Abstract
Lung cancer remains the most lethal cancer among men and women in the United States and worldwide. The majority of lung cancer cases are classified as non‐small cell lung cancer (NSCLC). Developing new therapeutics on the basis of better understanding of NSCLC biology is critical to improve the treatment of NSCLC. MicroRNAs (miRNAs or miRs) are a superfamily of genome‐derived, small noncoding RNAs that govern posttranscriptional gene expression in cells. Functional miRNAs are commonly dysregulated in NSCLC, caused by genomic deletion, methylation, or altered processing, which may lead to the changes of many cancer‐related pathways and processes, such as growth and death signaling, metabolism, angiogenesis, cell cycle, and epithelial to mesenchymal transition, as well as sensitivity to current therapies. With the understanding of miRNA biology in NSCLC, there are growing interests in developing new therapeutic strategies, namely restoration of tumor suppressive miRNAs and inhibition of tumor promotive miRNAs, to combat against NSCLC. In this article, we provide an overview on the molecular features of NSCLC and current treatment options with a focus on pharmacotherapy and personalized medicine. By illustrating the roles of miRNAs in the control of NSCLC tumorigenesis and progression, we highlight the latest efforts in assessing miRNA‐based therapies in animal models and discuss some critical challenges in developing RNA therapeutics.
Keywords: Cancer, miRNA, NSCLC, regulation, therapy, tumorigenesis
Abbreviations
- 3’UTR
3’‐untranslated region
- ABC
ATP‐binding cassette
- ABCB9
ATP‐binding cassette subfamily B member 9
- AGO
Argonaute
- ALK
anaplastic lymphoma kinase
- Bak
BCL2 agonist/killer
- Bax
BCL2 Associated X
- BCL
B‐cell leukemia/lymphoma
- BCL2L2
BCL2‐like 2
- Bid
BH3 interacting domain death agonist
- BRAF
v‐Raf murine sarcoma viral oncogene homolog B
- CCN
Cyclin
- Cdc
cell division cycle
- CDK
cyclin‐dependent kinase
- CTLA4
cytotoxic T‐lymphocyte‐associated protein 4
- DISC
death induced signaling complex
- E2F
E2 factor
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EML4
echinoderm microtubule‐associated protein‐like 4
- EMT
epithelial to mesenchymal transition
- ERK
extracellular‐signal‐related kinase
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- FIH
factor inhibiting HIF‐1
- FOXM
forkhead box M
- GLUT1
glucose transporter 1
- GTP
guanosine triphosphate
- HER
hormone epidermal growth factor receptor
- HGF
hepatocyte growth factor
- HGFR/MET
hepatocyte growth factor receptor
- HIF‐1
hypoxia‐inducible‐factor‐1
- HIF1AN
HIF‐1 subunit alpha inhibitor
- HK2
hexokinase 2
- HUVEC
human umbilical vein endothelial cell
- IFGR
insulin‐like growth factor receptor
- IGF
ligand insulin‐like growth factor
- KRAS
Kirsten rat sarcoma
- LDHA
lactate dehydrogenase A
- MAPK
mitogen‐activated protein kinase
- MEK
mitogen‐activated protein kinase kinase
- miRNAs or miRs
MicroRNAs
- MMP
matrix metallopeptidase
- NDUFA4
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4
- NRP
neuropilin
- NSCLC
non‐small cell lung cancer
- PD‐1
programmed cell death protein‐1
- PDK1
phosphoinositide‐dependent protein kinase‐1
- PD‐L1
PD‐1 ligand
- PI3K
phosphatidylinositol 3‐kinase
- PIGF
placenta growth factor
- PIK3CA
phosphatidylinositol‐4,5‐bisphosphate 3‐kinase catalytic subunit
- pre‐miRNA
precursor miRNA
- pri‐miRNA
primary micro‐RNA
- PTEN
phosphatase and tensin homolog
- RAC
RAS‐related C3 botulinum toxin substrate
- RAF
v‐Raf murine sarcoma
- RAS
rat sarcoma
- Rb
retinoblastoma tumor suppressor
- RhoA
Ras homolog family protein A
- RISC
RNA‐induced silencing complex
- ROCK
Rho‐associated protein kinase
- SDHD
dehydrogenase complex, subunit D
- SMAD
mothers against decapentaplegic
- SOX
Sry‐related HMG box
- STAT
signal transducer and activator of transcription
- TGFβ
transforming growth factor beta
- TGFβR
TGFβ receptor
- TIMP
tissue inhibitor of metalloproteases 3
- TNF
tumor necrosis factor
- TNFR
TNF receptors
- VEGF
vascular endothelial growth factor
- VEGFR
VEGF receptor
- vHL
von Hippel Lindau
- XPO5
Exportin‐5
- ZEB
zinc finger e‐box‐binding homeobox
1. INTRODUCTION
Lung and bronchus cancer is the second most commonly diagnosed cancer in the United States, with over 220,000 estimated new diagnoses in 2019, accounting for almost 13% of all cancer diagnoses.1 One in 15 men and 1 in 17 women will be diagnosed with lung cancer during their lifetime and the average age at diagnosis is 70 years.1 Lung cancer accounts for the highest cancer‐related deaths in the United States, causing 23% of all cancer‐related deaths which is more than colon, breast, and prostate cancers combined.1 The five‐year survival rate of all lung cancer diagnoses is 19%, which is lower than colon, breast, and prostate cancers. When the disease is detected while still localized to the lung, the five‐year survival rate is 56%; however, only 16% of cases at diagnosed at that stage. By contrast, the five‐year survival rate for metastatic lung cancer patients is only 5%. More than half of patients diagnosed with lung cancer will die within one year. It is also estimated that lung cancer care may be increased to 173 billion dollars in 2020 in the United States.2
The lung epithelium undergoes a series of morphological changes before becoming invasive, such as hyperplasia, metaplasia, and finally dysplasia and carcinoma in situ. Lung cancer is classified by the site of origin, and method of diagnosis, prognosis, and treatment. The two main types of lung cancer are small cell lung cancer, accounting for 15% of all cases, and non‐small cell lung cancer (NSCLC) which is any type of epithelial lung cancer, and accounts for 80% to 85% of all cases (Figure 1).3 The three most common histological forms of NSCLC are epidermoid or squamous cell carcinoma, large cell carcinoma, and adenoma; among them adenocarcinoma accounts for 40% of all lung cancer cases.4 Squamous cell carcinoma occurs inside the airways, adenocarcinoma occurs in the cells lining the alveoli located in the outer part of the lungs, and large cell carcinoma is in any other part of the lung. A major risk factor for NCSLC is smoking;5 other risks include secondhand smoke, radiation exposure, air pollution, family history, and human immunodeficiency virus infection. Lung cancer may present as a persistent cough, chest pain, weight loss, malaise, difficulty breathing, pleural effusion, pneumonia, chronic obstructive pulmonary disease or pulmonary fibrosis.6 Diagnostic procedures include sputum cytology, tissue biopsy and imaging tests such as bronchoscopy, X‐ray, MRI, positron emission tomography, or computed tomography scan.7 Lung cancer is staged based on the size of the primary tumor, the involvement of the lymph nodes, and the presence of distant metastasis.8
Figure 1.
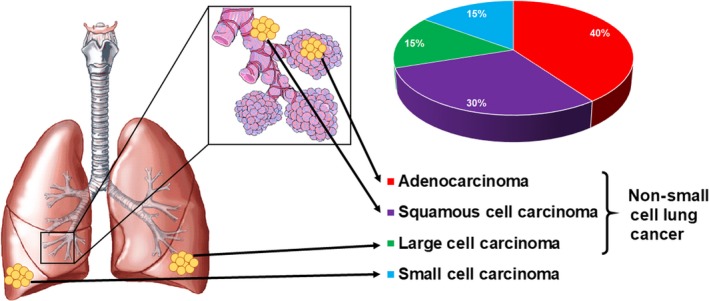
Lung cancer classifications and frequency of diagnosis. There are two main types of lung cancer: small‐cell lung cancer and non‐small cell lung cancer. Small‐cell carcinoma occurs in the outer edges of the lungs and accounts for about 15% of all cases. Non‐small cell lung cancer (NSCLC) makes up 85% of all lung cancer cases, and can be further classified into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. Adenocarcinoma, the most common NSCLC subtype, occurs in the cells lining the alveoli, squamous cell carcinoma is generally found in the airways or bronchi, and large cell carcinoma is in the edges of the lungs
2. MOLECULAR FEATURES OF NSCLC
Molecular features of NSCLC tumors may not only predict the prognosis and outcome the cancer but also serve as targets for therapies. The most frequent mutations in NSCLC occur in the TP53 gene, occurring in about 50% of NSCLC cases (Table 1).9 Mutations in EGFR, a tyrosine kinase receptor, account for 10%‐35% of cases and can cause dysfunction of the AKT and MAPK signaling which enhances cell survival and stimulates proliferation.10 The most common mutations of EGFR are in‐frame deletions of exon 19, and the second most common EGFR mutation is single nucleotide substitutions L858R in exon 21.11 The most common mutation detected after treatment with EGFR inhibitors is T790M in exon 20 which can confer drug resistance.12 The third most frequent mutations occur in KRAS, accounting for 15%‐25% of cases.13 Usually mutations in KRAS and EGFR are mutually exclusive and non‐overlapping. Another common molecular feature of NSCLC is the presence of ALK fusion gene, which encodes a receptor tyrosine kinase not normally expressed in the lung.13 At least nine different variants of fusion of ALK with an upstream partner EML4 have been identified causing constitutive activation of the kinase.13 The HER2 protein, a HER family receptor tyrosine kinase, is overexpressed in 20% of all NSCLC and gene amplification occurs in 2%.14, 15 These mutations commonly lead to constitutive activation of the HER2 signaling pathway.16 Mutations in the main catalytic subunit, PIK3CA, of phosphatidylinositol 3‐kinase occur in about 2% of NSCLC cases.17 These tumors can activate the protein kinase B signaling pathway without growth factors. Protein kinase B is encoded by AKT1, which is mutated from a glutamate to a lysine at position 17 in 1% of NSCLC cases and causes PI3K‐independent activation of protein kinase B.18, 19 BRAF is a member of the RAF kinase family which confers signaling of the MAPK family from the RAS GTPases to control cell proliferation.20 BRAF mutations are mostly found in adenocarcinomas and lead to higher kinase activity and constitutively active MAPK2 and MAPK321. MAPK2 and MAPK3 are downstream of BRAF and three mutations in the non‐kinase portion of these proteins have been found in cancer.22 Amplification of the gene MET, which codes hepatocyte growth factor receptor (HGFR) causes resistance to EGFR tyrosine kinase inhibitors.23 The molecular heterogeneity of NSCLC tumors increases the complexity of treatment for NSCLC patients.
Table 1.
Common genetic alterations and their prevalence in NSCLC
| Gene | Alteration | Prevalence in NSCLC |
|---|---|---|
| TP53 | Mutation | 50% |
| EGFR | Mutation | 10%‐35% |
| KRAS | Mutation | 15%‐27% |
| ALK | Fusion | 5%‐15% |
| HER2 | Overexpression/ Gene amplification | 20%/ 2% |
| PIK3CA | Mutation | 2%‐8% |
| AKT1 | Mutation | 1% |
| BRAF | Mutation | 5% |
| MET | Mutation | 5% |
3. CURRENT TREATMENTS FOR NSCLC PATIENTS
Lung cancer that is diagnosed at the early stages is commonly treated with resection surgery or lobectomy, chemotherapy, and radiation. Surgery may range from removing an entire lung to removing part of a lobe, depending on the size and location of the tumors. Radiation, alone or concurrent with surgery or chemotherapy is also commonly utilized. External beam radiation therapy, the most widely used form of radiation for NSCLC, consists of administering 1.5 to 2.5 Gray to the lungs usually 5 days a week for 5‐7 weeks, while stereotactic radiotherapy consists of a larger dose, around 22 Gray, in usually fewer than 5 doses.24 Brachytherapy, or internal radiation therapy, involves the inserting a radioactive pellet in or near the tumor for a short amount of time or permanently. The radiation stays localized and gets weaker over time. Radiofrequency ablation uses radio waves emitted from a probe guided by a computed tomography scan.
Molecular medicine or pharmacotherapy spans from chemotherapy to targeted therapy and the most recent immunotherapy, which utilize small‐molecule and protein or antibody drugs (Table 2). Commonly used chemotherapies for the treatment of NSCLC include cisplatin, carboplatin, docetaxel, paclitaxel, pemetrexed, and vinorelbine that usually interfere with DNA synthesis or replication to achieve the inhibition of cancer cell proliferation and growth (Table 2). Nevertheless, chemotherapy may not be effective for all patients and many cancers will eventually become resistant to the drugs. Furthermore, chemotherapy kills cancer cells less specifically and could cause some side effects like pain, nausea, vomiting, blood disorders, and hair‐loss.
Table 2.
List of drugs approved in the US for the treatment of NSCLC and their molecular targets or mechanistic actions
| Treatment | Classification | Target or Action | Approval | Overall Response Rate |
|---|---|---|---|---|
| Bevacizumab | antibody/protein | VEGF | Non‐squamous NSCLC | 35% with carboplatin and paclitaxel 150 |
| Ramucirumab | antibody/protein | VEGFR | Metastatic non‐squamous NSCLC | 23% with docetaxel 151 |
| Erlotinib | small molecule | EGFR | EGFR L858R mutation, metastatic NSCLC | 74.4% 152 |
| Necitumumab | antibody/protein | EGFR | Metastatic squamous NSCLC | 48.1% with cisplatin and gemcitabine 153 |
| Gefitinib | small molecule | EGFR | Advanced or metastatic NSCLC with L858R EGFR mutations | 76.9% 152 |
| Afatinib | small molecule | EGFR | Metastatic squamous NSCLC with non‐resistant EGFR mutations | 56% 154 |
| Osimertinibe | small molecule | EGFR T790M mutations | Advanced or metastatic NSCLC with T790M EGFR mutations | 77% 155 |
| Crizotinib | small molecule | ALK/CD246, ROS | Advanced or metastatic ALK‐positive NSCLC | 74% 156 |
| Ceritinib | small molecule | ALK/CD246 | Metastatic ALK‐positive NSCLC | 58% 157 |
| Brigatinib | small molecule | ALK/CD246 | Metastatic ALK‐positive NSCLC | 71% 158 |
| Alectinib | small molecule | ALK/CD246, RET | Metastatic ALK‐positive NSCLC | 82.9% 159 |
| Dabrafenib | small molecule | B‐Raf | Metastatic NSCLC with B‐Raf V600E mutation | 67% in combination with trametinib 160 |
| Trametinib | small molecule | MEK | Metastatic NSCLC with B‐Raf V600E mutation | See dabrafenib |
| Entrectinib | small molecule | ROS1/NTRK fusion | Metastatic, ROS1/NTRK‐positive NSCLC | 78% 161 |
| Nivolumab | antibody/protein | PD‐1/CD279 | Metastatic squamous NSCLC | 47% in patients with a high tumor‐mutation burden 162 |
| Pembrolizumab | antibody/protein | PD‐1/CD279 | Advance or metastatic squamous NSCLC | 44.8% 163 |
| Atezolizumab | antibody/protein | PD‐L1/CD274/B7‐H1 | Metastatic non‐squamous NSCLC | 63.5% with bevacizumab, carboplatin, and paclitaxel in patients with no EGFR or ALK alterations 164 |
| Ipilimumab | antibody/protein | CTLA4/CD152 | Metastatic NSCLC | 45.3% with nivolumab 165 |
| Carboplatin & cisplatin | small molecule | Inhibition of DNA replication | Advanced or metastatic NSCLC | 62% carboplatin with paclitaxel 166 |
| Irinotecan | small molecule | Topoisomerase I | Advanced NSCLC | 43.7% with cisplatin 167 |
| Etoposide | small molecule | Topoisomerase II | Metastatic NSCLC | 21.9% with cisplatin 168 |
| Docetaxel | small molecule | Microtubules; inhibition of mitosis | Advanced NSCLC | 9% in patients previously treated with chemotherapy 169 |
| Paclitaxel | small molecule | Tubulin; inhibition of mitosis | Advanced or metastatic NSCLC | See carboplatin & cisplatin |
| Vinorelbine | small molecule | Tubulin; inhibition of mitosis | Advanced NSCLC | 43% with cisplatin 170 |
| Vinblastine | small molecule | Microtubule; inhibition of mitosis | Advanced NSCLC | 41% with cisplatin 171 |
| Pemetrexed | small molecule | Thymidylate synthase, dihydrofolate reductase | Advanced NSCLC | 9.1 in patients previously treated with chemotherapy 172 |
| Gemcitabine | small molecule | Inhibition of DNA synthesis | Advanced or metastatic NSCLC | 40.6% with cisplatin 168 |
Pharmacotherapy for NSCLC has been benefited greatly by the development of targeted and personalized medications, either small molecules or antibodies, which act more selectively on particular molecular targets including transmembrane and cell surface proteins or receptors (eg, EGFR, PD‐1, PD‐L1, etc) as well as signal proteins (eg, cytokines, VEGF) and cytoplasmic kinases (eg, MEK) (Table 2). While all the therapies listed in Table 2 are approved in the United States, many are also approved in Europe and elsewhere. The response rate for most of the therapies is generally consistent across subtypes with higher rates for tumors with high mutational burden. The effectiveness of two antibody drugs, anti‐VEGF bevacizumab and anti‐VEGFR ramucirumab, is attributable to the inhibition of angiogenesis.25 NSCLC patients with an overexpression of EGFR mRNA or increased copy number have a 70% or higher response rate to small‐molecule EGFR inhibitors like gefitinib or EGFR antibodies like necitumumab.26 Furthermore, some targeted therapies are approved for specific subsets of NSCLC patients. Osimertinib is used to treat NSCLC patients with T790M mutations of EGFR. The EML4‐ALK tumors are mostly responsive to small‐molecule tyrosine inhibitors of ALK like crizotinib. Dabrafenib and trametinib, which target BRAF and MEK1/2, respectively, are prescribed for patients with BRAF V600E mutations.27 Immunotherapies such as PD‐1 and PD‐L1 antibodies (eg, nivolumab and atezolizumab) are also effective for the treatment of some NSCLC patients regardless of the subtype.28 While targeted and immunotherapies are generally less toxic and personalized for particular patients, some patients do exhibit primary or acquires resistance 29, 30 or show severe adverse effects such as diarrhea and pneumonitis.31, 32, 33 In addition, targeted therapies have the greatest response rate for patients with the indicated mutation, therefore, due to the high heterogeneity of mutations within NSCLC, targeted therapies may not work in every patient. Large efforts are underway to advance the understanding of NSCLC biology and assess novel therapies.
4. GENOME‐DERIVED MICRORNAS ARE DYSREGULATED IN NSCLC
As less than 5% of the human genome is processed to functional proteins in cells, the majority is transcribed into enormous numbers of functional noncoding RNAs. Among them, microRNAs (miRNAs or miRs) are a superfamily of short RNAs that act on corresponding transcripts via complementary binding to achieve mRNA degradation or translation inhibition 34 (Figure 2). The biogenesis of miRNAs starts with the transcription of miRNA‐coding genes into primary miRNA (pri‐miRNA) transcripts. The pri‐miRNA is thus processed by the Drosha‐DGCR8 complex within the nucleus to produce a precursor miRNA (pre‐miRNA) that can be exported into the cytoplasm by Ran‐GTP‐dependent Exportin‐5 (XPO5). The pre‐miRNA is cleaved into a miRNA duplex by the RNase Dicer in the cytoplasm 35 (Figure 2). The miRNA duplex is then unwound to offer two strands, among which the guide strand is preferably incorporated into the RNA‐induced silencing complex (RISC) consisting of the Argonaute family of proteins while the passenger strand is readily degraded.36, 37 The RISC proteins stabilize and aid the mature miRNA in binding to the 3’‐untranslated region (3’UTR) of a target transcript to accomplish the regulation of target gene expression (Figure 2).
Figure 2.
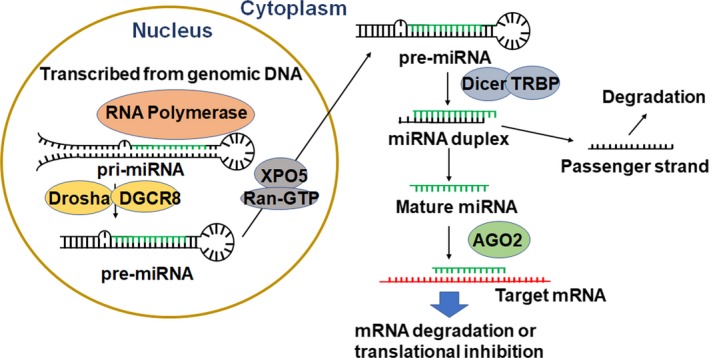
MicroRNAs are derived from the genome to control target gene expression through their actions on mRNAs. Transcribed from the genomic DNA by RNA polymerase as primary miRNA (pri‐miRNA) and subsequently processed by the Drosha/DGCR8 complex to a shorter form within the nucleus, the resultant precursor miRNA (pre‐miRNA) is transported into the cytoplasm by Ran‐GTP‐dependent Exportin‐5 (XPO5) and further processed by Dicer and TRBP to miRNA duplex. Unwinding of the duplex offers two strands, among which the passenger strand is readily degraded while the mature miRNA, guided by argonaut‐2 (AGO2), acts on target transcript through complementary binding and leads to mRNA degradation or translational inhibition
Many miRNAs are involved in the control of target gene expression behind various cancer cellular processes (see the following section), exhibiting tumor suppressive or promotive activities. Specifically, a miRNA that reduces the expression of tumor suppressors acts as a tumor promotor, and a miRNA that degrades oncogene transcripts functions as a tumor suppressor. Interestingly, some miRNAs are dysregulated in NSCLC (Table 3) that may be indicative of disease status or therapeutic outcome. With some exceptions, generally there is a decrease of tumor suppressive miRNAs (eg, miR‐34a‐5p and miR‐124‐3p) and increase of tumor promotive miRNAs (eg, miR‐21‐5p and miR‐183‐5p) in many human cancers including NSCLC (Table 3), as compared to normal tissues; however, the magnitude of dysregulation varies by case. Dysregulation of miRNA expression may be caused by different mechanisms such as chromosomal deletion or methylation, or dysregulation of their transcription factors, enzymes, or binding proteins involved in miRNA biogenesis. Dicer, the RNase responsible for the processing of pre‐miRNAs, is essential for mouse development and stem cell maintenance.38 Dicer was reported to be downregulated in some lung cancer patients, leading to a global decrease in miRNAs and associated with poor prognosis,39 and conditional deletion of Dicer led to increased lung tumorigenesis in mice.40 Actually, the role of Dicer in cancer remains contradictive.41 It has been suggested that a partial loss or downregulation of Dicer is oncogenic, while a complete loss is tumor protective. In addition to Dicer, the expression of some miRNAs is also modulated by the tumor suppressive transcription factor p53.42 During DNA damage, p53 associates with the Drosha/DGCR8 complex and facilitates processing of pri‐miRNA to pre‐miRNA. p53 is mutated and inactivated in many cancers, including lung,43 reducing the total levels of pre‐miRNAs. Taken together, a global decrease in miRNA levels, by either decreased Dicer expression or loss of p53, might be involved in tumor initiation and progression. Ultimately, restoration of tumor suppressive miRNAs and inhibition of tumor promotive miRNAs represent new anti‐cancer strategies.
Table 3.
List of miRNAs most commonly dysregulated in NSCLC and some of their corresponding targets validated by biological experiments
| miRNA | Expression | Direct Targets Verified | References |
|---|---|---|---|
| miR‐124‐3p | Decreased | STAT‐3, MYO10, SMAD4 | [173, 174, 175, 176] |
| miR‐126‐3p | Decreased | PIK3R2, VEGF‐A, Crk | [77, 120, 177, 178, 179] |
| miR‐143‐3p | Decreased | KRAS, NRAS, BCL2, HK2, PKCε, Limk1, ATG2B | [112, 180, 181, 182, 183, 184, 185, 186] |
| miR‐34a‐5p | Decreased | TGFβR2, Cyclin E1, PEBP4, Notch‐1, Axl | [187, 188, 189, 190, 191, 192] |
| let‐7c‐5p | Decreased | N/K‐RAS, ABCC2, Bcl‐XL, ITGB3, MAP4K3, HOXA1 | [122, 193, 194, 195, 196] |
| miR‐101‐3p | Decreased | ZEB1, ROCK2, MALAT‐1, EZH2 | [48, 180, 197, 198, 199] |
| miR‐100‐5p | Decreased | FGFR3, PLK1 | [67, 180, 200, 201] |
| miR‐181a‐5p | Decreased | BCL2, KRAS, VCAM‐1, CDK1 | [99, 180, 202, 203, 204] |
| miR‐145‐5p | Decreased | c‐Myc, SMAD3, AEG/MTDH, OCT4, SOX2, Fascin1 | [138, 202, 205, 206, 207] |
| miR‐486‐5p | Decreased | PIM‐1, ARHGAP5, IGF1, IGFR, p85α, CDK4 | [62, 63, 208, 209, 210] |
| miR‐451a‐5p | Decreased | PSMB8, RAB14 | [175, 180, 211] |
| miR‐21‐5p | Increased | PTEN | [180, 212] |
| miR‐210‐3p | Increased | E2F3, NDUFA4, SDHD | [79, 213] |
| miR‐205‐5p | Increased | PTEN, PHLPP2, ITGα5 | [131, 214, 215, 216] |
| miR‐31‐5p | Increased | ABCB9, hMLH1 | [121, 180, 217] |
| miR‐200b‐5p | Increased | FOXF2, IL‐8, CXCL1, FSCN1 | [129, 218, 219, 220] |
| miR‐182‐5p | Increased | PDCD4, RGS17 | [213, 221, 222, 223] |
| miR‐183‐5p | Increased | FOXO1, VIL2 | [134, 213, 224] |
5. MICRORNAS ARE INVOLVED IN THE CONTROL OF MULTIPLE NSCLC CELLULAR PROCESSES
5.1. Epithelial to Mesenchymal Transition
The epithelial to mesenchymal transition (EMT) is a process in which an epithelial‐like cell loses its attachment to the basal membrane and assumes mesenchymal characteristics like greater motility and invasiveness. EMT allows for cancer cells to metastasize by migrating from the primary tumor through the blood stream and invading other organs. Comprehensive reviews of the process of EMT have recently been published.44 Some miRNAs can affect cells’ ability or likelihood to undergo EMT by regulating the expression of EMT‐related genes. One important EMT signaling cascade involves TGFβ, is a signaling cytokine, that binds to its receptor, TGFβR1/2 to transduce a signal through RAS, PI3K, RhoA/ROCK, or Smad2/3 and activate transcription factors, like ZEB1/2, Twist and Snail. This ultimately results in the loss of epithelial attachment proteins, such as E‐cadherin, and gain of intermediate filament or cell‐cell adhesion proteins, such as vimentin and N‐cadherin.45 The EMT phenotype is reduced in NSCLC by the action of miR‐17‐5p, 20a‐5p, and 20b‐5p that directly target TGFβR2,46 miR‐148a‐3p and miR‐101‐3p that target ROCK1 and ROCK2 respectively,47, 48 and miR‐132‐3p, miR‐638‐5p, and miR‐338‐5p which target transcription factors ZEB2, SOX2, and SOX4, respectively 49, 50, 51 (Figure 3). Both miR‐149‐5p and miR‐509‐3p target the transcription factor FOXM1 to reduce invasion in H1299 cells as determined by Matrigel invasion assay.52 In addition, miR‐186‐5p targets CDC42 leading to the inhibition of migration and related EMT processes.53 Likewise, dysregulation of these miRNAs, as evident in NSCLC, leads to greater EMT and a more invasive, migratory, and potentially metastatic phenotype.
Figure 3.
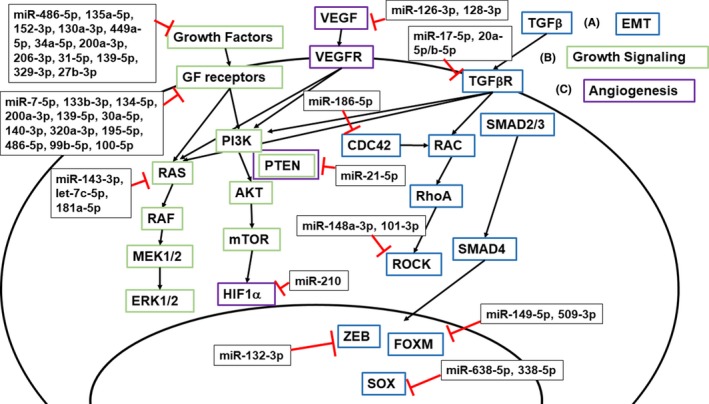
MicroRNAs modulate many cancer cellular processes important in tumor initiation, progression, and metastasis. (A) The epithelial to mesenchymal transition (EMT) is driven in part by transforming Growth Factor β (TGFβ) binding to the TGFβ Receptor (TGFβR) and signaling to RAC and activating RhoA and ROCK. SMAD2/3 signaling activates transcription factors ZEB, FOXM, and SOX to turn on transcription of other genes necessary for EMT. Some miRNAs that target TGFβR, CDC42, ROCK, or the downstream transcription factors are dysregulated in NSCLC, which can increase EMT signaling and therefore enhance cancer cell invasion and metastasis. (B) Growth factors (GF), such as epidermal growth factor (EGF), bind to corresponding growth factor receptors, such as EGF receptor (EGFR), to activate RAS or PI3K. This leads to a series of signal transductions that eventually enhance cancer cell proliferation and growth. Inhibition of growth factors and their receptors by miRNAs may inhibit tumor progression. (C) One mechanism behind angiogenesis involves vascular endothelial growth factor (VEGF) binding to VEGF Receptor (VEGFR) and activating hypoxia‐inducible factor‐1α (HIF1α). Those miRNAs that target VEGF or HIF1α may reduce angiogenesis essential for tumor progression
5.2. Signal transduction in lung cancer survival and proliferation
Oncogenesis is driven by an over‐expression or activation of growth signaling, such as growth factors, receptors, or downstream signaling molecules. Growth factor ligands bind to their corresponding receptors to relay a signal and induce proliferation. Cancer cells can hijack signaling by over‐expressing or mutating growth factor receptors to increase proliferative signals and miRNAs target certain receptor to modulate signaling. miR‐7‐5p,54 miR‐133b‐3p,55 miR‐134‐5p,56 and miR‐200a‐3p 57 target epidermal growth factor (EGFR) (Figure 3), which is commonly overexpressed in NSCLC (Table 1), to alter downstream signaling molecules such as AKT and ERK1/2 and decrease the growth phenotype. In addition, miR‐139‐5p,58 miR‐30a‐5p,59 miR‐140‐3p,60 miR‐320a‐3p 61 and miR‐195‐5p 62 all target insulin‐like growth factor 1 receptor (IGF1R), while miR‐486‐5p directly targets both IGF1R and its ligand insulin‐like growth factor 1 (IGF1) 63 and miR‐135a‐5p targets only IGF1.64 miRNA inhibition of the IGF pathway results in a lower proliferation as assayed by CCK‐8 kit among others.60 miR‐152‐3p targets fibroblast growth factor 2 (FGF2) 65 and miR‐99b‐5p 66 and miR‐100‐5p 67 target fibroblast growth factor receptor 3 (FGFR3). MET is a receptor for the hepatocyte growth factor (HGF) and is targeted by a number of miRNAs including miR‐130a‐3p,68 miR‐449a‐5p 69 miR‐34a‐5p,70 miR‐200a‐3p,57 miR‐206‐3p,71 miR‐31‐5p,72 miR‐139‐5p,73 miR‐329‐3p,74 miR‐27b‐3p 75 to decrease growth and proliferation. EGF, IGF, FGF, and HGF signaling results in activation of RAS or PI3K pathways and downstream growth signaling. A decrease in the miRNAs that target growth factors and their receptors, as well as downstream targets, as evident in NSCLC, results in an increase in growth signal transduction and an increase in cancer cell proliferation and growth.
5.3. Angiogenesis
Angiogenesis is the process of building new blood vessels for nutrients and gas exchange which is essential for cancer cells to survive and proliferate. Dysregulation of miRNAs in cancer cells can lead to increased angiogenesis through multiple pathways, including vascular endothelial growth factor (VEGF) or placenta growth factor (PIGF) binding to VEGF receptor (VEGFR) or neuropilin (NRP).76 During normal conditions, tumor suppressor von Hippel Lindau (vHL) mediates the degradation of hypoxia‐inducible‐factor‐1 (HIF1) through the ubiquitin‐proteasome pathway. By contrast, HIF‐1α associates with HIF1β during hypoxia and thus increases VEGF transcription by binding to the promoter.76 miRNAs regulate many important factors of angiogenesis (Figure 3). For example, miR‐126‐3p and miR‐128‐3p, both of which are commonly decreased in NSCLC, directly target VEGF‐A and VEGF‐C, respectively, to decrease angiogenesis and blood vessel formation, as measured by tube formation assay.77, 78 miR‐210‐3p is overexpressed in late‐stage NSCLC and protects against hypoxia induced apoptosis by indirectly stabilizing HIF1α to promote angiogenesis and increase glycolysis.79 miR‐21‐5p directly targets PTEN and activates AKT and ERK1/2 which leads to higher levels of HIF1α and VEGF expression.80 miR‐378‐5p is over‐expressed in NSCLC tumors in patients with brain metastasis and leads to increased VEGF expression and angiogenesis.81 miR‐206‐3p directly suppresses the expression of protein 14‐3‐3ζ which consequently decreases VEGF, HIF1α, and phosphorylated STAT3 and results in a lower degree of angiogenesis as assayed by HUVECs recruitment as well as inhibition of intratumoral capillary tube formation in vivo.82 In one study, coculture of NSCLC cell lines with vascular endothelial cells leads to higher levels of miR‐494‐3p in the vascular endothelial cells, in addition, a miR‐494 antagomir decreases tumor vascularization, suggesting that miRNAs may be transferred to vascular endothelial cells to control angiogenesis.83 Such miRNAs are important in the control and development of vascularization which is critical for tumorigenesis and metastasis.
5.4. Cell Cycle
The cell cycle is altered among almost all cancer cells to allow for uncontrolled growth. Cyclins and cyclin‐dependent kinases (CDKs) are partly responsible for entry into the different cell cycle stages. G1 begins with cyclins D1, D2, and D3 associating with CDK4 and 6 84 to phosphorylate Rb and repress the E2F transcription factor.85 G1/S transition is characterized by cyclin E complexing with CDK2 86 and cyclin A/CDK2 complex during S phase.87 Cyclin A complexes with CDK1 to transition to M phase, then cyclin B and CDK1 are complexed during M phase.88 Such proteins regulating G1 and S phases of many types of cancers, including NSCLC, are dysregulated, and some are direct targets of particular miRNAs (Figure 4 and Table 3). Tumor suppressive miR‐34a‐5p directly targets CCND1 and CDK6, leading to the arrest of the cell cycle in G1 phase.89 Furthermore, miR‐15a‐5p and miR‐16‐5p, down‐regulated in NSCLC, directly targets CCND1, CCND2, and CCNE1, and arrests the cell cycle in G1 to G0 in an Rb‐dependent manner.90 Combination of miR‐34a‐5p and miR‐15a/16 produces synergism in G1 cell cycle arrest in an Rb‐dependent manner due to an increase in miRNAs targeting more cell cycle related mRNAs.91 In addition, miR‐30d‐5p targets CCNE2,92 miR‐186‐5p targets CCND1, CDK2, and CDK6,93 and miR‐129‐5p targets CDK6 94 to arrest the cells in G1. Generally, cell cycle‐regulating miRNAs exhibit tumor suppressive actions by targeting cell cycle promotive genes to induce a cancer cell cycle arrest.
Figure 4.
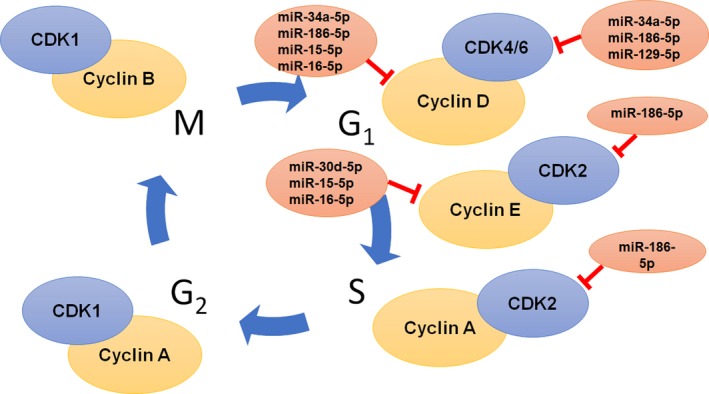
MicroRNAs directly target core cell cycle regulators. Some miRNAs, such as miR‐34a‐5p, miR‐186, and miR‐15, which regulate the expression of cell cycle regulators, are downregulated in NSCLC. This causes dysregulation of the cell cycle and ultimately increases cancer cell proliferation. Restoration of such tumor suppressive miRNAs may lead to cell cycle arrest to achieve anticancer effects
5.5. Evading Apoptosis
Cell death mechanisms, including apoptosis or necrosis, are important for cells to maintain homeostasis, and dysregulation of these pathways leads to an alteration of cell proliferation, including NSCLC cells. Some miRNAs regulate certain proteins involved in cell death (Figure 5), and dysregulation of such miRNAs may make the cells evade death signals and continue to proliferate. In brief, apoptosis occurs when a death ligand, such as tumor necrosis factor (TNF)‐related apoptosis‐inducing ligand (TRAIL), binds to a death receptor, including TNF receptors 1 and 2 (TNFR1/2), causing receptor multimerization and activation of the death induced signaling complex (DISC).95 This can result in direct activation of caspase‐8 mediated cleavage of effector caspases, like caspase‐3,96 or caspase‐8 cleavage of Bid which releases mitochondrial cytochrome c to associate with Bax and Bak and forms the apoptosome and cleaves effector caspases.97 BCL2, an anti‐apoptotic protein that mainly functions to inhibit release of cytochrome c from the mitochondria,98 is targeted by miR‐181‐5p,99 miR‐7‐5p,100 miR‐503‐5p,101 miR‐200bc/429 cluster,102 and miR‐497‐5p 103 (Figure 5). In addition, BCL2L2 and BCL6, also anti‐apoptotic proteins, are directly targeted by miR‐15a‐5p and miR‐187‐3p, respectively, to enhance apoptosis.104, 105 TRAIL expression induces apoptosis; however, NSCLC can confer resistance to TRAIL‐mediated apoptosis through many mechanisms including loss of PTEN and constitutive activation of AKT 106 or increased matrix metalloproteases.107 Therefore, over‐expression of miR‐148a‐3p can sensitize NSCLC to TRAIL by targeting MMP15.108 miR‐221‐3p and miR‐222‐3p can confer TRAIL resistance by targeting tumor suppressors PTEN and tissue inhibitor of metalloproteases 3 (TIMP3),109 while miR‐130a‐3p can reverse this effect by targeting miR‐221‐3p and miR‐222‐3p.68 miR‐21‐5p also targets PTEN and results in the inhibition of apoptosis, which can be reversed by the transfection of anti‐miR‐21‐5p.110
Figure 5.
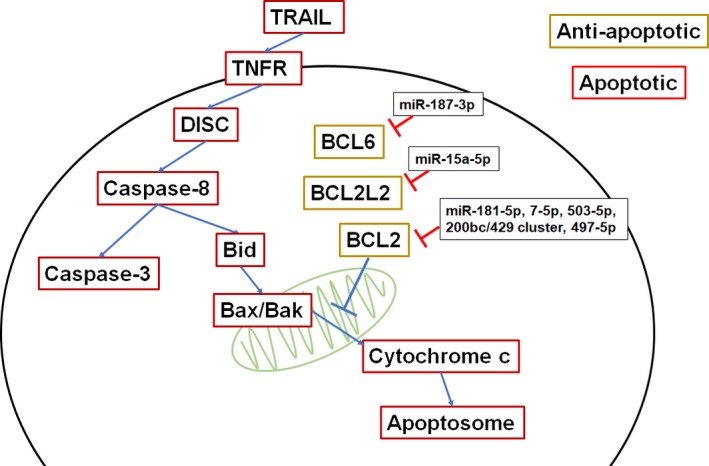
miRNAs affect the ability to evade of apoptosis. BCL6, BCL2L2, and BCL2 are anti‐apoptotic as they inhibit cytochrome c release from the mitochondria. Dysregulation or malfunction of miRNAs in NSCLC that inhibit the anti‐apoptotic cascade may reduce apoptotic capacity and enhance cancer progression. Therefore, restoration of such miRNA expression or function represents a novel therapeutic strategy
5.6. Metabolism
Some miRNAs can alter the metabolic potential of cancer cells. A higher metabolic rate may enhance the tumorigenesis and growth of NSCLC cells. miR‐155‐5p promotes aerobic glycolysis by indirectly upregulating HK2, as determined by a hexokinase colorimetric assay as well as glucose and L‐lactate test kits.111 The increase in glycolysis leads to greater degree of cell viability. miR‐143‐3p directly targets HK2, the first rate‐limiting enzyme in glycolysis, to decrease glycolysis and proliferation as well as tumorigenesis in vivo.112 miR‐124‐3p overexpression decreases glucose consumption, lactate production, and ATP content by decreasing HK2 and glucose transporter 1 (GLUT1), leading to a lower extent of cell proliferation.113 miR‐449‐5p directly targets lactate dehydrogenase A (LDHA) and suppresses glycolysis.114 miR‐182‐5p and miR‐31‐5p target HIF1AN and FIH, respectively, both of which are HIF‐1α inhibitors and lead to enhanced glycolysis.115, 116 miR‐210‐3p directly targets two genes important in the electron transport chain, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4 (NDUFA4), and succinate dehydrogenase complex subunit D (SDHD), which leads to alteration of the physical structure of the mitochondria, visualized by electron microscopy as well as alterations of the mitochondrial membrane potential that are phenotypic of mitochondria dysfunction.79 miR‐145‐5p and miR‐138‐5p directly target phosphoinositide‐dependent protein kinase‐1 (PDK1), an important enzyme in glucose and fatty acid metabolism.115, 117 Dysregulation or loss of those miRNAs that inhibit the metabolic potential of NSCLC cells may alter tumor progression.
6. MICRORNAS AFFECT THE SENSITIVITY OF NSCLC CELLS TO CURRENT THERAPIES
Dysregulation of miRNAs can confer the resistance to current therapies including chemotherapy and radiation therapy. For instance, upregulation of miR‐21‐5p leads to a reduction of apoptosis and decrease of sensitivity to two chemotherapeutics, docetaxel and cisplatin.110 Induced by radiation, miR‐155‐5p does confer resistance to radiation therapy by indirectly increasing HK2 to promote aerobic glycolysis.111 Furthermore, chronic treatment with EGFR inhibitor gefitinib reduces the expression of miR‐155‐5p and miR‐200c‐3p and may decrease the sensitivity to gefitinib.118 Therefore, miRNA profiles from tissue or body fluids may also be used as predictive biomarkers for the sensitivity of NSCLC tumors to certain therapies and to determine optimal therapies for the treatment of NSCLC. As an example, miR‐143‐3p is downregulated in NSCLC and suppresses NSCLC cell proliferation, migration, and invasion by regulating EGFR expression.119 NSCLC patients showing lower miR‐143‐3p levels and higher EGFR levels may benefit from anti‐EGFR therapy like gefitinib. PI3K or VEGF inhibitors may be beneficial for patients with decreased expression of miR‐126‐3p because dysregulation of miR‐126‐3p may lead to an increase in PI3K and VEGF‐A.77, 120 In addition, miRNAs can play an important role in drug uptake and efflux via direct targeting of drug transporters. For example, miR‐31‐5p is upregulated in cisplatin resistant cell lines and directly regulates the expression of ABCB9, a drug transporter, to confers cisplatin resistance.121 Let‐7c‐5p modulates the expression of ABCC2 transporter to sensitize cisplatin resistant cells.122 ABCC4 involved in the transport of many anti‐cancer drugs such as methotrexate and topotecan and is directly targeted by miR‐124‐3p and miR‐506‐3p.123 Excellent reviews on the potential of miRNAs as biomarkers in lung cancer have been recently published.124 Understanding miRNA‐controlled regulation may not only improve the understanding of multidrug resistance mechanisms but also offer clues to the development of new therapeutic strategies.125, 126
7. MICRORNAS IMPACT THE TUMORIGENESIS OF NSCLC CELLS
To determine the effect of a miRNA on tumorigenesis, investigators transiently or stably express the target miRNA in a cell line for implantation in vivo. While this does not necessarily model therapeutic potential of miRNAs, it does provide important information beyond cell‐based findings regarding the importance of miRNAs in the control of tumor initiation and development or tumorigenesis (Table 4). For example, subcutaneously or tail vein injected A549 cells expressing miR‐124‐3p, miR‐126‐3p, miR‐143‐3p, miR‐34a‐5p, Let‐7b‐5p, or miR‐182‐5p showed reduced tumor growth and in some cases reduce lung metastasis as compared to control cells. Lewis lung carcinoma cells transiently transfected with miR‐101‐3p displayed a smaller increase of tumor volume over time when subcutaneously injected into the flank of mice, as well as a reduction of metastasis to the lung when intraperitoneally injected.127 Cells overexpressing miR‐145‐5p or miR‐486‐5p subcutaneously implanted or tail vein injected in mice displayed a slower rate of tumor growth.62, 128 Lung cancer cells 344SQ transfected with miR‐200a‐3p or miR‐200b‐5p or both showed smaller tumor volume with transfection of both miRNAs having the greatest impact.129 By contrast, transgenic KRAS mutant mice with conditionally global overexpression of miR‐21‐5p showed much greater tumor burden as well as lower survival rate, as compared to KRAS mutant mice without miR‐21‐5p overexpression.130 After subcutaneous implantation, H460 cells overexpressing miR‐205‐5p grew faster and led to greater tumor volume and vascularization, as compared to control cells.131 Compared to corresponding controls, overexpression of miR‐31‐5p in three different lung cancer cell lines, H1993, H1437, and H460, led to an increase in subcutaneous tumor volume.132 The same study also demonstrated that transgenic mice with a doxycycline‐inducible miR‐31‐5p expression in the lung exhibited greater levels of hyperplasia and adenomas.132 In conclusion, ectopic or overexpression of certain functional miRNAs can largely influence the tumorigenesis of NSCLC cells, which may provide insights into development of new miRNA‐based therapies.
Table 4.
Some miRNAs shown to affect tumorogenesis of NSCLC cells in animal models
| miRNA | Cell line | Mouse strain | Finding | Reference |
|---|---|---|---|---|
| miR‐124‐3p | A549 | nude BALB/c | Reduced lung metastasis from tail vein injected cells | [176] |
| miR‐126‐3p | A549 | nude BALB/c | Reduced tumor wieght | [77] |
| miR‐143‐3p | A549 | nude BALB/c | Reduced tumor wieght | [225] |
| miR‐34a‐5p | A549 | nude BALB/c | Reduced tumor wieght, and lung tumor metastasis | [226] |
| let‐7b‐5p | A549, H460 | nod/scid | Reduce tumor growth | [227] |
| miR‐101‐3p | LLC | C57BL/6 | Reduced tumor wieght, metastasis from IP injected cells | [127] |
| miR‐100‐5p | SPC‐A1/DTX | nude | Reduced tumor volume in response to docetaxel | [201] |
| miR‐145‐5p | A549 CIC | nude | Reduced tumor volume | [128] |
| miR‐486‐5p | H460‐luc2 | athymic Swiss | Reduced lung metastasis from tail vein injected cells | [62] |
| miR‐451‐5p | A549 | nude BALB/c | Reduced tumor volume in respose to cisplatin | [228] |
| miR‐21‐5p | CAG‐miR‐21;K‐rasLA2 | Reduced tumor burden and increased survival | [130] | |
| miR‐205‐5p | H460 | BALB/c | Reduced tumor volume | [131] |
| miR‐31‐5p | H1993/ H1437/H460 | nude | Reduced tumor volume | [132] |
| miR‐200a/b | 344SQ | nude | Reduced tumor volume | [129] |
| miR‐182‐5p | A549 | nude | Reduced tumor volume and weight and increased survival | [229] |
8. THERAPEUTIC POTENTIAL OF MICRORNAS DEMONSTRATED IN NSCLC ANIMAL MODELS IN VIVO
Two miRNA‐based therapeutic strategies have been established, aiming to restore tumor suppressive miRNAs and inhibit tumor promotive miRNAs, respectively. Many studies were thus conducted to define the effectiveness of specific miRNA therapeutics for the treatment of NSCLC in animal models in vivo (Table 5). AntagomiRs were employed for the inhibition of tumor promotive miR‐21‐5p, miR‐183‐5p, and miR‐206‐3p and shown to inhibit subcutaneous A549 tumors.82, 133, 134 Whereas, tumor suppressive miRNAs let‐7b‐5p or miR‐34a‐5p reduced KRAS‐activated tumor burden in vivo.135 Let‐7c‐5p and let‐7a‐5p were both shown to decrease the progression of NSCLC in vivo.136 Synthetic miR‐34a‐5p injected either intratumorally or through the tail vein was effective in inhibiting the growth of subcutaneous xenograft NSCLC,137 and it did not show major impact on the cytokine profiles or liver or kidney functions. A specific type of chemical modification of miR‐145‐5p, namely locked nucleic acid, delivered with a polyurethane‐short branched‐polyethylenimine, led to significant inhibition of tumor growth and the effects were enhanced by radiation and cisplatin therapy.138 Orthotopic NSCLC tumor growth, metastasis, and vascularization were decreased in mice treated with miR‐200a/b.129 miR‐29b‐3p decreased cell proliferation and increased apoptosis in subcutaneous NSCLC tumors.139 Combination treatment with miR‐34a‐5p and let‐7c‐5pwas effective in improving overall survival and reducing tumor burden in KRAS mutant mice.140 Nevertheless, the majority of miRNAs used for in vivo therapies are made by chemical synthesis or in vitro transcription, or achieved through viral vectors or plasmids, where RNAs or mimics are often delivered with lipids or polymers. Very recently, a novel RNA bioengineering technology has been established for the production of new miRNA reagents for the assessment of miRNA therapies.141, 142, 143, 144, 145 Biologic or recombinant miR‐34a‐5p or miR‐124‐3p molecules produced in bacteria and delivered with in vivo‐jetPEI into tumor‐bearing mouse models decreased the growth of both subcutaneous and metastatic NSCLC tumors, with minimal influence on blood chemistry or cytokine profiles, in two different studies.144, 145 A variety of different delivery methods have been tested in vivo to deliver the above‐mentioned miRNAs. Many miRNAs were formulated with lipid‐based technologies, such as liposomes, lipoplex, siPORTamine, and MaxSuppressor, which surround the RNA and protect it from degradation.136, 137, 139, 140, 146 Viral vectors, such as lentivirus or adenovirus, as well is positively charged polyethylenimine, which associates with the negatively charged RNA, were also used.134, 136, 138, 144, 145 In most cases, miRNA therapeutics were administered through the tail vein or intra‐tumoral injection. These in vivo findings demonstrate the promise of miRNA‐based therapies for the treatment of NSCLC.
Table 5.
Some miRNA‐based therapies for the treatment of NSCLC assessed in animal models in vivo
| miR | Mouse model | Delivery | Findings | Reference |
|---|---|---|---|---|
| let‐7b‐5p and miR‐34a‐5p (synthetic) | Cre‐Kras mutant | Neutral lipid emulsion (NLE) | Lower tumor burden, increased apoptosis, decreased proliferation | [135] |
| let‐7b‐5p (synthetic) |
Subcutaneous H460 cell line |
siPORTamine (lipid based) | Decreased proliferation | [136] |
|
let‐7a‐5p (lenti‐let‐7) |
Cre‐Kras mutant | Lentiviral | Decreased proliferation | [136] |
| miR‐34a‐5p (synthetic) |
Subcutaneous H460 |
MaxSupressor in vivo RNALancerII (lipid based) | Decreased proliferation, increased apoptosis, minimal change in blood chemistry or cytokine profile | [137] |
| miR‐145‐5p (synthetic; LNA) | Subcutaneous, intrabronchial or intravenous patient derived primary lung adenocarcinoma CD133+ | Cationic polyurethane‐short branched polyethylenimine (PU‐PEI) | miR‐145‐5p alone showed moderate tumor inhibition, increased tumor inhibition and survival in combination with radiation and cisplatin | [138] |
| miR‐200a/b (synthetic) | intrapulmonary 344SQ (murine) cell line | 1,2‐dioleoyl‐sn‐glycero‐3‐phosphatidylcholine (DOPC) nanoliposomes | Reduced proliferation, metastasis, and tumor vasculature permeabilization | [129] |
| miR‐29b‐3p (synthetic) | Subcutaneous A549 cell line | Lipoplex | Suppressed target expression, reduced proliferation, increased apoptosis | [139] |
| miR‐34a‐5p & miR‐124‐3p (biologic RNA) | Intravenous A549 cell line, (metastatic) | In vivo‐jetPEI | Decreased lung lesions, minimal change on blood chemistry or cytokine release | [144] |
| miR‐34a‐5p (biologic RNA) | Subcutaneous A459 cell line | In vivo‐jetPEI | Decreased tumor size, minimal change on blood chemistry or cytokine release | [145] |
| miR‐34a‐5p (synthetic) |
Intramuscular H460 or H1299 cell line |
NOV340 (Liposomal nanoparticle) | Sensitized tumor to irradiation | [146] |
| miR‐34a‐5p and let‐7b‐5p (synthetic) | Kras/p53 mutant, Cre‐ adenoviral activated | NOV340 (liposomal nanoparticle) | Combination of miR‐34a‐5p and let‐7c‐5p reduced tumor burden, decreased proliferation, and increased survival with minimal cytokine induction | [140] |
| anti‐miR‐21‐5p (synthetic) | Subcutaneous A549 cell line | QTsome (cationic lipids) | Stable tumor growth or tumor regression after treatment, increased survival | [133] |
| anti‐miR‐183‐5p (synthetic) | Subcutaneous A549‐LUC‐GFP | Adenovirus (intra‐tumoral injection) | Decreased tumor growth as measured by luminescence | [134] |
| miR‐206‐3p‐agomir (synthetic) | Subcutaneous A549 cell line | No vehicle mentioned (intra‐tumoral injection) | Decreased tumor volume and formation of intra‐tumoral capillary tubes, and increased apoptosis | [82] |
One tumor suppressive miRNA, namely MRX34 or liposomal miR‐34a mimic, was also moved into first‐in‐human Phase I clinical trials.147 It was evaluated as dose‐escalating intravenous infusions under a regimen of twice a week in three‐week cycles. This study consisted of 47 patients with advanced solid tumors, including one patient with NSCLC who had stable disease for 8 cycles of treatment. While efficacy of MRX34 was obvious among some patients, 96% of all patients experienced immune‐related adverse effects where multiple deaths also occurred with complex and uncertain causes. The most common adverse effects were fever, fatigue, nausea, diarrhea, and vomiting, and laboratory abnormalities included lymphopenia, neutropenia, and increased AST among others. The study does not distinguish whether the toxicity resulted from the RNA or the liposomal carrier, however, both components have shown immune toxicities in previous studies.148, 149 The immune‐related toxicity suggests that more studies are warranted to understand the effect of miRNA therapies and the carriers on the immune system. The termination of this trial reiterates the importance of safety study in addition to efficacy during drug development.
9. CONCLUSIONS AND PERSPECTIVES
Functional miRNAs derived from the human genome are critical factors in posttranscriptional regulation of target gene expression underlying many cellular processes, including metabolism, proliferation, apoptosis, and disease initiation and progression. Uncontrolled NSCLC cell growth and tumor development is associated with dysregulated miRNA expression in addition to the alterations of proteins and signaling pathways, among which tumor suppressive miRNAs are generally downregulated and tumor promotive miRNAs are commonly upregulated. With the improved understanding of miRNA biology in NSCLC, new miRNA‐based therapies are under active investigations, in particular, the restoration of tumor suppressive miRNAs and inhibition of tumor promotive miRNAs. Nevertheless, many challenges remain for the development of new therapeutics. Although a number of RNA drugs have been approved for clinical practice,142 the pharmacokinetic and pharmacodynamic properties of RNA molecules are still of concern since RNA molecules are generally susceptible to serum RNases and cannot pass freely through cell membrane barriers. Chemical modifications and formulation with biocompatible lipids or polymers have proven useful for improving the metabolic stability and delivery of RNA therapeutics. As chemical modifications undoubtedly lead to different RNA folding, stability, biologic activity, and safety profiles, there are growing interests in producing biologic or recombinant RNA molecules in living cells for RNA research and drug development,141, 142 similar as the success of protein‐based therapeutic modalities. Improved formulations with lipid or polymer‐based drug delivery systems may aid in protecting the RNA from degradation or recognition by the immune system. In any case, evidence is required to address two fundamental questions: whether the drug is effective against the disease and whether the drug is safe for the patients, which warrants more extensive studies.
DISCLOSURES
The authors declare that there is no conflict of interest.
AUTHOR CONTRIBUTIONS
Research design/conducting experiments: n/a. Literature research & analysis: Petrek & Yu. Writing and revising the manuscript: Petrek & Yu.
ACKNOWLEDGEMENTS
This work was supported by National Cancer Institute (grant No. R01CA225958) and National Institute of General Medical Sciences (R01GM113888), National Institutes of Health.
Petrek H, Yu A‐M. MicroRNAs in non‐small cell lung cancer: Gene regulation, impact on cancer cellular processes, and therapeutic potential. Pharmacol Res Perspect. 2019;e00528 10.1002/prp2.528
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7‐34. [DOI] [PubMed] [Google Scholar]
- 2. Mariotto AB, Robin Yabroff K, Shao Y, Feuer EJ, Brown ML. Projections of the cost of cancer care in the United States: 2010–2020. J Natl Cancer Inst. 2011;103(2):117‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non‐small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol. 2015;10(9):1240‐1242. [DOI] [PubMed] [Google Scholar]
- 5. O’Keeffe LM, Taylor G, Huxley RR, Mitchell P, Woodward M, Peters S. Smoking as a risk factor for lung cancer in women and men: a systematic review and meta‐analysis. BMJ Open. 2018;8(10):e021611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooley ME. Symptoms in adults with lung cancer. A systematic research review. J Pain Symptom Manage. 2000;19(2):137‐153. [DOI] [PubMed] [Google Scholar]
- 7. Travis WD. Pathology of lung cancer. Clin Chest Med. 2002;23(1):65‐81 [DOI] [PubMed] [Google Scholar]
- 8. Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest 2017;151(1):193‐203. [DOI] [PubMed] [Google Scholar]
- 9. Consortium, A.P.G . AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7(8):818‐831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pao W, Miller VA. Epidermal growth factor receptor mutations, small‐molecule kinase inhibitors, and non‐small‐cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23(11):2556‐2568. [DOI] [PubMed] [Google Scholar]
- 11. Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118(2):257‐262. [DOI] [PubMed] [Google Scholar]
- 12. Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pao W, Girard N. New driver mutations in non‐small‐cell lung cancer. Lancet Oncol. 2011;12(2):175‐180. [DOI] [PubMed] [Google Scholar]
- 14. Heinmoller P, Gross C, Beyser K, et al. HER2 status in non‐small cell lung cancer: results from patient screening for enrollment to a phase II study of herceptin. Clin Cancer Res. 2003;9(14):5238‐5243. [PubMed] [Google Scholar]
- 15. Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431(7008):525‐526. [DOI] [PubMed] [Google Scholar]
- 16. Shimamura T, Ji H, Minami Y, et al. Non‐small‐cell lung cancer and Ba/F3 transformed cells harboring the ERBB2 G776insV_G/C mutation are sensitive to the dual‐specific epidermal growth factor receptor and ERBB2 inhibitor HKI‐272. Cancer Res. 2006;66(13):6487‐6491. [DOI] [PubMed] [Google Scholar]
- 17. Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. [DOI] [PubMed] [Google Scholar]
- 18. Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448(7152):439‐444. [DOI] [PubMed] [Google Scholar]
- 19. Malanga D, Scrima M, De Marco C, et al. Activating E17K mutation in the gene encoding the protein kinase AKT1 in a subset of squamous cell carcinoma of the lung. Cell Cycle. 2008;7(5):665‐669. [DOI] [PubMed] [Google Scholar]
- 20. Leicht DT, Balan V, Kaplun A, et al. Raf kinases: function, regulation and role in human cancer. Biochim Biophys Acta. 2007;1773(8):1196‐1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Naoki K, Chen TH, Richards WG, Sugarbaker DJ, Meyerson M. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62(23):7001‐7003. [PubMed] [Google Scholar]
- 22. Marks JL, Gong Y, Chitale D, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68(14):5524‐5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316(5827):1039‐1043. [DOI] [PubMed] [Google Scholar]
- 24. Foster CC, Sher DJ, Rusthoven CG, et al. Overall survival according to immunotherapy and radiation treatment for metastatic non‐small‐cell lung cancer: a National Cancer Database analysis. Radiat Oncol. 2019;14(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Villaruz LC, Socinski MA. The role of anti‐angiogenesis in non‐small‐cell lung cancer: an update. Curr Oncol Rep. 2015;17(6):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non‐small‐cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121‐128. [DOI] [PubMed] [Google Scholar]
- 27. Odogwu L, Mathieu L, Blumenthal G, et al. FDA approval summary: Dabrafenib and Trametinib for the Treatment of Metastatic Non‐Small Cell Lung Cancers Harboring BRAF V600E Mutations. Oncologist 2018;23(6):740‐745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rolfo C, Caglevic C, Santarpia M, et al. Immunotherapy in NSCLC: A Promising and Revolutionary Weapon. Adv Exp Med Biol. 2017;995:97‐125. [DOI] [PubMed] [Google Scholar]
- 29. Tseng D, Padda SK, Wakelee HA. Perspectives on acquired resistance to PD‐1 axis inhibitors in patients with non‐small cell lung cancer. J Thorac Oncol. 2018;13(6):741‐744. [DOI] [PubMed] [Google Scholar]
- 30. Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17(11):637‐658. [DOI] [PubMed] [Google Scholar]
- 31. Suresh K, Naidoo J, Lin CT, Danoff S. Immune checkpoint immunotherapy for non‐small cell lung cancer: benefits and pulmonary toxicities. Chest. 2018;154(6):1416‐1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rahouma M, Baudo M, Yahia M, et al. Pneumonitis as a complication of immune system targeting drugs?‐a meta‐analysis of anti‐PD/PD‐L1 immunotherapy randomized clinical trials. J Thorac Dis. 2019;11(2):521‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299‐311. [DOI] [PubMed] [Google Scholar]
- 34. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350‐355. [DOI] [PubMed] [Google Scholar]
- 35. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363‐366. [DOI] [PubMed] [Google Scholar]
- 36. Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22‐32. [DOI] [PubMed] [Google Scholar]
- 37. Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199‐208. [DOI] [PubMed] [Google Scholar]
- 38. Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215‐217. [DOI] [PubMed] [Google Scholar]
- 39. Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96(2):111‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673‐677. [DOI] [PubMed] [Google Scholar]
- 41. Swahari V, Nakamura A, Deshmukh M. The paradox of dicer in cancer. Mol Cell Oncol. 2016;3(3):e1155006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529‐533. [DOI] [PubMed] [Google Scholar]
- 43. Takahashi T, Nau M, Chiba I, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246(4929):491‐494. [DOI] [PubMed] [Google Scholar]
- 44. Kalluri R, Weinberg RA. The basics of epithelial‐mesenchymal transition. J Clin Invest. 2009;119(6):1420‐1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zaravinos A. The regulatory role of MicroRNAs in EMT and Cancer. J Oncol. 2015;2015:865816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiang Z, Yin J, Fu W, et al. MiRNA 17 family regulates cisplatin‐resistant and metastasis by targeting TGFbetaR2 in NSCLC. PLoS ONE. 2014;9(4):e94639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li J, Song Y, Wang Y, Luo J, Yu W. MicroRNA‐148a suppresses epithelial‐to‐mesenchymal transition by targeting ROCK1 in non‐small cell lung cancer cells. Mol Cell Biochem. 2013;380(1–2):277‐282. [DOI] [PubMed] [Google Scholar]
- 48. Ye Z, Yin S, Su Z, et al. Downregulation of miR‐101 contributes to epithelial‐mesenchymal transition in cisplatin resistance of NSCLC cells by targeting ROCK2. Oncotarget. 2016;7(25):37524‐37535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. You J, Li Y, Fang N, et al. MiR‐132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS ONE. 2014;9(3):e91827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xia Y, Wu Y, Liu B, Wang P, Chen Y. Downregulation of miR‐638 promotes invasion and proliferation by regulating SOX2 and induces EMT in NSCLC. FEBS Lett. 2014;588(14):2238‐2245. [DOI] [PubMed] [Google Scholar]
- 51. Li Y, Chen P, Zu L, Liu B, Wang M, Zhou Q. MicroRNA‐338‐3p suppresses metastasis of lung cancer cells by targeting the EMT regulator Sox4. Am J Cancer Res. 2016;6(2):127‐140. [PMC free article] [PubMed] [Google Scholar]
- 52. Ke Y, et al. miR‐149 inhibits non‐small‐cell lung cancer cells EMT by targeting FOXM1. Biochem Res Int. 2013;2013:506731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dong Y, Jin X, Sun Z, Zhao Y, Song X. MiR‐186 Inhibited Migration of NSCLC via Targeting cdc42 and Effecting EMT Process. Mol Cells. 2017;40(3):195‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA‐7. J Biol Chem. 2009;284(9):5731‐5741. [DOI] [PubMed] [Google Scholar]
- 55. Liu L, Shao X, Gao W, et al. MicroRNA‐133b inhibits the growth of non‐small‐cell lung cancer by targeting the epidermal growth factor receptor. FEBS J. 2012;279(20):3800‐3812. [DOI] [PubMed] [Google Scholar]
- 56. Qin Q, Wei F, Zhang J, Wang X, Li B. miR‐134 inhibits non‐small cell lung cancer growth by targeting the epidermal growth factor receptor. J Cell Mol Med. 2016;20(10):1974‐1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhen Q, Liu J, Gao L, et al. MicroRNA‐200a Targets EGFR and c‐Met to Inhibit Migration, Invasion, and Gefitinib resistance in non‐small cell lung cancer. Cytogenet Genome Res. 2015;146(1):1‐8. [DOI] [PubMed] [Google Scholar]
- 58. Xu W, Li J, Xu C, Zhang X. MicroRNA‐139‐5p inhibits cell proliferation and invasion by targeting insulin‐like growth factor 1 receptor in human non‐small cell lung cancer. Int J Clin Exp Pathol. 2015;8(4):3864‐3870. [PMC free article] [PubMed] [Google Scholar]
- 59. Wen XP, Ma HL, Zhao LY, Zhang W, Dang CX. MiR‐30a suppresses non‐small cell lung cancer progression through AKT signaling pathway by targeting IGF1R. Cell Mol Biol (Noisy‐le‐grand). 2015;61(2):78‐85. [PubMed] [Google Scholar]
- 60. Yuan Y, Shen Y, Xue L, Fan H. miR‐140 suppresses tumor growth and metastasis of non‐small cell lung cancer by targeting insulin‐like growth factor 1 receptor. PLoS ONE. 2013;8(9):e73604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang J, Shi C, Wang J, Cao LI, Zhong LI, Wang D. MicroRNA‐320a is downregulated in non‐small cell lung cancer and suppresses tumor cell growth and invasion by directly targeting insulin‐like growth factor 1 receptor. Oncol Lett. 2017;13(5):3247‐3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang J, Tian X, Han R, et al. Downregulation of miR‐486‐5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene. 2014;33(9):1181‐1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Peng Y, Dai Y, Hitchcock C, et al. Insulin growth factor signaling is regulated by microRNA‐486, an underexpressed microRNA in lung cancer. Proc Natl Acad Sci USA. 2013;110(37):15043‐15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou Y, Li S, Li J, Wang D, Li Q. Effect of microRNA‐135a on Cell Proliferation, Migration, Invasion, Apoptosis and Tumor Angiogenesis Through the IGF‐1/PI3K/Akt Signaling Pathway in Non‐Small Cell Lung Cancer. Cell Physiol Biochem. 2017;42(4):1431‐1446. [DOI] [PubMed] [Google Scholar]
- 65. Cheng Z, Ma R, Tan W, Zhang LI. MiR‐152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp Mol Med. 2014;46:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kang J, Lee SY, Lee SY, et al. microRNA‐99b acts as a tumor suppressor in non‐small cell lung cancer by directly targeting fibroblast growth factor receptor 3. Exp Ther Med. 2012;3(1):149‐153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Luo J, Chen B, Ji X‐X, Zhou S‐W, Zheng DI. Overexpression of miR‐100 inhibits cancer growth, migration, and chemosensitivity in human NSCLC cells through fibroblast growth factor receptor 3. Tumour Biol. 2015. [DOI] [PubMed] [Google Scholar]
- 68. Acunzo M, Visone R, Romano G, et al. miR‐130a targets MET and induces TRAIL‐sensitivity in NSCLC by downregulating miR‐221 and 222. Oncogene. 2012;31(5):634‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Luo W, Huang BO, Li Z, et al. MicroRNA‐449a is downregulated in non‐small cell lung cancer and inhibits migration and invasion by targeting c‐Met. PLoS ONE. 2013;8(5):e64759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhou J‐Y, Chen XI, Zhao J, et al. MicroRNA‐34a overcomes HGF‐mediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET. Cancer Lett. 2014;351(2):265‐271. [DOI] [PubMed] [Google Scholar]
- 71. Chen Q‐Y, Jiao D‐M, Yan LI, et al. Comprehensive gene and microRNA expression profiling reveals miR‐206 inhibits MET in lung cancer metastasis. Mol Biosyst. 2015;11(8):2290‐2302. [DOI] [PubMed] [Google Scholar]
- 72. Hou C, Sun BO, Jiang Y, et al. MicroRNA‐31 inhibits lung adenocarcinoma stem‐like cells via down‐regulation of MET‐PI3K‐Akt signaling pathway. Anticancer Agents Med Chem. 2016;16(4):501‐518. [DOI] [PubMed] [Google Scholar]
- 73. Sun C, Sang M, Li S, et al. Hsa‐miR‐139‐5p inhibits proliferation and causes apoptosis associated with down‐regulation of c‐Met. Oncotarget. 2015;6(37):39756‐39792. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74. Sun CC, Li SJ, Zhang F, et al. Hsa‐miR‐329 exerts tumor suppressor function through down‐regulation of MET in non‐small cell lung cancer. Oncotarget. 2016;7(16):21510‐21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhou H, Liu Y, Xiao L, Hu Z, Xia K. Overexpression of MicroRNA‐27b inhibits proliferation, migration, and invasion via suppression of MET expression. Oncol Res. 2017;25(1):147‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23(5):1011‐1027. [DOI] [PubMed] [Google Scholar]
- 77. Liu BO, Peng X‐C, Zheng X‐L, Wang J, Qin Y‐W. MiR‐126 restoration down‐regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66(2):169‐175. [DOI] [PubMed] [Google Scholar]
- 78. Hu J, Cheng Y, Li Y, et al. microRNA‐128 plays a critical role in human non‐small cell lung cancer tumourigenesis, angiogenesis and lymphangiogenesis by directly targeting vascular endothelial growth factor‐C. Eur J Cancer. 2014;50(13):2336‐2350. [DOI] [PubMed] [Google Scholar]
- 79. Puisségur M‐P, Mazure NM, Bertero T, et al. miR‐210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF‐1 activity. Cell Death Differ. 2011;18(3):465‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu LZ, Li C, Chen Q, et al. MiR‐21 induced angiogenesis through AKT and ERK activation and HIF‐1alpha expression. PLoS ONE. 2011;6(4):e19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen L‐T, Xu S‐D, Xu H, Zhang J‐F, Ning J‐F, Wang S‐F. MicroRNA‐378 is associated with non‐small cell lung cancer brain metastasis by promoting cell migration, invasion and tumor angiogenesis. Med Oncol. 2012;29(3):1673‐1680. [DOI] [PubMed] [Google Scholar]
- 82. Xue D, Yang Y, Liu Y, et al. MicroRNA‐206 attenuates the growth and angiogenesis in non‐small cell lung cancer cells by blocking the 14‐3‐3zeta/STAT3/HIF‐1alpha/VEGF signaling. Oncotarget. 2016;7(48):79805‐79813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mao G, Liu Y, Fang XI, et al. Tumor‐derived microRNA‐494 promotes angiogenesis in non‐small cell lung cancer. Angiogenesis. 2015;18(3):373‐382. [DOI] [PubMed] [Google Scholar]
- 84. Sherr CJ. G1 phase progression: cycling on cue. Cell. 1994;79(4):551‐555. [DOI] [PubMed] [Google Scholar]
- 85. Buchkovich K, Duffy LA, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell‐cycle. Cell. 1989;58(6):1097‐1105. [DOI] [PubMed] [Google Scholar]
- 86. Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1‐to‐S phase transition. Mol Cell Biol. 1995;15(5):2612‐2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Girard F, Strausfeld U, Fernandez A, Lamb N. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67(6):1169‐1179. [DOI] [PubMed] [Google Scholar]
- 88. Arellano M, Moreno S. Regulation of CDK/cyclin complexes during the cell cycle. Int J Biochem Cell Biol. 1997;29(4):559‐573. [DOI] [PubMed] [Google Scholar]
- 89. Sun F, Fu H, Liu Q, et al. Downregulation of CCND1 and CDK6 by miR‐34a induces cell cycle arrest. FEBS Lett. 2008;582(10):1564‐1568. [DOI] [PubMed] [Google Scholar]
- 90. Bandi N, Zbinden S, Gugger M, et al. miR‐15a and miR‐16 are implicated in cell cycle regulation in a Rb‐dependent manner and are frequently deleted or down‐regulated in non‐small cell lung cancer. Cancer Res. 2009;69(13):5553‐5559. [DOI] [PubMed] [Google Scholar]
- 91. Bandi N, Vassella E. miR‐34a and miR‐15a/16 are co‐regulated in non‐small cell lung cancer and control cell cycle progression in a synergistic and Rb‐dependent manner. Mol Cancer. 2011;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chen DI, Guo W, Qiu Z, et al. MicroRNA‐30d‐5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non‐small cell lung cancer. Cancer Lett. 2015;362(2):208‐217. [DOI] [PubMed] [Google Scholar]
- 93. Cai J, Wu J, Zhang H, et al. miR‐186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell‐cycle regulation. Cancer Res. 2013;73(2):756‐766. [DOI] [PubMed] [Google Scholar]
- 94. Wu J, Qian J, Li C, et al. miR‐129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle. 2010;9(9):1809‐1818. [DOI] [PubMed] [Google Scholar]
- 95. Degterev A, Boyce M, Yuan J. A decade of caspases. Oncogene. 2003;22(53):8543‐8567. [DOI] [PubMed] [Google Scholar]
- 96. Scaffidi C, Fulda S, Srinivasan A, et al. Two CD95 (APO‐1/Fas) signaling pathways. EMBO J. 1998;17(6):1675‐1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Korsmeyer SJ, Wei MC, Saito M, Weiler S, Oh KJ, Schlesinger PH. Pro‐apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 2000;7(12):1166‐1173. [DOI] [PubMed] [Google Scholar]
- 98. Kluck RM, Bossy‐Wetzel E, Green DR, Newmeyer DD The release of cytochrome c from mitochondria: a primary site for Bcl‐2 regulation of apoptosis. Science. 1997;275(5303):1132‐1136. [DOI] [PubMed] [Google Scholar]
- 99. Huang P, Ye BO, Yang YU, Shi J, Zhao H. MicroRNA‐181 functions as a tumor suppressor in non‐small cell lung cancer (NSCLC) by targeting Bcl‐2. Tumour Biol. 2015;36(5):3381‐3387. [DOI] [PubMed] [Google Scholar]
- 100. Xiong S, Zheng Y, Jiang P, Liu R, Liu X, Chu Y. MicroRNA‐7 inhibits the growth of human non‐small cell lung cancer A549 cells through targeting BCL‐2. Int J Biol Sci. 2011;7(6):805‐814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Qiu T, Zhou LI, Wang T, et al. miR‐503 regulates the resistance of non‐small cell lung cancer cells to cisplatin by targeting Bcl‐2. Int J Mol Med. 2013;32(3):593‐598. [DOI] [PubMed] [Google Scholar]
- 102. Zhu W, Xu H, Zhu DanXia, et al. miR‐200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother Pharmacol. 2012;69(3):723‐731. [DOI] [PubMed] [Google Scholar]
- 103. Zhu W, Zhu DanXia, Lu S, et al. miR‐497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2012;29(1):384‐391. [DOI] [PubMed] [Google Scholar]
- 104. Yang T, Thakur A, Chen T, et al. MicroRNA‐15a induces cell apoptosis and inhibits metastasis by targeting BCL2L2 in non‐small cell lung cancer. Tumour Biol. 2015;36(6):4357‐4365. [DOI] [PubMed] [Google Scholar]
- 105. Sun C, Li S, Yang C, et al. MicroRNA‐187‐3p mitigates non‐small cell lung cancer (NSCLC) development through down‐regulation of BCL6. Biochem Biophys Res Commun. 2016;471(1):82‐88. [DOI] [PubMed] [Google Scholar]
- 106. Whang YE, Yuan XJ, Liu Y, Majumder S, Lewis TD. Regulation of sensitivity to TRAIL by the PTEN tumor suppressor. Vitam Horm. 2004;67:409‐426. [DOI] [PubMed] [Google Scholar]
- 107. Abraham R, Schäfer J, Rothe M, Bange J, Knyazev P, Ullrich A. Identification of MMP‐15 as an anti‐apoptotic factor in cancer cells. J Biol Chem. 2005;280(40):34123‐34132. [DOI] [PubMed] [Google Scholar]
- 108. Joshi P, Jeon Y‐J, Laganà A, et al. MicroRNA‐148a reduces tumorigenesis and increases TRAIL‐induced apoptosis in NSCLC. Proc Natl Acad Sci USA. 2015;112(28):8650‐8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Garofalo M, Di Leva G, Romano G, et al. miR‐221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498‐509. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110. Liu Z‐L, Wang HE, Liu J, Wang Z‐X. MicroRNA‐21 (miR‐21) expression promotes growth, metastasis, and chemo‐ or radioresistance in non‐small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372(1–2):35‐45. [DOI] [PubMed] [Google Scholar]
- 111. Lv X, Yao LI, Zhang J, Han P, Li C. Inhibition of microRNA‐155 sensitizes lung cancer cells to irradiation via suppression of HK2‐modulated glucose metabolism. Mol Med Rep. 2016;14(2):1332‐1338. [DOI] [PubMed] [Google Scholar]
- 112. Fang R, Xiao T, Fang Z, et al. MicroRNA‐143 (miR‐143) regulates cancer glycolysis via targeting hexokinase 2 gene. J Biol Chem. 2012;287(27):23227‐23235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhao X, Lu C, Chu W, et al. MicroRNA‐124 suppresses proliferation and glycolysis in non‐small cell lung cancer cells by targeting AKT‐GLUT1/HKII. Tumour Biol. 2017;39(5):1010428317706215. [DOI] [PubMed] [Google Scholar]
- 114. Li L, Liu H, Du L, et al. miR‐449a suppresses LDHA‐mediated glycolysis to enhance the sensitivity of non‐small cell lung cancer cells to ionizing radiation. Oncol Res. 2018;26(4):547‐556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chen G‐M, Zheng A‐J, Cai J, Han P, Ji H‐B, Wang L‐L. microRNA‐145‐3p inhibits non‐small cell lung cancer cell migration and invasion by targeting PDK1 via the mTOR signaling pathway. J Cell Biochem. 2018;119(1):885‐895. [DOI] [PubMed] [Google Scholar]
- 116. Zhu B, Cao X, Zhang W, et al. MicroRNA‐31‐5p enhances the Warburg effect via targeting FIH. FASEB J. 2019;33(1):545‐556. [DOI] [PubMed] [Google Scholar]
- 117. Ye X‐W, Yu H, Jin Y‐K, et al. miR‐138 inhibits proliferation by targeting 3‐phosphoinositide‐dependent protein kinase‐1 in non‐small cell lung cancer cells. Clin Respir J. 2015;9(1):27‐33. [DOI] [PubMed] [Google Scholar]
- 118. Narita M, Shimura E, Nagasawa A, et al. Chronic treatment of non‐small‐cell lung cancer cells with gefitinib leads to an epigenetic loss of epithelial properties associated with reductions in microRNA‐155 and ‐200c. PLoS ONE. 2017;12(2):e0172115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhang H‐B, Sun L‐C, Ling L, Cong L‐H, Lian R. miR‐143 suppresses the proliferation of NSCLC cells by inhibiting the epidermal growth factor receptor. Exp Ther Med. 2016;12(3):1795‐1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Song L, Li D, Gu Y, et al. MicroRNA‐126 Targeting PIK3R2 Inhibits NSCLC A549 cell proliferation, migration, and invasion by regulation of PTEN/PI3K/AKT Pathway. Clin Lung Cancer. 2016;17(5):e65‐e75. [DOI] [PubMed] [Google Scholar]
- 121. Dong Z, Zhong Z, Yang L, Wang S, Gong Z. MicroRNA‐31 inhibits cisplatin‐induced apoptosis in non‐small cell lung cancer cells by regulating the drug transporter ABCB9. Cancer Lett. 2014;343(2):249‐257. [DOI] [PubMed] [Google Scholar]
- 122. Zhan M, Qu Q, Wang G, Zhou H. Let‐7c sensitizes acquired cisplatin‐resistant A549 cells by targeting ABCC2 and Bcl‐XL. Pharmazie. 2013;68(12):955‐961. [PubMed] [Google Scholar]
- 123. Markova SM, Kroetz DL. ABCC4 is regulated by microRNA‐124a and microRNA‐506. Biochem Pharmacol. 2014;87(3):515‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Inamura K, Ishikawa Y. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment. J Clin Med. 2016;5(3):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yu AM, Pan YZ. Noncoding microRNAs: small RNAs play a big role in regulation of ADME? Acta Pharmaceutica Sinica B. 2012;2(2):93‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Yu A‐M, Tian Y, Tu M‐J, Ho PY, Jilek JL. MicroRNA pharmacoepigenetics: posttranscriptional regulation mechanisms behind variable drug disposition and strategy to develop more effective therapy. Drug Metab Dispos. 2016;44(3):308‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Yan F, Shen N, Pang J, et al. Restoration of miR‐101 suppresses lung tumorigenesis through inhibition of DNMT3a‐dependent DNA methylation. Cell Death Dis. 2014;5:e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hu J, Qiu M, Jiang F, et al. MiR‐145 regulates cancer stem‐like properties and epithelial‐to‐mesenchymal transition in lung adenocarcinoma‐initiating cells. Tumour Biol. 2014;35(9):8953‐8961. [DOI] [PubMed] [Google Scholar]
- 129. Pecot CV, Rupaimoole R, Yang D, et al. Tumour angiogenesis regulation by the miR‐200 family. Nat Commun. 2013;4:2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K‐Ras‐dependent lung tumorigenesis by MicroRNA‐21. Cancer Cell. 2010;18(3):282‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Cai J, Fang L, Huang Y, et al. miR‐205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non‐small cell lung cancer. Cancer Res. 2013;73(17):5402‐5415. [DOI] [PubMed] [Google Scholar]
- 132. Edmonds MD, Boyd KL, Moyo T, et al. MicroRNA‐31 initiates lung tumorigenesis and promotes mutant KRAS‐driven lung cancer. J Clin Invest. 2016;126(1):349‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Yung BC, Li J, Zhang M, et al. Lipid Nanoparticles Composed of Quaternary Amine‐Tertiary Amine Cationic Lipid Combination (QTsome) for Therapeutic Delivery of AntimiR‐21 for Lung Cancer. Mol Pharm. 2016;13(2):653‐662. [DOI] [PubMed] [Google Scholar]
- 134. Zhang L, Quan H, Wang S, Li XueHui, Che X. MiR‐183 promotes growth of non‐small cell lung cancer cells through FoxO1 inhibition. Tumour Biol. 2015;36(10):8121‐8126. [DOI] [PubMed] [Google Scholar]
- 135. Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116‐1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Trang P, Medina PP, Wiggins JF, et al. Regression of murine lung tumors by the let‐7 microRNA. Oncogene. 2010;29(11):1580‐1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA‐34. Cancer Res. 2010;70(14):5923‐5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Chiou G‐Y, Cherng J‐Y, Hsu H‐S, et al. Cationic polyurethanes‐short branch PEI‐mediated delivery of Mir145 inhibited epithelial‐mesenchymal transdifferentiation and cancer stem‐like properties and in lung adenocarcinoma. J Control Release. 2012;159(2):240‐250. [DOI] [PubMed] [Google Scholar]
- 139. Wu Y, Crawford M, Mao Y, et al. Therapeutic delivery of MicroRNA‐29b by cationic lipoplexes for lung cancer. Mol Ther Nucleic Acids. 2013;2:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non‐small cell lung cancer. Oncogene. 2015;34(27):3547‐3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ho PY, Yu AM. Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip Rev RNA. 2016;7(2):186‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Yu AM, Jian C, Allan HY, Tu MJ. RNA therapy: are we using the right molecules? Pharmacol Ther. 2019;196:91‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Chen Q‐X, Wang W‐P, Zeng SU, Urayama S, Yu A‐M. A general approach to high‐yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucleic Acids Res. 2015;43(7):3857‐3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ho PY, Duan Z, Batra N, et al. Bioengineered noncoding RNAs selectively change cellular miRNome profiles for cancer therapy. J Pharmacol Exp Ther. 2018;365(3):494‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wang W‐P, Ho PY, Chen Q‐X, et al. Bioengineering novel chimeric microRNA‐34a for prodrug cancer therapy: high‐yield expression and purification, and structural and functional characterization. J Pharmacol Exp Ther. 2015;354(2):131‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Cortez MA, Valdecanas D, Niknam S, et al. In vivo delivery of miR‐34a sensitizes lung tumors to radiation through RAD51 regulation. Mol Ther Nucleic Acids. 2015;4:e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Beg MS, Brenner AJ, Sachdev J, et al. Phase I study of MRX34, a liposomal miR‐34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35(2):180‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Szebeni J, Muggia F, Gabizon A, Barenholz Y. Activation of complement by therapeutic liposomes and other lipid excipient‐based therapeutic products: prediction and prevention. Adv Drug Deliv Rev. 2011;63(12):1020‐1030. [DOI] [PubMed] [Google Scholar]
- 149. Snove O Jr, Rossi JJ. Toxicity in mice expressing short hairpin RNAs gives new insight into RNAi. Genome Biol. 2006;7(8):231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Sandler A, Gray R, Perry MC, et al. Paclitaxel‐carboplatin alone or with bevacizumab for non‐small‐cell lung cancer. N Engl J Med. 2006;355(24):2542‐2550. [DOI] [PubMed] [Google Scholar]
- 151. Garon EB, Ciuleanu T‐E, Arrieta O, et al. Ramucirumab plus docetaxel versus placebo plus docetaxel for second‐line treatment of stage IV non‐small‐cell lung cancer after disease progression on platinum‐based therapy (REVEL): a multicentre, double‐blind, randomised phase 3 trial. Lancet. 2014;384(9944):665‐673. [DOI] [PubMed] [Google Scholar]
- 152. Wang Y, Schmid‐Bindert G, Zhou C. Erlotinib in the treatment of advanced non‐small cell lung cancer: an update for clinicians. Ther Adv Med Oncol. 2012;4(1):19‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. di Noia V, D’Argento E, Pilotto S, et al. Necitumumab in the treatment of non‐small‐cell lung cancer: clinical controversies. Expert Opin Biol Ther. 2018;18(9):937‐945. [DOI] [PubMed] [Google Scholar]
- 154. Wu Y‐L, Sequist LV, Tan E‐H, et al. Afatinib as first‐line treatment of older patients with EGFR mutation‐positive non‐small‐cell lung cancer: subgroup analyses of the LUX‐Lung 3, LUX‐Lung 6, and LUX‐Lung 7 Trials. Clin Lung Cancer. 2018;19(4):e465‐e479. [DOI] [PubMed] [Google Scholar]
- 155. Ramalingam SS, et al. Osimertinib as first‐line treatment of EGFR mutation‐positive advanced non‐small‐cell lung cancer. J Clin Oncol. 2018;36(9):841‐849. [DOI] [PubMed] [Google Scholar]
- 156. Chuang JC, Neal JW. Crizotinib as first line therapy for advanced ALK‐positive non‐small cell lung cancers. Transl Lung Cancer Res. 2015;4(5):639‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK‐rearranged non‐small‐cell lung cancer. N Engl J Med. 2014;370(13):1189‐1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Camidge DR, Kim HR, Ahn MJ, et al. Brigatinib versus crizotinib in ALK‐positive non‐small‐cell lung cancer. N Engl J Med. 2018;379(21):2027‐2039. [DOI] [PubMed] [Google Scholar]
- 159. Peters S, Camidge DR, Shaw AT, et al. Alectinib versus crizotinib in untreated ALK‐positive non‐small‐cell lung cancer. N Engl J Med. 2017;377(9):829‐838. [DOI] [PubMed] [Google Scholar]
- 160. Planchard D, Smit EF, Groen H, et al. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)‐mutant metastatic non‐small‐cell lung cancer: an open‐label, phase 2 trial. Lancet Oncol. 2017;18(10):1307‐1316. [DOI] [PubMed] [Google Scholar]
- 161. Liu D, Offin M, Harnicar S, Li BT, Drilon A. Entrectinib: an orally available, selective tyrosine kinase inhibitor for the treatment of NTRK, ROS1, and ALK fusion‐positive solid tumors. Ther Clin Risk Manag. 2018;14:1247‐1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Carbone DP, Reck M, Paz‐Ares L, et al. First‐line nivolumab in stage IV or recurrent non‐small‐cell lung cancer. N Engl J Med. 2017;376(25):2415‐2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Reck M, Rodríguez‐Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD‐L1‐positive non‐small‐cell lung cancer. N Engl J Med. 2016;375(19):1823‐1833. [DOI] [PubMed] [Google Scholar]
- 164. Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for first‐line treatment of metastatic nonsquamous NSCLC. N Engl J Med. 2018;378(24):2288‐2301. [DOI] [PubMed] [Google Scholar]
- 165. Hellmann MD, Ciuleanu T‐E, Pluzanski A, et al. Nivolumab plus Ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093‐2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Langer CJ, Leighton JC, Comis RL, et al. Paclitaxel and carboplatin in combination in the treatment of advanced non‐small‐cell lung cancer: a phase II toxicity, response, and survival analysis. J Clin Oncol. 1995;13(8):1860‐1870. [DOI] [PubMed] [Google Scholar]
- 167. Negoro S, Masuda N, Takada Y, et al. Randomised phase III trial of irinotecan combined with cisplatin for advanced non‐small‐cell lung cancer. Br J Cancer. 2003;88(3):335‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Cardenal F, López‐Cabrerizo MP, Antón A, et al. Randomized phase III study of gemcitabine‐cisplatin versus etoposide‐cisplatin in the treatment of locally advanced or metastatic non‐small‐cell lung cancer. J Clin Oncol. 1999;17(1):12‐18. [DOI] [PubMed] [Google Scholar]
- 169. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N Engl J Med. 2015;373(2):123‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Depierre A, Chastang CL, Quoix E, et al. Vinorelbine versus vinorelbine plus cisplatin in advanced non‐small cell lung cancer: a randomized trial. Ann Oncol. 1994;5(1):37‐42. [DOI] [PubMed] [Google Scholar]
- 171. Kris MG, et al. Randomized trial comparing vindesine plus cisplatin with vinblastine plus cisplatin in patients with non‐small cell lung cancer, with an analysis of methods of response assessment. Cancer Treat Rep. 1985;69(4):387‐395. [PubMed] [Google Scholar]
- 172. Hanna N, Shepherd FA, Fossella FV, et al. Randomized phase III trial of pemetrexed versus docetaxel in patients with non‐small‐cell lung cancer previously treated with chemotherapy. J Clin Oncol. 2004;22(9):1589‐1597. [DOI] [PubMed] [Google Scholar]
- 173. Li X, Yu Z, Li Y, et al. The tumor suppressor miR‐124 inhibits cell proliferation by targeting STAT3 and functions as a prognostic marker for postoperative NSCLC patients. Int J Oncol. 2015;46(2):798‐808. [DOI] [PubMed] [Google Scholar]
- 174. Sun Y, Ai X, Shen S, Lu S. NF‐kappaB‐mediated miR‐124 suppresses metastasis of non‐small‐cell lung cancer by targeting MYO10. Oncotarget. 2015;6(10):8244‐8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Yin P, Peng R, Peng H, et al. MiR‐451 suppresses cell proliferation and metastasis in A549 lung cancer cells. Mol Biotechnol. 2015;57(1):1‐11. [DOI] [PubMed] [Google Scholar]
- 176. Zu L, Xue Y, Wang J, et al. The feedback loop between miR‐124 and TGF‐beta pathway plays a significant role in non‐small cell lung cancer metastasis. Carcinogenesis. 2016;37(3):333‐343. [DOI] [PubMed] [Google Scholar]
- 177. Yang J, Lan H, Huang X, Liu B, Tong YU. MicroRNA‐126 inhibits tumor cell growth and its expression level correlates with poor survival in non‐small cell lung cancer patients. PLoS ONE. 2012;7(8):e42978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Zhu X, Li H, Long L, et al. miR‐126 enhances the sensitivity of non‐small cell lung cancer cells to anticancer agents by targeting vascular endothelial growth factor A. Acta Biochim Biophys Sin (Shanghai). 2012;44(6):519‐526. [DOI] [PubMed] [Google Scholar]
- 179. Crawford M, Brawner E, Batte K, et al. MicroRNA‐126 inhibits invasion in non‐small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373(4):607‐612. [DOI] [PubMed] [Google Scholar]
- 180. Gao W, Yu Y, Cao H, et al. Deregulated expression of miR‐21, miR‐143 and miR‐181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64(6):399‐408. [DOI] [PubMed] [Google Scholar]
- 181. Chen X, Guo X, Zhang H, et al. Role of miR‐143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28(10):1385‐1392. [DOI] [PubMed] [Google Scholar]
- 182. Wang L, Shi Z‐M, Jiang C‐F, et al. MiR‐143 acts as a tumor suppressor by targeting N‐RAS and enhances temozolomide‐induced apoptosis in glioma. Oncotarget. 2014;5(14):5416‐5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Li W‐H, Wu H‐J, Li Y‐X, Pan H‐G, Meng T, Wang X. MicroRNA‐143 promotes apoptosis of osteosarcoma cells by caspase‐3 activation via targeting Bcl‐2. Biomed Pharmacother. 2016;80:8‐15. [DOI] [PubMed] [Google Scholar]
- 184. Zhang X, Dong Y, Ti H, et al. Down‐regulation of miR‐145 and miR‐143 might be associated with DNA methyltransferase 3B overexpression and worse prognosis in endometrioid carcinomas. Hum Pathol. 2013;44(11):2571‐2580. [DOI] [PubMed] [Google Scholar]
- 185. Xia H, Sun S, Wang BO, et al. miR‐143 inhibits NSCLC cell growth and metastasis by targeting Limk1. Int J Mol Sci. 2014;15(7):11973‐11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wei J, Ma Z, Li Y, et al. miR‐143 inhibits cell proliferation by targeting autophagy‐related 2B in non‐small cell lung cancer H1299 cells. Mol Med Rep. 2015;11(1):571‐576. [DOI] [PubMed] [Google Scholar]
- 187. Ma ZL, Hou PP, Li YL, et al. MicroRNA‐34a inhibits the proliferation and promotes the apoptosis of non‐small cell lung cancer H1299 cell line by targeting TGFbetaR2. Tumour Biol. 2015;36(4):2481‐2490. [DOI] [PubMed] [Google Scholar]
- 188. Hong JH, Roh KS, Suh S‐S, et al. The expression of microRNA‐34a is inversely correlated with c‐MET and CDK6 and has a prognostic significance in lung adenocarcinoma patients. Tumour Biol. 2015;36(12):9327‐9337. [DOI] [PubMed] [Google Scholar]
- 189. Han Z, Zhang Y, Yang Q, et al. miR‐497 and miR‐34a retard lung cancer growth by co‐inhibiting cyclin E1 (CCNE1). Oncotarget. 2015;6(15):13149‐13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Yu G, Zhong N, Chen G, Huang B, Wu S. Downregulation of PEBP4, a target of miR‐34a, sensitizes drug‐resistant lung cancer cells. Tumour Biol. 2014;35(10):10341‐10349. [DOI] [PubMed] [Google Scholar]
- 191. Kang JiHoon, Kim EunGi, Kim W, et al. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial‐mesenchymal transition (EMT) by miR‐34a‐mediated suppression of Notch‐1 expression in non‐small cell lung cancer cell lines. J Biol Chem. 2013;288(38):27343‐27357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H. Regulation of Axl receptor tyrosine kinase expression by miR‐34a and miR‐199a/b in solid cancer. Oncogene. 2011;30(25):2888‐2899. [DOI] [PubMed] [Google Scholar]
- 193. Dou H, Wang Y, Su G, Zhao S. Decreased plasma let‐7c and miR‐152 as noninvasive biomarker for non‐small‐cell lung cancer. Int J Clin Exp Med. 2015;8(6):9291‐9298. [PMC free article] [PubMed] [Google Scholar]
- 194. Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let‐7 microRNA family. Cell. 2005;120(5):635‐647. [DOI] [PubMed] [Google Scholar]
- 195. Zhan M, Qu Q, Wang G, et al. Let‐7c inhibits NSCLC cell proliferation by targeting HOXA1. Asian Pac J Cancer Prev. 2013;14(1):387‐392. [DOI] [PubMed] [Google Scholar]
- 196. Zhao B, Han H, Chen J, et al. MicroRNA let‐7c inhibits migration and invasion of human non‐small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342(1):43‐51. [DOI] [PubMed] [Google Scholar]
- 197. Han L, Chen W, Xia Y, et al. MiR‐101 inhibits the proliferation and metastasis of lung cancer by targeting zinc finger E‐box binding homeobox 1. Am J Transl Res. 2018;10(4):1172‐1183. [PMC free article] [PubMed] [Google Scholar]
- 198. Zhang X, He X, Liu Y, et al. MiR‐101‐3p inhibits the growth and metastasis of non‐small cell lung cancer through blocking PI3K/AKT signal pathway by targeting MALAT‐1. Biomed Pharmacother. 2017;93:1065‐1073. [DOI] [PubMed] [Google Scholar]
- 199. Zhang J‐G, Guo J‐F, Liu D‐L, Liu Q, Wang J‐J. MicroRNA‐101 exerts tumor‐suppressive functions in non‐small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6(4):671‐678. [DOI] [PubMed] [Google Scholar]
- 200. Liu J, Lu KH, Liu ZL, Sun M, De W, Wang ZX. MicroRNA‐100 is a potential molecular marker of non‐small cell lung cancer and functions as a tumor suppressor by targeting polo‐like kinase 1. BMC Cancer. 2012;12:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Feng B, Wang R, Chen LB. MiR‐100 resensitizes docetaxel‐resistant human lung adenocarcinoma cells (SPC‐A1) to docetaxel by targeting Plk1. Cancer Lett. 2012;317(2):184‐191. [DOI] [PubMed] [Google Scholar]
- 202. Ma Z, Qiu X, Wang D, et al. MiR‐181a‐5p inhibits cell proliferation and migration by targeting Kras in non‐small cell lung cancer A549 cells. Acta Biochim Biophys Sin (Shanghai). 2015;47(8):630‐638. [DOI] [PubMed] [Google Scholar]
- 203. Cao Y, Zhao D, Li P, et al. MicroRNA‐181a‐5p Impedes IL‐17‐Induced Nonsmall Cell Lung Cancer Proliferation and Migration through Targeting VCAM‐1. Cell Physiol Biochem. 2017;42(1):346‐356. [DOI] [PubMed] [Google Scholar]
- 204. Shi Q, Zhou Z, Ye N, Chen Q, Zheng X, Fang M. MiR‐181a inhibits non‐small cell lung cancer cell proliferation by targeting CDK1. Cancer Biomark. 2017;20(4):539‐546. [DOI] [PubMed] [Google Scholar]
- 205. Chen Z, Zeng H, Guo Y, et al. miRNA‐145 inhibits non‐small cell lung cancer cell proliferation by targeting c‐Myc. J Exp Clin Cancer Res. 2010;29:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Hu H, Xu Z, Li C, et al. MiR‐145 and miR‐203 represses TGF‐beta‐induced epithelial‐mesenchymal transition and invasion by inhibiting SMAD3 in non‐small cell lung cancer cells. Lung Cancer. 2016;97:87‐94. [DOI] [PubMed] [Google Scholar]
- 207. Wang M, Wang J, Deng J, Li X, Long W, Chang Y. MiR‐145 acts as a metastasis suppressor by targeting metadherin in lung cancer. Med Oncol. 2015;32(1):344. [DOI] [PubMed] [Google Scholar]
- 208. Pang W, Tian X, Bai F, et al. Pim‐1 kinase is a target of miR‐486‐5p and eukaryotic translation initiation factor 4E, and plays a critical role in lung cancer. Mol Cancer. 2014;13:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Shao Y, Shen Y‐Q, Li Y‐L, et al. Direct repression of the oncogene CDK4 by the tumor suppressor miR‐486‐5p in non‐small cell lung cancer. Oncotarget. 2016;7(23):34011‐34021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Zhu J, Zeng Y, Xu C, et al. Expression profile analysis of microRNAs and downregulated miR‐486‐5p and miR‐30a‐5p in non‐small cell lung cancer. Oncol Rep. 2015;34(4):1779‐1786. [DOI] [PubMed] [Google Scholar]
- 211. Wang R, Wang Z‐X, Yang J‐S, Pan X, De W, Chen L‐B. MicroRNA‐451 functions as a tumor suppressor in human non‐small cell lung cancer by targeting ras‐related protein 14 (RAB14). Oncogene. 2011;30(23):2644‐2658. [DOI] [PubMed] [Google Scholar]
- 212. Zhang J‐G, Wang J‐J, Zhao F, Liu Q, Jiang KE, Yang G‐H. MicroRNA‐21 (miR‐21) represses tumor suppressor PTEN and promotes growth and invasion in non‐small cell lung cancer (NSCLC). Clin Chim Acta. 2010;411(11–12):846‐852. [DOI] [PubMed] [Google Scholar]
- 213. Zhu WangYu, Zhou KaiYu, Zha Y, et al. Diagnostic value of serum miR‐182, miR‐183, miR‐210, and miR‐126 levels in patients with early‐stage non‐small cell lung cancer. PLoS ONE. 2016;11(4):e0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Lei L, Huang Y, Gong W. miR‐205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol Rep. 2013;30(6):2897‐2902. [DOI] [PubMed] [Google Scholar]
- 215. Larzabal L, et al. TMPRSS4 regulates levels of integrin alpha5 in NSCLC through miR‐205 activity to promote metastasis. Br J Cancer. 2014;110(3):764‐774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Jiang M, Zhang P, Hu G, et al. Relative expressions of miR‐205‐5p, miR‐205‐3p, and miR‐21 in tissues and serum of non‐small cell lung cancer patients. Mol Cell Biochem. 2013;383(1–2):67‐75. [DOI] [PubMed] [Google Scholar]
- 217. Zhong Z, Dong Z, Yang L, Chen X, Gong Z. MicroRNA‐31‐5p modulates cell cycle by targeting human mutL homolog 1 in human cancer cells. Tumour Biol. 2013;34(3):1959‐1965. [DOI] [PubMed] [Google Scholar]
- 218. Kundu ST, Byers LA, Peng DH, et al. The miR‐200 family and the miR‐183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2016;35(2):173‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8(9):1156‐1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Võsa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta‐analysis of microRNA expression in lung cancer. Int J Cancer. 2013;132(12):2884‐2893. [DOI] [PubMed] [Google Scholar]
- 221. Wang M, Wang Y, Zang W, et al. Downregulation of microRNA‐182 inhibits cell growth and invasion by targeting programmed cell death 4 in human lung adenocarcinoma cells. Tumour Biol. 2014;35(1):39‐46. [DOI] [PubMed] [Google Scholar]
- 222. Xiao P, Liu W, Zhou H. miR‐200b inhibits migration and invasion in non‐small cell lung cancer cells via targeting FSCN1. Mol Med Rep. 2016;14(2):1835‐1840. [DOI] [PubMed] [Google Scholar]
- 223. Sun Y, Fang R, Li C, et al. Hsa‐mir‐182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem Biophys Res Commun. 2010;396(2):501‐507. [DOI] [PubMed] [Google Scholar]
- 224. Wang G, Mao W, Zheng S. MicroRNA‐183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008;582(25–26):3663‐3668. [DOI] [PubMed] [Google Scholar]
- 225. Cheng TL, Hu C, Yang H, Cao L, An J. Transforming growth factor‐beta‐induced miR‐143 expression in regulation of non‐small cell lung cancer cell viability and invasion capacity in vitro and in vivo. Int J Oncol. 2014;45(5):1977‐1988. [DOI] [PubMed] [Google Scholar]
- 226. Li Y‐L, Liu X‐M, Zhang C‐Y, et al. MicroRNA‐34a/EGFR axis plays pivotal roles in lung tumorigenesis. Oncogenesis. 2017;6(8):e372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Esquela‐Kerscher A, Trang P, Wiggins JF, et al. The let‐7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759‐764. [DOI] [PubMed] [Google Scholar]
- 228. Bian HB, Pan X, Yang JS, Wang ZX, De W. Upregulation of microRNA‐451 increases cisplatin sensitivity of non‐small cell lung cancer cell line (A549). J Exp Clin Cancer Res. 2011;30:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Zhang L, Liu T, Huang Y, Liu J. microRNA‐182 inhibits the proliferation and invasion of human lung adenocarcinoma cells through its effect on human cortical actin‐associated protein. Int J Mol Med. 2011;28(3):381‐388. [DOI] [PubMed] [Google Scholar]


