Abstract
Epithelial ovarian cancer (EOC) is the most lethal gynecological malignancy, with the presence of chemoresistance contributing to the poor prognosis. Heat Shock Proteins (HSPs) genes are activated in response to pathophysiological stress and serve a role in a variety of stages in carcinogenesis, acting primarily as anti-apoptotic agents and in chemotherapy resistance in a variety of tumor types. The current study evaluated the HSP gene expression profile in women with ovarian cancer (OC) and their correlation with clinical and pathological aspects of patients with OC. A total of 51 patients included in the current study were divided into four groups: Primary Epithelial Ovarian Cancer (EOC; n=14), metastatic EOC (n=11), ovarian serous cystadenoma (n=7) and no evidence of ovarian malignancy or control groups (n=19). RNA extraction and reverse transcription-quantitative (RT-q) PCR was then performed on the samples obtained. RT-qPCR was performed to compare TNF receptor associated protein 1 (TRAP1), heat shock protein family (HSP) HSPB1, HSPD1, HSPA1A and HSPA1L expression in primary and metastatic EOCs. TRAP1, HSPB1, HSPD1, HSPA1A and HSPA1L gene expression did not differ among groups. HSPA1A, HSPA1L and TRAP1 were revealed to be underexpressed in the primary and metastatic EOC groups, with HSPA1L exhibiting the lowest expression. TRAP1 expression was higher in tumors at stages I/II compared with those at stages III/IV. No correlation was exhibited between HSP expression and age, menarche, menopause, parity, period after menopause initiation, cytoreduction, CA-125 or overall and disease-free survival. HSPA1A was negatively correlated with the risk of mortality from OC. The results indicated that the downregulation of HSPA1A, HSPA1L and TRAP1 could be associated with the clinical prognostic features of women with EOC.
Keywords: ovarian cancer, prognosis, resistance to chemotherapy, heat shock proteins, gene expression
Introduction
Epithelial ovarian cancer (EOC) causes around 125,000 deaths globally per year (1). Approximately 70% of women with ovarian cancer (OC) are diagnosed with locally advanced or metastatic disease (stages III/IV), of whom only ~30% will survive more than 5 years. By contrast, women diagnosed with earlier stage (stage I) disease have a 5-year survival rate >90%. Unfortunately, signs and symptoms of OC are usually absent or too subtle to be easily detected in the early stages of the disease. Despite of high initial response rates to chemotherapy, approximately 80% of women with advanced OC relapse within 2 years after initial drug treatment (2,3).
The standard treatment for EOC involves maximal cytoreductive surgery followed by platinum and taxane-based chemotherapy. At first, most patients with advanced stage (III/IV) EOC respond well to surgery and chemotherapy; however, within two years after initial treatment, cancer frequently relapses with a drug-resistant phenotype and most patients die of the disease (4). Age and disease staging at diagnosis, tumor histology, and performance status (PS) are the best known prognostic factors (5), albeit limited by our restricted understanding of EOC's biology and complicated by disease heterogeneity.
Currently, there is a growing interest in finding specific molecular markers that could function both in the prognosis of the disease and the patient's response to chemotherapy. Good candidates include heat shock proteins (HSPs) because of their role in facilitating malignant transformation, tumor progression, and tumor survival (6,7). These evolutionarily conserved proteins are classified according to their molecular weight and, in mammalian cells, are grouped into six main classes: HSP27, HSP40, HSP60, HSP70, HSP90, and HSP110 (7). The contribution of HSPs to tumorigenesis can be attributed to their activities governing folding/unfolding, turn-over, and transport of client proteins as well as assembly of multiprotein complexes. As a result, various crucial and clinically important cell responses are vitally influenced and modulated by HSPs, e.g., cell growth, apoptosis, metastasis, and treatment resistance (6,7).
Although the existing data for HSP's function in OC progression and drug resistance is appealing, it is still limited and conflicting at times. For instance, the cytosolic HSPB1(HSP27), HSP70 (HSPA1A, HSPA1L), and HSP90(TRAP1) as well as the mitochondrial HSP60 proteins and the tumor necrosis factor receptor-associated protein 1 (TRAP-1) have all been shown to be induced by drug treatment and frequently associated with cross-resistance to anticancer compounds of different classes in ovarian and other cancer types (6–10). Moreover, the levels of circulating HSP27 protein were decreased after chemotherapy treatment in metastatic OC patients (11). Thus, drug-mediated regulation of HSPs in OC may follow differentially controlled stress signaling pathways. Due to our limited understanding of HSP's role in OC biology, studies elucidating their potential to help the prognostic evaluation of patients and the therapeutic strategy upon relapse after platinum-based chemotherapy are warranted.
In the present study, we correlated the expression of TRAP1, HSPB1, HSPA1, HSPAl, and HSPD1 genes and the clinical and pathological aspects of patients with OC. To this end, we compared the expression of these genes in the primary and metastatic ovarian tumor and investigated the relationship between the observed expression profile with other known prognostic factors and with the patients' response to chemotherapy and relapse-free survival.
Materials and methods
Ethics
This study was approved by the Research Ethics Committee of Vera Cruz Hospital (Belo Horizonte, Minas Gerais, Brazil), under the protocol CAAE: 01242212.2.0000.5135. All participants voluntarily signed an informed consent form.
Patients and tumor tissue samples
We collected ovarian tissue from 51 women divided into four groups: Primary Epithelial Ovarian Cancer EOC (n=14), metastatic EOC (n=11), ovarian serous cystadenoma (n=7) and normal ovary (n=19). The patients were recruited to our study using convenience sampling and they did not match any of the following exclusion criteria: Previously treated with chemotherapy and/or radiotherapy; HIV positive; presenting any infectious process diagnosed or not during laparotomy; present or previous history of other malignant neoplasms; using or with a previous history of use of immunosuppressives, systemic corticosteroids or non-steroidal anti-inflammatory drugs in the three months prior to the study. All cases were reevaluated blindly by a senior consultant subspecialized in gynecologic pathology and a representative portion of each tumor containing >80% tumor cells were selected for storage until analysis. Clinical and pathologic information documented at the time of surgery included disease stage, tumor grade and histotype, residual tumor size and debulking success.
In the EOC patients, samples were collected from primary tumors and of metastatic tumors, when extra pelvic disease above 1 cm was observed. Tumor staging was performed according to the FIGO recommendations (12). Normal ovarian epithelial tissue samples were taken from postmenopausal women who required a bilateral oophorectomy. After excision, the samples were immediately frozen in liquid nitrogen and stored at −80°C until use.
RNA extraction, cDNA synthesis and gene expression analysis
Total RNA was extracted from 50 to 100 mg of each ovarian tumor sample using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The RNA yield and A260/280 ratio were determined by a NanovueTM Plus Spectrophotometer (GE Healthcare Biosciences). RNA integrity and quality were characterized through 1% agarose gel electrophoresis. Subsequently, the samples were treated with RNAse-Free DNAse Set® (Qiagen) to remove possible traces of genomic DNA.
cDNAs were synthesized using M-MLV Reverse transcriptase (Promega Corporation) according to the manufacturer's recommendations and were subjected to RT-qPCR using TaqMan® Universal PCR master mix and inventoried TaqMan® Assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to manufacturer's recommendation. Taqman assays were selected for each target gene: TRAP1 (Hs00212476_m1), HSPB1 (Hs00356629_g1), HSPA1A (Hs00359163_s1), HSPA1L (Hs00271466_s1), HSPD1 (Hs01036753_g1) and for TBP (Hs00427620_m1) used as endogenous control. A sample without a template was included as a control in each assay. Each 40-cycle reaction was performed in duplicate using a Step OnePlus detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Two technical replicates were adopted for each sample. Relative gene expression was determined using the 2−ΔΔCq method (13).
Gene functional and Network pathway analysis
The differentially expressed genes determined using the 2−ΔΔCq method and for the pathway analysis of gene-associated proteins, the Kyoto Encyclopedia of Genes and Genomes (KEGG) was investigated by using STRING database, version 10.5 (14). STRING was also used to evaluate protein-protein interactions (PPI) among the associated genes.
Statistics
Student's t-test and ANOVA were used to compare gene expression and qualitative variables (15). To detect correlation between the genes and to compare their expression with the quantitative variables, we used Pearson's correlation and Spearman's correlation (16), respectively. To compare disease free time curves and survival curves with gene expression, the log-rank test (17) was used. It is worth mentioning that the gene expressions were recoded as greater than 1, less than 1, or equal to 1. To analyze the factors influencing survival and the disease-free interval, a univariate analysis was performed using the Cox Regression Model and the Risk Ratio was computed (17). Different from logistic regression, the Cox model has the advantage of including the effect of time up to the death and relapse besides allowing the interpretation through the risk ratio and not the odds ratio. The probability of survival and significance were calculated using the Kaplan-Meier method (18). All statistical analyzes were performed using the statistical software package R (version 3.4.1) (http://www.r-project.org/), using stats package (19) to quantitative variables and survival package to analyze the survival rates (20). P<0.05 was considered to indicate a statistically significant difference.
Results
The general characteristics of the patients are shown in Table I. The parity was 2.07 births with a range between 0 and 8 deliveries. The clinicopathologic characteristics of the tumor samples are shown in Table II. The stage was I in 7 patients (29.17%) and III/IV in 17 patients in the EOC group (70.83%). All samples were identified as high-grade serous carcinoma by histopathological evaluation.
Table I.
General characteristics of patients.
| Variables | Cystadenoma mean ± standard error | Primary EOC mean ± standard error | Metastatic EOC mean ± standard error | Normal ovary mean ± standard error | P-valuea |
|---|---|---|---|---|---|
| Age (years) | 50.00±16.54 | 57.93±10.54 | 59.55±10.76 | 47.68±8.33 | 0.017 |
| Menarche | 12.57±1.13 | 12.79±1.37 | 13.10±1.45 | – | – |
| Parity (births) | 2.14±3.08 | 2.00±1.24 | 2.10±1.20 | – | – |
| Period after Menopause | 6.86±11.19 | 8.62±9.03 | 10.13±9.03 | – | – |
Kruskal-Wallis. EOC, epithelial ovarian carcinoma.
Table II.
Clinicopathologic characteristics in ovarian sample.
| Variables | Cystadenoma N (%) | Primary EOC N (%) | Metastatic EOC N (%) | Normal ovary N (%) | P-valuea |
|---|---|---|---|---|---|
| Stage | |||||
| I | 0 (0.0) | 5 (35.7) | 2 (20.0) | – | – |
| III | 0 (0.0) | 6 (42.9) | 6 (60.0) | – | |
| IV | 0 (0.0) | 3 (21.4) | 2 (20.0) | – | |
| Menopause | |||||
| No | 4 (57.1) | 3 (21.4) | 2 (20.0) | 17 (89.5) | <0.001 |
| Yes | 3 (42.9) | 11 (78.6) | 8 (80.0) | 2 (10.5) | |
| Ascites | |||||
| No | 7 (100.0) | 4 (30.8) | 3 (37.5) | – | – |
| Yes | 0 (0.0) | 9 (69.2) | 5 (62.5) | – | |
| Tumor differentiation grade | |||||
| G2 | 0 (0.0) | 5 (35.7) | 4 (40.0) | – | – |
| G3 | 0 (0.0) | 9 (64.3) | 6 (60.0) | – | |
| CA-125 | |||||
| <35 U/ml | 3 (42.9) | 4 (28.6) | 1 (9.10) | – | – |
| >35 U/ml | 4 (57.1) | 10 (71.4) | 10 (90.9) | – |
Fisher exact test. EOC, epithelial ovarian carcinoma.
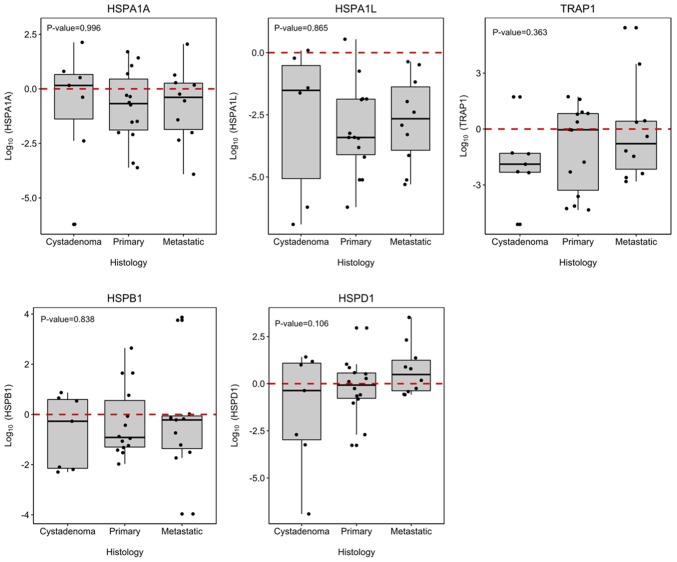
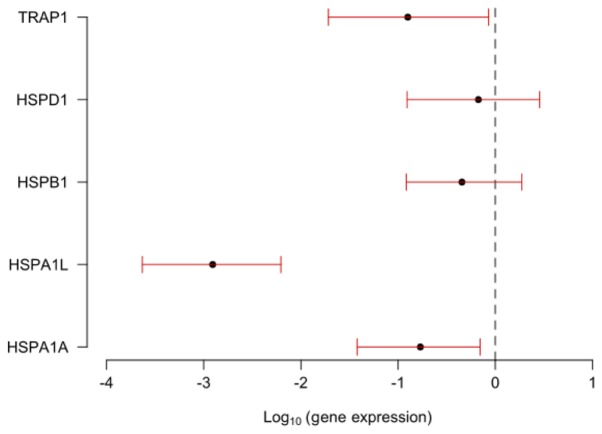
TRAP1, HSPA1A, HSPA1L, HSPD1 and HSPB1 genes showed differential expression between tumor samples of the EOC group and samples from the cystadenoma, primary and metastatic EOC samples. Although, no significantly differ among the groups (P>0.050; Fig. 1). When the groups were compared singly, HSPA1A, HSPA1L and TRAP1 were significantly under-expressed in the primary and metastatic EOC groups in comparison to the expression profile presented in normal ovarian tissues, with HSPA1L showing the lowest expression in both carcinoma groups (Fig. 2).
Figure 1.
The association between TRAP1, HSPD1, HSPB1, HSPA1L and HSPA1A expression in ovarian tumors. The values represent TRAP1, HSPD1, HSPB1, HSPA1L and HSPA1A expression. The horizontal line indicates the median expression ratio, and the box plots demonstrate the interquartile range (25–75%). The 10 to 90th percentile ranges are also presented. The differences between groups were evaluated using a Man-Whitney U test and a Wilcoxon test. TRAP1, TNF receptor associated protein 1; HSP, heat shock protein family.
Figure 2.
Comparison of the expression of the TRAP1, HSPD1, HSP1, HSPA1 and HSPA1A genes in patients diagnosed with primary and metastatic EOC compared with patients with normal ovaries. Patients with normal ovary were considered as a control group (value of the expression equals 1, logarithm equals 0). The confidence intervals (red stems) of the genes intersecting the dashed line do not differ from each other. Intervals with confidence below the dashed line (less than 0) indicate that the gene is underexpressed. TRAP1, TNF receptor associated protein 1; HSP, heat shock protein family; EOC, epithelial ovarian carcinoma.
There was no correlation between the expression levels of the analyzed genes and age, menarche, parity or period after menopause initiation as well as between the seric levels of the CA125 tumor marker and the expression of the HSP genes analyzed (Table III).
Table III.
Association between clinicopathologic characteristics of EOC patients and TRAP1, HSPD1, HSPB1, HSPA1L and HSPA1A gene expression profile.
| HSP gene expression profile correlation | |||||
|---|---|---|---|---|---|
| Clinicopathologic characteristics | HSPA1A | HSPA1L | HSPB1 | HSPD1 | TRAP1 |
| Histopathology | 0.996 | 0.865 | 0.838 | 0.106 | 0.363 |
| Age (years) | 0.797 | 0.309 | 0.723 | 0.287 | 0.451 |
| Menarche | 0.782 | 0.713 | 0.68 | 0.554 | 0.351 |
| Parity (births) | 0.119 | 0.061 | 0.852 | 0.152 | 0.594 |
| Period after Menopause | 0.804 | 0.643 | 0.486 | 0.632 | 0.409 |
| CA-125 | 0.222 | 0.806 | 0.539 | 0.842 | 0.315 |
| Stage | 0.962 | 0.327 | 0.075 | 0.193 | 0.040 |
| Menopause | 0.927 | 0.664 | 0.786 | 0.600 | 0.492 |
| Ascites | 0.562 | 0.573 | 0.585 | 0.174 | 0.798 |
| Tumor differentiation grade | 0.397 | 0.305 | 0.035 | 0.080 | 0.163 |
| Cytoreduction | 0.797 | 0.772 | 0.239 | 0.824 | 0.422 |
| Risk of dying | 0.73 | 1.09 | 0.92 | 1.02 | 0.94 |
| (P-value) | (0.048) | (0.491) | (0.527) | (0.892) | (0.511) |
TRAP1, TNF receptor associated protein 1; HSP, heat shock protein family; EOC, epithelial ovarian carcinoma.
A comparison between the expression profile of the HSP genes and the OC staging showed that TRAP1 expression was significantly greater in tumors at stage I than in tumors at stages III and IV of EOC patients (P=0.040; Table III).
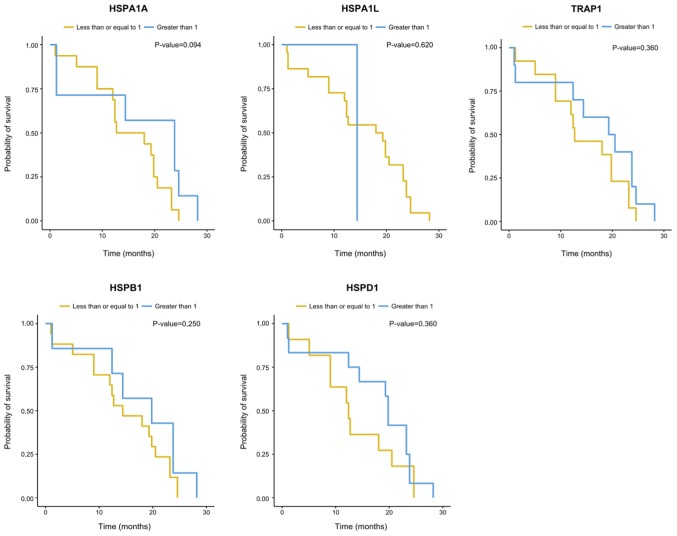
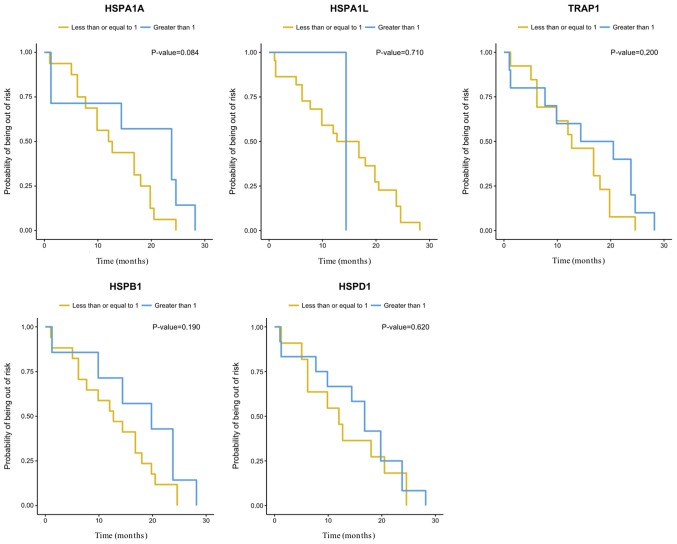
There was no correlation between cytoreduction and the expression of the HSP genes analyzed herein (Table III). There were no significant differences (P=0.05) between the expression of the HSP genes evaluated and overall survival (OS) or disease-free survival (DFS) (Figs. 3 and 4). However, the gene expression analysis in relation to OS suggested influence of HSPA1A expression levels on the risk of dying of EOC (P=0.048). An increase of one unit in the gene log decreased the risk of dying by 0.73 times [0.53; 0.99] (Table III).
Figure 3.
Kaplan-Meier analysis of overall survival among women with primary and metastatic EOC according to their TRAP1, HSPD1, HSPB1, HSPA1L and HSPA1A gene expression. TRAP1, TNF receptor associated protein 1; HSP, heat shock protein family; EOC, epithelial ovarian carcinoma.
Figure 4.
Kaplan-Meier analysis of disease-free survival among women with primary and metastatic EOC according to their TRAP1, HSPD1, HSPB1, HSPA1L and HSPA1A gene expression. TRAP1, TNF receptor associated protein 1; HSP, heat shock protein family; EOC, epithelial ovarian carcinoma.
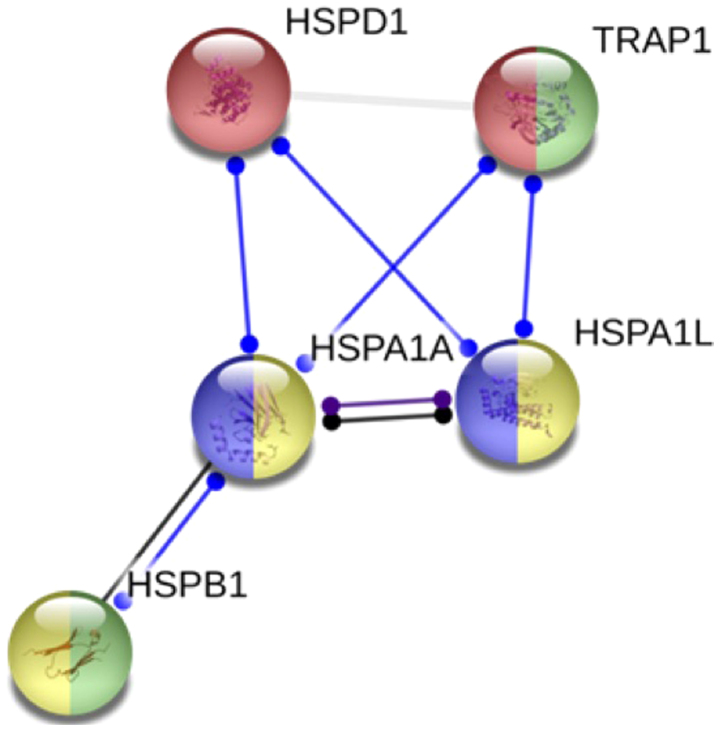
In silico network protein analysis made on STRING database revealed the protein-protein interactions between the proteins codified by the genes analyzed by us (Fig. 5).
Figure 5.
Gene functional and Network pathway analysis of TRAP1, HSPD1, HSPB1, HSPA1L and HSPA1A. The Gene functional and Network pathway analysis evaluated in silico by STRING database. The circles colors indicate the HSP functions in network. Chaperone mediated protein folding, negative regulated of oxidative stress-induced intrinsic apoptotic signaling pathway (green), MAPK signaling pathway (yellow), and spliceossoma (purple). TRAP1, TNF receptor associated protein 1; HSP, heat shock protein family.
Discussion
EOC is a very heterogeneous disease and the most lethal gynecological neoplasia (21). Despite extensive effort, EOC continues to be a poorly understood disease and patients survival rates remain low. Therefore, new strategies for early diagnosis, prognostic markers for clinical assessment and a better understanding of the mechanisms related to ovarian carcinogenesis are of extreme importance in order to obtain better outcomes for the affected patients. In this study, we investigated whether a gene signature among patients with and without EOC could be identified. To this end, we evaluated the level of expression of the genes TRAP1, HSPD1, HSPB1, HSPA1A, and HSPA1L in tumor samples obtained from patients with cystadenoma, primary and metastatic EOC in relation to baseline expression of these genes in normal ovary (NO) tissues. Therefore, expression of each gene in NO was assigned an arbitrary quantity of ‘1’ and their expression in the tumor samples were expressed in terms of their fold difference to NO (21). We found that these five genes were differentially expressed between the groups, but the prediction of EOC metastasis with gene expression profiling was not better than chance alone. The comparison between the expression levels of the studied genes in the tumor groups with the NO group showed that HSPA1A, HSPA1L and TRAP1 were significantly under-expressed in the EOC groups.
The under-expression of HSP70 isoforms was previously observed in OC (22). According to these authors, the genes HSPA1A and HSPA1L reside on a particularly vulnerable CpG island, which is subject to methylation and boosts the immune response. In addition, the copy number variation (CNV) of the HSP genes described for different tumors may also explain the under-expression of the observed HSPA1A, HSPA1L and TRAP1 genes in our study. TRAP1 expression is correlated with the copy number, suggesting this could be one of the driving mechanisms for the loss of TRAP1 expression in OC (23).
Several gene expression studies identifying molecular markers related to cancer progression have been published. Overall, there is a considerable overlap between previous studies and our study in terms of differentially expressed genes between normal and tumor tissues. Furthermore, all studies demonstrate the great diversity of tumor pathobiology, a feature that makes cancer a difficult disease to treat effectively (24–26).
TRAP1, HSPD1, HSPB1, HSPA1A, and HSPA1L belong a stress or HSPs family of highly conserved genes that are expressed in response to a wide variety of physiological and environmental insults in order to maintain cellular homeostasis or to contribute to cell survival to lethal conditions. The stresses involving HSPs include such as hypoxia, exposure to UV light and chemicals, viral agents, nutritional deficiencies (e.g., glucose deprivation), surgical, emotional and mechanical stress, among other stresses (27–30). Beyond that, biological processes of proteins among HSP associated genes analyzed by the Gene functional and Network pathway analysis performed herein revealed their function in chaperone mediated protein folding.
TRAP1 encodes a mitochondrial chaperone protein that is a member of the heat shock family 90 (HSP90). The protein has ATPase activity and interacts with tumor necrosis factor type I (31). Interestingly, alternate splicing results in multiple transcript variants (32) and other study suggested that TRAP1 has an oncogenic role in a variety of cancer types (33). In colorectal carcinoma, increased expression of TRAP1 was correlated with increased lymph node involvement, more advanced stages of the disease, and reduction in overall survival. TRAP1 is currently a marker predicting worse outcomes in colorectal cancer (34). However, low levels of TRAP1 has been related to high tumor grade, more advanced stage and resistance to platinum in OC (35). In OC cells lines and tissues, TRAP1 was shown to be associated with a metabolic shift, ultimately causing the onset of resistance to cisplatin-based chemotherapy (36). Furthermore, in OC clinical samples, TRAP1 is often deleted in high-grade serous OC patients and it is correlated directly with epithelial-mesenchymal transition, which is an important determinant of the invasive potential of tumor cells (23). Therefore, TRAP1 downregulation is linked to tumor progression in OC patients. Similarly, TRAP1 was under-expressed in the EOC group compared with the NO group and higher expression was showed in tumors at stages I/II than at III/IV (P=0.040), a finding that could be correlate with a negative impact on the response to chemotherapy and survival of patients with OC.
The HSPA1A, HSPA1L, HSPD1, and HSPB1 genes are expressed either constitutively or regulated inductively. High molecular weight HSPs are ATP-dependent chaperones (HSPA1A, HSPA1L, HSPD1), whereas small HSPs act in an ATP-independent fashion (HSPB1). As molecular chaperones, the function of HSPs is to regulate protein folding, transport, translocation and assembly, particularly to refold misfolded proteins or assist their elimination (27,30). The literature relates the overexpression of these genes as a possible marker of worse prognosis in other tumor types (37). It is known that in cancer there is a need for ambiguous signal transduction, hence there are greater demand for chaperones. The phenomenon is probably linked to the drastic changes in protein homeostasis caused by the accumulation of mutated proteins in cancer cells (27).
In our study, HSPA1A was the gene that presented the lowest level of expression in relation to the NO group. In addition, it showed a significant influence on the overall survival of patients with EOC, who showed a decrease in their risk of death by 0.73 times for every increase in one unit in HSPA1A expression, suggesting a protective role for this gene. This result highlights the potential of this gene as a possible genetic marker to assist the clinical evaluation of the prognosis of the disease.
The association between HSPB1 expression and high-grade OC primary tumors and metastases was described previously (38) and similar results were already observed in cell lines (39). The immune response to HSPB1 is also increased in women with OC and other gynecological tumors and some studies suggested the use of anti-HSP27 antibody concentrations for early diagnosis of relapse or disease progression (40). The association of HSPB1 with early disease staging and longer survival of patients with OC, most studies suggest an association between HSPB1 overexpression and worse prognosis (41). There appears to be a co-expression of HSPB1 or a positive correlation between HSPB1 expression and resistance to chemotherapy and expression of MDR1 (gene for resistance to multiple drugs) (42). Thus, there is evidence that the overexpression and activity of HSP27 is associated with increased carcinogenesis, metastatic potential, and resistance to chemotherapy. However, in our series the expression of HSPB1 was not significantly different in the EOC group in comparison to the control group.
This study has some limitations which have to be considered. The small number of patients and controls do not allow us to draw any meaningful conclusions regarding the relation between gene expression and anatomic site or clinicopathologic parameters. It is worth mentioning that, differently from tumors studies in other sites, the low prevalence of ovarian tumors, along with the ethical and biological determinants for control group selection imposes a limiting factor of patient numbers in EOC cohorts. So, our study was led with convenience samples, also known as availability sampling, a specific type of nonprobability sampling method that relies on data collection from a population who are conveniently available to participate in study. Because of that, it observed the unbalanced number of samples of each group. Furthermore, the lack of immunohistochemistry to confirm the expressions of HSP genes could be considered other limitation to our study. However, it was designed to evaluate gene expression by tracking messenger RNA using RT-qPCR, due access to standardized protocols and automation ensures an accurate performance and fast turn-around. Our findings highlight the importance of understanding the role of HSP genes in the ovarian carcinogenesis process and future investigations should be performed cloning HSP genes into OC cell lines or using CRISPR gene editing to verify if chemoresistance and others prognosis features can be altered in OC by HSPs gene expression and confirm our results.
We can hypothesis that HSPA1A, HSPA1L and TRAP1 downregulation seems to enhance the ability of the cancer cells to die in a range of lethal conditions. Further studies with a larger number of patients and longer follow-up are necessary to assess the accuracy of the prognostic impact of these results.
Acknowledgements
Not applicable.
Funding
The current study was financially supported from grants from Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG; grant. no. CDS-APQ-02373-17), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
Regarding the authorship of the manuscript, AWP, SAL, BLC, GNG and SLM gave individual contribution in the concept and design/analysis and interpretation of data, drafting the manuscript or revised it critically for important intellectual content and final approval. AWP and SAL were responsible for patient recruitment. BLC, GNG and SLM were responsible for performing the experimental assays.
Ethics approval and consent to participate
The current study was approved by the Research Ethics Committee of Vera Cruz Hospital, Belo Horizonte, Brazil, under the protocol CAAE: 01242212.2.0000.5135. Informed consent forms were obtained when the patients were accepted for the study by the hospital.
Patient consent for publication
Consent for publication was obtained from all participants of the current study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Corrado G, Salutari V, Palluzzi E, Distefano MG, Scambia G, Ferrandina G. Optimizing treatment in recurrent epithelial ovarian cancer. Expert Rev Anticancer Ther. 2017;17:1147–1158. doi: 10.1080/14737140.2017.1398088. [DOI] [PubMed] [Google Scholar]
- 4.du Bois A, Luck HJ, Meier W, Adams HP, Möbus V, Costa S, Bauknecht T, Richter B, Warm M, Schröder W, et al. A randomized clinical trial of cisplatin/paclitaxel versus carboplatin/paclitaxel as first-line treatment of ovarian cancer. J Natl Cancer Inst. 2003;95:1320–1329. doi: 10.1093/jnci/djg036. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal R, Kaye SB. Prognostic factors in ovarian cancer: How close are we to a complete picture? Ann Oncol. 2005;16:4–6. doi: 10.1093/annonc/mdi104. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Liu T, Rios Z, Mei Q, Lin X, Cao S. Heat shock proteins and cancer. Trends Pharmacol Sci. 2017;38:226–256. doi: 10.1016/j.tips.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Ciocca DR, Arrigo AP, Calderwood SK. Heat shock proteins and heat shock factor 1 in carcinogenesis and tumor development: An update. Arch Toxicol. 2013;87:19–48. doi: 10.1007/s00204-012-0918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song TF, Zhang ZF, Liu L, Yang T, Jiang J, Li P. Small interfering RNA-mediated silencing of heat shock protein 27 (HSP27) Increases chemosensitivity to paclitaxel by increasing production of reactive oxygen species in human ovarian cancer cells (HO8910) J Int Med Res. 2009;37:1375–1388. doi: 10.1177/147323000903700512. [DOI] [PubMed] [Google Scholar]
- 9.Landriscina M, Amoroso MR, Piscazzi A, Esposito F. Heat shock proteins, cell survival and drug resistance: The mitochondrial chaperone TRAP1, a potential novel target for ovarian cancer therapy. Gynecol Oncol. 2010;117:177–182. doi: 10.1016/j.ygyno.2009.10.078. [DOI] [PubMed] [Google Scholar]
- 10.Elstrand MB, Stavnes HT, Trope CG, Davidson B. Heat shock protein 90 is a putative therapeutic target in patients with recurrent advanced-stage ovarian carcinoma with serous effusions. Hum Pathol. 2012;43:529–535. doi: 10.1016/j.humpath.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 11.Zhao M, Ding JX, Zeng K, Zhao J, Shen F, Yin YX, Chen Q. Heat shock protein 27: A potential biomarker of peritoneal metastasis in epithelial ovarian cancer? Tumour Biol. 2014;35:1051–1056. doi: 10.1007/s13277-013-1139-7. [DOI] [PubMed] [Google Scholar]
- 12.Kim HS, Song YS. International federation of gynecology and obstetrics (FIGO) staging system revised: What should be considered critically for gynecologic cancer? J Gynecol Oncol. 2009;20:135–136. doi: 10.3802/jgo.2009.20.3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database Issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montgomery DC, Peck EA, Vining GG. John Wiley & Sons; New York: 2012. Introduction to linear regression analysis 5th edition. [Google Scholar]
- 16.Myles H, Wolfe DA. John Wiley & Sons; New York: 1999. Nonparametric Statistical Methods. [Google Scholar]
- 17.Colosimo EA, Giolo SR. Edgard Blücher; 2006. ABE-Projeto Fisher. [Google Scholar]
- 18.Kaplan EL, Meyer P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 19.R Core Team, corp-author. R Foundation for Statistical Computing; Vienna: 2018. A language and environment for statistical computing. [Google Scholar]
- 20.Therneau TM, Grambsch PM. Springer; New York, NY: 2000. Modeling survival data: Extending the Cox Model. [Google Scholar]
- 21.Kurman RJ, Shih Ie M. The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am J Surg Pathol. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh MK, Sharma B, Tiwari PK. The small heat shock protein Hsp27: Present understanding and future prospects. J Therm Biol. 2017;69:149–154. doi: 10.1016/j.jtherbio.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Amoroso MR, Matassa DS, Agliarulo I, Avolio R, Lu H, Sisinni L, Lettini G, Gabra H, Landriscina M, Esposito F. TRAP1 downregulation in human ovarian cancer enhances invasion and epithelial-mesenchymal transition. Cell Death Dis. 2016;7:e2522. doi: 10.1038/cddis.2016.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfister K, Radons J, Busch R, Tidball JG, Pfeifer M, Freitag L, Feldmann HJ, Milani V, Issels R, Multhoff G. Patient survival by Hsp70 membrane phenotype: Association with different routes of metastasis. Cancer. 2007;110:926–935. doi: 10.1002/cncr.22864. [DOI] [PubMed] [Google Scholar]
- 25.Braga Lda C, Silva LM, Ramos AP, Piedade JB, Vidigal PV, Traiman P, da Silva Filho AL. Single CpG island methylation is not sufficient to maintain the silenced expression of CASPASE-8 apoptosis-related gene among women with epithelial ovarian cancer. Biomed Pharmacother. 2014;68:87–91. doi: 10.1016/j.biopha.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 26.de Lima AB, Silva LM, Goncales NG, Carvalho MRS, da Silva Filho AL, da Conceicao Braga L. Three-dimensional cellular arrangement in epithelial ovarian cancer cell lines TOV-21G and SKOV-3 is associated with apoptosis-related miRNA expression modulation. Cancer Microenviron. 2018;11:85–92. doi: 10.1007/s12307-017-0203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arrigo AP, Gibert B. HspB1, HspB5 and HspB4 in human cancers: Potent oncogenic role of some of their client proteins. Cancers (Basel) 2014;6:333–365. doi: 10.3390/cancers6010333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beere HM. Stressed to death: Regulation of apoptotic signaling pathways by the heat shock proteins. Sci STKE. 2001;2001:re1. doi: 10.1126/stke.2001.93.re1. [DOI] [PubMed] [Google Scholar]
- 29.Khalil AA, Kabapy NF, Deraz SF, Smith C. Heat shock proteins in oncology: Diagnostic biomarkers or therapeutic targets? Biochim Biophys Acta. 2011;1816:89–104. doi: 10.1016/j.bbcan.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Chen M, Zhou J, Zhang X. HSP27, 70 and 90, anti-apoptotic proteins, in clinical cancer therapy (Review) Int J Oncol. 2014;45:18–30. doi: 10.3892/ijo.2014.2399. [DOI] [PubMed] [Google Scholar]
- 31.Matassa DS, Amoroso MR, Maddalena F, Landriscina M, Esposito F. New insights into TRAP1 pathway. Am J Cancer Res. 2012;2:235–248. [PMC free article] [PubMed] [Google Scholar]
- 32.Costantino E, Maddalena F, Calise S, Piscazzi A, Tirino V, Fersini A, Ambrosi A, Neri V, Esposito F, Landriscina M. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009;279:39–46. doi: 10.1016/j.canlet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 33.Aust S, Bachmayr-Heyda A, Pateisky P, Tong D, Darb-Esfahani S, Denkert C, Chekerov R, Sehouli J, Mahner S, Van Gorp T, et al. Role of TRAP1 and estrogen receptor alpha in patients with ovarian cancer-a study of the OVCAD consortium. Mol Cancer. 2012;11:69. doi: 10.1186/1476-4598-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han JJ, Baek SK, Lee JJ, Kim GY, Kim SY, Lee SH. Combination of TRAP1 and ERCC1 expression predicts clinical outcomes in metastatic colorectal cancer treated with Oxaliplatin/5-Fluorouracil. Cancer Res Treat. 2014;46:55–64. doi: 10.4143/crt.2014.46.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cappello F, Di Stefano A, David S, Rappa F, Anzalone R, La Rocca G, D'Anna SE, Magno F, Donner CF, Balbi B, Zummo G. Hsp60 and Hsp10 down-regulation predicts bronchial epithelial carcinogenesis in smokers with chronic obstructive pulmonary disease. Cancer. 2006;107:2417–2424. doi: 10.1002/cncr.22265. [DOI] [PubMed] [Google Scholar]
- 36.Matassa DS, Amoroso MR, Lu H, Avolio R, Arzeni D, Procaccini C, Faicchia D, Maddalena F, Simeon V, Agliarulo I, et al. Oxidative metabolism drives inflammation-induced platinum resistance in human ovarian cancer. Cell Death Differ. 2016;23:1542–1554. doi: 10.1038/cdd.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vidyasagar A, Wilson NA, Djamali A. Heat shock protein 27 (HSP27): Biomarker of disease and therapeutic target. Fibrogenesis Tissue Repair. 2012;5:7. doi: 10.1186/1755-1536-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elstrand MB, Kleinberg L, Kohn EC, Trope CG, Davidson B. Expression and clinical role of antiapoptotic proteins of the bag, heat shock, and Bcl-2 families in effusions, primary tumors, and solid metastases in ovarian carcinoma. Int J Gynecol Pathol. 2009;28:211–221. doi: 10.1097/PGP.0b013e31818b0f5e. [DOI] [PubMed] [Google Scholar]
- 39.Langdon SP, Rabiasz GJ, Hirst GL, King RJ, Hawkins RA, Smyth JF, Miller WR. Expression of the heat shock protein HSP27 in human ovarian cancer. Clin Cancer Res. 1995;1:1603–1609. [PubMed] [Google Scholar]
- 40.Olejek A, Damasiewicz-Bodzek A, Bodzek P, Wielkoszyński T, Zamłyński J, Stołtny P, Skutil M. Concentrations of antibodies against heat shock protein 27 in the sera of women with ovarian carcinoma. Int J Gynecol Cancer. 2009;19:1516–1520. doi: 10.1111/IGC.0b013e3181bf425b. [DOI] [PubMed] [Google Scholar]
- 41.Geisler JP, Tammela JE, Manahan KJ, Geisler HE, Miller GA, Zhou Z, Wiemann MC. HSP27 in patients with ovarian carcinoma: Still an independent prognostic indicator at 60 months follow-up. Eur J Gynaecol Oncol. 2004;25:165–168. [PubMed] [Google Scholar]
- 42.Schneider J, Jimenez E, Marenbach K, Marx D, Meden H. Co-expression of the MDR1 gene and HSP27 in human ovarian cancer. Anticancer Res. 1998;18:2967–2971. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.