharolytic capabilities for more complex carbon sources, as observed for bifidobacteria.28,29 Strains of LAB often grew to a limited extent on pGOS compared to their positive controls, with OD600nm values ranging between 32 and 49% of Glc controls (Figure 1B). L. acidophilus W37 was an exception and reached an OD600nm over 70% of its positive control. Growth results on pGOS were supported by the final culture pH, close to (for bifidobacteria) or above (for most lactobacilli) their positive controls. These results show that LAB strains grew on pGOS only to a limited extent and that these strains thus differ from most Bifidobacterium strains in their metabolic capabilities to use pGOS.
Structural Identity of pGOS Compounds Consumed by Probiotic Strains
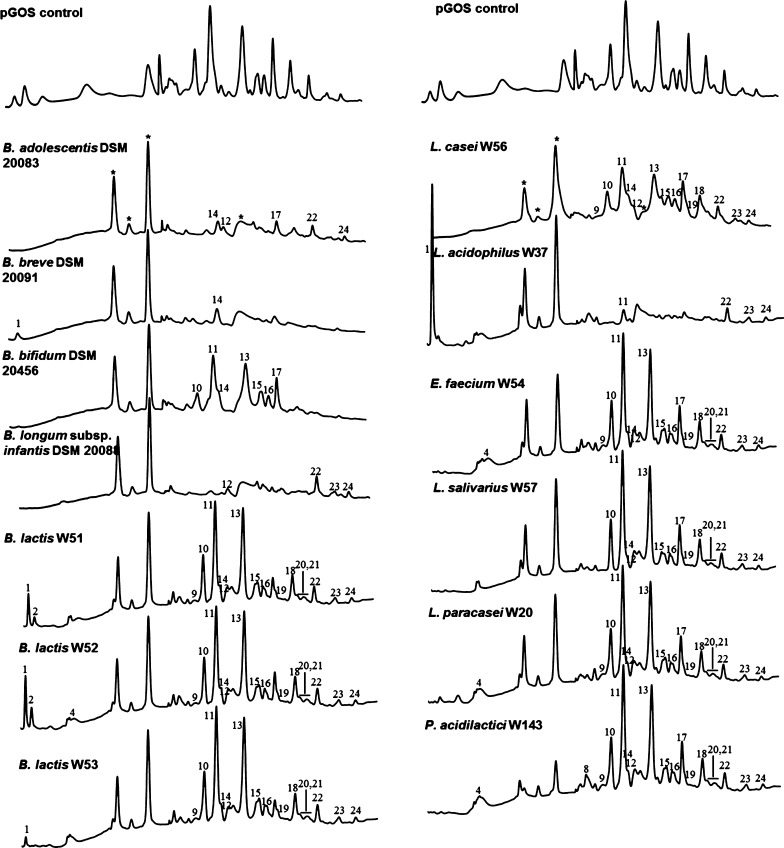
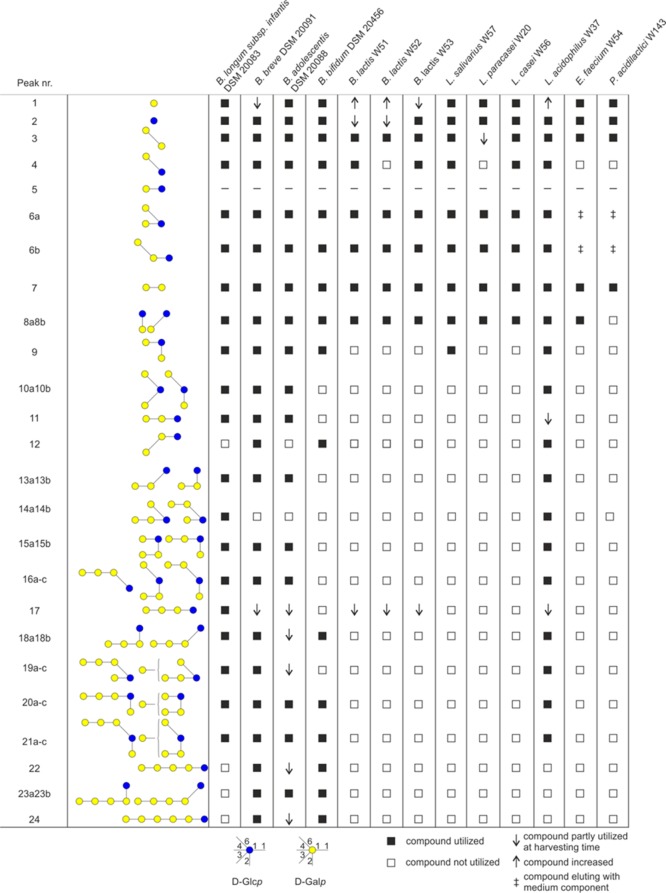
The pGOS mixture contained 40 different GOS compounds (Figures 2 and 3) as identified by peak height differences above the baseline. Analysis of GOS compounds remaining at the stationary growth phase (harvesting time points for Bifidobacterium strains, 25 or 32 h, depending on the time required to reach the stationary phase; for LAB strains 18 h, see Figure 1) revealed strain-dependent GOS consumption profiles. The three B. lactis strains W51–W53 completely utilized the DP2 compounds β-d-Galp-(1 → 2)-d-Glc, β-d-Galp-(1 → 3)-d-Glc (peak 8), and partially utilized the linear DP4 β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-d-Glc (peak 17) (Figure 2A). In addition, HPAEC–PAD chromatograms of strains W51 and W52 showed an increase in galactose (peak 1, Figure 2A) while strain W53 had consumed virtually all galactose at the time the cultures were harvested, as previously observed for various lactobacilli.21 As the pH in these cultures remained above controls, this effect is most likely linked to a lactose–galactose antiporter involved in pGOS metabolism present in the B. lactis strains (further described below).30,31 The B. breve strain DSM 20091 showed the broadest utilization of GOS compounds amongst all the strains tested. It consumed 38 out of the 40 compounds in pGOS; the remaining compounds were the DP4-branched compounds β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-[β-d-Galp-(1 → 6)]-d-Glc and β-d-Galp-(1 → 4)-[β-d-Galp-(1 → 4)-β-d-Galp-(1 → 6)]-d-Glc (peak 14a and b). Partially consumed compounds (detected with the reduced peak height) represented the two linear elongated DP4 and DP6 structures (peaks 17 and 24). In the chromatogram of this strain, the galactose peak was found to be reduced compared to the pGOS standard. Previously, Watson et al.21 studied the growth and GOS consumption by various other B. breve strains, revealing that strain-dependent differences may occur. Similar to B. breve DSM 20091, the B. adolescentis strain DSM 20083 did not use the two DP4-branched compounds eluting under peak 14 nor DP3 β-d-Galp-(1 → 3)-β-d-Galp-(1 → 4)-d-Glc (peak 12). Interestingly, the latter structure has been linked to cross-reactive allergies in Japan.32,33 Partial utilization was observed with the β-(1 → 4) linear elongated products of 4′-galactosyllactose (peak 11), that is, DP4 (peak 17), DP5 (peak 22), and DP6 (peak 24). The latter two compounds were preferentially utilized by the B. bifidum strain DSM 20456. Furthermore, this strain selectively left the DP3-branched compounds β-d-Galp-(1 → 6)-[β-d-Galp-(1 → 2)]-d-Glc and β-d-Galp-(1 → 6)-[β-d-Galp-(1 → 2)]-d-Glc (peaks 10 a and b) while the linear β-d-Galp-(1 → 3)-β-d-Galp-(1 → 4)-d-Glc compound (peak 12) was consumed. B. bifidum DSM 20456 also did not utilize the DP3 compounds β-d-Galp-(1 → 4)-β-d-Galp-(1 → 2)-d-Glc, β-d-Galp-(1 → 4)-β-d-Galp-(1 → 3)-d-Glc, and β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-d-Glc (peaks 13 and 11, Figure 2A). The DP4-branched compounds eluting as peaks 14, 15, and 16 were not utilized, neither was the β-(1 → 4) linear elongation of 4′-galactosyllactose (peak 17). Similar to B. breve, the B. longum infantis strain DSM 20088 consumed the majority of GOS molecules. Only DP 5+ and 3′-galacosyllactose (peak 12) were left by this strain. Overall these results show that most Bifidobacterium strains grew well on pGOS and consumed GOS molecules with a distinct DP and different glycosidic linkage compositions. Most of the branched structures (peaks 10, 13, 15, and 16) were only consumed by a limited number of strains, particularly by B. infantis, B. breve, B. adolescentis, and L. acidophilus. Peak 14 is only consumed by B. infantis DSM 20088 and L. acidophilus W37. Branched structure 6a was fermented by all the strains tested (Figures 2 and 3).
Figure 2.
HPAEC–PAD chromatograms of pGOS (control, first line) and pGOS after the growth of probiotic strains. For each strain, pGOS composition was analyzed in n = 3 biological replicates (numbers 1, 2, and 4 indicate Gal, Glc, and lactose, respectively). Other numbers indicate single pGOS compounds that were not utilized by the strains at the stationary growth phase (Figures 1 and 3). Bifidobacterium strains were grown in a carbon source-free Bifidobacterium medium with 5 mg/mL pGOS added for 25–32 h. LAB strains were grown in modified MRS-medium with 5 mg/mL pGOS added for 18 h. Peaks marked * are non-GOS peaks stemming from the growth medium.
Figure 3.
Differential utilization of pGOS components for growth as observed for Bifidobacterium and LAB strains highlighting their diverse capabilities to consume GOS of a specific DP level and different glycosidic linkages present.
Rather opposite effects were seen for the LAB strains that we tested. These strains mostly utilized a narrow and specific range of GOS molecules. P. acidilactici W143 hardly used the GOS molecules, only the disaccharides β-d-Galp-(1 → 6)-d-Gal (peak 3) and β-d-Galp-(1 → 4)-d-Gal (peak 7; Figures 2B and 3). E. faecium W54, L. salivarius W57, L. paracasei W20, and L. casei W56 utilized in addition to these structures also the disaccharides β-d-Galp-(1 → 2)-d-Glc and β-d-Galp-(1 → 3)-d-Glc (peak 8). Strains B. lactis W51, B. lactis W53, L. salivarius W57, and L. casei W56 also consumed allolactose (peak 4). L. salivarius W57 also consumed branched trisaccharide peak 9. L. acidophilus W37 was exceptional amongst the LAB strains in utilizing a broad range of GOS molecules, similar to what was observed for the B. infantis strain DSM 20088. However, residual levels of the linear components β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-d-Glc and β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-d-Glc were still found, while B. infantis completely utilized these compounds (Figure 2A,B). The results show that LAB strains often utilized only the DP2-3 GOS molecules. This is in line with the results reported in a recent study.16 In our work, we were further able to identify the precise molecules consumed from this DP2-3 fraction by the individual LAB strains. These results show that LAB grow to a limited extent on pGOS and often only utilize the DP2-3 GOS molecules. Strain-dependent differences may occur however: Watson et al.21 studied L. casei DN-144-001 and reported the use of virtually all Vivinal GOS-derived components.
Genome Analysis
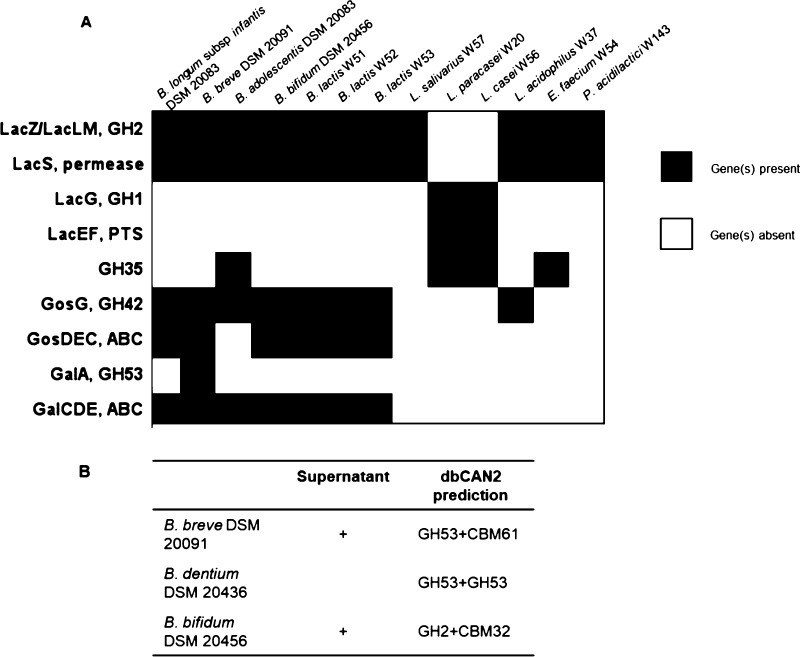
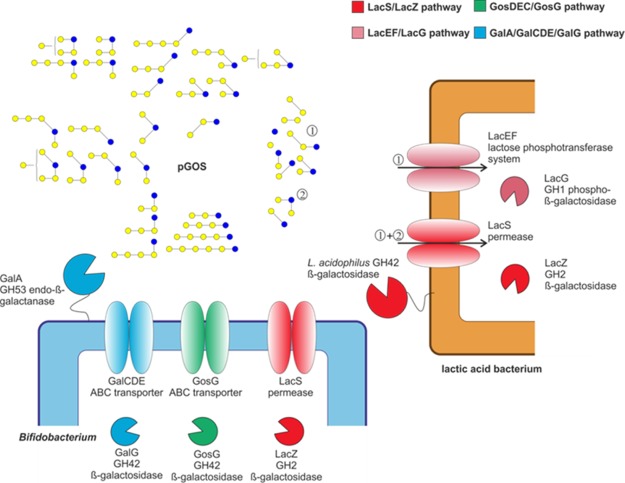
Bacterial strains employ specific catabolic pathways to utilize GOS and several possible routes were identified in bifidobacterial and lactic acid bacterial strains (Figures 4 and 5). The most common route observed for lactobacilli involves a lactose permease transporter (LacS, identified in L. acidophilus NCFM) in combination with one or more (intracellular) β-galactosidases of the family GH2 (LacZ/LacLM) (Figure 4).34 This pathway is often found in genomes of (probiotic) lactobacilli.35 A second route was identified in L. casei using a lactose phosphotransferase system (Lac_PTS, LacEF) and an intracellular family GH1 phospho-β-galactosidase (LacG).36 We searched the available genomes of the probiotic LAB strains studied here to identify the catabolic pathways that may be involved in degradation of pGOS compounds. We found that most of the LAB strains encoded the genes for the LacS/LacZ pathway; however, they differ in the total number of candidate genes for LacZ (GH2 β-galactosidase) (Figure 4). As an exception, L. paracasei W20 encoded genes for the lactose phosphotransferase system (Lac_PTS, LacEF). In the growth experiments, L. paracasei W20 reached an OD600nm clearly below the values of other strains (Figure 1B), and the structural analysis of pGOS compounds utilized by this strain revealed specific utilization of only 7 out of 40 pGOS components, mostly from the DP2 fraction. It is known that this lactose PTS is specific for lactose uptake and therefore, may explain the low number of pGOS compounds used by L. paracasei W20.14 While all other LAB strains employ genes for the LacS/LacZ pathway, the common presence of this pathway in these strains does not explain the individual differences observed in growth and pGOS utilization (Figures 1B and 3). For example, L. salivarius W57 utilized DP2 GOS, but also metabolized the branched DP3 component β-d-Galp-(1 → 4)-[β-d-Galp-(1 → 2)]-d-Glc (Figure 3). It was previously shown that the GH2 β-galactosidases associated with the LacZ/LacLM system cleaved GOS with a wide variety of glycosidic linkages including β(1 → 2,3,4,6) and DP (2–6).37 Therefore, we suggest that the GH2 β-galactosidase or the LacS transporter is the limiting factor in utilization of GOS. The substrate specificity and (3-dimensional) structural organization of this transporter needs to be further characterized within the probiotic strains in order to identify the differences that may explain differential utilization of pGOS components among probiotic LAB strains.
Figure 4.
Candidate genes involved in GOS catabolism in probiotic bacterial strains. (A) Candidate genes in known pathways for LacEF/LacG, LacS/LacZ, GosDEC/GosG, and GalA/GalCDE/GalG retrieved from BLAST searches of reference genes against bacterial genomes. The family and number of GHs annotated with dbCAN2. (B) Signal sequences for extracellular secretion searched by dbCAN2, PSORTb 3.0 (1, 2).
Figure 5.
Catabolic routes identified in Bifidobacterium and LAB to degrade β-galactooligosaccharide compounds from pGOS. Lactobacillus strains employ genes of (i) LacEF/LacG pathway to utilize mostly pGOS compounds with a similar structure to lactose (labelled as fraction “1”) or (ii) LacS/LacZ pathway to utilize mostly DP2 compounds of pGOS and certain DP3 compounds (labelled as fraction 1 and 2, respectively). Bifidobacterial strains employ (i) also the LacS/LacZ pathway to utilize DP2 compounds from pGOS and in addition (ii) GosDEC/GosG and/or the GalA/GalCDE/GalG pathway(s) to utilize GOS compounds with a higher DP.
In comparison to lactobacilli, bifidobacteria employ diverse catabolic systems to degrade GOS. In B. breve UCC2003 (and in Bacteroides thetaiotaomicron), it was demonstrated that efficient GOS utilization, particularly of higher DP compounds, correlated with the expression of a membrane-associated GH53 endo-galactanase (GalA).38,39 The gene encoding this enzyme was part of a galactan utilization operon encoding also an ABC transporter (GalCDE) and an intracellular GH42 β-galactosidase (GalG). Another GOS utilization system was identified in B. lactis Bl-04 which employs the LacS/LacZ pathway and in addition the GosDEC (ABC transporter) with GosG (intracellular GH42 β-galactosidase).40 Other strains with GH53 GalA homologues were found in B. longum but were lacking among the B. longum subsp. infantis strains.41 The strains in the present study showed variation in the set of genes potentially involved in the abovementioned pathways. The B. lactis strains W51–W53 studied here encode an enzyme and a transporter of the GosG/GosDEC pathway, in addition to the LacS pathway.40 The genome of the B. infantis strain DSM 20088 contained multiple candidate genes for the GosDEC and GalDEC transporter, but no extracellular GH53 enzyme. The lack of a GH53 enzyme, as reported for B. infantis strains,41 may explain why the strain DSM 20088 used in this study was unable to use larger GOS (Figure 3). The B. breve strain DSM 20091 encoded the highest number of genes possibly expressed as GosDEC and GalCDE transporters among all the strains tested (8 and 9, respectively, Figures 4 and 5) as well as an extracellular GH53 enzyme. Our structural analysis of the pGOS components utilized by B. breve showed that the strain DSM 20091 used 38 out of the 40 identified components and only left the DP4-branched molecules β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-[β-d-Galp-(1 → 6)]-d-Glc and β-d-Galp-(1 → 4)-[β-d-Galp-(1 → 4)-β-d-Galp-(1 → 6)]-d-Glc (Figure 3). As shown in B. breve UCC2003, the GH53 enzyme and not the GalCDE transporter was essential for GOS utilization;42 this most likely shows that the B. breve GH53 galactanase is inactive on these two components. Interestingly, the genome of B. adolescentis DSM 20083 encoded almost no genes for ABC transporters, although the strain used a broad range of pGOS molecules. We do not know whether the presence of LacS and GalDEC genes is responsible for GOS uptake in B. adolescentis DSM 20083 or that another (unidentified) system is active in this strain. B. adolescentis was the only Bifidobacterium strain in this study that encoded one family GH35 gene. Another extracellular enzyme (lacZ β-galactosidase, Acc. Nr. WP_021648433) was found in B. bifidum DSM 20456. This extracellular galactosidase may allow the strain to degrade larger GOS molecules of DP5 and DP6 (specifically utilized by this strain, Figure 3). At the same time, B. bifidum DSM 20456 did not utilize the DP4- and DP3-branched GOS (compounds nr. 10, 14–16). Together with the linearly elongated structures, β-d-Galp-(1 → 4)-β-d-Galp-(1 → 2)-d-Glc, β-d-Galp-(1 → 4)-β-d-Galp-(1 → 3)-d-Glc, and β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-d-Glc (nr. 11 and 13, Figure 3), these branched molecules may be unsuitable substrates for the extracellular GH2 enzyme of B. bifidum. The genetic organization of this strain shows similarity to that of the B. lactis strains W51–W53 (Figure 5). An additional extracellular GH2 enzyme may enable the strain to access larger DP GOS compounds that comprise linear elongated galactose residues at the nonreducing end while these molecules are not accessible for the B. lactis strains.
Overall, the diversity of utilization of GOS compounds with different DP and glycosidic linkages is reflected in the variable catabolic systems encoded in bacterial genomes. The presence or absence of a particular system together with different numbers of candidate genes reflects the differences that are observed between strains in terms of consumption of GOS molecules. We need further data to show why strains that encode the same system differ in utilization of particular GOS molecules. Further characterization (biochemically and/or 3-dimensional structure) may identify what differences at the protein level result in the use of (or inability to use) certain components.
Moreover, the role of strains encoding extracellular enzymes during potential cross-feeding on GOS should be further characterized. These strains may together with their (purified) extracellular enzymes find application in synergistic synbiotics, comprising a mixture of probiotic strains and GOS.
Conclusions
This study has characterized the GOS consumption profiles of 13 probiotic bifidobacteria and LAB strains, using a purified Vivinal GOS derived sample (pGOS), with 40 different GOS molecules, as the carbon source for anaerobic growth. For this purpose, we identified GOS compounds remaining in the medium at the stationary growth phase of these bacteria when incubated with pGOS, not only identifying the DP of the individual GOS compounds left but also their glycosidic linkage composition.
The results revealed bacterial species-dependent profiles of GOS compound utilization: the different bacterial strains examined, selectively consumed a variable number of GOS molecules. The DP4-branched compounds β-d-Galp-(1 → 4)-β-d-Galp-(1 → 4)-[β-d-Galp-(1 → 6)]-d-Glc and β-d-Galp-(1 → 4)-[β-d-Galp-(1 → 4)-β-d-Galp-(1 → 6)]-d-Glc were hardly utilized by these strains and apparently are poorly accessible for the GOS catabolic systems encoded in their bacterial genomes. GOS mixtures comprise mostly compounds that at the molecular level mimic structures close to lactose and β-galactan; thus, the GOS utilization correlated well with the different lactose uptake/degradation and/or β-galactan degradation systems genomically encoded by these strains. Dietary supplementation with this GOS mixture is likely to result in enrichment for bacterial strains encoding such catabolic systems.
At the nonreducing end, pGOS majorly comprises compounds with β(1 → 4) linked galactose. Only three strains studied were able to use the GOS DP3+ fraction, providing interesting potential selectivity. In future work we aim to characterize whether GOS mixtures enriched in β(1 → 3) or β(1 → 6) elongations at the nonreducing end are degraded similarly, involving the catabolic systems described here or whether other pathways are involved in the selective metabolism of these prebiotic GOS compounds.
The results also show that pGOS, applied as a functional ingredient in infant nutrition, stimulates growth of a broad range of probiotic bacteria. This may be advantageous in generation of a diversified gut microbiota close or beyond the composition of breast-fed infants. These data also show that synbiotic mixtures of this type of GOS with B. breve, B. infantis, and/or L. acidophilus are most likely to be successful. Although beyond the scope of this paper, such data may also be used to prepare GOS mixtures with a tailored composition to restore dysfunctional microbiome composition in patients with gut-related diseases.
Acknowledgments
Research of MCLB was performed in the public-private partnership CarboHealth coordinated by the Carbohydrate Competence Center (CCC, www.cccresearch.nl) and financed by participating partners and allowances of the TKI Agri&Food program, Ministry of Economic Affairs. ALVB was a recipient of an NWO VENI Grant.
Author Present Address
† Department of Laboratory Medicine, University Medical Center Groningen, University of Groningen, 9713 GZ, Groningen, The Netherlands.
Author Present Address
‡ CarbExplore Research BV, Zernikepark 12, 9747 AN Groningen, The Netherlands.
The authors declare no competing financial interest.
References
- Krogsgaard L. R.; Andersen L. O.; Johannesen T. B.; Engsbro A. L.; Stensvold C. R.; Nielsen H. V.; Bytzer P. Characteristics of the bacterial microbiome in association with common intestinal parasites in irritable bowel syndrome. Clin. Transl. Gastroenterol. 2018, 9, e161 10.1038/s41424-018-0027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochhar S.; Martin F.. Metabonomics and Gut Microbiota in Nutrition and Disease; Springer: London, 2015. [Google Scholar]
- McIlroy J.; Ianiro G.; Mukhopadhya I.; Hansen R.; Hold G. L. Review article: the gut microbiome in inflammatory bowel disease-avenues for microbial management. Aliment. Pharmacol. Ther. 2018, 47, 26–42. 10.1111/apt.14384. [DOI] [PubMed] [Google Scholar]
- Roychowdhury S.; Selvakumar P.; Cresci G. The role of the gut microbiome in nonalcoholic fatty liver disease. Med. Sci. 2018, 6, 47. 10.3390/medsci6020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. R.; Hutkins R.; Sanders M. E.; Prescott S. L.; Reimer R. A.; Salminen S. J.; Scott K.; Stanton C.; Swanson K. S.; Cani P. D.; Verbeke K.; Reid G. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- Bindels L. B.; Delzenne N. M.; Cani P. D.; Walter J. Towards a more comprehensive concept for prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 303–310. 10.1038/nrgastro.2015.47. [DOI] [PubMed] [Google Scholar]
- Bäckhed F.; Roswall J.; Peng Y.; Feng Q.; Jia H.; Kovatcheva-Datchary P.; Li Y.; Xia Y.; Xie H.; Zhong H.; Khan M. T.; Zhang J.; Li J.; Xiao L.; Al-Aama J.; Zhang D.; Lee Y. S.; Kotowska D.; Colding C.; Tremaroli V.; Yin Y.; Bergman S.; Xu X.; Madsen L.; Kristiansen K.; Dahlgren J.; Wang J. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Bode L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oozeer R.; van Limpt K.; Ludwig T.; Ben Amor K.; Martin R.; Wind R. D.; Boehm G.; Knol J. Intestinal microbiology in early life: Specific prebiotics can have similar functionalities as human-milk oligosaccharides. Am. J. Clin. Nutr. 2013, 98, 561S–571S. 10.3945/ajcn.112.038893. [DOI] [PubMed] [Google Scholar]
- Fanaro S.; Boehm G.; Garssen J.; Knol J.; Mosca F.; Stahl B.; Vigi V. Galacto-oligosaccharides and long-chain fructo-oligosaccharides as prebiotics in infant formulas: a review. Acta Paediatr. 2005, 94, 22–26. 10.1080/08035320510043538. [DOI] [PubMed] [Google Scholar]
- Lamsal B. P. Production, health aspects and potential food uses of dairy prebiotic galactooligosaccharides. J. Sci. Food Agric. 2012, 92, 2020–2028. 10.1002/jsfa.5712. [DOI] [PubMed] [Google Scholar]
- Fischer C.; Kleinschmidt T. Synthesis of galactooligosaccharides in milk and whey: a review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 678–697. 10.1111/1541-4337.12344. [DOI] [PubMed] [Google Scholar]
- Lombard V.; Golaconda Ramulu H.; Drula E.; Coutinho P. M.; Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal P. K.; Sullivan P. A.; Smart J. B. Utilisation of galacto-oligosaccharides as selective substrates for growth by lactic acid bacteria including Bifidobacterium lactis DR10 and Lactobacillus rhamnosus DR20. Int. Dairy J. 2001, 11, 19–25. 10.1016/s0958-6946(01)00026-7. [DOI] [Google Scholar]
- Watson D.; O’Connell Motherway M.; Schoterman M. H. C.; van Neerven R. J. J.; Nauta A.; van Sinderen D. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J. Appl. Microbiol. 2013, 114, 1132–1146. 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- Thongaram T.; Hoeflinger J. L.; Chow J.; Miller M. J. Prebiotic galactooligosaccharide metabolism by probiotic lactobacilli and bifidobacteria. J. Agric. Food Chem. 2017, 65, 4184–4192. 10.1021/acs.jafc.7b00851. [DOI] [PubMed] [Google Scholar]
- Peacock K. S.; Ruhaak L. R.; Tsui M. K.; Mills D. A.; Lebrilla C. B. Isomer-specific consumption of galactooligosaccharides by bifidobacterial species. J. Agric. Food Chem. 2013, 61, 12612–12619. 10.1021/jf403789r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittibunchakul S.; Maischberger T.; Domig K.; Kneifel W.; Nguyen H.-M.; Haltrich D.; Nguyen T.-H. Fermentability of a Novel Galacto-Oligosaccharide Mixture by Lactobacillus spp. and Bifidobacterium spp. Molecules 2018, 23, 3352. 10.3390/molecules23123352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen S. S.; Kuipers B. J. H.; Dijkhuizen L.; Kamerling J. P. Development of a 1 H NMR structural-reporter-group concept for the analysis of prebiotic galacto-oligosaccharides of the [β- d -Gal p -(1→ x)] n - d -Glc p type. Carbohydr. Res. 2014, 400, 54–58. 10.1016/j.carres.2014.08.011. [DOI] [PubMed] [Google Scholar]
- van Leeuwen S. S.; Kuipers B. J. H.; Dijkhuizen L.; Kamerling J. P. Comparative structural characterization of 7 commercial galacto-oligosaccharide (GOS) products. Carbohydr. Res. 2016, 425, 48–58. 10.1016/j.carres.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Watson D.; O’Connell Motherway M.; Schoterman M. H. C.; van Neerven R. J. J.; Nauta A.; van Sinderen D. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J. Appl. Microbiol. 2013, 114, 1132–1146. 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- Ryan S. M.; Fitzgerald G. F.; van Sinderen D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 2006, 72, 5289–5296. 10.1128/aem.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels L.; Zeikus J. G. Improved culture flask for obligate anaerobes. Appl. Microbiol. 1975, 29, 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boger M. C. L.; Lammerts van Bueren A.; Dijkhuizen L. Cross-feeding among probiotic bacterial strains on prebiotic inulin involves the extracellular exo-inulinase of Lactobacillus paracasei strain W20. Appl. Environ. Microbiol. 2018, 84, e01539 10.1128/aem.01539-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K.; Bartels D.; Best A. A.; DeJongh M.; Disz T.; Edwards R. A.; Formsma K.; Gerdes S.; Glass E. M.; Kubal M.; Meyer F.; Olsen G. J.; Olson R.; Osterman A. L.; Overbeek R. A.; McNeil L. K.; Paarmann D.; Paczian T.; Parrello B.; Pusch G. D.; Reich C.; Stevens R.; Vassieva O.; Vonstein V.; Wilke A.; Zagnitko O. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Yohe T.; Huang L.; Entwistle S.; Wu P.; Yang Z.; Busk P. K.; Xu Y.; Yin Y. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N. Y.; Wagner J. R.; Laird M. R.; Melli G.; Rey S.; Lo R.; Dao P.; Sahinalp S. C.; Ester M.; Foster L. J.; Brinkman F. S. L. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F.; Milani C.; Duranti S.; Mahony J.; van Sinderen D.; Ventura M. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol. 2018, 26, 339–350. 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Goh Y. J.; Klaenhammer T. R. Genetic mechanisms of prebiotic oligosaccharide metabolism in probiotic microbes. Annu. Rev. Food Sci. Technol. 2015, 6, 137–156. 10.1146/annurev-food-022814-015706. [DOI] [PubMed] [Google Scholar]
- Silvestroni A.; Connes C.; Sesma F.; de Giori G. S.; Piard J.-C. Characterization of the melA Locus for -Galactosidase in Lactobacillus plantarum. Appl. Environ. Microbiol. 2002, 68, 5464–5471. 10.1128/aem.68.11.5464-5471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucaud C.; Poolman B. Lactose transport system of Streptococcus thermophilus. Functional reconstitution of the protein and characterization of the kinetic mechanism of transport. J. Biol. Chem. 1992, 267, 22087–22094. [PubMed] [Google Scholar]
- Kaneko K.; Watanabe Y.; Kimura K.; Matsumoto K.; Mizobuchi T.; Onoue M. Development of hypoallergenic galacto-oligosaccharides on the basis of allergen analysis. Biosci., Biotechnol., Biochem. 2014, 78, 100–108. 10.1080/09168451.2014.877819. [DOI] [PubMed] [Google Scholar]
- Jyo T.; Kuwabara M.; Kodommari Y.; Tanemori N.; Asaoku Y.; Katsutani T.; Otsuka T.; Tsuboi S.; Ono K.; Shigeta S. Cases of immediate-type allergy in oyster shuckers due to galactooligosaccharide. J. Hiroshima Med. Assoc. 1993, 25, 19–26. [Google Scholar]
- Andersen J. M.; Barrangou R.; Abou Hachem M.; Lahtinen S.; Goh Y. J.; Svensson B.; Klaenhammer T. R. Transcriptional and functional analysis of galactooligosaccharide uptake by lacS in Lactobacillus acidophilus. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 17785–17790. 10.1073/pnas.1114152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gänzle M. G.; Follador R. Metabolism of oligosaccharides and starch in lactobacilli: a review. Front. Microbiol. 2012, 3, 340. 10.3389/fmicb.2012.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos W.; Vaughan E. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 1994, 15, 217–237. 10.1016/0168-6445(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. The structure of E. coli β-galactosidase. C. R. Biol. 2005, 328, 549–556. 10.1016/j.crvi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- O’Connell Motherway M.; Kinsella M.; Fitzgerald G. F.; van Sinderen D. Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003. Microb. Biotechnol. 2013, 6, 67–79. 10.1111/1751-7915.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böger M.; Hekelaar J.; van Leeuwen S. S.; Dijkhuizen L.; Lammerts van Bueren A. Structural and functional characterization of a family GH53 β-1,4-galactanase from Bacteroides thetaiotaomicron that facilitates degradation of prebiotic galactooligosaccharides. J. Struct. Biol. 2019, 205, 1–10. 10.1016/j.jsb.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Andersen J. M.; Barrangou R.; Abou Hachem M.; Lahtinen S. J.; Goh Y. J.; Svensson B.; Klaenhammer T. R. Transcriptional analysis of oligosaccharide utilization by Bifidobacterium lactis Bl-04. BMC Genomics 2013, 14, 312. 10.1186/1471-2164-14-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M.; Fitzgerald G. F.; van Sinderen D. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb. Biotechnol. 2011, 4, 403–416. 10.1111/j.1751-7915.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M.; Kinsella M.; Fitzgerald G. F.; van Sinderen D. Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003. Microb. Biotechnol. 2013, 6, 67–79. 10.1111/1751-7915.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]